Paramecium bursaria—A Complex of Five Cryptic Species: Mitochondrial DNA COI Haplotype Variation and Biogeographic Distribution †
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
2.2. Methods
2.2.1. Culturing and Identification of P. bursaria Strains
2.2.2. Molecular Techniques
2.2.3. Data Analysis
3. Results
3.1. Paramecium bursaria Species Complex
3.2. Analysis of COI mtDNA Sequences of the Paramecium bursaria Complex
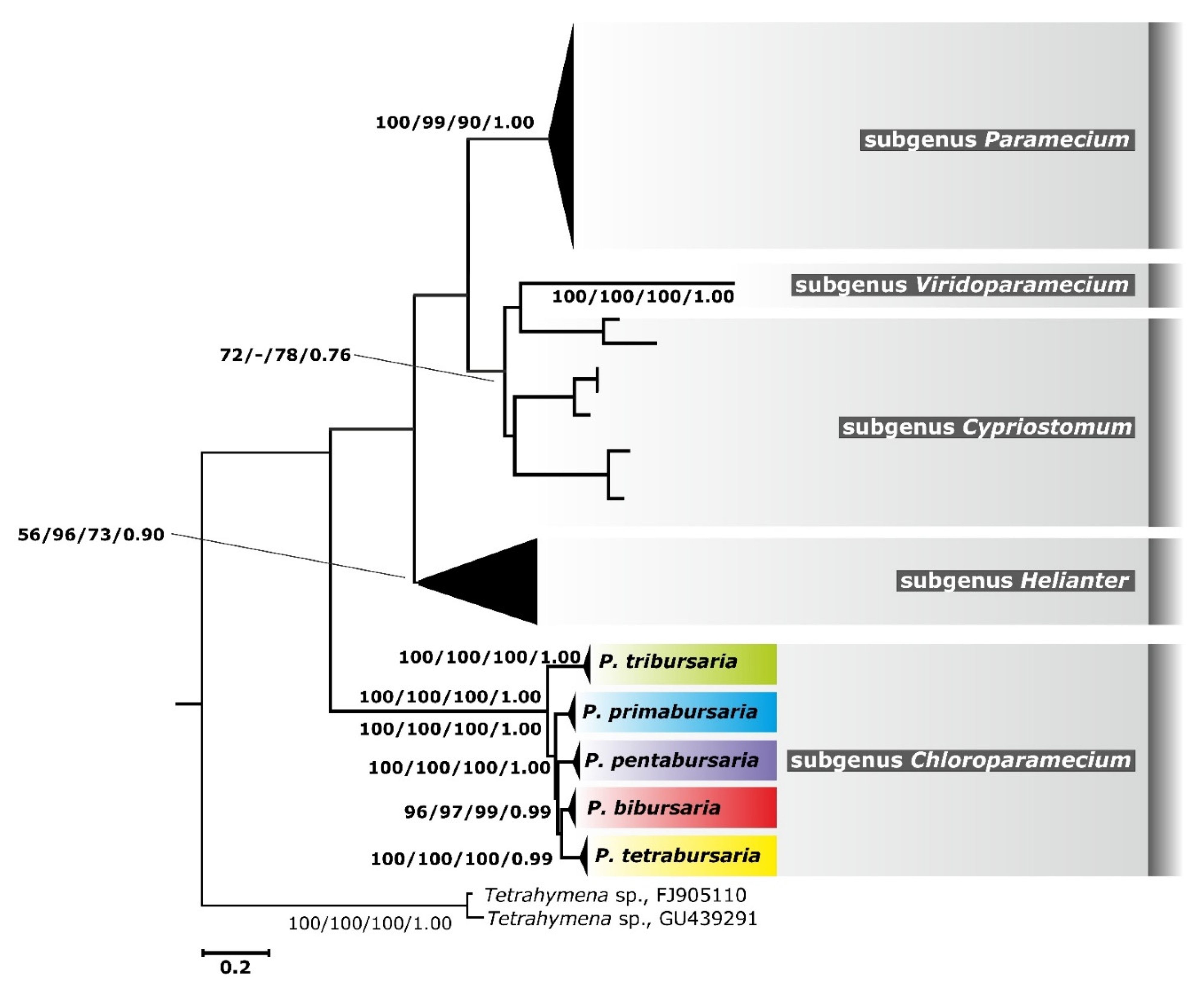
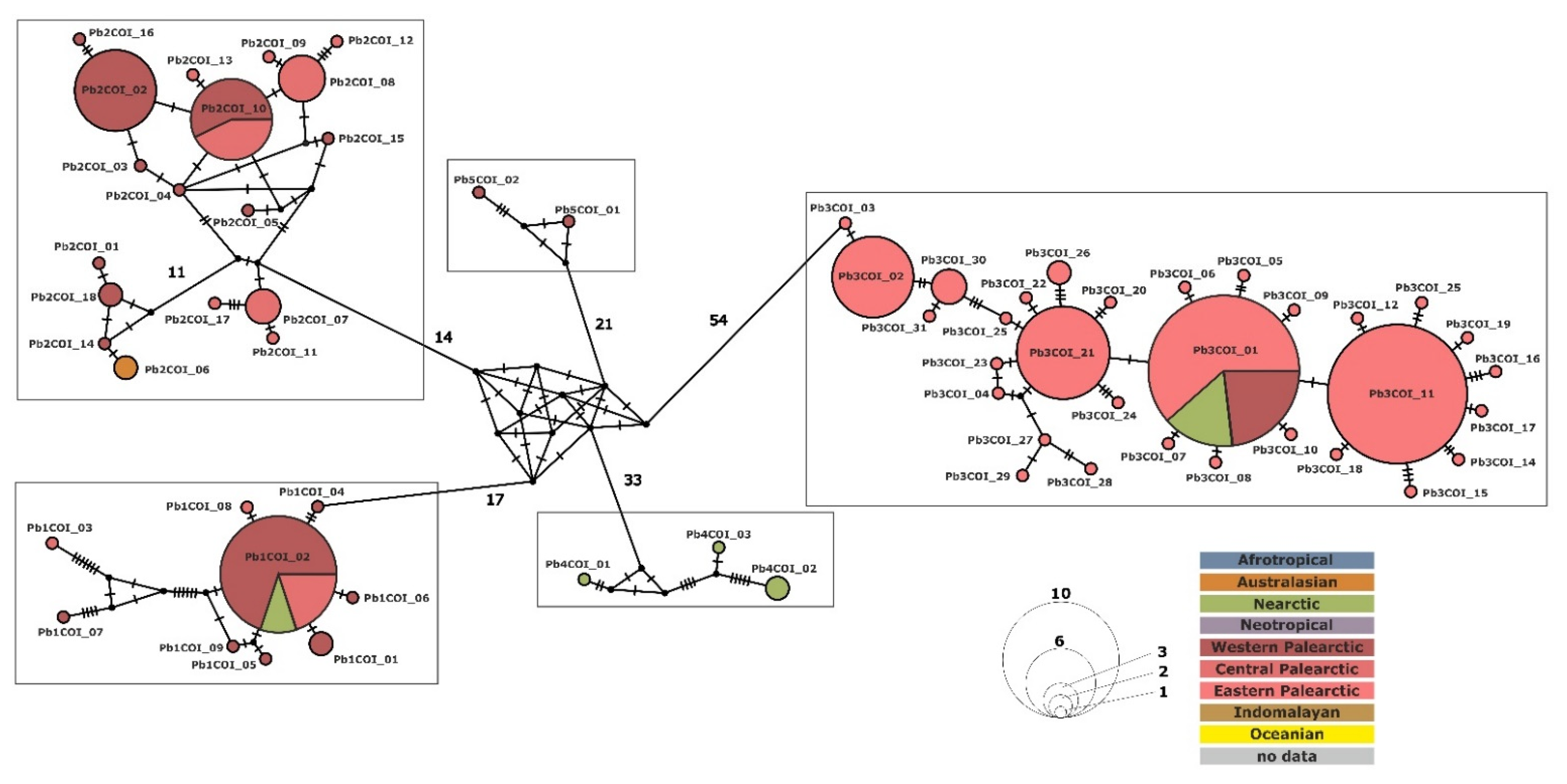
3.3. Phylogenetic Relationship and Haplotype Analysis
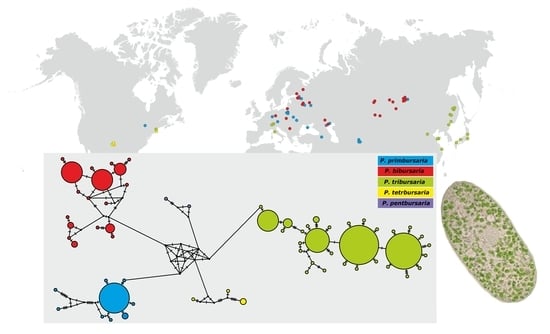
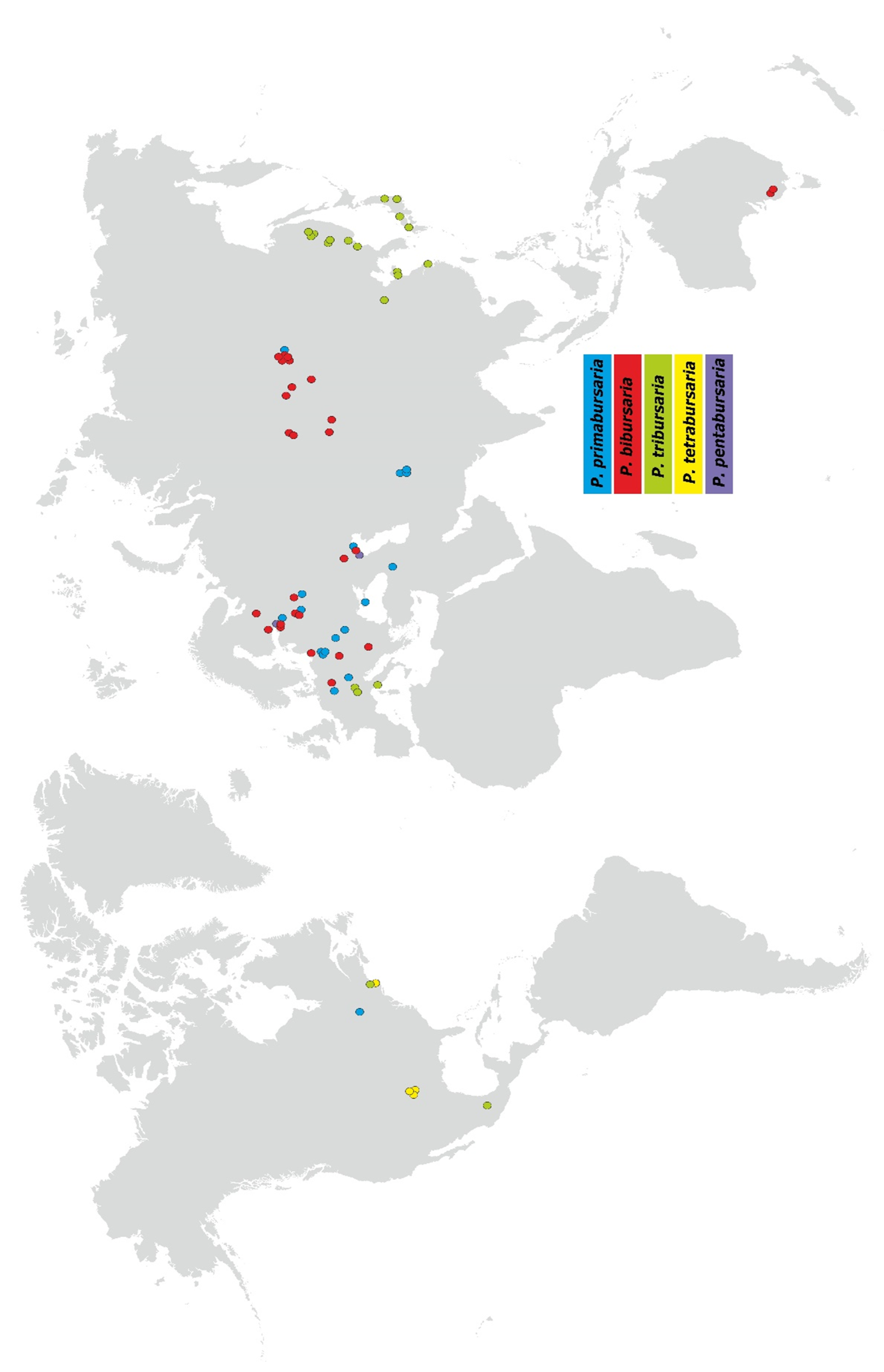
3.4. Paramecium bursaria Complex Biogeography—Analysis of Haplotype Variability
4. Discussion
4.1. COI mtDNA as an Appropriate DNA Marker for Ciliate Species Delimitation
4.2. Cryptic Species in the Genus Paramecium
4.3. Paramecium bursaria Species Complex—Wide or Narrow Range Species?
4.4. Taxonomic Summary
- Class Oligohymenophorea de Puytorac et al., 1974 [87]
- Order Peniculida Fauré-Fremiet (in Corliss), 1956 [88]
- Family Parameciidae Dujardin, 1841 [89]
- Genus Paramecium O.F. Müller, 1773 [90]
- Subgenus Chloroparamecium Fokin et al., 2004 [26]
- Paramecium bursaria Focke, 1836 [39]
- The Paramecium bursaria species complex Greczek-Stachura et al., 2021 [42]
