Abstract
MADS-box transcription factors play a crucial role in plant growth and development. Although previous genome-wide analyses have investigated the MADS-box family in cucumber, this study provides the first comprehensive reannotation of the MADS-box gene family in Cucumis sativus using updated Cucurbitaceae genome data, offering novel insights into the gene family’s evolution and functional diversity. The results show that a total of 48 CsMADS-box genes were identified in the V3 version of cucumber, while 3 of the 43 genes identified in the V1 version were duplicated. The V1 version actually has only 40 genes. Additionally, we analyzed the variability in protein sequences and found that the amino acid sequences of 14 genes showed no differences between the two versions of the database, while the amino acid sequences of 29 genes exhibited significant differences. The further analysis of conserved motifs revealed that although the amino acid lengths of 15 genes had changed, their conserved motifs remained unchanged; however, the conserved motifs of 12 genes had altered. Furthermore we found that motif1 and motif2 were present in most proteins, indicating that they are highly conserved. Gene structure analysis revealed that most type I (Mα, Mβ) MADS-box genes lack introns, whereas type II (MIKC) genes exhibit a similar structure with a higher number of introns. Chromosomal localization analysis indicated that CsMADS-box genes are unevenly distributed across the seven chromosomes of cucumber. Promoter region analysis showed that the promoter regions of CsMADS-box genes contain response elements related to plant growth and development, suggesting that CsMADS-box genes may be extensively involved in plant growth and development. Different CsMADS-box genes exhibit specific high expression in roots, stems, leaves, tendrils, male flowers, female flowers, and ovaries, suggesting that these genes play crucial roles in the growth, development, reproduction and morphogenesis of cucumber. Moreover, 26, 18, 8, and 10 CsMADS-box genes were differentially expressed under high temperature, NaCl and/or silicon, downy mildew, and powdery mildew treatments, respectively. Interestingly, CsMADS07 and CsMADS16 responded to all tested stress conditions. These findings provide a reference and basis for further investigation into the function and mechanisms of the MADS-box genes for resistance breeding in cucumber.
1. Introduction
The MADS-box gene family is one of the most widely studied transcription factor genes in plants. It is characterized by a highly conserved DNA-binding MADS domain at the N-terminus, containing 56–60 amino acids [1,2]. Plant MADS-box genes can be divided into two categories according to their evolutionary lineages: type I and type II. Type II genes are also known as MIKC genes because they share a common structure of four domains. In addition to the MADS (M) domain, MIKC-type genes also contain three other conserved domains: I, K and C domains [3]. The I domain is responsible for the specificity of DNA binding dimer formation, the K domain is involved in protein-protein interaction, and the C domain is involved in transcriptional activation [4,5,6]. Based on its structural characteristics, MIKC can be further divided into two types: MIKC* and MIKCC [7]. Compared with MIKCC-type proteins, MIKC* -type proteins tend to have longer I domains and fewer conserved K domains. MIKCC-type MADS-box genes are the most representative class of MADS-box genes, which play important and diverse roles in plant growth and development [3,8]. MIKCC-type MADS-box genes can be divided into 12 subclasses according to their phylogenetic relationships in Arabidopsis thaliana [9]. Compared with the type II lineage group, the type I genes have a simpler gene structure and lacks the K domain. They are thought to share a common ancestor with type I genes from animals and fungi, but their functions are generally not well understood [3,10]. Type I MADS-box genes can be further subdivided into Mα, Mβ, Mγ and Mδ in plants [11,12].
MADS-box genes play an important role in plant development. The most important role is as a major component of the well-known ABCDE model, which performs its function in regulating floral organs. Different combinations of A, B, C, D and E functions of MADS-box genes determine different floral organ characteristics: (sepals (A + E), petals (A + B + E), stamens (B + C + E), carpels (C + E) and ovules (D + E)). Class A genes mainly control the development of calyx, corolla and floral organs. B-type genes control the development of corolla and stamens, and also affect the development of calyx in a few plants. Class C genes control the development of three-wheeled floral organs of stamens, pistils and ovules, while only two-wheeled structures of stamens and pistils are controlled in some plants. Class D genes control the development of ovules, and class E genes are involved in the formation of floral organs during each round of flower development, and form a tetramer model complex with class A, B, and C genes. In Arabidopsis, the corresponding functional genes are class A, APETALA 1 (AP 1); class B, PISTILATA (PI) and AP 3; class C, AGAMOUS (AG); class D, SEEDSTICK/AGAMOUS-LIKE 11(STK/AGL 11); and E, SEPALLATA (SEP 1, SEP 2, SEP 3 and SEP 4) [3,13,14,15].
In addition to its important role in determining floral organ traits, MADS-box genes have also been found to be involved in the regulation of flowering time and flower initiation, such as SOC1, FLC, AGL and SVP [16,17,18,19,20,21,22,23]; as well as in fruit formation (FUL) [24,25] and root development (AGL12 and AGL17) [26,27].
MADS-box genes have been demonstrated to participate in the nutrient growth processes and various stress responses in different plants such as Arabidopsis [28], rice [29], wheat [30,31], and cabbage [32]. Therefore, the MADS-box protein family is an important transcription factor family, influencing almost the entire process of plant growth and development. Model plant species of the MADS-box family have been extensively studied, including rice (Oryza sativa), cabbage (Brassica rapa), poplar (Populus trichocarpa), wheat (Triticum aestivum L.), banana (Musa acuminata), and others.
Cucumber (Cucumis sativus L.) is an economically and nutritionally important vegetable crop cultivated worldwide. It is loved by people because of its good taste, high nutritional value and economic value. Although MADS-box genes exist as a superfamily, little is known about MADS-box genes in cucumber. Therefore, it is of great significance to identify the MADS-box gene family of cucumber. In previous studies, 43 MADS-box genes were identified in the V1 genome of cucumber [33]. With the update of the Cucurbitaceae genome database, the cucumber genome version has been updated to the V3 version. Therefore, it is necessary to re-identify and modify the members of the MADS-box gene family in cucumber. This work is helpful in the context of a more comprehensive study of cucumber MADS-box gene family members and provides a basis for further functional analysis to elucidate their roles in development.
