Cloning and Functional Analysis of Glyoxalase I Gene BrGLYI 13 in Brassica rapa L.
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of BrGLYI Gene Family Members
2.2. Phylogenetic Analysis and Classification of the BrGLYI Gene Family
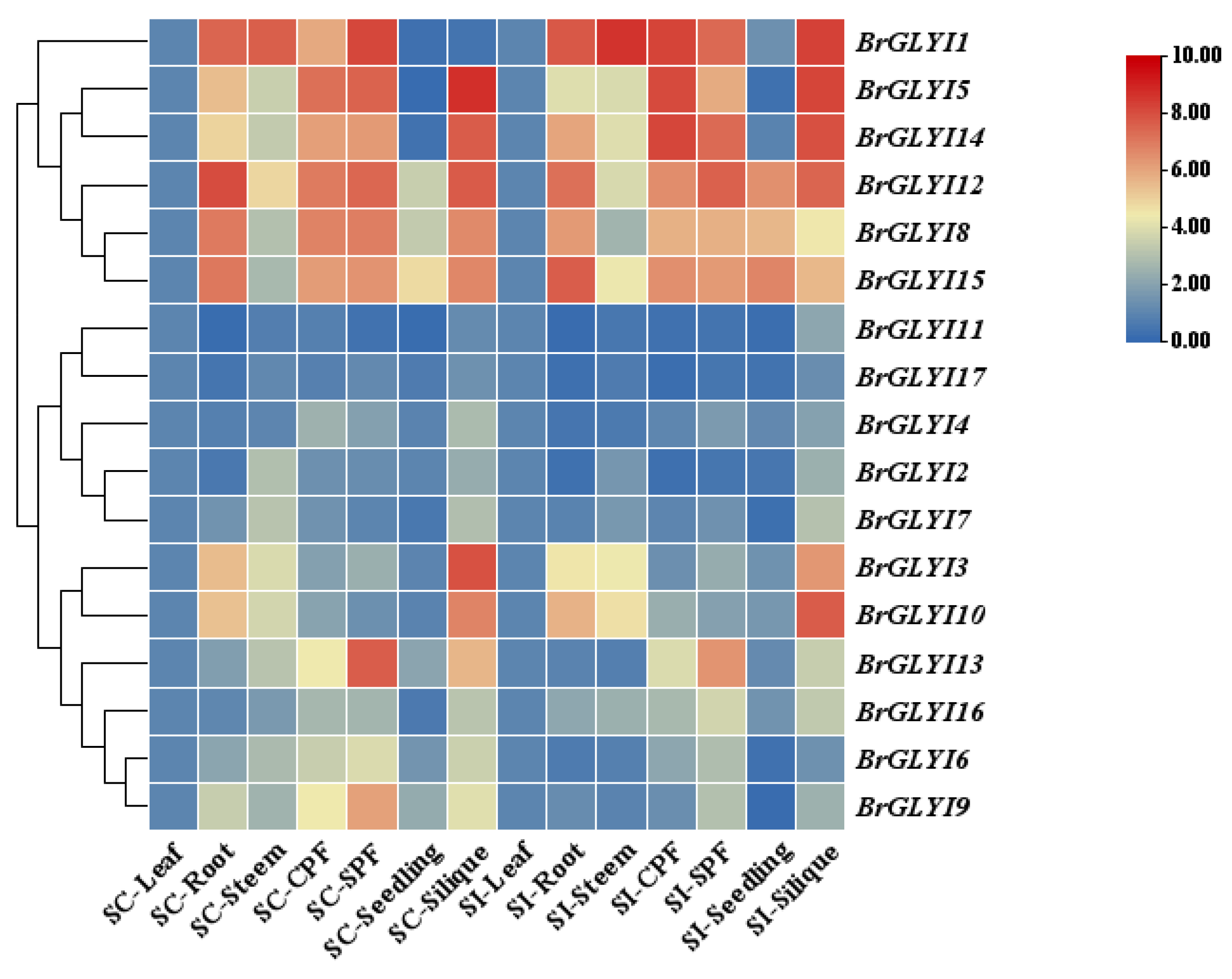
2.3. Expression Analysis of BrGLYIs in B. rapa
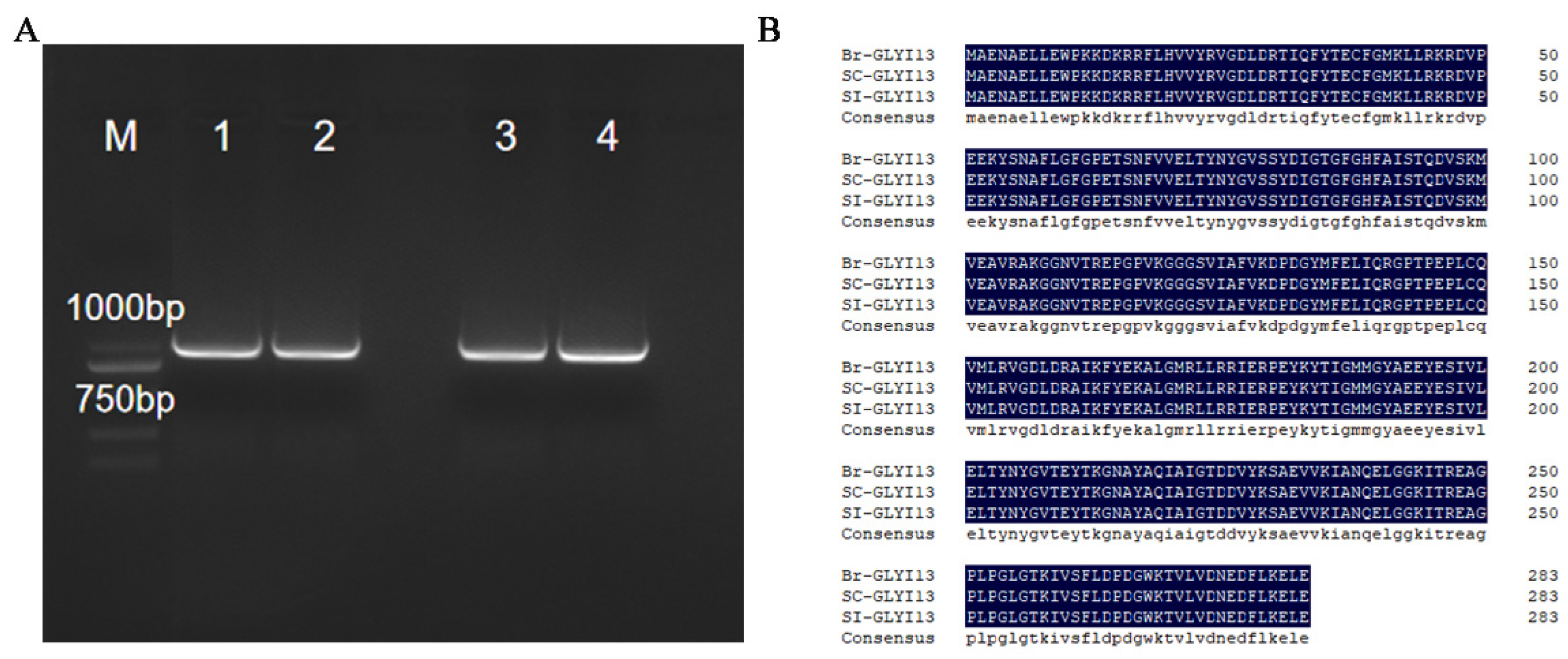
2.4. Cloning and Sequence Alignment of BrGLYI13
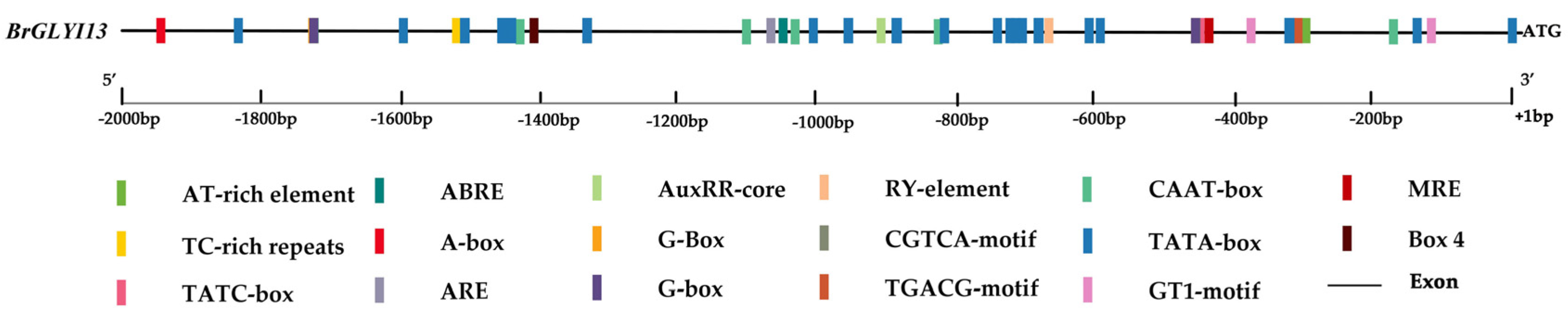
2.5. Identification and Analysis of Cis-Acting Elements in the Promoter Region of BrGLYI13
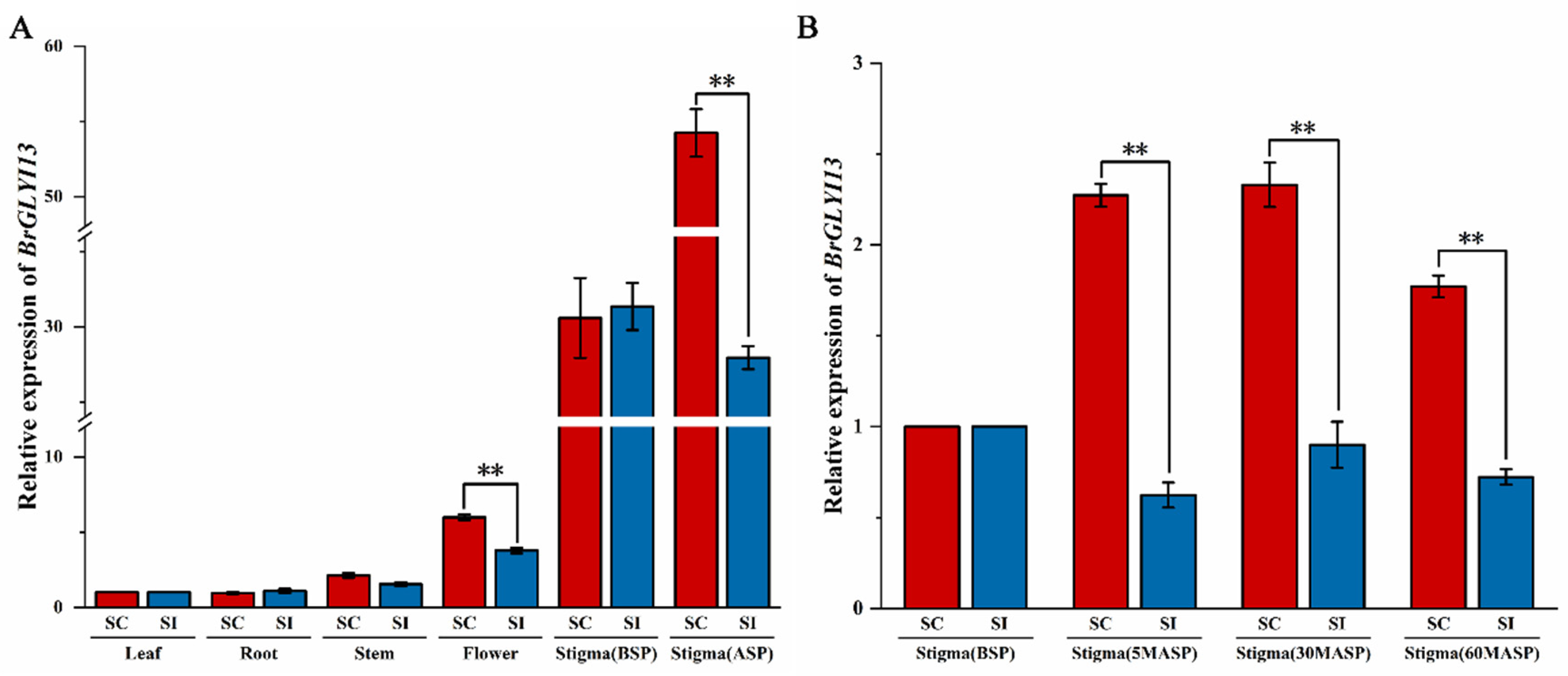
2.6. Expression Analysis of BrGLYI13 in Different Tissues of B. rapa
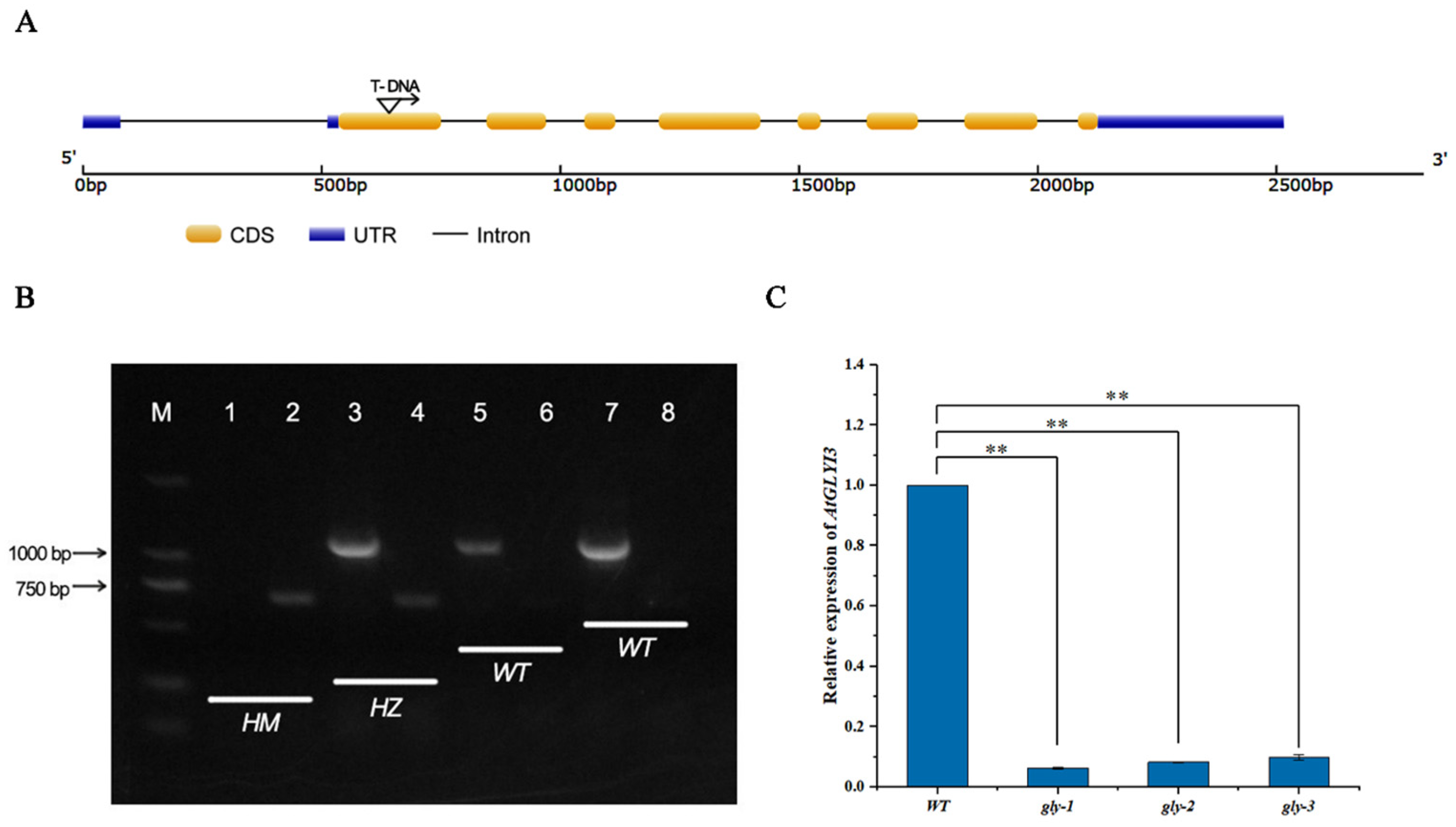
2.7. Effect of T-DNA Insertion Homozygous Mutant of AtGLYI3 on Self-Compatibility in A. thaliana
2.7.1. Identification of DNA and Transcript Levels in Homozygous Mutants
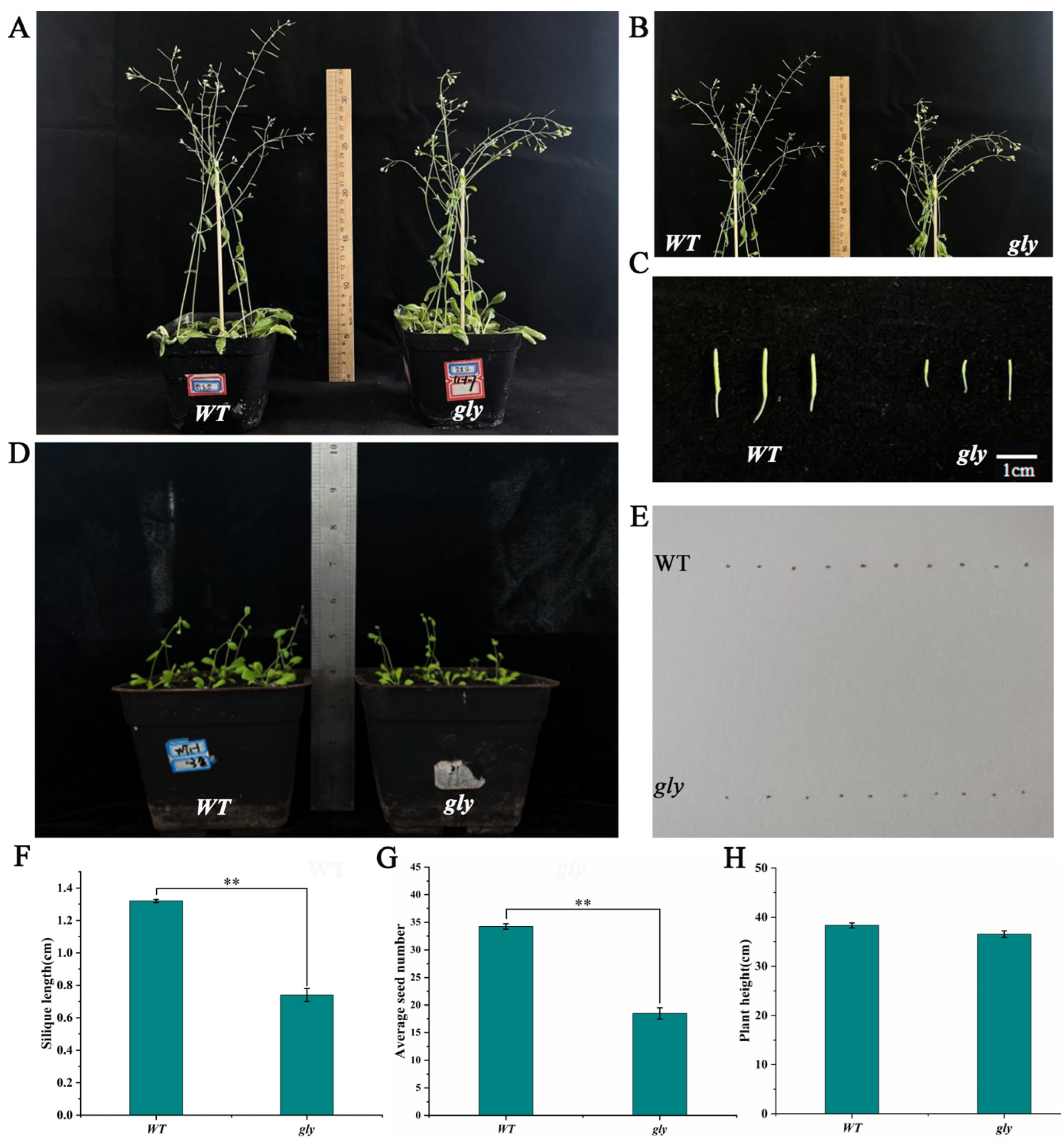
2.7.2. Impact of Homozygous Mutant on A. thaliana Self-Compatibility
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Genome-Wide Identification and Physicochemical Property Analysis of the BrGLYI Gene Family
4.3. Real-Time Quantitative Reverse Transcription PCR (RT-qPCR)
4.4. Cloning of BrGLYI13
4.5. Analysis of Cis-Regulatory Elements in the BrGLYI13 Gene Promoter
4.6. Identification and Phenotypic Observations of T-DNA Insertion Pure Mutants of A. thaliana AtGLYI3
4.6.1. Identification of Homozygous Mutants at the DNA Level
4.6.2. RT-qPCR Verification of AtGLYI3 Expression in Homozygous Mutants