- Desciption of the morphospecies Paramecium bursaria
- Etymology. The name is derived from the Latin word bursa (purse), which refers to the shape of the cell.
- Holotype: 87 MS-1.
- Type locality: St Petersburg vicinity, Russia (59°52′ N 29°54′ E), a pond in park.
- Distribution: widespread in Europe, several locations in Asia, including the Baikalarea (easternmost location).
- Occurrence/number of mating types: eight mating types.
- May correspond to Bomford’s syngen B6.
- Gene sequence: cytochrome c oxidase subunit I sequence of the holotype specimen. Has been deposited in GenBank under OK356526 accession number.
- Holotype: Ek.
- Type locality: St Petersburg, Russia (59°58′ N 30°14′ E), a pond in a public garden.
- Distribution: widespread in Europe, and central part of in Asia, two localities in South Australia.
- Occurrence/number of mating types: eight mating types.
- Demonstrate very good intersyngenic mating reactions with P. tetrabursaria, which suggests that it corresponds to Bomford’s syngen B4.
- Gene sequence: cytochrome c oxidase subunit I sequence of the holotype specimen. Has been deposited in GenBank under JF708911 accession number.
- Holotype: T316.
- Type locality: Tsukuba, Japan (36°03′ N 140°07′ E) no habitat details.