2. Results
2.1. Characterization of MADS-Box from Chinese Long 9930 (V3 Version)
In previous studies, we identified 43 MADS-box genes in the cucumber V1 genome [33]. With the update of the cucumber family genome database, the cucumber genome version has been updated to V3. Therefore, we re-identified and revised the MADS-box gene family members in cucumber. In the cucumber V3 version, we identified a total of 48 MADS-box genes, and found that Csa014213 and Csa025232 aligned to the gene CsaV3_6G006010 in the cucumber V3 version database; similarly, Csa014249 and Csa026408 aligned to CsaV3_1G009750; while Csa014140 and Csa025231 aligned to CsaV3_6G006020. Csa014213 and Csa025232 have the same CDS sequence. Csa014140 and Csa025231 also have the same CDS sequence. The CDS sequence similarity between Csa014140 and Csa025231 is as high as 96%. These results indicate that there are some errors in the MADS-box genes identified in the V1 version. Therefore, there are actually 40 MADS-box genes in the V1 version, 48 in the cucumber V3 version, and the 48 MADS-box genes are divided into 14 subfamilies (Figure 1). The eight newly added genes are CsaV3_6G052910, CsaV3_6G051220, CsaV3_5G040310, CsaV3_5G040370, CsaV3_6G051590, CsaV3_3G009400, CsaV3_3G016620 and CsaV3_UNG063480 (Table 1).
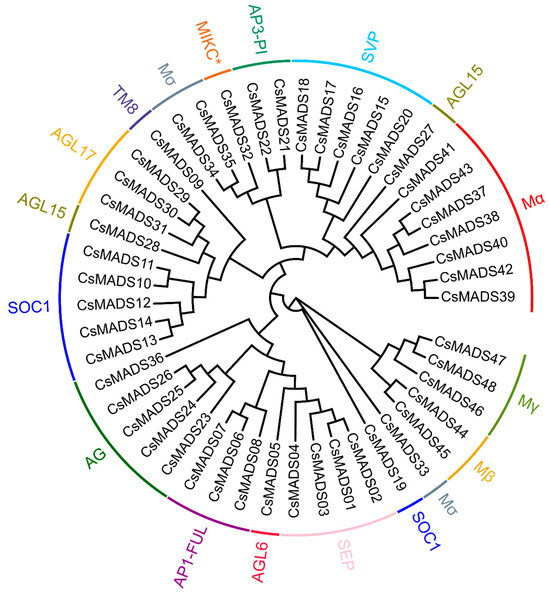
Figure 1.
The phylogenetic tree of the MADS-box proteins from cucumber. These proteins were phylogenetically analyzed using MEGA7 software with 1000 bootstrap tests. Different colors represent different subgroups of the MADS-box family (AG (AGAMOUS) and SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO 1)) “*” represents a type of MICK subfamily used to emphasize specific changes and specific amino acid sequence variations.

Table 1.
Comparison of the number of MADS-box families in the Cucumber V1 and V3 versions.
2.2. Analysis of the Differences of Amino Acid Sequences of MADS-Box Family Members Between V1 and V3 Versions
Due to the difference in the number of members of the MADS-box gene family in the V1 and V3 versions, we analyzed the differences in the protein sequences of the 43 genes found in the V1 version compared to the V3 versions. The results show that there were no differences in the amino acid sequences of 14 genes in the two versions of the database. They were Csa004117, Csa008448, Csa014140, Csa025231, Csa012879, Csa012099, Csa017355, Csa000939, Csa021069, Csa017317, Csa020265, Csa017909, Csa002566 and Csa001552. The amino acid sequences of 29 genes were significantly different between the two versions of the data (Table 2). See Dataset S1 for amino acid sequence information.

Table 2.
Comparison of the amino acid number of MADS-box in the cucumber V1 and V3 versions.
2.3. Analysis of Protein Motif Difference of MADS-Box
In order to explore whether differences in amino acid sequence lead to the changes in protein conserved motifs, we compared and analyzed the protein conserved motifs of MADS-box family members in the V1 and V3 versions. The results show that the amino acid length of 15 family members changed, but their protein motifs did not change. They were CsMADS05, CsMADS06, CsMADS07, CsMADS09, CsMADS10, CsMADS16, CsMADS21, CsMADS22, CsMADS28, CsMADS32, CsMADS33, CsMADS39, CsMADS44, CsMADS45 and CsMADS48, respectively. The protein motifs of 12 family members changed, including CsMADS03, CsMADS04, CsMADS08, CsMADS11, CsMADS17, CsMADS18, CsMADS23, CsMADS27, CsMADS29, CsMADS30, CsMADS31 and, CsMADS35. Similarly, we analyzed the conserved motifs of the eight newly identified proteins, and found that CsMADS42 contains only two motifs (motif1, motif2), and CsMADS43 contains three motifs (motif1, motif2, motif6) (Figure 2). In addition, the conserved motifs of the remaining six proteins numbered five to six. Moreover, we found that motif1 and motif2 are present in most proteins, indicating that motif1 and motif2 are highly conserved (Figure 2). The amino acid sequence for each motif is presented in Figure S1.
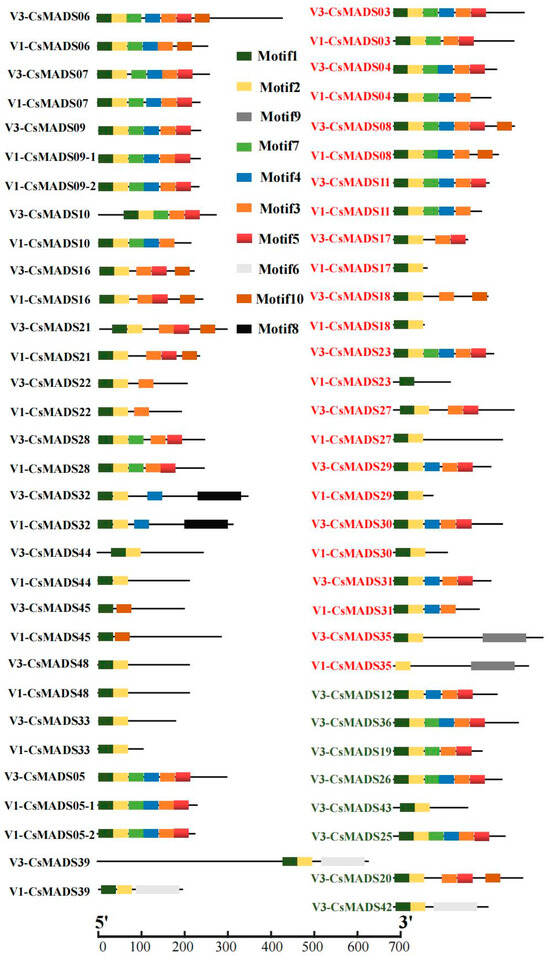
Figure 2.