4.6.3. Phenotypic Observation of Homozygous Mutant Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ye, X.Y.; Qiu, X.M.; Wang, Y.; Li, Z.G. Glyoxalase system and its role in response and adaptation of plants to environmental stress. Plant Physiol. J. 2019, 55, 401–410. [Google Scholar] [CrossRef]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 2005, 337, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Parvin, K.; Hasanuzzaman, M.; Bhuyan, M.; Mohsin, S.M.; Fujita, A.M. Quercetin Mediated Salt Tolerance in Tomato through the Enhancement of Plant Antioxidant Defense and Glyoxalase Systems. Plants 2019, 8, 247. [Google Scholar] [CrossRef] [PubMed]
- Welchen, E.; Schmitz, J.; Fuchs, P.; García, L.; Wagner, S.; Wienstroer, J.; Schertl, P.; Braun, H.P.; Schwarzländer, M.; Gonzalez, D.H.; et al. D-Lactate Dehydrogenase Links Methylglyoxal Degradation and Electron Transport through Cytochrome c. Plant Physiol. 2016, 172, 901–912. [Google Scholar] [CrossRef]
- Li, Z.G. Methylglyoxal and Glyoxalase System in Plants: Old Players, New Concepts. Bot. Rev. 2016, 82, 183–203. [Google Scholar] [CrossRef]
- Klebanoff, S.J. The effect of X-radiation on the glutathione metabolism of intact erythrocytes in vitro. J. Gen. Physiol. 1958, 41, 725–736. [Google Scholar] [CrossRef]
- Klebanoff, S.J. The relationship of hydrogen peroxide to the inhibition of the glyoxalase activity of intact erythrocytes by X-radiation. J. Gen. Physiol. 1958, 41, 737–753. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Jamshed, M.; Kumar, A.; Skori, L.; Scandola, S.; Wang, T.; Spiegel, D.; Samuel, M.A. Glyoxalase Goes Green: The Expanding Roles of Glyoxalase in Plants. Int. J. Mol. Sci. 2017, 18, 898. [Google Scholar] [CrossRef]
- Guo, Y.L. Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 2013, 73, 941–951. [Google Scholar] [CrossRef]