- Distribution: widespread in eastern Asia (Russia, Japan, China), three locations in southern Europe, and one in North America (Boston, MA, USA).
- Occurrence/number of mating types: eight mating types.
- It is still maintained in several laboratories in Japan, and these strains were once used for the identification of strains from the current collection. That makes it the only link to the lost Bomford’s collection.
- Corresponds to Bomford’s syngen B1.
- Gene sequence: cytochrome c oxidase subunit I sequence of the holotype specimen. Has been deposited in GenBank under JF708900 accession number.
- Holotype: AB2-32.
- Type locality: Boston, USA (42°21′ N 71°04′ W), a pond in public garden.
- Distribution: restricted to USA area.
- Occurrence/number of mating types: six mating types.
- Demonstrate very good intersyngenic mating reactions with P. bibursaria which suggests that it correspond to Bomford’s syngens B2.
- Gene sequence: cytochrome c oxidase subunit I sequence of the holotype specimen. Has been deposited in GenBank under JF708916 accession number.
- Holotype: AZ20-1.
- Type locality: Astrakhan Nature Reserve, Russia (45°53′ N 48°35′ E), backwaters in the delta of the Volga river.
- Distribution: restricted to two location in Europe.
- Occurrence/number of mating types: four mating types.
- Corresponds to Bomford’s syngen B3 or B5.
