Abstract
Gold nanoparticles (AuNPs) have found widespread applications in life sciences. While synthesis of monodispersed AuNPs has been fairly convenient by using chemical reduction of chloroauric acid by sodium citrate, we found that AuNPs of high quality and high concentrations were not readily obtained via this method. As an example, we showed that monodispersed 13-nm AuNPs were readily synthesized at relatively low concentrations (e.g. 3.5 nM); in contrast, 13-nm AuNPs of 17 nM obtained by the direct reduction method were irregularly shaped and not well dispersed. In this work, we demonstrated that AuNPs of high concentration could be prepared by a two-step approach, i.e. chemical reduction at low concentrations and subsequent centrifugation. Compared to the direct reduction method, this new two-step method led to AuNPs with high salt resistance and high stability, which are essential for the preparation of DNA-AuNPs conjugates for DNA biodetection.
1. Introduction
Gold nanoparticles have found widespread applications in life sciences and attracted significant research interest. In fact, the use of AuNPs has a long history in biology, dating back to the application of “immunogold” in biological imaging in the 1970’s [1]. In 1996, Mirkin’s group and Alivisatos’ group independently reported that AuNPs-DNA conjugates could serve as scaffolds for nanostructures [2, 3]. By exploiting the unique optical and electronic properties of AuNPs, Mirkin and coworkers later developed a series of novel methods for ultrasensitive detection of DNA and proteins [4–8]. Their approaches relied on the modification of AuNPs with thiolated DNA probes, which underwent characteristic red-to-blue color changes during the process of target DNA-mediated aggregation of AuNPs. Very recently, Li et al. reported an even simpler method for colorimetric detection of target DNA by using unmodified AuNPs [9,10]. The mechanism lying behind their strategy is that single-stranded (ss-) DNA, while not double-stranded (ds-) DNA, spontaneously binds to unmodified AuNPs, and effectively stabilizes AuNPs in solutions of high ionic strength. Interestingly, the differentiation ability of AuNPs between ss- and ds- DNA also forms the basis of our recently developed “nanoparticle PCR”, a novel AuNPs-enhanced PCR strategy with high selectivity [11].
In view of the wide applicability of AuNPs, it is important to synthesize high-quality AuNPs in DNA-based biodetection. In certain applications, it is also important to prepare AuNPs of high concentrations [12]. Therefore, it is the goal of this work to develop a new approach to synthesize AuNPs of high quality and high concentration that are appropriate for preparation of DNA-AuNPs conjugates.
2. Results and Discussion
We evaluated the size distribution of synthesized 13-nm AuNPs via Atomic Force Microscopy (AFM). As shown in Figure 1, AuNPs of 3.5 nM synthesized by direct reduction method were almost monodispersed, with a size of 12.5 ± 2.3 nm. In spite of this, we still observed a small amount of particles of smaller size as indicated in Figure 1a. In contrast, AuNPs of 17 nM prepared from the direct reduction method (method I) were poorly dispersed in size (14 ± 5 nm), containing approximately 30 % large particles. We then used the two-step approach (method II), and found that the size distribution of thus prepared AuNPs was as good as or even better than that of AuNPs of 3.5 nM. In fact, even the small particles observed in 3.5-nM AuNPs samples were absent in this centrifugation method. This clearly showed that centrifugation significantly improved the quality of high-concentration AuNPs. We then measured the UV-Vis spectra of AuNPs. As shown in Figure 2, all these AuNPs showed characteristic absorption at approx. 520 nm, corresponding to the strong surface plasomon resonance of nanosized gold.