- Xu, M.; Zuo, D.; Wang, Q.; Lv, L.; Zhang, Y.; Jiao, H.; Zhang, X.; Yang, Y.; Song, G.; Cheng, H. Identification and molecular evolution of the GLX genes in 21 plant species: A focus on the Gossypium hirsutum. BMC Genom. 2023, 24, 474. [Google Scholar] [CrossRef]
- Mustafiz, A.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genom. 2011, 11, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Islam, T. Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 2016, 16, 87. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Xiao, X.; Wang, N.; Zhang, F.; Gao, G.; Xu, K.; Chen, B.; Qiao, J.; Wu, X. Genome-wide analysis and expression profiles of glyoxalase gene families in Chinese cabbage (Brassica rapa L.). PLoS ONE 2018, 13, e0191159. [Google Scholar] [CrossRef]
- Yan, G.; Zhang, M.; Guan, W.; Zhang, F.; Dai, W.; Yuan, L.; Gao, G.; Xu, K.; Chen, B.; Li, L.; et al. Genome-Wide Identification and Functional Characterization of Stress Related Glyoxalase Genes in Brassica napus L. Int. J. Mol. Sci. 2023, 24, 2130. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Kaur, C.; Kumar, B.; Pareek, A.; Sopory, S.K. Reassessing plant glyoxalases: Large family and expanding functions. New Phytol. 2020, 227, 714–721. [Google Scholar] [CrossRef]
- Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. Glyoxalase and Methylglyoxal as Biomarkers for Plant Stress Tolerance. Crit. Rev. Plant Sci. 2014, 33, 429–456. [Google Scholar] [CrossRef]
- Schmitz, J.; Dittmar, I.C.; Brockmann, J.D.; Schmidt, M.; Hüdig, M.; Rossoni, A.W.; Maurino, V.G. Defense against Reactive Carbonyl Species Involves at Least Three Subcellular Compartments Where Individual Components of the System Respond to Cellular Sugar Status. Plant Cell 2017, 29, 3234–3254. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Jamshed, M.; Samuel, M.A. Degradation of glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat. Plants 2015, 1, 15185. [Google Scholar] [CrossRef]