- Gene sequence: cytochrome c oxidase subunit I sequence of the holotype specimen. Has been deposited in GenBank under JF708905 accession number.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Debroas, D.; Domaizon, I.; Humbert, J.-F.; Jardillier, L.; Lepère, C.; Oudart, A.; Taïb, N. Overview of freshwater microbial eukaryotes diversity: A first analysis of publicly available metabarcoding data. FEMS Microbiol. Ecol. 2017, 93, fix023. [Google Scholar] [CrossRef]
- Finlay, B.J. Global dispersal of free-living microbial eucaryote species. Environ. Microbiol. 2002, 296, 1061–1064. [Google Scholar]
- Katz, L.A.; McManus, G.B.; Snoeyenbos-West, O.; Griffin, A.; Pirog, K.; Costas, B.; Foissner, W. Reframing the ‘Everything is everywhere’ debate: Evidence for high gene flow and diversity in ciliate morphospecies. Aquat. Microb. Ecol. 2005, 41, 55–65. [Google Scholar] [CrossRef][Green Version]
- Schlegel, M.; Meisterfeld, R. The species problem in protozoa revisited. Eur. J. Protistol. 2003, 39, 349–355. [Google Scholar] [CrossRef]
- Abraham, J.S.; Sripoorna, S.; Maurya, S.; Makhija, S.; Gupta, R.; Toteja, R. Techniques and tools for species identification in ciliates: A review. Int. J. Syst. Evol. Microbiol. 2019, 69, 877–894. [Google Scholar] [CrossRef] [PubMed]
- Clamp, J.C.; Lynn, D.H. Investigating the biodiversity of ciliates in the ‘Age of Integration’. Eur. J. Protistol. 2017, 61, 314–322. [Google Scholar] [CrossRef]
- Doerder, F.P. Barcodes Reveal 48 New Species of Tetrahymena, Dexiostoma, and Glaucoma: Phylogeny, Ecology, and Biogeography of New and Established Species. J. Eukaryot. Microbiol. 2019, 66, 182–208. [Google Scholar] [CrossRef] [PubMed]
- Lind, A.L.; Pollard, K.S. Accurate and sensitive detection of microbial eukaryotes from whole metagenome shotgun sequencing. Microbiome 2021, 9, 58. [Google Scholar] [CrossRef]
- Schrallhammer, M. Biodiversity of Ciliates and Their Symbionts: A Special Issue. Diversity 2020, 12, 441. [Google Scholar] [CrossRef]
- Barth, D.; Tischer, K.; Berger, H.; Schlegel, M.; Berendonk, T.U. High mitochondrial haplotype diversity of Coleps sp. (Ciliophora: Prostomatida). Environ. Microbiol. 2008, 10, 626–634. [Google Scholar] [CrossRef]
- Gentekaki, E.; Lynn, D.H. Evidence for Cryptic Speciation in Carchesium polypinum Linnaeus, 1758 (Ciliophora: Peritrichia) Inferred from Mitochondrial, Nuclear, and Morphological Markers. J. Eukaryot. Microbiol. 2010, 57, 508–519. [Google Scholar] [CrossRef]
- Katz, L.A.; DeBerardinis, J.; Hall, M.S.; Kovner, A.M.; Dunthorn, M.; Muse, S.V. Heterogeneous Rates of Molecular Evolution Among Cryptic Species of the Ciliate Morphospecies Chilodonella uncinata. J. Mol. Evol. 2011, 73, 266–272. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boscaro, V.; Syberg-Olsen, M.J.; Irwin, N.A.T.; del Campo, J.; Keeling, P.J. What Can Environmental Sequences Tell Us About the Distribution of Low-Rank Taxa? The Case of Euplotes (Ciliophora, Spirotrichea), Including a Description of Euplotes enigma sp. nov. J. Eukaryot. Microbiol. 2019, 66, 281–293. [Google Scholar] [CrossRef]
- Cai, X.; Wang, C.; Pan, X.; El-Serehy, H.A.; Mu, W.; Gao, F.; Qiu, Z. Morphology and systematics of two freshwater Frontonia species (Ciliophora, Peniculida) from northeastern China, with comparisons among the freshwater Frontonia spp. Eur. J. Protistol. 2018, 63, 105–116. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Pan, X.; Ding, W.; Rasheid, K.A.; Qiu, Z. Morphology and Phylogeny of a New Frontonia Ciliate, F. paramegna spec. nov. (Ciliophora, Peniculida) from Harbin Northeast China. Zootaxa 2014, 3827, 375–386. [Google Scholar] [CrossRef]
- Krenek, S.; Berendonk, T.; Fokin, S. New Paramecium (Ciliophora, Oligohymenophorea) congeners shape our view on its biodiversity. Org. Divers. Evol. 2015, 15, 215–233. [Google Scholar] [CrossRef]
- Liang, Z.; Shen, Z.; Zhang, Y.; Ji, D.; Li, J.; Warren, A.; Lin, X. Morphology and Phylogeny of Four New Vorticella Species (Ciliophora: Peritrichia) from Coastal Waters of Southern China. J. Eukaryot. Microbiol. 2019, 66, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.H.; Doerder, F.P.; Gillis, P.L.; Prosser, R.S. Tetrahymena glochidiophila n. sp., a new species of Tetrahymena (Ciliophora) that causes mortality to glochidia larvae of freshwater mussels (Bivalvia). Dis. Aquat. Org. 2018, 127, 125–136. [Google Scholar] [CrossRef]