Analysis of differences in protein motifs of MADS-box in versions V1 and V3 (comparison of protein motifs with amino acid sequence differences between the two genomes).
2.4. CsMADS Multiple Sequence Alignment
MADS-box genes have a highly conserved DNA binding domain, namely MADS box. By analyzing the amino acid sequence, we found that CsMADS contains a highly conserved MADS box (Figure 3).

Figure 3.
The multiple protein sequence alignment of the domains of MADS from cucumber.Conserved sequences are highlighted in black and grey shading; the black shading represents completely conserved sequences, while the grey shading represents incompletely conserved sequences. Black region: similarity over 80%. Grey region: similarity over 60–80%. “*” from left to right represents 430, 450, 470.
2.5. Phylogenetic Relationship and Gene Structure Analysis of MADS-Box Genes
The genome annotation information of the V1 cannot be obtained. Therefore, it is impossible to analyze the differences in the gene structures of the MADS-box family in the V1 and V3 versions, so only the structure of the CsMADS in the V3 version is analyzed. The results show that the numbers of introns in CsMADS varied greatly, ranging from 0 to 10. CsMADS38, CsMADS40, CsMADS41, CsMADS44, and CsMADS46 have no introns, and these all belong to type I genes in the Mα and Mβ subfamilies. Type II genes, also known as MIKC genes, contain a large number of introns (CsMADS01-CsMADS32) and have similar structures (Figure 4).
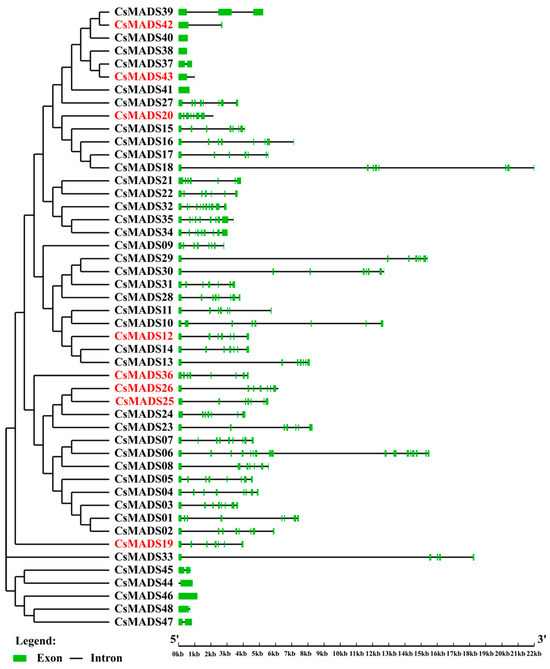
Figure 4.
Phylogenetic tree and gene structure of MADS family members in C. sativus. The phylogenetic tree was constructed using the neighbor-joining (NJ) method with 1000 bootstrap replicates, based on the alignment of the identified MADS proteins in C. sativus. The gene structures of the 48 MADS genes identified in C. sativus were generated utilizing the Gene Structure Display Server v.2.0. In the structures, red represents the new genes in version of V3, the green box represents the exon, and the black line represents the intron.
2.6. Chromosome Distribution Analysis of CsMADS-Box Genes
The chromosome distribution analysis of the cucumber CsMADS gene shows that the distribution of the CsMADS gene on seven chromosomes was uneven (Figure 5). Specifically, there are nine CsMADS genes on chromosome 1 and chromosome 3, three CsMADS genes on chromosome 2, eight CsMADS genes on chromosome 4, and five CsMADS genes on chromosome 5. The number of CsMADS genes on chromosome 6 is the largest, with 12 CsMADS genes, and the number of CsMADS genes on chromosome 7 is the smallest, with only one CsMADS gene (Figure 5).
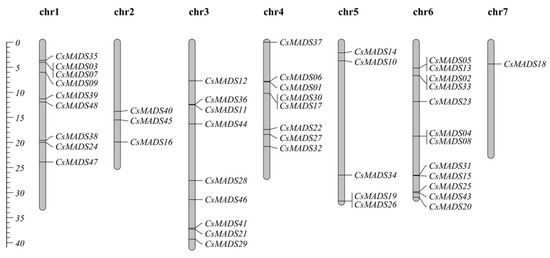
Figure 5.
Chromosomal location of CsMADS genes.
2.7. Analysis of Cis-Acting Elements in Gene Promoter Region
The analysis of the 1500 bp upstream sequence of the CsMADS gene revealed a variety of cis-acting elements (Figure 6). For example, there were hormone response elements (ABRE, CGTCA-motif, GARE-motif, TCA-element, TATC~box, TCA-element, TGACG motif, etc.), stress corresponding components (MBS, MBSI, etc.), and other important response elements in plant growth and development (ARE, CAT-box, LTR, GC-motif, O2-site, RE-element, TC~rich). This shows that the MADS-box family is widely involved in the growth and development of plants (Figure 6).

Figure 6.
Predicted cis-elements in promoter regions of CsMADS genes. The promoter region was defined as a 1.5 kb sequence upstream of the translation initiation codon of the MADS gene. Identification of cis-acting elements using the online tool Plant CARE. Different types of cis-acting elements are represented by closed boxes of different colors.
2.8. Expression Patterns of CsMADS in Different Tissues
This part aims to study the role of MADS genes in cucumber development by analyzing public RNA-seq data of different tissues. The results show that, compared with other tissues, the expression of most CsMADS genes in cucumber stems was relatively low (Figure 7). In contrast, CsMADS10, CsMADS12, CsMADS16 and CsMADS31 were highly expressed in tendrils. CsMADS03, CsMADS22, CsMADS32, CsMADS34, CsMADS36 and CsMADS40 were highly expressed in female flowers, indicating their potential roles in flower differentiation and development. CsMADS14, CsMADS19, CsMADS29, CsMADS30, CsMADS37, CsMADS38, CsMADS41, CsMADS44, CsMADS45 and CsMADS46 were mainly expressed in roots, indicating that they may be involved in regulating ion transport in the underground part of cucumber (Figure 7).