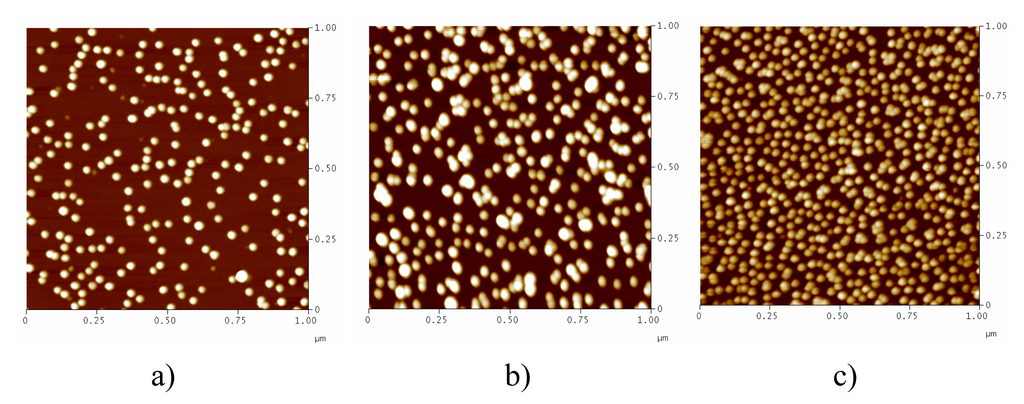
Figure 1.
AFM images of 13-nm AuNPs. a) 3.5 nM, direct reduction b) 17 nM, direct reduction; c) 17 nM, centrifuging.
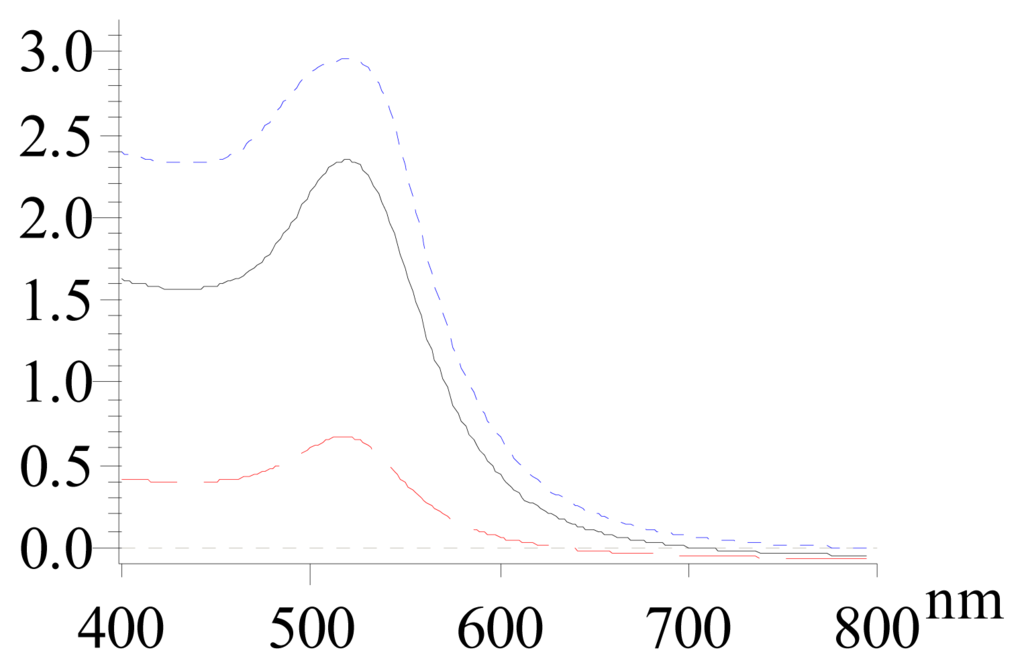
Figure 2.
UV spectra of 13-nm AuNPs. a) 3.5 nM, direct reduction; b) 17 nM, direct reduction; c) 17 nM, centrifuging (from bottom to top).
AuNPs prepared by method I and II showed distinctly different stability upon salt addition. We found that the addition of 10 μl of 0.1 M PBS to 100 μl of AuNPs prepared from method I resulted in a color change from red to purple (Figure 3). At higher concentrations of salts, AuNPs became blue colored, suggesting the formation of large aggregation and the shift of SPR absorption. The UV-Vis absorption also correlated well with this color change. As shown in Figure 4, the absorption at 520 nm gradually decreased along with the addition of salts. In contrast, AuNPs prepared from method II were much more resistant to salt-induced aggregation. Even with 25 μl of 0.1 M PBS, AuNPs remained to be red colored. In fact, the characteristic color change was only found after the addition of 80 μl of 0.1 M PBS. This significantly increased stability might arise due to the absence of large particles in solution.

Figure 3.