- Xu, Y.; Pan, H.; Miao, M.; Hu, X.; Al-Farraj, S.A.; Al-Rasheid, K.A.S.; Song, W. Morphology and Phylogeny of Two Species of Loxodes (Ciliophora, Karyorelictea), with Description of a New Subspecies, Loxodes striatus orientalis subsp. n. J. Eukaryot. Microbiol. 2015, 62, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Rataj, M.; Vd’ačný, P. Cryptic host-driven speciation of mobilid ciliates epibiotic on freshwater planarians. Mol. Phylogenet. Evol. 2021, 161, 107174. [Google Scholar] [CrossRef]
- Bieliavskaia, A.; Kiselev, A.; Rautian, M. New Paramecium species “Candidatus Paramecium ossipovi”. Protistology 2016, 10, 8. [Google Scholar]
- Paiva, T.S.; Borges, B.N.; Harada, M.L.; Silva-Nero, I.D. Description and molecular phylogeny of Paramecium grohmannae sp. nov. (Ciliophora, Peniculida) from a wastewater treatment plant in Brazil. Revista Brasileira de Zoociências Ciliophora 2016, 17, 7–19. [Google Scholar]
- Shakoori, F.R.; Tasneem, F.; Al-Ghanim, K.; Mahboob, S.; Al-Misned, F.; Jahan, N.; Shakoori, A.R. Variability in Secondary Structure of 18S Ribosomal RNA as Topological Marker for Identification of Paramecium species. J. Cell. Biochem. 2014, 115, 2077–2088. [Google Scholar] [CrossRef]
- Przyboś, E.; Tarcz, S. Paramecium jenningsi complex: Existence of three cryptic species confirmed by multi-locus analysis and strain crosses. Syst. Biodivers. 2016, 14, 140–154. [Google Scholar] [CrossRef]
- Potekhin, A.; Mayén-Estrada, R. Paramecium Diversity and a New Member of the Paramecium aurelia Species Complex Described from Mexico. Diversity 2020, 12, 197. [Google Scholar] [CrossRef]
- Fokin, S.; Przybos, E.; Chivilev, S.M.; Beier, C.L.; Horn, M.; Skotarczak, B.; Wodecka, B.; Fujishima, M. Morphological and molecular investigations of Paramecium schewiakoffi sp. nov. (Ciliophora, Oligohymenophorea) and current status of distribution and taxonomy of Paramecium spp. Eur. J. Protistol. 2004, 40, 22–243. [Google Scholar] [CrossRef]
- Kreutz, M.; Stoeck, T.; Foissner, W. Morphological and Molecular Characterization of Paramecium (Viridoparamecium nov. subgen.) chlorelligerum Kahl (Ciliophora). J. Eukaryot. Microbiol. 2012, 59, 548–563. [Google Scholar] [CrossRef] [PubMed]
- Sonneborn, T.M. The Paramecium aurelia complex of fourteen sibling species. Trans. Am. Microsc. Soc. 1975, 94, 155–178. [Google Scholar] [CrossRef]
- Greczek-Stachura, M.; Potekhin, A.; Przyboś, E.; Rautian, M.; Skoblo, I.; Tarcz, S. Identification of Paramecium bursaria syngens through molecular markers--comparative analysis of three loci in the nuclear and mitochondrial DNA. Protist 2012, 163, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Tarcz, S.; Rautian, M.; Potekhin, A.; Sawka, N.; Beliavskaya, A.; Kiselev, A.; Nekrasova, I.; Przyboś, E. Paramecium putrinum (Ciliophora, Protozoa): The first insight into the variation of two DNA fragments—Molecular support for the existence of cryptic species. Mol. Phylogenet. Evol. 2014, 73, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Przyboś, E.; Rautian, M.; Beliavskaia, A.; Tarcz, S. Evaluation of the molecular variability and characteristics of Paramecium polycaryum and Paramecium nephridiatum, within subgenus Cypriostomum (Ciliophora, Protista). Mol. Phylogenet. Evol. 2019, 132, 296–306. [Google Scholar] [CrossRef]
- Mayr, E. Systematics and the Origin of Species; Columbia University Press: New York, NY, USA, 1942. [Google Scholar]
- Caron, D.A. Towards a Molecular Taxonomy for Protists: Benefits, Risks, and Applications in Plankton Ecology. J. Eukaryot. Microbiol. 2013, 60, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Fokin, S. Paramecium genus: Biodiversity, some morphological features and the key to the main morphospecies discrimination. Protistology 2010, 6, 227–235. [Google Scholar]
- Zhan, Z.; Li, J.; Xu, K. Ciliate Environmental Diversity Can Be Underestimated by the V4 Region of SSU rDNA: Insights from Species Delimitation and Multilocus Phylogeny of Pseudokeronopsis (Protist, Ciliophora). Microorganisms 2019, 7, 493. [Google Scholar] [CrossRef]
- Strüder-Kypke, M.C.; Lynn, D.H. Comparative analysis of the mitochondrial cytochrome c oxidase subunit I (COI) gene in ciliates (Alveolata, Ciliophora) and evaluation of its suitability as a biodiversity marker. Syst. Biodivers. 2010, 8, 131–148. [Google Scholar] [CrossRef]
- Park, M.-H.; Jung, J.-H.; Jo, E.; Park, K.-M.; Baek, Y.-S.; Kim, S.-J.; Min, G.-S. Utility of mitochondrial CO1 sequences for species discrimination of Spirotrichea ciliates (Protozoa, Ciliophora). Mitochondrial DNA Part A 2019, 30, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Przyboś, E.; Tarcz, S. Global molecular variation of Paramecium jenningsi complex (Ciliophora, Protista): A starting point for further, detailed biogeography surveys. Syst. Biodivers. 2019, 17, 527–539. [Google Scholar] [CrossRef]