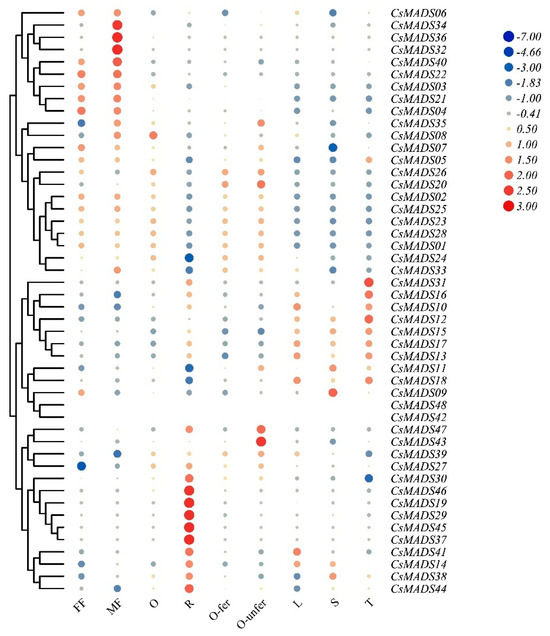
Figure 7.
Temporal–spatial expression of cucumber MADS genes. Heatmap displaying the expression profiles of CsMADS genes in nine different cucumber tissues. The RNA-seq datasets with accession number PRJNA80169 were obtained from the Cucurbit Genomics Data website. The color scale represents Log2(FPKM) values, where blue and red indicate low and high expression levels, respectively. The FPKM values of CsMADS genes can be found in Table S1. R, root; S, stem; L, leaf; FF, female flower; MF, male flower; O, unexpanded ovary; O-fer, expanded fertilized ovary; O-unfer, expanded unfertilized ovary; T, tendril.
2.9. Expression Profiles of CsMADS Genes Under Abiotic and Biotic Stresses
Although MADS genes in many species have been identified to be involved in a variety of stress responses, they have not been studied in cucumber. In this study, based on the public transcriptome information, the expression patterns of CsMADS genes under different stress conditions such as salt, heat, downy mildew (DM, Pseudoperonospora cubensis) and powdery mildew (PM, Podosphaera fusca) were analyzed to explore the role of CsMADS genes under different stress conditions.
Firstly, the role of the CsMADS gene under salt stress was analyzed. The results are presented in the form of heat maps (Figure 8). Under NaCl treatment, most genes were up-regulated; only a small number of genes were down-regulated, including CsMADS13, CsMADS25 and CsMADS40. Silicon (Si) is considered to be an essential element for plant growth and development, and plays a significant role in promoting plant growth and development and enhancing stress resistance. We observed that gene expression undergoes significant changes exclusively under the Si treatment condition. The expressions of CsMADS06, CsMADS09, CsMADS16, CsMADS19, CsMADS21, CsMADS37 and CsMADS40 were up-regulated (Figure 8). In contrast, the expression of CsMADS12, CsMADS13, CsMADS28 and CsMADS30 were down-regulated. It is worth noting that the expressions of CsMADS06, CsMADS07, CsMADS09, CsMADS16, CsMADS29 and CsMADS40 were up-regulated under the treatment of NaCl and silicon(Si), suggesting that they play an important role in the process of plant resistance to salt stress (Figure 8).
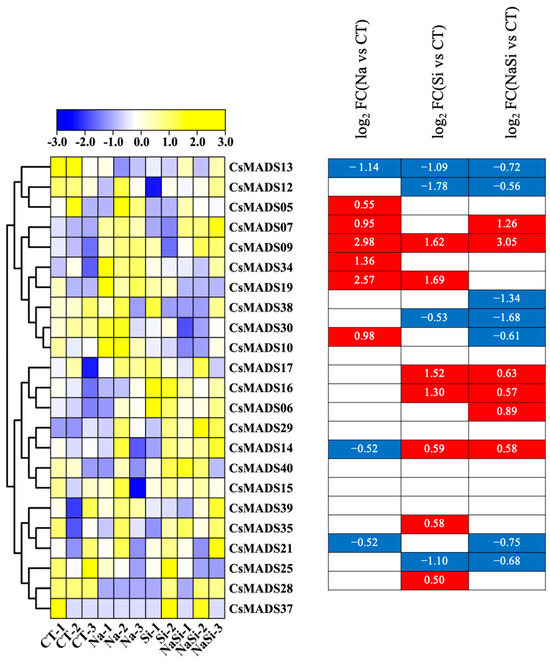
Figure 8.
Expression profiles of CsMADS genes in response to salt stress treatments: A range of −3.00 to 3.00 was artificially set with the color scale limits according to the normalized values. The color scale shows increasing expression levels from blue to yeollow. Blue region: Down-regulated Red region: Up-regulated. The FPKM value of CsMADS genes under salt are listed in Table S2.
We also analyzed the responses of CsMADS genes to heat stress. The expressions of CsMADS04, CsMADS06, CsMADS16, CsMADS29, CsMADS30, CsMADS35, CsMADS39, CsMADS41, CsMADS42 and CsMADS43 were significantly up-regulated in both three-hour and six-hour heat and high temperature treatments. The expressions of CsMADS08, CsMADS10, CsMADS12, CsMADS15, CsMADS18, CsMADS19, CsMADS21, CsMADS28, CsMADS33 and CsMADS46 were significantly down-regulated. In addition, the results show that the expression of CsMADS07 was significantly down-regulated after 3 h of high-temperature treatment, but significantly up-regulated after 6 h of high-temperature treatment (Figure 9).
