Molecular Characterization and Pathogenicity of Watermelon Isolates of Begomovirus cucurbitachinaense
Abstract
1. Introduction
2. Results
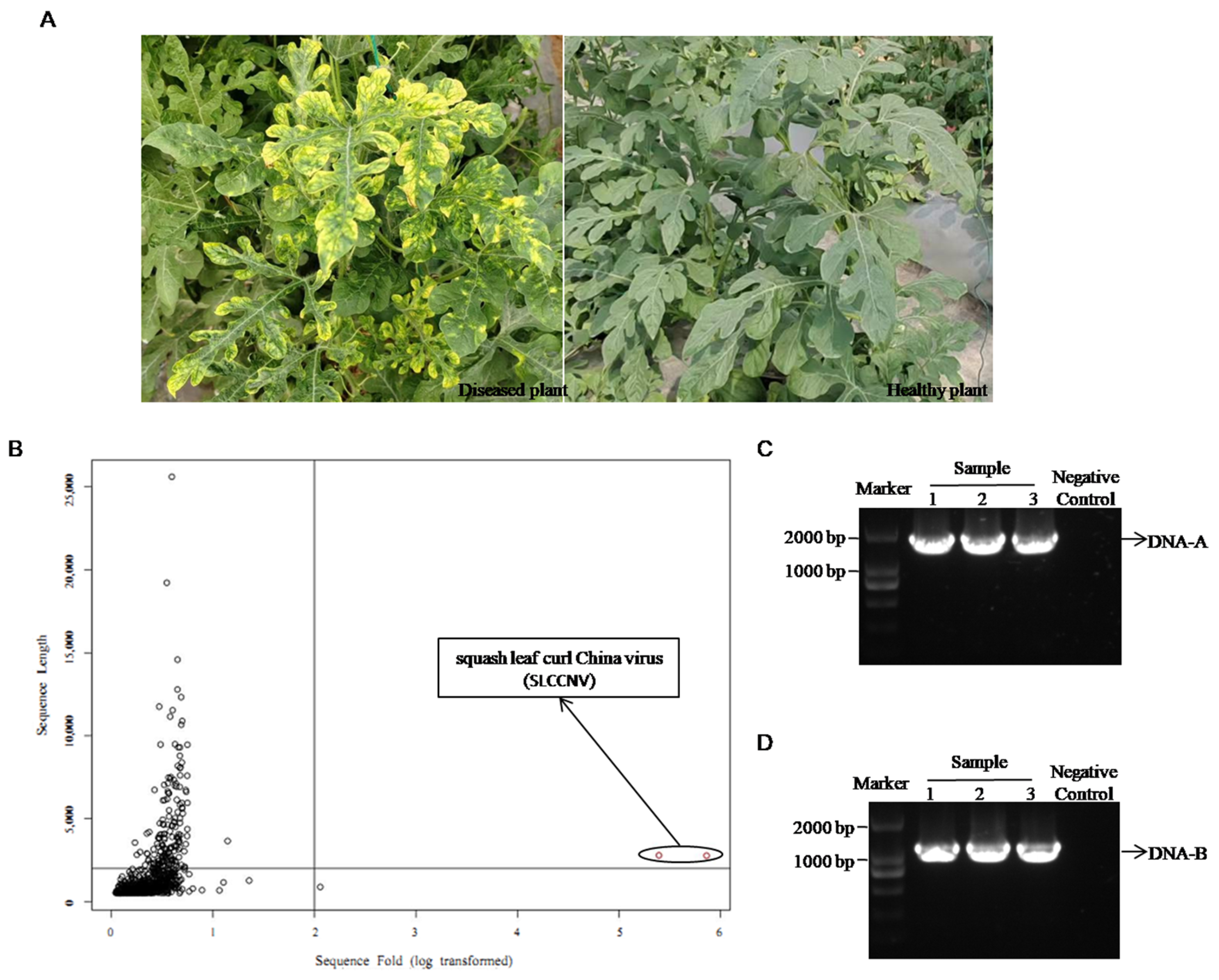
2.1. Discovery of Squash Leaf Curl China Virus in Watermelon Diseased Leaves Through Next-Generation Sequencing (NGS) and PCR Identification
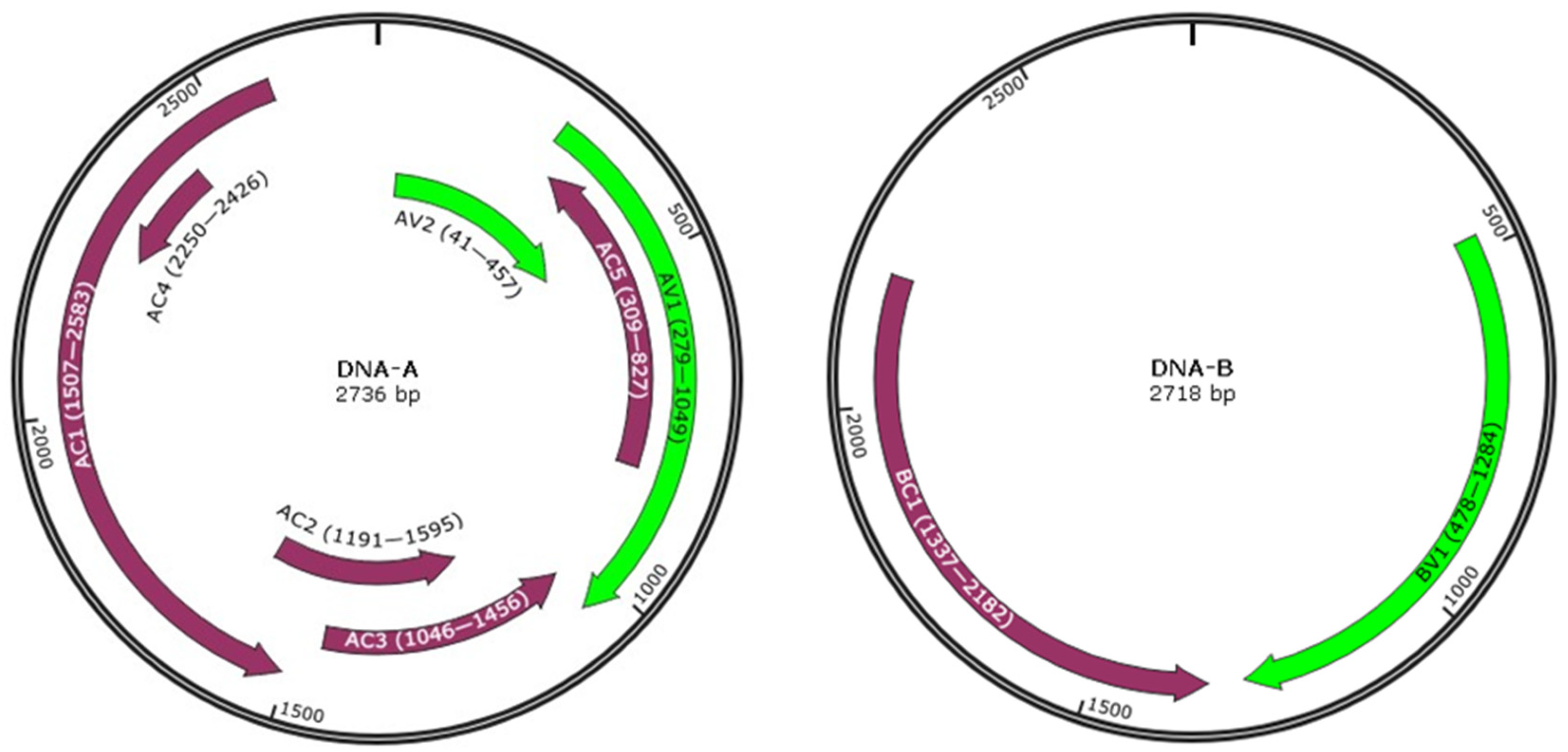
2.2. Complete Genome Sequences of SLCCNV Watermelon Isolates and Sequence Identity Analysis
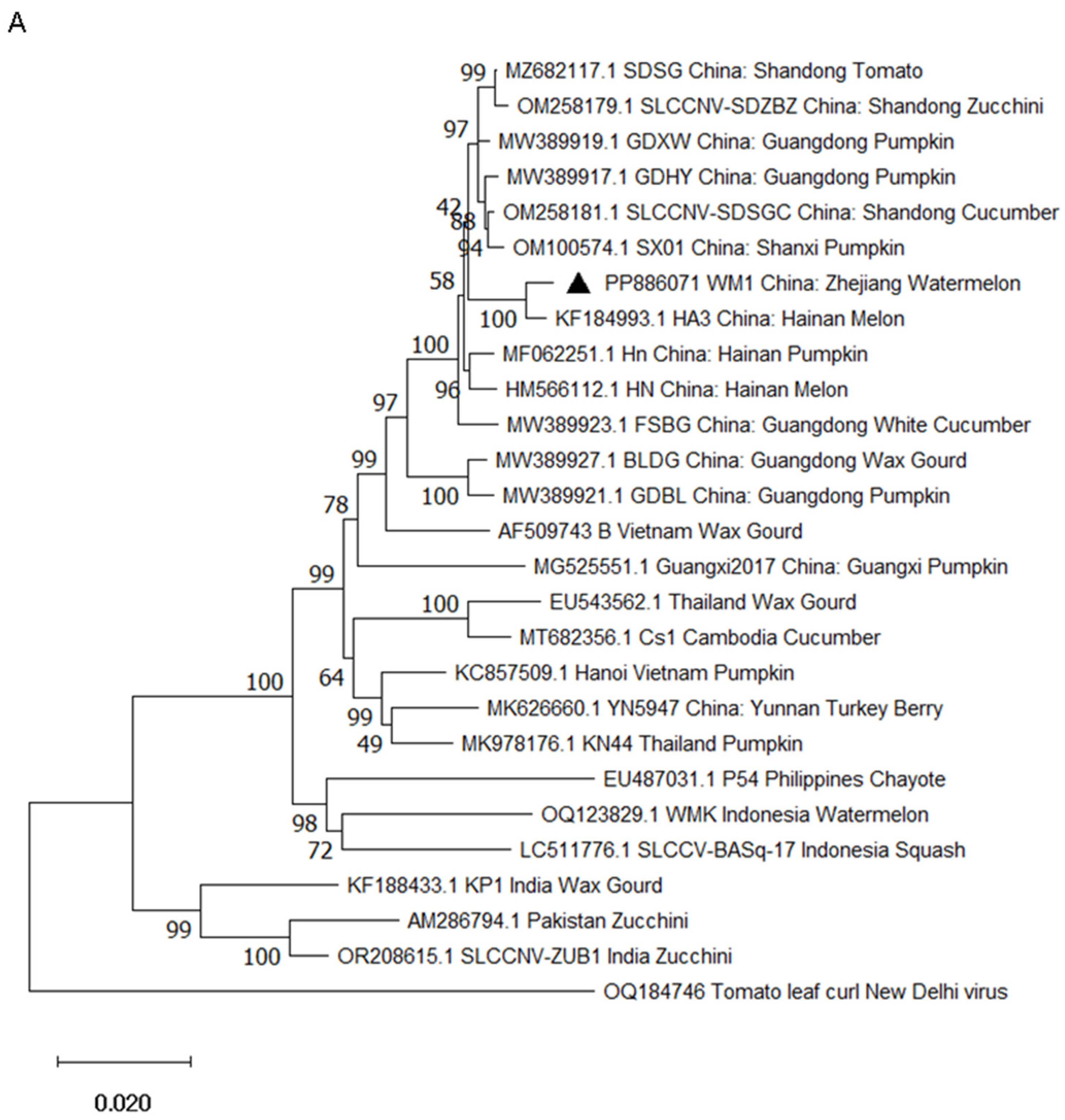
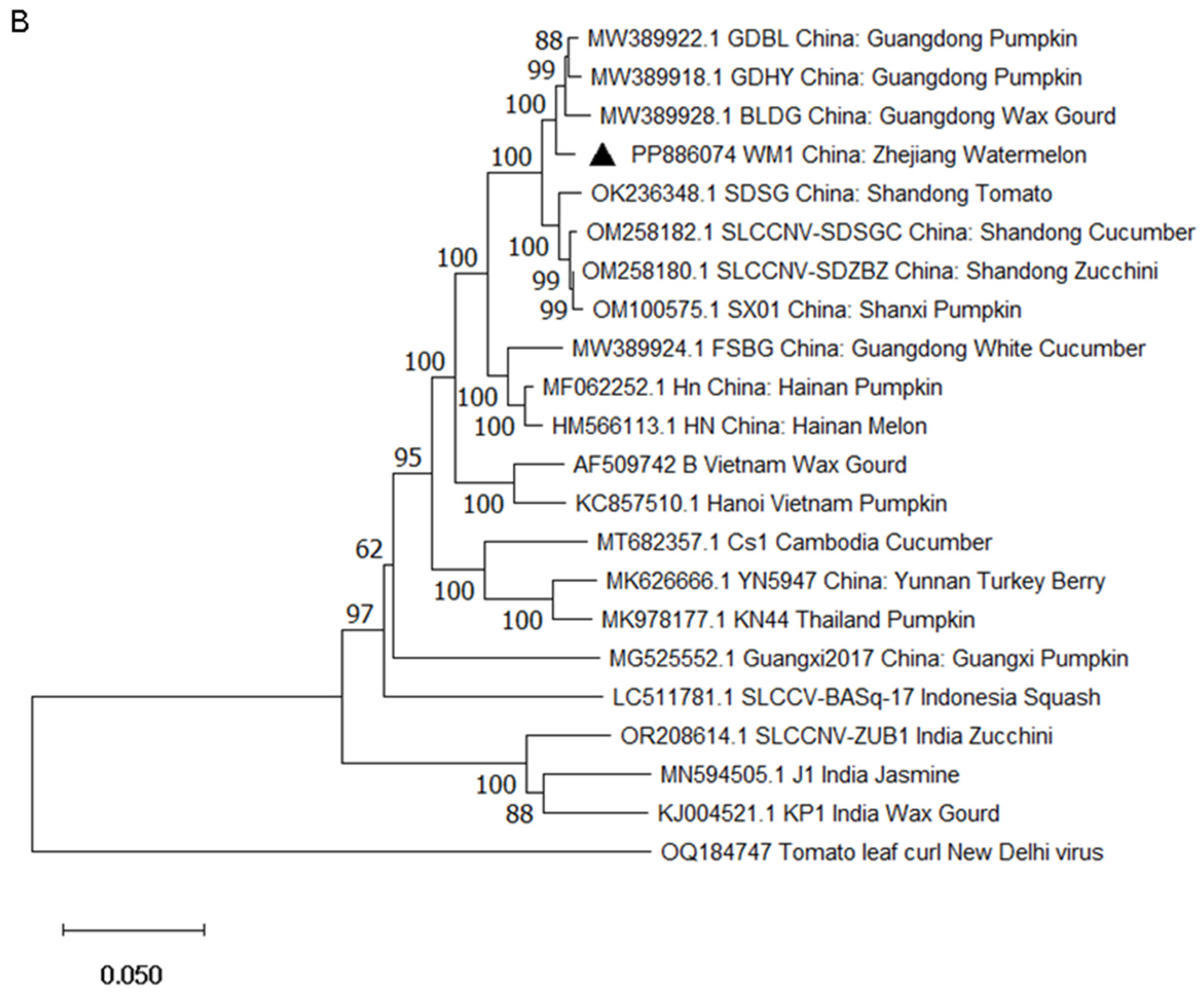
2.3. Phylogenetic Analysis
2.4. The Infectious Clone of Isolate WM1 Can Systematically Infect Watermelon
2.5. The Infectious Clone of Isolate WM1 Can Also Systematically Infect Melon, Squash, Pumpkin, Wax Gourd, Cucumber, and N. benthamiana
2.6. Progeny Virions Derived from the Cloned WM1 Isolate Can Be Transmitted by Whiteflies Rather than Mechanical
2.7. The Infectious Clone of Melon Isolate SLCCNV-HN Can Systemically Infect Melon and Watermelon, but Exhibits Lower Pathogenicity in Watermeon Compared to the WM1 Isolate
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Next-Generation Sequencing (NGS) and Sequence Assembly
4.3. PCR Identification
4.4. Amplification of Complete Genome Sequences of SLCCNV Isolates
4.5. Sequence Analysis
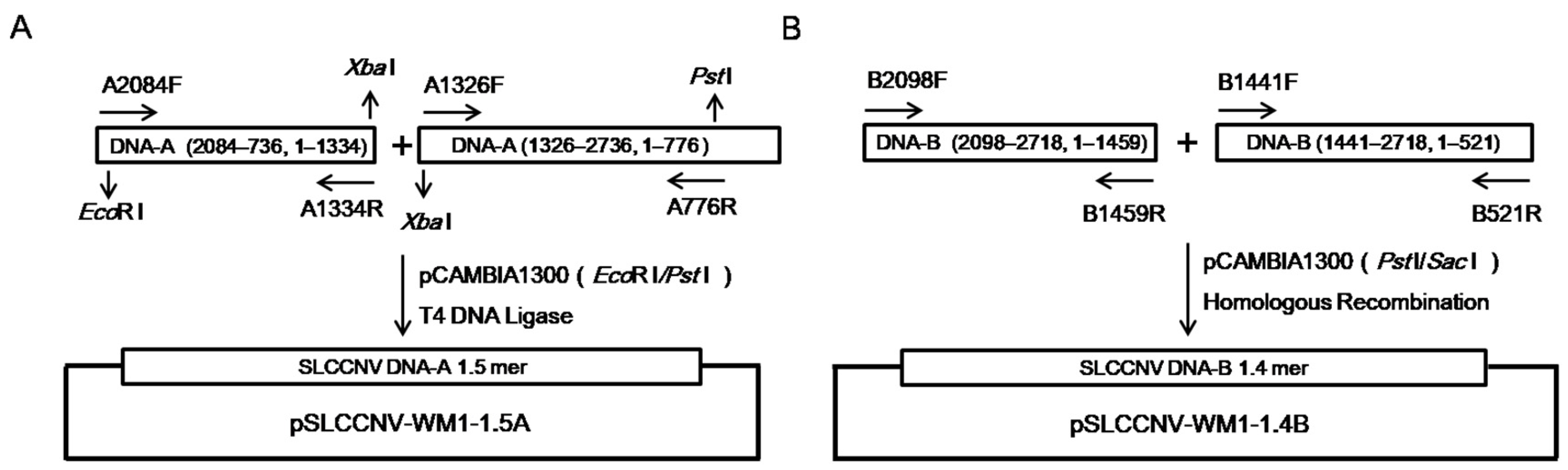
4.6. Construction of Infectious Clone
4.7. Agrobacterium-Mediated Inoculation
4.8. Whitefly Transmission and Mechanical Inoculation
4.9. Southern Blot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fiallo-Olive, E.; Lett, J.M.; Martin, D.P.; Roumagnac, P.; Varsani, A.; Zerbini, F.M.; Navas-Castillo, J. ICTV Report Consortium. ICTV virus taxonomy profile: Geminiviridae 2021. J. Gen. Virol. 2021, 102, 001696. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Qiao, R.; Wang, Z.; Yang, X.; Zhou, X. Occurrence and distribution of geminiviruses in China. Sci. China Life Sci. 2022, 65, 1498–1503. [Google Scholar] [CrossRef] [PubMed]