- Focke, G.W. Ueber einige organisations-verhaltnisse bei polygastrischen infusorien. Oken. Isis 1836, 10, 785–787. [Google Scholar]
- Vivier, E. Morphology, Taxonomy and General Biology of the Genus Paramecium. In Paramecium, A Current Survey; van Wagtendonk, W.J., Ed.; Elsevier: Amsterdam, The Netherlands, 1974; pp. 1–89. [Google Scholar]
- Wichterman, R. The Biology of Paramecium, 2nd ed.; Plenum Press: New York, NY, USA, 1986. [Google Scholar]
- Greczek-Stachura, M.; Leśnicka, P.Z.; Tarcz, S.; Rautian, M.; Możdżeń, K. Genetic Diversity of Symbiotic Green Algae of Paramecium bursaria Syngens Originating from Distant Geographical Locations. Plants 2021, 10, 609. [Google Scholar] [CrossRef]
- Pröschold, T.; Darienko, T.; Silva, P.C.; Reisser, W.; Krienitz, L. The systematics of Zoochlorella revisited employing an integrative approach. Environ. Microbiol. 2011, 13, 350–364. [Google Scholar] [CrossRef]
- Bomford, R. The Syngens of Paramecium bursaria: New Mating Types and Intersyngenic Mating Reactions. J. Protozool. 1966, 13, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Sonneborn, T.M. Breeding systems, reproductive methods and species problem in Protozoa. In The Species Problem; Mayer, E., Ed.; AAAS: Washington, DC, USA, 1957; pp. 155–324. [Google Scholar]
- Sonneborn, T.M. Chapter 12 Methods in Paramecium Research. In Methods in Cell Biology; Prescott, D.M., Ed.; Academic Press: New York, NY, USA, 1970; Volume 4, pp. 241–339. [Google Scholar]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Page, R.D.M. Tree View: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Moritz, C.; Cicero, C. DNA Barcoding: Promise and Pitfalls. PLoS Biol. 2004, 2, e354. [Google Scholar] [CrossRef]
- Pawlowski, J.; Audic, S.; Adl, S.; Bass, D.; Belbahri, L.; Berney, C.; Bowser, S.S.; Cepicka, I.; Decelle, J.; Dunthorn, M.; et al. CBOL Protist Working Group: Barcoding Eukaryotic Richness beyond the Animal, Plant, and Fungal Kingdoms. PLoS Biol. 2012, 10, e1001419. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, T.; Przybos, E.; Dunthorn, M. The D1-D2 region of the large subunit ribosomal DNA as barcode for ciliates. Mol. Ecol. Resour. 2014, 14, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. Paramecium aurelia Revisited. J. Eukaryot. Microbiol. 2005, 52, 68–77. [Google Scholar] [CrossRef]
- Boscaro, V.; Fokin, S.I.; Verni, F.; Petroni, G. Survey of Paramecium duboscqui using three markers and assessment of the molecular variability in the genus Paramecium. Mol. Phylogenet. Evol. 2012, 65, 1004–1013. [Google Scholar] [CrossRef]
- Tarcz, S.; Potekhin, A.; Rautian, M.; Przyboś, E. Variation in ribosomal and mitochondrial DNA sequences demonstrates the existence of intraspecific groups in Paramecium multimicronucleatum (Ciliophora, Oligohymenophorea). Mol. Phylogenet. Evol. 2012, 63, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Barth, D.; Krenek, S.; Fokin, S.I.; Berendonk, T.U. Intraspecific Genetic Variation in Paramecium Revealed by Mitochondrial Cytochrome c Oxidase I Sequences. J. Eukaryot. Microbiol. 2006, 53, 20–25. [Google Scholar] [CrossRef]
- Brown, W.M.; George, M., Jr.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef]
- De Souza, B.A.; Dias, R.J.P.; Senra, M.V.X. Intrageneric evolutionary timing and hidden genetic diversity of Paramecium lineages (Ciliophora: Oligohymenophorea). Syst. Biodivers. 2020, 18, 662–674. [Google Scholar] [CrossRef]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Grundt, H.H.; Kjølner, S.; Borgen, L.; Rieseberg, L.H.; Brochmann, C. High biological species diversity in the arctic flora. Proc. Natl. Acad. Sci. USA 2006, 103, 972–975. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Amato, A.; Kooistra, W.H.C.F.; Levialdi Ghiron, J.H.; Mann, D.G.; Pröschold, T.; Montresor, M. Reproductive Isolation among Sympatric Cryptic Species in Marine Diatoms. Protist 2007, 158, 193–207. [Google Scholar] [CrossRef]
- Smirnov, A.V. Cryptic freshwater amoeba species in the bottom sediments of Nivå Bay (Øresund, Baltic Sea). Europ. J. Protistol. 2007, 43, 87–94. [Google Scholar] [CrossRef]
- Chantangsi, C.; Lynn, D.H.; Brandl, M.T.; Cole, J.C.; Hetrick, N.; Ikonomi, P. Barcoding ciliates: A comprehensive study of 75 isolates of the genus Tetrahymena. Int. J. Syst. Evol. Microbiol. 2007, 57, 2412–2423. [Google Scholar] [CrossRef] [PubMed]