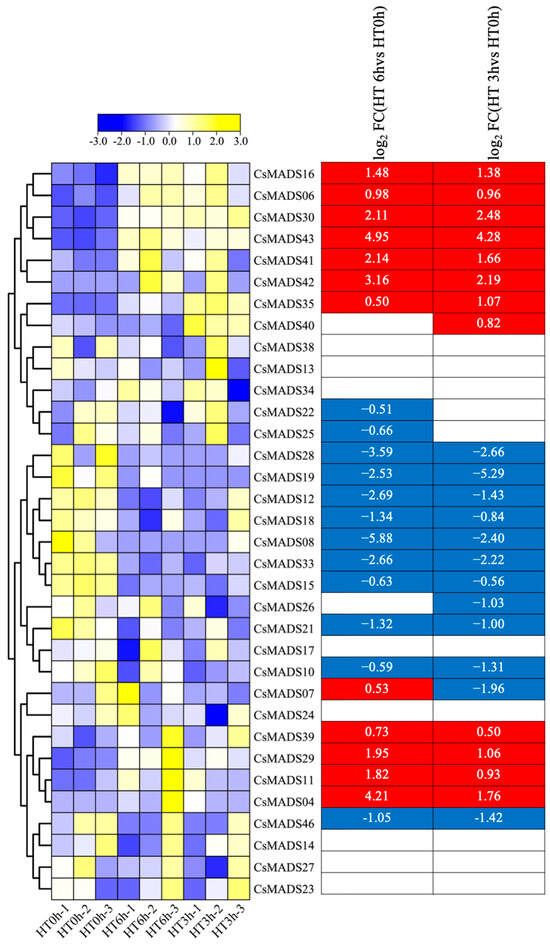
Figure 9.
Expression profiles of CsMADS genes in response to heat stress treatments: A range of −3.00 to 3.00 was artificially set with the color scale limits according to the normalized values. The color scale shows increasing expression levels from blue to yeollow. Blue region: Down-regulated Red region: Up-regulated.The FPKM value of CsMADS genes under heat stress are listed in Table S3.
In order to explore the role of CsMADS in biological stress resistance, we used the RNA-Seq database to analyze the expression of CsMADS. The results show that CsMADS17, CsMADS39 and CsMADS48 were significantly up-regulated in susceptible and resistant cucumber lines after inoculation with powdery mildew (PM). On the contrary, the expression of CsMADS09 was down-regulated (Figure 10A). In addition, we found that the expression of CsMADS25 was significantly up-regulated in susceptible cucumber lines, indicating that the gene may play an important role in the mechanism of cucumber resistance to powdery mildew.
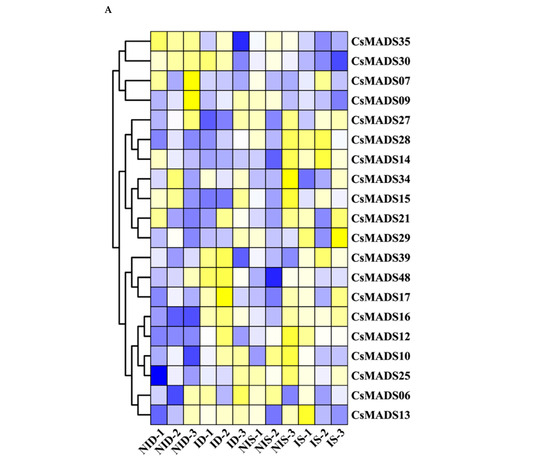
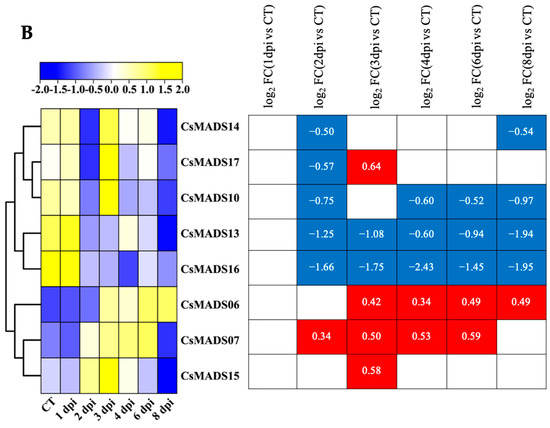
Figure 10.
Expression analysis of CsMADS under biotic stresses: The transcriptional levels of CsMADS genes after infection with powdery mildew (PM) for 48 h (A) and with downy mildew (DM) for 1–8 days post-inoculation (B) are shown on the heatmaps. A range of −3.00 to 3.00 was artificially set with the color scale limits according to the normalized values. The color scale shows increasing expression levels from blue to red. ID, PM-inoculated susceptible cucumber line D8 leaves; NID, non-inoculated D8 leaves; IS, PM-inoculated resistant cucumber line SSL508–28 leaves; NIS, non-inoculated SSL508–28 leaves; CT, without inoculation; DPI, days post inoculation; FC, fold-change. Blue region: Down-regulated Red region: Up-regulated. The FPKM value of CsMADS genes under powdery mildew (PM) and downy mildew (DM) are listed in Table S4.
In the transcriptome data of cucumber seedlings inoculated with DM, only eight MADS genes were detected. CsMADS06 and CsMADS07 were up-regulated at most treatment time points, while CsMADS10, CsMADS13 and CsMADS16 were down-regulated at most treatment time points (Figure 10B).
3. Discussion
The MADS-box gene is an important transcriptional regulator in eukaryotes, which is involved in the regulation of growth and development and signal transduction processes. It has been widely identified in many species [32,33]. Although the MADS-box gene of cucumber has been previously identified, its comprehensive identification and characterization are limited due to the low quality of the genome. In addition, the function of MADS-box family in signal transduction and response to different stress conditions is also relatively insufficient. With the update of the Cucurbitaceae genome database, the cucumber genome has been upgraded. Therefore, it is necessary to re-identify and modify the members of the MADS-box gene family in cucumber to further explore its variation and potential functions, and to elucidate its role in cucumber development.
In previous studies, 43 CsMADS-box genes were identified in the cucumber V1 database [33], while 48 CsMADS-box genes were identified in the cucumber V3 database. By analyzing the 43 CsMADS-box genes in the V1 version, we found that Csa014213 and Csa025232 had the same CDS sequence, as well as Csa014249 and Csa026408. In addition, the CDS sequence similarity of Csa014140 and Csa025231 was as high as 96%, indicating that they may actually be the same gene (Table 1). These results indicate that there are some errors in previous studies, so it is necessary to re-identify and correct the members of the MADS-box gene family in cucumber.