- Venkataravanappa, V.; Ashwathappa, K.V.; Reddy, C.N.L.; Shankarappa, K.S.; Reddy, M.K. Characterization of Tomato leaf curl New Delhi virus associated with leaf curl and yellowing disease of watermelon and development of LAMP assay for its detection. 3 Biotech 2020, 10, 282. [Google Scholar] [CrossRef]
- Moriones, E.; Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000, 71, 123–134. [Google Scholar] [CrossRef]
- Li, F.; Xu, X.; Huang, C.; Gu, Z.; Cao, L.; Hu, T.; Ding, M.; Li, Z.; Zhou, X. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 2015, 208, 555–569. [Google Scholar] [CrossRef]
- Gutierrez, C. Strategies for geminivirus DNA replication and cell cycle interference. Physiol. Mol. Plant Pathol. 2002, 60, 219–230. [Google Scholar] [CrossRef]
- Jeske, H.; Lütgemeier, M.; Preiss, W. DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic virus. Embo J. 2001, 20, 6158–6167. [Google Scholar] [CrossRef]
- Sanderfoot, A.A.; Lazarowitz, S.G. Getting it together in plant virus movement: Cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 1996, 6, 353–358. [Google Scholar] [CrossRef]
- Pascal, E.; Goodlove, P.E.; Wu, L.C.; Lazarowitz, S.G. Transgenic tobacco plants expressing the geminivirus BL1 protein exhibit symptoms of viral disease. Plant Cell 1993, 5, 795–807. [Google Scholar] [CrossRef]
- Jose, J.; Usha, R. Bhendi yellow vein mosaic disease in India is caused by association of a DNA β satellite with a begomovirus. Virology 2003, 305, 310–317. [Google Scholar] [CrossRef]
- Saunders, K.; Bedford, I.D.; Yahara, T.; Stanley, J. Aetiology: The earliest recorded plant virus disease. Nature 2003, 422, 831. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Pinner, M.S.; Stanley, J.; Markham, P.G. Geminivirus coat protein gene replacement alters insect specificity. Virology 1990, 177, 85–94. [Google Scholar] [CrossRef]