Salt-resistance for AuNPs from method II (top) and method I (bottom).(From left to right: addition of 0, 5, 10, 15, 20, 25 μl of 0.1 M PBS)
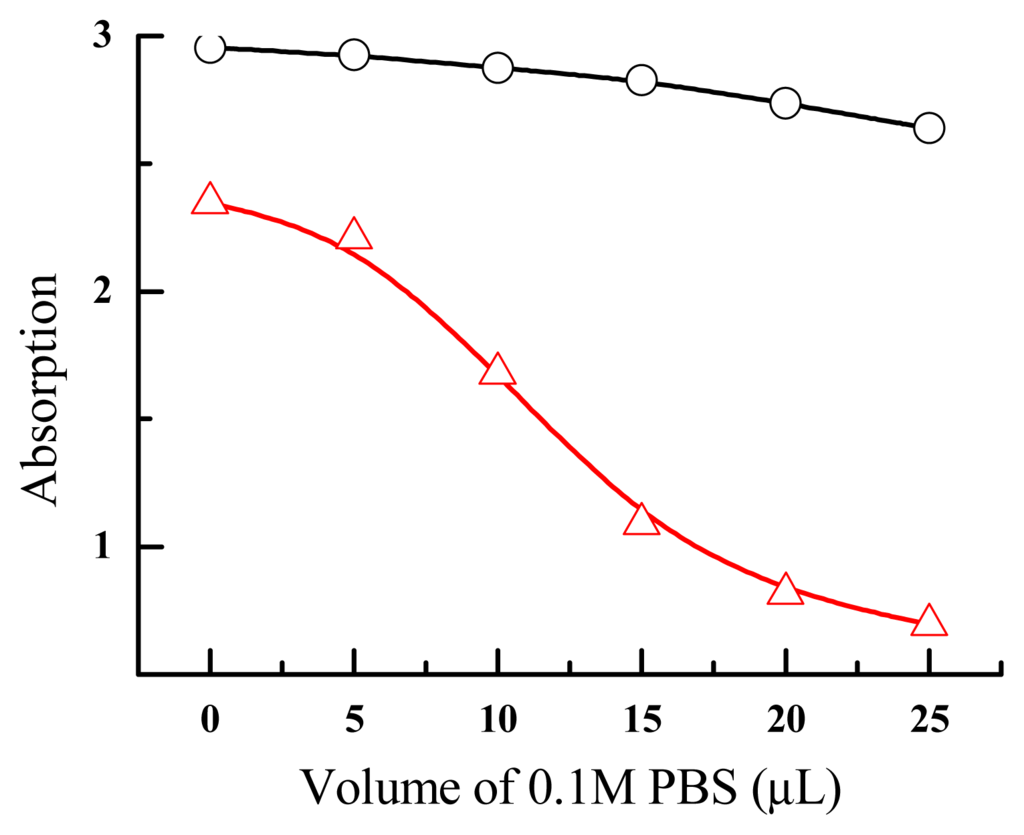
Figure 4.
UV-Vis analysis of AuNPs from method II (top) and method I (bottom).
High-concentration AuNPs are useful for the preparation of DNA-AuNPs conjugates. As shown in Figure 5, after DNA self-assembly and centrifuging, DNA absorption at 260 nm largely decreased, suggesting the removal of unbound DNA, while the thiolated DNA remained at the surface of AuNPs. We again found that it was better to use AuNPs prepared from method II than from method I. After the formation of DNA-AuNPs, the precipitates from method II were readily dispersed while those from method I required repetitive tapping. In addition, AuNPs from method I showed slight absorption shift from 520 nm to 522 nm, suggesting the formation of some aggregation. In contrast, AuNPs from method II did not exhibit such changes.