- Takayama, S.; Shimosato, H.; Shiba, H.; Funato, M.; Che, F.S.; Watanabe, M.; Iwano, M.; Isogai, A. Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 2001, 413, 534–538. [Google Scholar] [CrossRef]
- Dixit, R.; Nasrallah, J.B. Recognizing self in the self-incompatibility response. Plant Physiol. 2001, 125, 105–108. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.C.; Ma, W.G.; Lei, J.M.; Liu, Q.; Yang, R.Y.; Wu, J.Y.; Wang, X.F.; Ye, J.; Zeng, J.; Zhang, Y.H.; et al. Study on Adaptation and Introduction Possibility of Winter Rapeseed to Dry and Cold Areas in Northwest China. Sci. Agric. Sin. 2007, 40, 2716–2726. [Google Scholar]
- Liu, H.Q.; Sun, W.C.; Liu, Z.G.; Wang, Z.J.; Fang, Y.; Wu, J.Y.; Li, X.C.; Fang, Y. Safe wintering and economic and ecological benefit of winter rapeseed in dry and cold areas of northern China. Ying Yong Sheng Tai Xue Bao 2015, 26, 3035–3044. [Google Scholar]
- Luo, Y.X. Study on Self-incompatibility and Pollinations of Brassica compestris L. Acta Agric. Boreali-Occident. 2007, 2, 56–58. [Google Scholar]
- Zhang, Y.; Xue, Y. Molecular Biology of S-Rnase-Based Self-Incompatibility. In Self-Incompatibility in Flowering Plants: Evolution, Diversity, and Mechanisms; Franklin-Tong, V.E., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 193–215. [Google Scholar]
- Sassa, H. Molecular mechanism of the S-RNase-based gametophytic self-incompatibility in fruit trees of Rosaceae. Breed. Sci. 2016, 66, 116–121. [Google Scholar] [CrossRef]