- He, Y.Z.; Wang, Y.M.; Yin, T.Y.; Fiallo-Olive, E.; Liu, Y.Q.; Hanley-Bowdoin, L.; Wang, X.W. A plant DNA virus replicates in the salivary glands of its insect vector via recruitment of host DNA synthesis machinery. Proc. Natl. Acad. Sci. USA 2020, 117, 16928–16937. [Google Scholar] [CrossRef]
- Wu, H.; Liu, M.; Kang, B.; Liu, L.; Hong, N.; Peng, B.; Gu, Q. AC5 protein encoded by squash leaf curl China virus is an RNA silencing suppressor and a virulence determinant. Front. Microbiol. 2022, 13, 980147. [Google Scholar] [CrossRef]
- Venkataravanappa, V.; Reddy, C.N.L.; Nandan, M.; Hiremath, S.; Ashwathappa, K.V.; Shankarappa, K.S.; Kumar, H.D.V.; Reddy, M.K. Transmission, characterization and occurrence of recombination in Indian strain of squash leaf curl China virus associated with yellow mosaic and leaf curl disease of Summer squash. 3 Biotech 2021, 11, 265. [Google Scholar] [CrossRef]
- Ito, T.; Ogawa, T.; Samretwanich, K.; Sharma, P.; Ikegami, M. Yellow leaf curl disease of pumpkin in thailand is associated with Squash leaf curl china virus. Plant Pathol. 2010, 57, 766. [Google Scholar] [CrossRef]
- Muniyappa, V.; Maruthi, M.N.; Babitha, C.R.; Colvin, J.; Briddon, R.W.; Rangaswamy, K.T. Characterisation of pumpkin yellow vein mosaic virus from india. Ann. Appl. Biol. 2003, 142, 323–331. [Google Scholar] [CrossRef]
- Riyaz, S.U.M.; Deepan, S.; Dharanivasan, G.; Jesse, M.I.; Muthuramalingam, R.; Kathiravan, K. First report on a variant of Squash leaf curl China virus (SLCCNV) infecting Benincasa hispida in India. New Dis. Rep. 2013, 28, 20. [Google Scholar] [CrossRef]
- Wu, H.; Li, M.; Hong, N.; Peng, B.; Gu, Q. Molecular and biological characterization of melon-infecting Squash leaf curl China virus in China. J. Integr. Agric. 2020, 19, 570–577. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhang, H.; Tian, W.; Fan, L.; Du, M.; Yuan, G.; Wang, D.; Wen, C.; Xu, X. First report of squash leaf curl China virus infecting tomato in China. Plant Dis. 2022, 106, 2539. [Google Scholar] [CrossRef]