- McManus, G.B.; Xu, D.; Costas, B.A.; Katz, L.A. Genetic Identities of Cryptic Species in the Strombidium stylifer/apolatum/oculatum Cluster, Including a Description of Strombidium rassoulzadegani n. sp. J. Eukaryot. Microbiol. 2010, 57, 369–378. [Google Scholar] [CrossRef]
- Foissner, W.; Chao, A.; Katz, L.A. Diversity and geographic distribution of ciliates (Protista: Ciliophora). Biodivers. Conserv. 2008, 17, 345–363. [Google Scholar] [CrossRef]
- Allen, S.; Farrow, S.W.; Golembiowski, P.A. Esterase variation between the 14 syngens of Paramecium aurelia under axenic growth. Genetics 1973, 73, 561–573. [Google Scholar] [CrossRef]
- Tait, A. Enzyme variation between syngens in Paramecium aurelia. Biochem. Genet. 1970, 4, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Catania, F.; Wurmser, F.; Potekhin, A.A.; Przyboś, E.; Lynch, M. Genetic Diversity in the Paramecium aurelia Species Complex. Mol. Biol. Evol. 2008, 26, 421–431. [Google Scholar] [CrossRef]
- Zagata, P.; Greczek-Stachura, M.; Tarcz, S.; Rautian, M. Molecular Identification of Paramecium bursaria Syngens and Studies on Geographic Distribution using Mitochondrial Cytochrome C Oxidase Subunit I (COI). Folia Biol. 2015, 63, 77–83. [Google Scholar] [CrossRef]
- Olefeld, J.L.; Bock, C.; Jensen, M.; Vogt, J.C.; Sieber, G.; Albach, D.; Boenigk, J. Centers of endemism of freshwater protists deviate from pattern of taxon richness on a continental scale. Sci. Rep. 2020, 10, 14431. [Google Scholar] [CrossRef]
- Finlay, B.J. The global diversity of protozoa and other small species. Int. J. Parasitol. 1998, 28, 29–48. [Google Scholar] [CrossRef]
- Foissner, W. Biogeography and Dispersal of Micro-organisms: A Reviev Emphasizing Protists. Acta Protozool. 2006, 45, 111–136. [Google Scholar]
- Segawa, T.; Matsuzaki, R.; Takeuchi, N.; Akiyoshi, A.; Navarro, F.; Sugiyama, S.; Yonezawa, T.; Mori, H. Bipolar dispersal of red-snow algae. Nat. Commun. 2018, 9, 3094. [Google Scholar] [CrossRef]
- Dunthorn, M.; Stoeck, T.; Wolf, K.; Breiner, H.-W.; Foissner, W. Diversity and endemism of ciliates inhabiting Neotropical phytotelmata. Syst. Biodivers. 2012, 10, 195–205. [Google Scholar] [CrossRef]
- Przyboś, E.; Tarcz, S.; Dusi, E. New Paramecium quadecaurelia strains (P. aurelia spp. complex, Ciliophora) identified by molecular markers (rDNA and mtDNA). Europ. J. Protistol. 2013, 49, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Przyboś, E.; Tarcz, S.; Rautian, M.; Lebedeva, N. The first European stand of Paramecium sonneborni (P. aurelia complex), a species known only from North America (Texas, USA). Europ. J. Protistol. 2014, 50, 236–247. [Google Scholar] [CrossRef]
- Tarcz, S.; Przyboś, E.; Surmacz, M. An assessment of haplotype variation in ribosomal and mitochondrial DNA fragments suggests incomplete lineage sorting in some species of the Paramecium aurelia complex (Ciliophora, Protozoa). Mol. Phylogenet. Evol. 2013, 67, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Salsbery, M.E.; DeLong, J.P. The benefit of algae endosymbionts in Paramecium bursaria is temperature dependent. Evol. Ecol. Res. 2018, 19, 669–678. [Google Scholar]
- Możdżeń, K.; Leśnicka, P.Z.; Burnecki, T.; Śliwińska-Wilczewska, S.; Skoczowski, A.; Greczek-Stachura, M. Photosynthetic efficiency of endosymbiotic algae of Paramecium bursaria originating from locations with cold and warm climates. Oceanol. Hydrobiol. Stud. 2018, 47, 202–210. [Google Scholar] [CrossRef]
- Tarcz, S.; Sawka-Gądek, N.; Przyboś, E. Worldwide sampling reveals low genetic variability in populations of the freshwater ciliate Paramecium biaurelia (P. aurelia species complex, Ciliophora, Protozoa). Org. Divers. Evol. 2018, 18, 39–50. [Google Scholar] [CrossRef]
- de Puytorac, P.; Batisse, A.; Bohatier, J.; Corliss, J.O.; Deroux, G.; Didier, P.; Dragesco, J.; Fryd-Versavel, G.; Grain, J.; Grolière, C.; et al. Proposition dúne classification du phylum Ciliophora Doflein, 1901 (réunion de systématique, Clermont-Ferrand). C. r. hebd. Séanc. Acad. Sci. Paris 1974, 278, 2799–2802. [Google Scholar]
- Corliss, J.O. On the evolution and systematics of ciliated protozoa. Syst. Zool. 1956, 5, 68–91. [Google Scholar] [CrossRef]
- Dujardin, F. Histoire Naturelle Des Zoophytes; Suies a Buffon; Infusoires: Paris, France, 1841. [Google Scholar]
- Müller, O.F. Vermium Terrestrium et Fluviatilium, seu Animalium Infusorium, Helminthicorum et Testaceorum, non Marinorum, Succincta Historia. Havniae et Lipsiae 1773, 1, 1–135. [Google Scholar]