By analyzing the differences in protein sequences and motifs, we found that more than half of the amino acid sequences were significantly different between the two versions of the data (Table 2). Specifically, there were no differences in the amino acid sequences of 14 genes, which were Csa004117, Csa008448, Csa014140, Csa025231, Csa012879, Csa012099, Csa017355, Csa000939, Csa021069, Csa017317, Csa020265, Csa017909, Csa002566 and Csa001552. In addition, the amino acid length of 15 family members changed, but their protein motifs did not change (CsMADS05, 06, 07, 09, 10, 16, 21, 22, 28, 32, 33, 39, 44, 45 and 48). Besides this, we found that motif1 and motif2 are highly conserved in most CsMADS genes (Figure 2). Type I (Mα, Mβ and Mγ) MADS-box genes usually lack introns or have only one intron, and their gene structure is very simple (Figure 4). In contrast, the gene structure of type II genes (MIKC and Mδ) seems to be more complex, including multiple exons and introns. Studies have shown that genes containing multiple introns are usually more conserved [34], so type I genes may not be as conserved as type II MADS-box genes. In addition, studies have shown that a small number of introns help genes respond quickly to various stresses and activate down-regulated genes [35]. The presence of introns may lead to alternative splicing, thereby delaying the response to stress, and type I genes may respond to stress earlier. Chromosomal localization analysis showed that CsMADS-box genes were unevenly distributed on seven chromosomes of cucumber (Figure 5).
We also identified 10 important cis-acting elements of the MADS-box family, most of which are related to plant hormones. Hormone response elements (ABRE, CGTCA-motif, GARE-motif, TCA-element, TATC-box, TCA-element, TGACG motif) were highly enriched in the promoter region of CsMADS gene, indicating that the CsMADS gene may be involved in the regulation of various hormone responses (Figure 6). Meristem response elements (ARE, CAT-box, LTR, GC-motif, O2-site, RE-element, TC-rich) are mainly identified in type II genes, suggesting that type II MADS-box genes have a function in determining meristem and floral organ identity in cucumber, which is in accordance with Callicarpa americana [36].
Understanding gene expression is essential to reveal the molecular mechanism of biological development [37]. MADS-box genes are widely thought to be associated with floral organ development and identity determination in plants. Previous studies have shown that AOAMOUS (AG) and APETALA1 (AP1) gene family members are mainly expressed in flowers, fruits and buds in species such as tomato [38], cotton [39], watermelon [40] and soybean [41]. The results of this study are consistent with previous observations. AG gene family members (CsMADS23, 24, 25, 26 and 36) and AP1 gene family members (CsMADS06, 07 and 08) are mainly expressed in cucumber flowers. Moreover, we found that 10 other MADS genes (CsMADS14, 19, 29, 30, 37, 38, 41, 44, 45 and 46) were highly expressed in roots, indicating that they may play an important role in plant growth and ion transport (Figure 7).
In addition to regulating the characteristics of floral organs and their meristems in plant flower development, MADS-box genes have also been found to be involved in a variety of stress responses [42]. For example, DgMADS114 and DgMADS115 can enhance the resistance of transgenic Arabidopsis to PEG, NaCl, ABA and high-temperature stress [43]. Under ABA treatment, the expression levels of MsMADS001 and MsMADS075 gradually increased with time, while MsMADS075 showed a trend of increasing first and then decreasing under drought treatment, which is consistent with the results of RNA-Seq and qRT-PCR analysis, indicating that they may play an important role in stress response [44]. In addition, the expression level of TaMADS19 was significantly increased after inoculation with wheat stripe rust. After inoculation with powdery mildew, the expression of TaMADS117 was significantly reduced. The expression levels of TaMADS121, 93 and 21 were significantly increased under phosphorus deficiency stress. Under high temperature stress, the expression levels of TaMADS63 and 41 were significantly reduced [45]. In this study, we found that the expression levels of 26,18,8 and 10 CsMADS-box genes showed significant change after high temperature, NaCl, silicon, downy mildew and powdery mildew treatments (Figure 8, Figure 9 and Figure 10). Only two genes, CsMADS07 and CsMADS16, showed responses to all tested stress conditions, suggesting their pivotal role in conferring cucumber’s resistance to multiple environmental stresses. These findings imply that CsMADS07 and CsMADS16 might function as key regulatory factors in the plant’s stress tolerance mechanisms, possibly by modulating pathways involved in stress signal transduction, reactive oxygen species (ROS) regulation, or hormonal responses. Given their broad response to various abiotic stresses, these genes could be valuable targets for genetic improvement programs aimed at enhancing cucumber’s resilience to climate change-induced stressors. In summary, this study highlights the significant contribution of CsMADS genes to cucumber growth and development, while providing new genetic resources for future breeding efforts focused on stress resistance in cucumber.
4. Material and Methods
4.1. Identification of MADS-Box Genes in Cucumber
The cucumber genome data were downloaded from the Cucurbit Genomics Database (http://cucurbitgenomics.org/) (accessed on 1 March 2024) and NCBI (https://www.ncbi.nlm.nih.gov/). The hidden Markov model (HMM) profile files of the MADS-box conserved domain (PF00319) were downloaded from the Pfam database (http://pfam.xfam.org/). The MADS-box genes of cucumber were identified from the genome database using HMMER 3.0 with the default parameters and a cutoff value of 0.01. All CsMADS-boxs were further examined to confirm the MADS-box conserved domain through the CDD, Pfam, and SMART online tools. All CsMADS-box genes were named according to their locations on seven cucumber chromosomes.
4.2. Gene Structure and Motif Analysis
The CDS sequences and genomic data for CsMADS-box genes retrieved from the C. sativus genome database (http://cucurbitgenomics.org/organism/20) (accessed on 8 April 2025) were visualized using the Gene Structure Display Server online tool (http://gsds.cbi.pku.edu.cn/) [46]. The conserved motifs of CsMADS-box proteins were then identified with MEME 4.9.1 (http://meme-suite.org/) [47] and visualized with WebLogo (http://weblogo.berkeley.edu/logo.cgi) (accessed on 8 April 2025) [48]. The total number of motifs (nmotifs) is 10, the minimum length of motifs (minw) is 6 amino acids, and the maximum length of motifs (maxw) is 10 amino acids.
4.3. Phylogenetic Analysis and Multiple Sequence Alignment
The protein sequences of MADS-box in cucumber were uploaded to the MEGA software (v7.0) to be aligned using ClustalW 2.x, and the phylogenetic relationships among all MADS-box proteins were examined via the neighbor-joining method with 1000 bootstrap replicates. Then, the phylogenetic trees were landscaped in Evolview (https://evolgenius.info//evolview-v2/#login, accessed on 8 April 2025).