- Zhao, L.; Zhong, J.; Shi, Z.; Li, T.; Ding, M.; Zhang, Z. Characterization of the genome organization of two begomoviruses mixedly infecting wild eggplant Solanum torvum. J. Plant Prot. 2020, 47, 355–364. [Google Scholar] [CrossRef]
- Du, J.; Zhang, L.; Cui, L.; Ma, Z.; Wang, C.; Wang, D. Molecular characteristic of squash leaf curl china virus (SLCCNV) infecting Bolbostemma paniculatum in HeBei Province. Acta Phytopathol. Sin. 2023, 53, 539–545. [Google Scholar] [CrossRef]
- Subiastuti, A.S.; Hartono, S.; Daryono, B.S. Detection and identification of Begomovirus infecting Cucurbitaceae and Solanaceae in Yogyakarta, Indonesia. Biodiversitas J. Biol. Divers. 2019, 20, 738–744. [Google Scholar] [CrossRef]
- Guzman, P.; Sudarshana, M.R.; Seo, Y.S.; Rojas, M.R.; Natwick, E.; Turini, T.; Mayberry, K.; Gilbertson, R.L. A new bipartite Geminivirus (Begomovirus) causing leaf curl and crumpling in cucurbits in the imperial valley of California. Plant Dis. 2000, 84, 488. [Google Scholar] [CrossRef]
- Al-Musa, A.; Anfoka, G.; Al-Abdulat, A.; Misbeh, S.; Ahmed, F.H.; Otri, I. Watermelon chlorotic stunt virus (WmCSV): A serious disease threatening watermelon production in Jordan. Virus Genes 2011, 43, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Romay, G.; Chirinos, D.; Geraud-Pouey, F.; Desbiez, C. Association of an atypical alphasatellite with a bipartite New World begomovirus. Arch. Virol. 2010, 155, 1843–1847. [Google Scholar] [CrossRef]
- Abudy, A.; Sufrin-Ringwald, T.; Dayan-Glick, C.; Guenoune-Gelbart, D.; Livneh, O.; Zaccai, M.; Lapidot, M. Watermelon chlorotic stunt and Squash leaf curl begomoviruses—New threats to cucurbit crops in the Middle East. Isr. J. Plant Sci. 2010, 58, 33–42. [Google Scholar] [CrossRef]
- Shahid, M.S.; Al-Sadi, A.M.; Briddon, R.W. First report of Chilli leaf curl virus and tomato leaf curl betasatellite infecting watermelon (Citrullus lanatus) in Oman. Plant Dis. 2017, 101, 1063. [Google Scholar] [CrossRef]