- Takayama, S.; Isogai, A. Self-incompatibility in plants. Annu. Rev. Plant Biol. 2005, 56, 467–489. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, Y.; Zhang, H.; Song, Y.; Zhao, F.; Zhang, Y.; Zhu, S.; Zhang, H.; Zhou, Z.; Guo, H.; et al. Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell 2022, 34, 579–596. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, K.; Dou, S.; Gao, E.; Hussey, P.J.; Lin, Z.; Wang, P. Exo84c-regulated degradation is involved in the normal self-incompatible response in Brassicaceae. Cell Rep. 2024, 43, 113913. [Google Scholar] [CrossRef]
- Hiscock, S.J.; McInnis, S.M. Pollen recognition and rejection during the sporophytic self-incompatibility response: Brassica and beyond. Trends Plant Sci. 2003, 8, 606–613. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Z.; Xue, Y. Roles of proteolysis in plant self-incompatibility. Annu. Rev. Plant Biol. 2009, 60, 21–42. [Google Scholar] [CrossRef]
- He, Y.T.; Cheng, B.Y.; Fu, T.D.; Li, D.R.; Tu, J.X. Origins and Evolution of Brassica campestris L. in China. J. Genet. Genom. 2003, 30, 1003–1012. [Google Scholar]
- Lei, J.M.; Wu, J.Y.; Zhang, Y. Analysis on the self-compatibility of winter rapeseed (Brassica rapa) in China. In Proceedings of the 12th International Rapeseed Congress, Wuhan, China, 26–30 March 2007; p. 3. [Google Scholar]
- Yamamoto, M.; Nishio, T. Commonalities and differences between Brassica and Arabidopsis self-incompatibility. Hortic. Res. 2014, 1, 14054. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Wu, J.; Xu, X.-D.; Liu, L.J.; Ma, L.; Pu, Y.; Wang, W.; Hua, X.-Y.; Song, J.-M.; Liu, K.; Lu, G.; et al. A Chromosome Level Genome Assembly of a Winter Turnip Rape (Brassica rapa L.) to Explore the Genetic Basis of Cold Tolerance. Front. Plant Sci. 2022, 13, 936958. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 Years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s Conserved Domain Database and Tools for Protein Domain Analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liang, X.; Peng, L.; Baek, C.H.; Katzen, F. Single step BP/LR combined Gateway reactions. Biotechniques 2013, 55, 265–268. [Google Scholar] [CrossRef]
- Xiong, H.; Lü, G.; Li, D.B.; Li, S.S. Molecular Identification and Phenotype of Arabidopsis thaliana Mutant erd15. J. Trop. Subtrop. Bot. 2020, 28, 505–512. [Google Scholar] [CrossRef]

| Gene Name | Sequence ID | Number of Amino Acids /aa | Molecular Weight /Da | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization Prediction |
|---|---|---|---|---|---|---|---|---|
| BrGLYI1 | Brapa02T002698.1 | 167 | 18,791.65 | 5.81 | 62.94 | 89.22 | −0.193 | Cytoplasm |
| BrGLYI2 | Brapa03T001219.1 | 193 | 21,797.96 | 8.43 | 37.75 | 90.41 | −0.461 | Chloroplast |
| BrGLYI3 | Brapa04T002215.1 | 185 | 13,878.76 | 6.08 | 42.68 | 85.61 | −0.601 | Nucleus |
| BrGLYI4 | Brapa05T001273.1 | 137 | 15,252.22 | 5.46 | 58.99 | 76.86 | −0.226 | Cytoplasm |
| BrGLYI5 | Brapa06T000491.1 | 171 | 19,274.70 | 7.77 | 36.81 | 78.60 | −0.405 | Cytoplasm |
| BrGLYI6 | Brapa06T000534.1 | 235 | 26,352.75 | 6.32 | 32.14 | 69.74 | −0.460 | Cytoplasm |
| BrGLYI7 | Brapa06T000824.1 | 1044 | 115,660.97 | 8.27 | 44.05 | 82.90 | −0.298 | Cytoplasm |
| BrGLYI8 | Brapa06T001102.1 | 174 | 19,778.79 | 5.68 | 65.91 | 89.54 | −0.209 | Nucleus |
| BrGLYI9 | Brapa06T001420.1 | 266 | 30,367.20 | 5.24 | 43.82 | 70.38 | −0.575 | Cytoplasm |
| BrGLYI10 | Brapa07T002043.1 | 185 | 21,015.55 | 4.84 | 37.80 | 73.78 | −0.709 | Cytoplasm |
| BrGLYI11 | Brapa07T003470.1 | 406 | 45,190.31 | 8.85 | 34.98 | 89.09 | −0.111 | Chloroplast |
| BrGLYI12 | Brapa08T003039.1 | 167 | 19,784.93 | 5.88 | 49.09 | 95.69 | −0.217 | Cytoplasm |