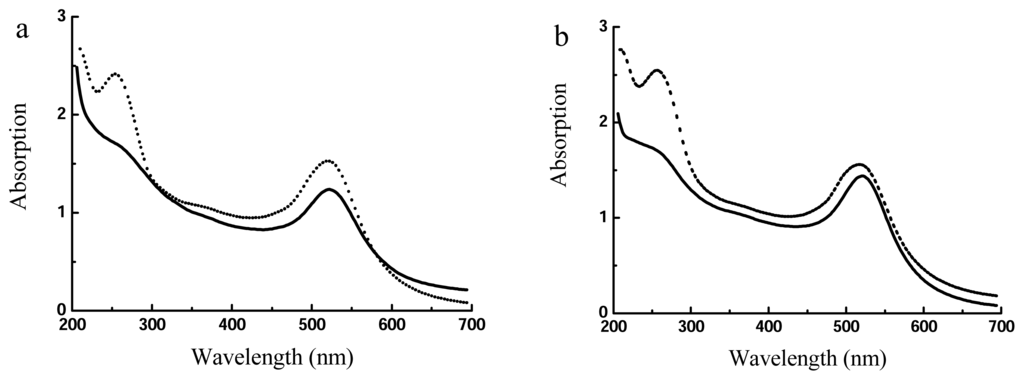
Figure 5.
UV-Vis spectra of AuNPs-DNA before (dashed line) and after (solid line) centrifugation. a) method I; b) method II.
We also tested the salt-resistance of DNA-AuNPs conjugates. We found that the conjugates from method II were highly stable, which could tolerate salt concentration as high as 2.6 M, while those from method I could only tolerate a solution of 1.5 M NaCl. More importantly, the storage stability of DNA-AuNPs conjugates prepared from method II was also much better. As a comparison, the conjugates from method I showed some precipitates only after 7-day storage at 4 °C, while those from method II were stable for at least 30 days. The Table 1 summarized the major differences of AuNPs prepared from either method.

Table 1.
Comparison of AuNPs preparation from method I and II.
Conclusion
In summary, we showed that monodispersed 13-nm AuNPs were readily synthesized at relatively low concentrations (e.g. 3.5 nM) while not at high concentrations (17 nM). We then proposed a two-step approach, i.e. chemical reduction at low concentrations and subsequent centrifugation. Compared to the direct reduction method, this new two-step method led to AuNPs with high salt resistance and high stability, which are essential for the preparation of DNA-AuNPs conjugates for biodetection. Indeed, DNA-AuNPs conjugates prepared from the two-step approach showed much improved stability toward salt-induced aggregation, as well as much longer storage time. These advantages are highly useful for AuNPs-based DNA biodetection.
3. Experimental Section
3.1 Reagents and instrumentation
The thiolated oligonucleotide was obtained from Sangon, Shanghai, with a sequence of 5′-HS-TTTTTTTTTTAGGGTCGGAACAGGAGA-3′. HAuCl4.4H2O (Au content > 47.8 %) was from National Pharmaceutical, China; sodium citrate was from Sangon, Shanghai, and all other reagents were of analytical grade. The water employed in this work was Milli Q (18.2 MΩ). Centrafuging was performed at a Himac CF16RX (Hitachi). UV-Vis spectra was performed at a U-3010 UV-Vis spectroscopy (Hitachi). Atomic force microscopy was a DI NanoScope IIIa from Veeco Instrument Co.
3.2 Synthesis of 13-nm AuNPs
The synthesis of AuNPs follows the work of Natan and coworkers [13]. For 3.5 nM AuNPs, 100 ml of 0.01 % HAuCl4 was added to a 250 ml round bottle, and boiled. Under rapid stirring, 3.5 ml of sodium citrate (1 %) was added and further rapidly stirred for 15 min. After stirred for 30-min, the solution was then gradually cooled to room temperature, and was filtered by 0.22 μm filter paper.
For 17 nM AuNPs, we used two methods, that is, direct reduction method (method I) and centrafuging method (method II). Method I: 100 ml of 1mM HAuCl4 was added to a 250 ml round bottle, and boiled. Under rapid stirring, 5 ml of 38.8 mM sodium citrate was added and further rapidly stirred for 15 min. After stirred for 30 min, the solution was then gradually cooled to room temperature, and was filtered by 0.22 μm filter paper. Method II: AuNPs of 3.5 nM was first synthesized by reduction, and then centrifuged to a concentrated solution of 17 nM. We experimentally found that the optimum conditions were centrifuging for 20 min at 4 °C, with a RCF of 11930 g.
Shape, concentration and size-distribution of syntheized AuNPs were characterized by UV-Vis spectroscopy and AFM.