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.; Fiallo-Olivé, E.; Briddon, R.W.; Hernández-Zepeda, C.; Idris, A.; et al. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Saritha, R.K.; Bag, T.K.; Loganathan, M.; Rai, A.B.; Rai, M. First report of Squash leaf curl china virus causing mosaic symptoms on summer squash (Cucurbita pepo) grown in Varanasi district of India. Arch. Phytopathol. Plant 2011, 44, 179–185. [Google Scholar] [CrossRef]
- Riyaz, S.U.M.; Deepan, S.; Jesse, M.I.; Dharanivasan, G.; Kathiravan, K. New record of bipartite Squash leaf curl China virus (SLCCNV) and Croton yellow vein mosaic beta satellite associated with yellow vein disease of ash gourd in India. New Dis. Rep. 2015, 31, 3. [Google Scholar] [CrossRef]
- Lu, S.; Li, J.; Wang, X.; Song, D.; Bai, R.; Shi, Y.; Gu, Q.; Kuo, Y.W.; Falk, B.W.; Yan, F. A semipersistent plant virus differentially manipulates feeding behaviors of different sexes and biotypes of its whitefly vector. Viruses 2017, 9, 4. [Google Scholar] [CrossRef]
- Celix, A.; Lopez-Sese, A.; Almarza, N.; Gomez-Guillamon, M.L.; Rodriguez-Cerezo, E. Characterization of cucurbit yellow stunting disorder virus, a Bemisia tabaci-transmitted closterovirus. Phytopathology 1996, 86, 1370–1376. [Google Scholar]
- Tahir, M.; Haider, M.S.; Briddon, R.W. First report of Squash leaf curl china virus in Pakistan. Australas. Plant Dis. 2010, 5, 21–24. [Google Scholar] [CrossRef]
- Kon, T.; Dolores, L.M.; Bajet, N.B.; Hase, S.; Takahashi, H.; Ikegami, M. Molecular characterization of a strain of Squash leaf curl china virus from the Philippines. J. Phytopathol. 2003, 151, 535–539. [Google Scholar] [CrossRef]
- Maina, S.; Edwards, O.R.; de Almeida, L.; Ximenes, A.; Jones, R.A.C. First complete Squash leaf curl China virus genomic segment DNA-A sequence from East Timor. Genome Announc. 2017, 5, e00483-17. [Google Scholar] [CrossRef] [PubMed]