4.4. Gene Duplication Analysis and Genome Distribution
CsMADS-box loci were extracted from the cucumber genome database (http://cucurbitgenomics.org/organism/20) and their locations on chromosomes were visualized using MapChart 3.x [49].
4.5. Analysis of Promoter Regions of CsMADS-Box Genes
The 1.5-kb sequences upstream of the initiation codons (ATG) of CsMADS-box genes were obtained from the cucurbit genomics data website (http://cucurbitgenomics.org/organism/20), and analyzed for cis-elements in the promoter region using the online tool PlantCARE [50].
4.6. Transcriptome Analysis of CsMADS-Box Genes in Cucumber
The expression patterns of the CsMADS genes were analyzed using the transcriptomic data of the roots, stems, leaves, flowers, ovaries, and tendrils of cucumber. The published RNA-Seq data (SRA046916) [51] were downloaded from the Cucurbit Genomics Database (http://cucurbitgenomics.org/). The remapped clean tags and the recalculated FPKM values were cited to analyze the expression patterns of the CsMADS. The genome-wide expression of the CsMADS genes was shown on a heatmap using TBtools v2.x [52]. The heatmap values were calculated according to the following steps: the fold change values of the FPKM value of the treatment group and the control group were calculated first, and then the logarithm based on two of the fold change values was taken.
4.7. Transcriptome Analysis of CsMADS in Response to Abiotic and Biotic Stresses
The publicly available transcriptomic data of cucumber treated with salt (GSE116265) [53], heat (GSE151055) [54], DM (SRP009350) [55], and PM (GSE81234) [56] were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) to analyze the expression patterns of CsMADS under different stresses. After aligning the gene IDs to the cucumber genome, the genome-wide expression of the CsMADS genes was shown on a heatmap using TBtools V2.056. For the transcriptome analysis of the CsMADS, a threshold of FDR (or p-value) ≤ 0.05 and an absolute value of log2 (fold-change) ≥ 1 or log2 (fold-change) ≤ −1 were used to define DEGs.
4.8. The Methodology Was Involved in the Development of the V1 and V3 Versions
Development Methodology of V1 Version: The V1 version of the cucumber genome was primarily developed using early sequencing technologies, including Sanger sequencing and early Illumina short-read sequencing. The assembly process in this version relied on lower coverage and fewer assembly strategies, which resulted in some issues such as incomplete gene lengths, inaccurate gene spacing, and assembly problems in repetitive regions. Although the V1 version provided an initial genomic framework, it had limitations in gene annotation and sequence completeness, particularly for longer genes and complex genomic regions.
Development Methodology of V3 Version: In contrast, the V3 version of the cucumber genome was developed using a more advanced combination of Illumina short-read sequencing and PacBio long-read sequencing technologies. These advancements provided higher coverage and longer read lengths, allowing for better handling of repetitive regions and structural variations within the genome. The V3 version also utilized more sophisticated gene prediction and annotation tools, incorporating extensive transcriptomic data and experimental validation to significantly improve the accuracy of gene annotations.
5. Conclusions
In this study, we re-identified and revised the members of the MADS-box gene family. In the updated version (V3), 48 CsMADS-box genes were identified—8 more than in the V1 version. In different versions, most amino acid sequences are significantly different. The further analysis of conserved motifs revealed that the conserved motifs of 12 genes were changed. In addition, we found that motif1 and motif2 are highly conserved. Gene structure analysis showed that most of the type I genes did not contain introns. The gene structure of type II is conserved, with a notably high number of introns. The CsMADS-box gene is distributed across seven chromosomes in cucumber. The promoter region of the CsMADS-box gene contains response elements and other elements related to plant growth and development. The expression pattern analysis of CsMADS-box in different tissues has shown that they were likely to be involved in the growth and morphogenesis of cucumber. Furthermore, transcriptome data under different stress conditions reveal the expression of CsMADS-box under abiotic and biotic stresses, and show that CsMADS07 and CsMADS16 responded to all four stresses. This study provides data and a theoretical reference for the potential role of CsMADS in cucumber stress resistance breeding.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms26083800/s1.
Author Contributions
Conceptualization, Z.R.; methodology, Z.W. and M.Y.; software, Z.W., X.W. and J.C.; validation, Z.W.; formal analysis, Z.W., M.Y. and J.H.; investigation, Z.W. and J.H.; resources, Z.R. and L.W.; data curation, Z.W.; writing—original draft preparation, Z.W. and Z.R.; writing—review and editing, L.W. and Z.R.; visualization, L.W. and Z.R.; supervision, L.W. and Z.R.; project administration, L.W. and Z.R.; funding acquisition, Z.W. and Z.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31972419 and 32172605), the Agricultural Variety Improvement Project of Shandong Province (2022LZGCQY001), and the “Taishan Scholar” Foundation of the People’s Government of Shandong Province (ts20130932).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available in this article and Supplementary Materials.