- Fokin, S.I. Morphology of the contractile vacuoles in ciliated protozoa of the genus Paramecium (Hymenostomatida, Peniculina) as a species-specific character. Zool. Zhurnal 1986, 65, 5–15. [Google Scholar]
- Engelmann, T.W. Über Licht- und Farbenperception niederster Organismen. Archiv. Physiol. 1882, 2, 387–400. [Google Scholar] [CrossRef]
- Pado, R. Spectral activity of light and phototaxis in Paramecium bursaria. Acta Protozool. 1972, 11, 387–393. [Google Scholar]
- Nakaoka, Y. Localization of photosensitivity in Paramecium. J. Comp. Physiol. A 1989, 165, 637–641. [Google Scholar] [CrossRef]
- Barth, D.; Berendonk, T.U. The mitochondrial genome sequence of the ciliate Paramecium caudatum reveals a shift in nucleotide composition and codon usage within the genus Paramecium. BMC Genom. 2011, 12, 272. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Gentekaki, E.; Yi, Z.; Lin, X. Genetic differentiation of the mitochondrial cytochrome oxidase c subunit I gene in genus Paramecium (Protista, Ciliophora). PLoS ONE 2013, 8, e77044. [Google Scholar] [CrossRef] [PubMed]
- Przyboś, E.; Tarcz, S.; Pothekin, A.; Rautian, M.; Prajer, M. A two-locus molecular characterisation of Paramecium calkinsi. Protist 2012, 163, 263–273. [Google Scholar] [CrossRef]
- Przyboś, E.; Tarcz, S.; Rautian, M.; Sawka, N. Delimiting species boundaries within a paraplyletic species complex: Insights from morphological, genetic, and molecular data on Paramecium sonneborni (Paramecium aurelia species complex, Ciliophora, Protozoa). Protist 2015, 166, 438–456. [Google Scholar] [CrossRef]
- Tarcz, S. Intraspecific differentiation of Paramecium novaurelia strains (Ciliophora, Protozoa) inferred from phylogenetic analysis of ribosomal and mitochondrial DNA variation. Europ. J. Protistol. 2013, 49, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Przyboś, E.; Tarcz, S. Three-locus analysis in conjunction with strain crosses confirms the existence of reproductively isolated populations in Paramecium jenningsi (Diller and Earl 1958). Syst. Biodivers. 2013, 11, 507–523. [Google Scholar] [CrossRef]
- Snoke, M.S.; Berendonk, T.U.; Barth, D.; Lynch, M. Large Global Effective Population Sizes in Paramecium. Mol. Biol. Evol. 2006, 23, 2474–2479. [Google Scholar] [CrossRef] [PubMed]
- Krenek, S.; Petzoldt, T.; Berendonk, T.U. Coping with Temperature at the Warm Edge—Patterns of Thermal Adaptation in the Microbial Eukaryote Paramecium caudatum. PLoS ONE 2012, 7, e30598. [Google Scholar] [CrossRef]
- Kher, C.P.; Doerder, F.P.; Cooper, J.; Ikonomi, P.; Achilles-Day, U.; Kupper, F.C.; Lynn, D.H. Barcoding Tetrahymena: Discriminating species and identifying unknowns using the cytochrome c oxidase subunit I (cox-1) barcode. Protist 2011, 162, 2–13. [Google Scholar] [CrossRef]
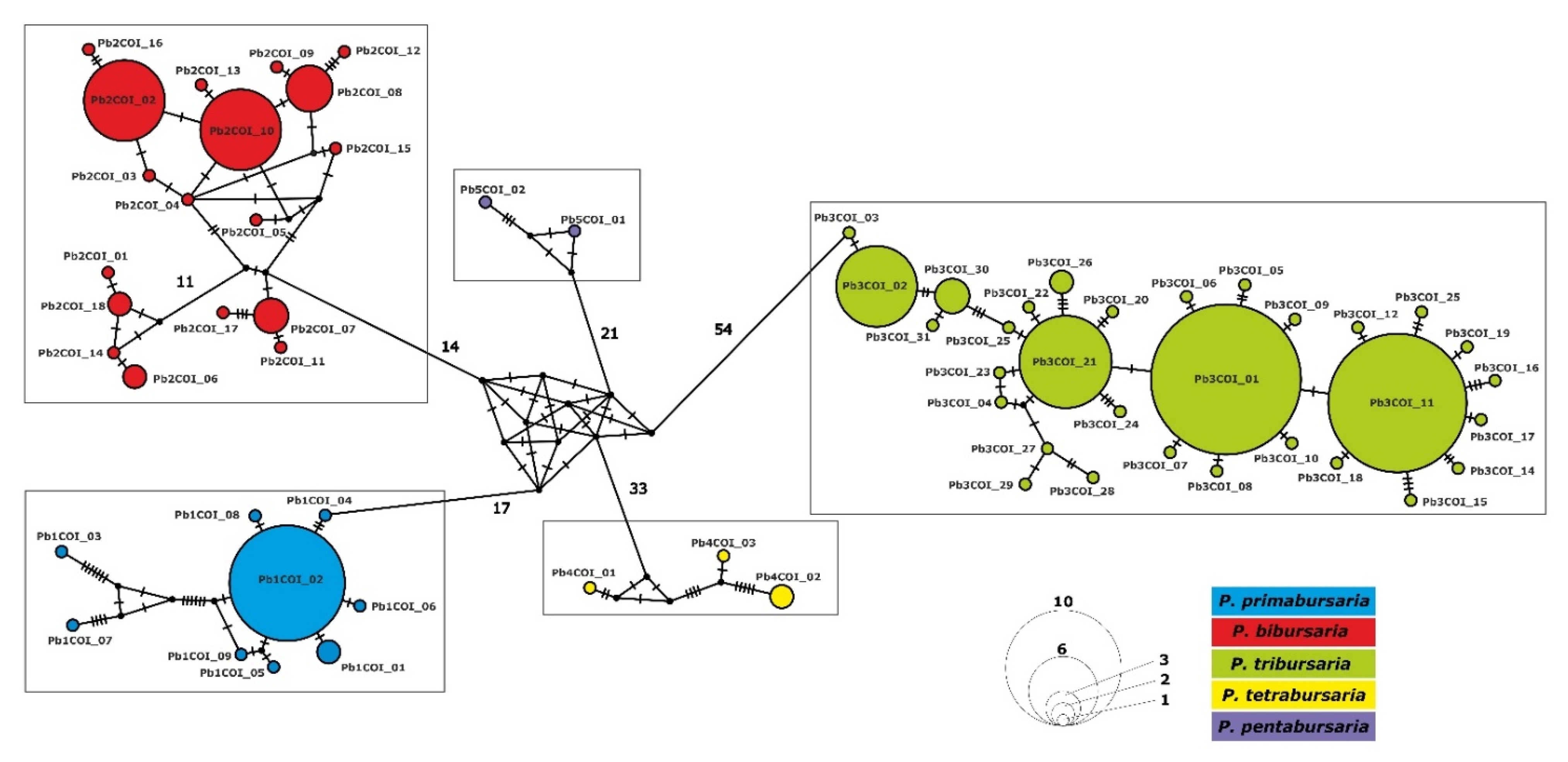
| Paramecium Species | Number of Sequences | Mean Uncorrected | Number of Haplotypes | Haplotype Diversity | Nucleotide Diversity |
|---|---|---|---|---|---|
| N | p-Distance | h | Hd | π | |
| P. primabursaria | 19 | 0.01 | 9 | 0.731 | 0.00552 |
| P. bibursaria | 37 | 0.01 | 18 | 0.920 | 0.0000029 |
| P. tribursaria | 70 | 0.01 | 31 | 0.918 | 0.00533 |
| P. tetrabursaria | 4 | 0.01 | 3 | 0.833 | 0.01133 |
| P. pentabursaria | 2 | 0.01 | 2 | 1.000 | 0.00618 |
| P. bursaria complex | 132 | 0.09 | 63 | 0.966 | 0.06351 |
| P. jenningsi complex * | 13 | 0.013 | 13 | 1.000 | 0.01779 |
| P. aurelia complex * | 15 | 0.084 | 15 | 1.000 | 0.07812 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greczek-Stachura, M.; Rautian, M.; Tarcz, S. Paramecium bursaria—A Complex of Five Cryptic Species: Mitochondrial DNA COI Haplotype Variation and Biogeographic Distribution. Diversity 2021, 13, 589. https://doi.org/10.3390/d13110589
Greczek-Stachura M, Rautian M, Tarcz S. Paramecium bursaria—A Complex of Five Cryptic Species: Mitochondrial DNA COI Haplotype Variation and Biogeographic Distribution. Diversity. 2021; 13(11):589. https://doi.org/10.3390/d13110589
Chicago/Turabian StyleGreczek-Stachura, Magdalena, Maria Rautian, and Sebastian Tarcz. 2021. "Paramecium bursaria—A Complex of Five Cryptic Species: Mitochondrial DNA COI Haplotype Variation and Biogeographic Distribution" Diversity 13, no. 11: 589. https://doi.org/10.3390/d13110589
APA StyleGreczek-Stachura, M., Rautian, M., & Tarcz, S. (2021). Paramecium bursaria—A Complex of Five Cryptic Species: Mitochondrial DNA COI Haplotype Variation and Biogeographic Distribution. Diversity, 13(11), 589. https://doi.org/10.3390/d13110589