- Revill, P.A.; Ha, C.V.; Porchun, S.C.; Vu, M.T.; Dale, J.L. The complete nucleotide sequence of two distinct geminiviruses infecting cucurbits in Vietnam. Arch. Virol. 2003, 148, 1523–1541. [Google Scholar] [CrossRef]
- Feng, R.; Du, M.; Chen, C.; Ma, Z.; Zhou, X.; Yang, X. Identification and complete genome sequence analysis of squash leaf curl China virus infecting squash in Shaanxi. Plant Prot. 2023, 49, 121–128. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Stanley, J. Revising the way we conceive and name viruses below the species level: A review of geminivirus taxonomy calls for new standardized isolate descriptors. Arch. Virol. 2005, 150, 2151–2179. [Google Scholar] [CrossRef] [PubMed]
- Sawangjit, S. The complete nucleotide sequence of Squash leaf curl China virus-[Wax gourd] and its phylogenetic relationship to other geminiviruses. ScienceAsia 2009, 35, 131–136. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Liu, L.; Peng, B.; Zhang, Z.; Wu, Y.; Miras, M.; Aranda, M.A.; Gu, Q. Exploring different mutations at a single amino acid position of cucumber green mottle mosaic virus replicase to attain stable symptom attenuation. Phytopathology 2017, 107, 1080–1086. [Google Scholar] [CrossRef]


| 1. Isolate | 2. Nucleotide Sequence Identity/% | 3. Amino Acid Sequence Identity/% | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA-A (GenBank Accession) | DNA-B (GenBank Accession) | AV1 | AV2 | AC1 | AC2 | AC3 | AC4 | AC5 | BC1 | BV1 | |
| BLDG | 96.7 (MW389927.1) | 97.9 (MW389928.1) | 99.2 | 95.6 | 96.9 | 92.5 | 94.8 | - | 92.4 | 82.7 | 98.5 |
| FSBG | 97.9 (MW389923.1) | 93.9 (MW389924.1) | 97.6 | 97.8 | 98.0 | 94.0 | 94.1 | - | 95.9 | 88.5 | 91.7 |
| GDBL | 96.6 (MW389921.1) | 98.1 (MW389922.1) | 98.8 | 88.0 | 91.9 | 66.4 | 94.1 | - | 91.2 | 95.3 | 99.6 |
| GDXW | 98.3 (MW389919.1) | - | 99.6 | 91.3 | 98.3 | 94.7 | 94.1 | - | 98.8 | - | - |
| GDHY | 98.2 (MW389917.1) | 98.3 (MW389918.1) | 99.2 | 97.8 | 98.0 | 94.0 | 94.1 | - | 98.2 | 79.3 | 98.8 |