Acknowledgments
We extend our appreciation to the anonymous reviewers for their valuable suggestions that helped improve this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Riechmann, J.L.; Meyerowitz, E.M. MADS domain proteins in plant development. Biol. Chem. 1997, 378, 1079–1101. [Google Scholar] [PubMed]
- Messenguy, F.; Dubois, E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 2003, 316, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Melzer, R.; Theissen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.; Egea-Cortines, M.; de Andrade Silva, E.; Saedler, H.; Sommer, H. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 1996, 15, 4330–4343. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Krizek, B.A.; Meyerowitz, E.M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGA-MOUS. Proc. Natl. Acad. Sci. USA 1996, 93, 4793–4798. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.M.; Dorit, R.L.; Irish, V.F. Molecular evolution of genes controlling petal and stamen development: Duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 1998, 149, 765–783. [Google Scholar] [CrossRef] [PubMed]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Munster, T.; Theissen, G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Liljegren, S.J.; Pelaz, S.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Vergara-Silva, F.; Yanofsky, M.F. MADS-box gene evolution beyond flowers: Expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 2000, 24, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Riquelme, J.; Lijavetzky, D.; Martínez-Zapater, J.M.; Carmona, M.J. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiol. 2009, 149, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Ribas de Pouplana, L.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef] [PubMed]
- De Bodt, S.; Raes, J.; Florquin, K.; Rombauts, S.; Rouze, P.; Theissen, G.; Van de Peer, Y. Genomewide structural annotation and evolutionary analysis of the type I MADS-box genes in plants. J. Mol. Evol. 2003, 56, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Pařenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Meyerowitz, E.M. The ABCs of floral homeotic genes. Cell 1994, 78, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; de Pamphilis, C. The ABCs of floral evolution. Cell 2000, 101, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Hepworth, S.R.; Valverde, F.; Ravenscroft, D.; Mouradov, A.; Coupland, G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002, 21, 4327–4337. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, H.; Er, H.L.; Soo, H.M.; Kumar, P.P.; Han, J.-H.; Liou, Y.C.; Yu, H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 2008, 135, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Krober, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Xu, Y.; Tan, E.L.; Kumar, P.P. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. USA 2002, 99, 16336–16341. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Ditta, G.; Gustafson-Brown, C.; Pelaz, S.; Yanofsky, M.; Amasino, R.M. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 2003, 33, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, U.; Hohmann, S.; Nettesheim, K.; Wisman, E.; Saedler, H.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000, 21, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Ferrándiz, C.; Yanofsky, M.F.; Martienssen, R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 1998, 125, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Ferrándiz, C.; Liljegren, S.J.; Yanofsky, M.F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 2000, 289, 436–438. [Google Scholar] [CrossRef] [PubMed]
- Rounsley, S.D.; Ditta, G.S.; Yanofsky, M.F. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 1995, 7, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Tapia-López, R.; García-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Pérez-Ruíz, R.V.; Kim, S.-H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.X.; Zhang, J.; Chen, S.Y.; Wu, J.; Xia, J.Q.; Sun, L.Q.; Ma, S.S.; Xiang, C.B. Arabidopsis MADS-box factor AGL16 is a negative regulator of plant response to salt stress by downregulating salt-responsive genes. New Phytol. 2021, 232, 2418–2439. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, B.; Wu, Q.; Min, Q.; Zeng, R.; Xie, Z.; Huang, J. OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PLoS Genet. 2021, 17, e1009699. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tardif, G.; Kane, N.A.; Adam, H.; Labrie, L.; Major, G.; Gulick, P.; Sarhan, F.; Laliberté, J.F. Interaction network of proteins associated with abiotic stress response and development in wheat. Plant Mol. Biol. 2007, 63, 703–718. [Google Scholar] [CrossRef]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Song, X.; Liu, T.; Huang, Z.; Ren, J.; Hou, X.; Li, Y. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet. Genom. 2015, 290, 239–255. [Google Scholar] [CrossRef]
- Hu, L.; Liu, S. Genome-wide analysis of the MADS-box gene family in cucumber. Genome 2012, 55, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Dong, Q.; Ji, Z.; Chi, F.; Cong, P.; Zhou, Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 2015, 555, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Alhindi, T.; Al-Abdallat, A.M. Genome-wide identification and analysis of the MADS-box gene family in American beautyberry (Callicarpa americana). Plants 2021, 10, 1805. [Google Scholar] [CrossRef]
- Bar-Joseph, Z.; Gitter, A.; Simon, I. Studying and modelling dynamic biological processes using time-series gene expression data. Nat. Rev. Genet. 2012, 13, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Pan, I.L.; McQuinn, R.; Giovannoni, J.J.; Irish, V.F. Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 2010, 61, 1795–1806. [Google Scholar] [CrossRef]
- Nardeli, S.M.; Artico, S.; Aoyagi, G.M.; de Moura, S.M.; da Franca Silva, T.; Grossi-de-Sa, M.F.; Romanel, E.; Alves-Ferreira, M. Genome-wide analysis of the MADS-box gene family in polyploid cotton (Gossypium hirsutum) and in its diploid parental species (Gossypium arboreum and Gossypium raimondii). Plant Physiol. Biochem. 2018, 127, 169–184. [Google Scholar] [CrossRef]
- Wang, P.; Wang, S.; Chen, Y.; Xu, X.; Guang, X.; Zhang, Y. Genome-wide analysis of the MADS-box gene family in watermelon. Comput. Biol. Chem. 2019, 80, 341–350. [Google Scholar] [CrossRef]
- Shu, Y.; Yu, D.; Wang, D.; Guo, D.; Guo, C. Genome-wide survey and expression analysis of the MADS-box gene family in soybean. Mol. Biol. Rep. 2013, 40, 3901–3911. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef]
- Yang, Z.; Nie, G.; Feng, G.; Xu, X.; Li, D.; Wang, X.; Huang, L.; Zhang, X. Genome-wide identification of MADS-box gene family in orchardgrass and the positive role of DgMADS114 and DgMADS115 under different abiotic stress. Int. J. Biol. Macromol. 2022, 223 Pt A, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Deng, H.; Ma, W.; Zhou, Q.; Liu, Z. Genome-wide identification of the MADS-box transcription factor family in autotetraploid cultivated alfalfa (Medicago sativa L.) and expression analysis under abiotic stress. BMC Genom. 2021, 22, 603. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ma, J.; Yang, Y.; Luo, W.; Yang, C.; Ding, P.; Liu, Y.; Qiao, L.; Chang, Z.; Geng, H.; Wang, P.; et al. Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0181443. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yin, J.; Liang, Y.; Liu, J.; Jia, J.; Huo, H.; Wu, Z.; Yang, R.; Gong, H. Transcriptomic dynamics provide an insight into the mechanism for silicon-mediated alleviation of salt stress in cucumber plants. Ecotoxicol. Environ. Saf. 2019, 174, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Z.; Tang, R.; Wang, L.; Chen, C.; Ren, Z. Genome-Wide identification and expression analysis of Hsf and Hsp gene families in cucumber (Cucumis sativus L.). Plant Growth Regul. 2021, 95, 223–239. [Google Scholar] [CrossRef]
- Adhikari, B.N.; Savory, E.A.; Vaillancourt, B.; Childs, K.L.; Hamilton, J.P.; Day, B.; Buell, C.R. Expression Profiling of Cucumis sativus in Response to Infection by Pseudoperonospora cubensis. PLoS ONE 2012, 7, e34954. [Google Scholar] [CrossRef]
- Xu, Q.; Xu, X.; Shi, Y.; Qi, X.; Chen, X. Elucidation of the molecular responses of a cucumber segment substitution line carrying Pm5.1 and its recurrent parent triggered by powdery mildew by comparative transcriptome profiling. BMC Genom. 2017, 18, 21. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).