3.3 Evaluation of salt-resistance of AuNPs
100 μl of AuNPs (17 nM) from either method I or method II were challenged by PBS (10 mM PB, pH 7.0, 0.1 M NaCl) of appropriate volumes, respectively. Instead, DNA-AuNPs conjugates were challenged by a solution of 5 M NaCl of appropriate volumes. Then the colors and UV-Vis spectra of these solutions were recorded.
3.4 Synthesis of DNA-AuNPs conjugates
The protocol for the synthesis of DNA-AuNPs conjugates was similar to previously reported [4,14]. AuNPs of 17 nM were added by thiolate DNA, leading to a final concentration of 10 nM. After 20-min, a solution of 1 M PBS (100 mM PB, pH 7.0, 1 M NaCl) was gradually added to the solution until the salt concentration reached 0. 1 M. This solution was aged for 16 hours at room temperature. After that, the solution was centrifuged, and the precipitate was washed by 100 μl of 0.1 M PBS. This washing process was repeated for three times, and the final product was diluted in 100 μl of 0.1 M PBS.
Acknowledgements
We thank the financial support from National Natural Science Foundation (60537030, 20404016 and 30570401), Ministry of Science and Technology (2006CB933000), Shanghai Municipal Commission for Science and Technology (0652nm006, 0652nm016 and 06ZR14106), Shanghai Rising-Star Program and Chinese Academy of Sciences.
References and Notes
- Hacker, G.W.; Gu, J. Gold and Silver Staining: Techniques in Molecular Morphology; CRC Press: Boca Raton, 2002. [Google Scholar]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [Google Scholar]
- Alivisatos, A.P.; Johnsson, K.P.; Peng, X.G.; Wilson, T.E.; Loweth, C.J.; Bruchez, M.P.; Schultz, P.G. Organization of 'nanocrystal molecules' using DNA. Nature 1996, 382, 609–611. [Google Scholar]
- Taton, T.A.; Mirkin, C.A.; Letsinger, R.L. Scanometric DNA Array Detection with Nanoparticle Probes. Science 2000, 289, 1757–1760. [Google Scholar]
- Cao, Y.W.C.; Jin, R.C.; Mirkin, C.A. Nanoparticles with Raman Spectroscopic Fingerprints for DNA and RNA Detection. Science 2002, 297, 1536–1540. [Google Scholar]
- Rosi, N.L.; Mirkin, C.A. Nanostructures in biodiagnostics. Chem. Rev 2005, 105, 1547–1562. [Google Scholar]
- Nam, J.M.; Thaxon, C.S.; Mirkin, C.A. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science 2003, 301, 1884–1886. [Google Scholar]
- Oh, B.K.; Nam, J.M.; Lee, S.W.; Mirkin, C.A. A fluorophore-based bio-barcode amplification assay for proteins. Small 2006, 2, 103–108. [Google Scholar]
- Li, H.; Rothberg, L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl. Acad. Sci., U.S.A 2004, 101, 14036–14039. [Google Scholar]
- Li, H.; Rothberg, L.J. Label-free colorimetric detection of specific sequences in genomic DNA amplified by the polymerase chain reaction. J. Am. Chem. Soc 2004, 126, 10958–10961. [Google Scholar]
- Li, H.; Huang, J.; Lv, J.; An, H.; Zhang, X.; Zhang, Z.; Fan, C.; Hu, J. Nanoparticle PCR: Nanogold-assisted PCR with enhanced specificity. Angew. Chem. Int. Ed 2005, 44, 5100–5103. [Google Scholar]
- Elghanian, R.; Storhoff, J.J.; Mucic, R.C.; Letsinger, R.L.; Mirkin, C.A. DNA-Directed Synthesis of Binary Nanoparticle Network Materials. Science 1997, 277, 1078–1081. [Google Scholar]
- Grabar, K.C.; Freeman, R.G.; Hommer, M.B.; Natan, M.J. Preparation and Characterization of Au Colloid Monolayers. Anal. Chem 1995, 67, 735–743. [Google Scholar]
- Rosi, N.L.; Giljohann, D.A.; Thaxton, C.S.; Lytton-Jean, A.K.R.; Han, M.S.; Mirkin, C.A. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science 2006, 312, 1027–1030. [Google Scholar]
© 2007 by MDPI Reproduction is permitted for noncommercial purposes.