| Guangxi2017 | 94.8 (MG525551.1) | 86.3 (MG525552.1) | 98.4 | - | 94.7 | 88.8 | 92.6 | - | 87.7 | 94.3 | 90.6 |
| HA3 | 99.2 (KF184993.1) | - | 98.8 | 99.2 | 98.8 | 97.7 | 98.5 | - | 97.6 | - | - |
| Hn | 98.1 (MF062251.1) | 93.1 (MF062252.1) | 99.6 | 97.8 | 98.0 | 94.7 | 94.1 | 91.3 | 98.2 | 98.2 | 95.1 |
| HN | 98.1 (HM566112.1) | 94.7 (HM566113.1) | 99.6 | 99.2 | 97.2 | 92.5 | 93.3 | - | 99.4 | 98.9 | 94.7 |
| SDSG | 98.3 (MZ682117.1) | 97.3 (OK236348.1) | 99.6 | 97.8 | 98.0 | 94.7 | 94.1 | 96.5 | 98.8 | 99.2 | 96.6 |
| SLCCNV-SDSGC | 98.2 (OM258181.1) | 97.4 (OM258182.1) | 99.6 | 97.8 | 98.3 | 93.2 | 94.1 | 96.5 | 98.8 | 99.6 | 97.0 |
| SLCCNV-SDZBZ | 98.0 (OM258179.1) | 97.6 (OM258180.1) | 99.6 | 97.1 | 98.0 | 93.2 | 92.6 | 94.8 | 98.8 | 99.6 | 97.7 |
| SX01 | 98.2 (OM100574.1) | 97.3 (OM100575.1) | 99.6 | 97.1 | 98.3 | 94.0 | 94.1 | 96.5 | 97.6 | 99.6 | 97.3 |
| YN5947 | 94.4 (MK626660.1) | 89.5 (MK626666.1) | 80.4 | - | 72.6 | 89.5 | 92.6 | 89.6 | 71.4 | 95.3 | 89.1 |
| J1 | 88.8 (MN594504.1) | 81.5 (MN594505.1) | 93.3 | 55.7 | 93.3 | 76.1 | 81.6 | 81.0 | - | 92.9 | 86.1 |
| KP1 | 91.3 (KF188433.1) | 81.6 (KJ004521.1) | 95.3 | 66.6 | 94.4 | 83.5 | 88.9 | 82.7 | - | 90.4 | 83.2 |
| SLCCNV-[PumIARI] | 89.6 (JN587811.1) | 84.3 (JN624306.1) | 93.7 | 66.6 | 91.9 | 80.5 | 86.2 | - | 68.0 | 97.1 | 87.6 |
| SLCCNV-ZUB1 | 91.4 (OR208615.1) | 82.9 (OR208614.1) | 96.0 | 66.6 | 93.6 | 81.3 | 88.9 | 79.3 | 70.3 | 97.1 | 88.0 |
| Cs1 | 94.7 (MT682356.1) | 89.8 (MT682357.1) | 96.8 | 88.6 | 95.2 | 88.8 | 92.6 | - | 87.2 | 95.7 | 90.2 |
| Hanoi | 95.6 (KC857509.1) | 91.9 (KC857510.1) | 98.4 | 97.1 | 97.2 | 90.2 | 93.3 | - | 91.8 | 98.2 | 92.9 |
| KN44 | 95.8 (MK978176.1) | 89.5 (MK978177.1) | 98.4 | 97.1 | 95.8 | 91.0 | 91.9 | - | - | 95.7 | 88.4 |
| WMK | 93.5 (OQ123829.1) | - | 98.4 | 81.3 | 95.8 | 82.8 | 89.7 | 93.1 | - | - | - |
| SLCCV-[BASq-17] | 93.3 (LC511776.1) | 84.6 (LC511781.1) | 96.8 | 75.3 | 93.3 | 86.5 | 90.4 | 84.4 | - | 95.3 | 89.5 |