| BrGLYI13 | Brapa08T003191.1 | 283 | 31,886.44 | 5.27 | 28.95 | 82.61 | −0.322 | Cytoplasm |
| BrGLYI14 | Brapa09T000323.1 | 137 | 15,394.22 | 5.84 | 33.70 | 71.09 | −0.464 | Cytoplasm |
| BrGLYI15 | Brapa09T000758.1 | 172 | 19,692.81 | 6.30 | 48.25 | 87.79 | −0.252 | Cytoplasm |
| BrGLYI16 | Brapa09T005172.1 | 118 | 12,967.91 | 6.40 | 45.49 | 100.76 | 0.069 | Nucleus |
| BrGLYI17 | Brapa10T001850.1 | 195 | 21,959.00 | 6.71 | 43.16 | 89.54 | −0.449 | Chloroplast |
| Variety (Line) | Source | Compatibility | Relative Compatibility Index |
|---|---|---|---|
| 039-1 | Gansu Agricultural University | Self-compatibility | 1.00 |
| GAU-28-5 | Gansu Agricultural University | Self-incompatibility | 0.07 |
| Gene Name | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) |
|---|---|---|
| BrGLYI1 | TGAGAGCAGTGGTGGAAGAAGG | AAGAGGCACGACGGGAAGG |
| BrGLYI2 | AGATTCTGGATAAAGCTGGGATTGC | TTGTGTCTGGATCACGAGTGAATATC |
| BrGLYI3 | CGTTAAACCATGTCTCAAGACTTTGC | ACGCTCTGTCTCCACGAATCC |
| BrGLYI4 | ACTCCTTCCTTCGCTCTCTCAAG | GCTACCTCTAATCCGTTTCCATCAG |
| BrGLYI5 | ATGGTGCGGTGGCTGTGAG | GTGGCTTCCGATGCGTACAAC |
| BrGLYI6 | CCACTGCTCTCGGTCGGATC | GCTGCTGATTCCTTCGCTTCC |
| BrGLYI7 | AGGATGAGGAGGAGAAGAAGAAGAAG | AAGTAGCGGCGTGACAGAGG |
| BrGLYI8 | GGACATGGAATTGGAATACATCTCTTG | TGACATACTCTCGCACTGGAAGG |
| BrGLYI9 | GAAGGAGAAGCAGTGGCGATTC | CGAGGGTTAAGGATGGGATGGG |
| BrGLYI10 | CGGTGCGTGGCTATTCAACTAC | TGTGATTATCCATCGGGTCCAAATG |
| BrGLYI11 | GTTATGCTCCGTGTTGGTGATCTC | AACTTGTACTCTGGATTGTCTCTTGTG |
| BrGLYI12 | AGTATGGGAGCAGTGGAGAAGAAG | ATCAGGGTCGTGGAAGAAGAGC |
| BrGLYI13 | CTGAAGTAGTGAAGATAGCCAACCAAG | GGTGCCGAGTCCAGGAAGAG |
| BrGLYI14 | TCAGCGAGCCAGAGGACAAG | ACCAATGCGGACAACGATGC |
| BrGLYI15 | GGAATTGGAATACATCTCTTGCGTTC | TCTCCACTGCTGCCATACTCTC |
| BrGLYI16 | AGCTTCAGTCTGGTCCTCTCAAG | GTGAACGACAGTAGCGAAGAGTATC |
| BrGLYI17 | GCGAGAACTTAGAACGGTCACTG | TACCCATAACCATGCTCCTCTATACG |
| Actin | TGTGCCAATCTACGAGGGTTT | TTTCCCGCTCTGCTGTTGT |
| Gene Name | Sequence (5′~3′) |
|---|---|
| gly-LP | TGTAACGCGTCTGTGATGATC |
| gly-RP | CCCAAAGCCAGTTCCAATATC |
| LBb1.3 | ATTTTGCCGATTTCGGAAC |
| AtGLYI3-F | CAGTCAAAGGTGGAGGCAGTGTC |
| AtGLYI3-R | TTGGCAGAAAGGTTCAGGAGTTGG |
| Actin2-F | GTTGGGATGAACCAGAAGGA |
| Actin2-R | GCTCTTCAGGAGCAATACGAAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Zhang, F.; Tao, X.; Li, Y.; Fan, T.; Wu, J.; Ma, L.; Liu, L.; Pu, Y.; Wang, W.; et al. Cloning and Functional Analysis of Glyoxalase I Gene BrGLYI 13 in Brassica rapa L. Int. J. Mol. Sci. 2025, 26, 2737. https://doi.org/10.3390/ijms26062737
Song X, Zhang F, Tao X, Li Y, Fan T, Wu J, Ma L, Liu L, Pu Y, Wang W, et al. Cloning and Functional Analysis of Glyoxalase I Gene BrGLYI 13 in Brassica rapa L. International Journal of Molecular Sciences. 2025; 26(6):2737. https://doi.org/10.3390/ijms26062737
Chicago/Turabian StyleSong, Xiaojia, Feng Zhang, Xiaolei Tao, Yapeng Li, Tingting Fan, Junyan Wu, Li Ma, Lijun Liu, Yuanyuan Pu, Wangtian Wang, and et al. 2025. "Cloning and Functional Analysis of Glyoxalase I Gene BrGLYI 13 in Brassica rapa L." International Journal of Molecular Sciences 26, no. 6: 2737. https://doi.org/10.3390/ijms26062737
APA StyleSong, X., Zhang, F., Tao, X., Li, Y., Fan, T., Wu, J., Ma, L., Liu, L., Pu, Y., Wang, W., Yang, G., & Sun, W. (2025). Cloning and Functional Analysis of Glyoxalase I Gene BrGLYI 13 in Brassica rapa L. International Journal of Molecular Sciences, 26(6), 2737. https://doi.org/10.3390/ijms26062737