| Isolate | Crop | Repeat 1 | Repeat 2 | Repeat 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Incidence Rate a | Infection Rate b | Incidence Rate | Infection Rate | Incidence Rate | Infection Rate | |||||
| 14 dpi | 21 dpi | PCR | 14 dpi | 21 dpi | PCR | 14 dpi | 21 dpi | PCR | ||
| SLCCNV-HN | Melon | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) |
| Watermelon | 1/8 (12.5%) | 3/8 (37.5%) | 8/8 (100%) | 3/6 (50%) | 3/6 (50%) | 6/6 (100%) | 1/6 (16.7%) | 1/6 (16.7%) | 6/6 (100%) | |
| WM1 | Melon | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) |
| Watermelon | 8/8 (100%) | 8/8 (100%) | 8/8 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | 6/6 (100%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Wang, Y.; Geng, Y.; Yu, B.; Yan, L.; Hao, F.; Wu, H.; Wang, P.; Gu, Q.; Kang, B. Molecular Characterization and Pathogenicity of Watermelon Isolates of Begomovirus cucurbitachinaense. Int. J. Mol. Sci. 2025, 26, 4289. https://doi.org/10.3390/ijms26094289
Liu L, Wang Y, Geng Y, Yu B, Yan L, Hao F, Wu H, Wang P, Gu Q, Kang B. Molecular Characterization and Pathogenicity of Watermelon Isolates of Begomovirus cucurbitachinaense. International Journal of Molecular Sciences. 2025; 26(9):4289. https://doi.org/10.3390/ijms26094289
Chicago/Turabian StyleLiu, Liming, Yanhui Wang, Yanfei Geng, Bo Yu, Leiyan Yan, Fangmin Hao, Huijie Wu, Pingyong Wang, Qinsheng Gu, and Baoshan Kang. 2025. "Molecular Characterization and Pathogenicity of Watermelon Isolates of Begomovirus cucurbitachinaense" International Journal of Molecular Sciences 26, no. 9: 4289. https://doi.org/10.3390/ijms26094289
APA StyleLiu, L., Wang, Y., Geng, Y., Yu, B., Yan, L., Hao, F., Wu, H., Wang, P., Gu, Q., & Kang, B. (2025). Molecular Characterization and Pathogenicity of Watermelon Isolates of Begomovirus cucurbitachinaense. International Journal of Molecular Sciences, 26(9), 4289. https://doi.org/10.3390/ijms26094289





