The Current Status of T Cell Receptor (TCR) Repertoire Analysis in Colorectal Cancer
Abstract
1. Introduction
2. TCR Basic Structure and Function
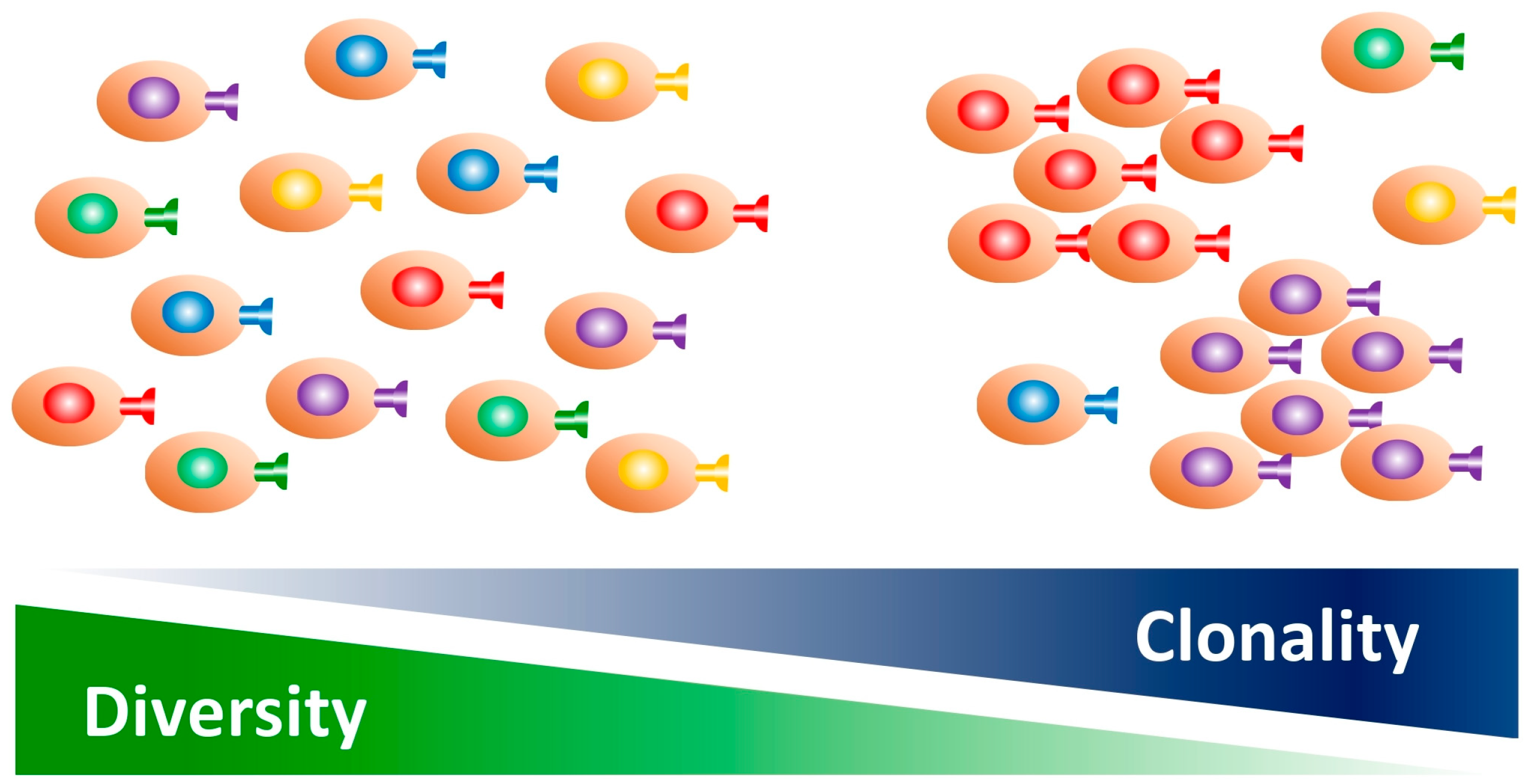
3. Clonality and Diversity of the TCR Repertoire
4. TCR Repertoire Differences Between CRC and Healthy Tissues
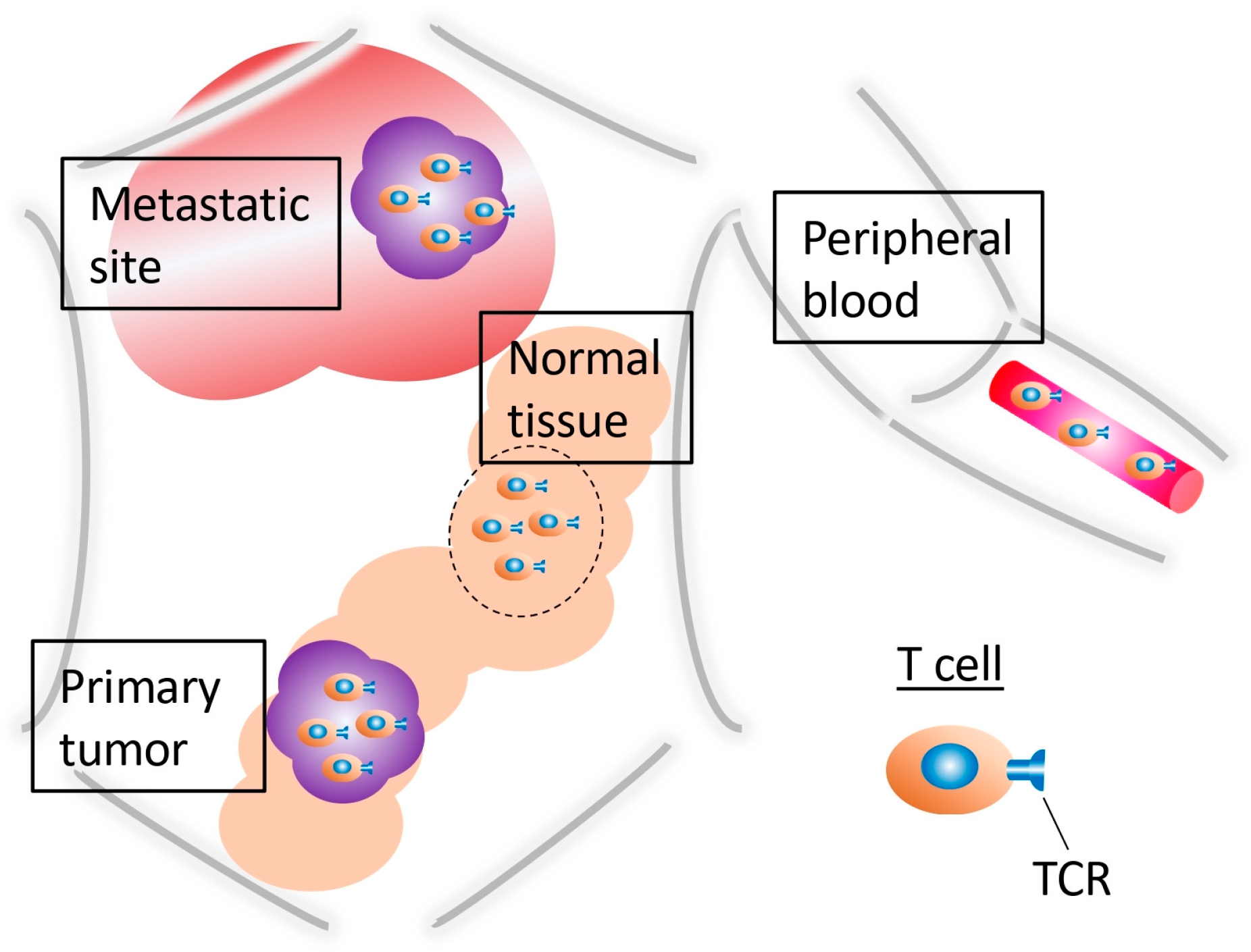
5. TCR Repertoire Differences Between Intratumoral and Peripheral T Cells
6. The TCR Repertoire in CRC Metastatic Sites
7. The Treg TCR Repertoire
8. TCR Repertoire Differences Between dMMR and pMMR Patients
9. Can the TCR Repertoire Serve as a Predictor of ICI Efficacy?
10. The TCR Repertoire as a Predictor of CRC Prognosis
11. The TCR Repertoire Sequence as a Predictor of Efficacy Using Conventional Multimodal Treatment Methods
12. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Kojima, D.; Wada, H.; Hanaoka, K.; Watanabe, T.; Nagano, H.; Yamakado, J.; Matsuda, A.; Irie, H.; Maki, T.; et al. A Bridge to Curative Surgery for Obstructive Colorectal Cancer: Self-expandable Metallic Stent Versus Decompression Tube. Anticancer Res. 2024, 44, 3427–3441. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef]
- Cercek, A.; Lumish, M.; Sinopoli, J.; Weiss, J.; Shia, J.; Lamendola-Essel, M.; El Dika, I.H.; Segal, N.; Shcherba, M.; Sugarman, R.; et al. PD-1 Blockade in Mismatch Repair-Deficient, Locally Advanced Rectal Cancer. N. Engl. J. Med. 2022, 386, 2363–2376. [Google Scholar] [CrossRef] [PubMed]
- Chalabi, M.; Verschoor, Y.L.; Tan, P.B.; Balduzzi, S.; Van Lent, A.U.; Grootscholten, C.; Dokter, S.; Buller, N.V.; Grotenhuis, B.A.; Kuhlmann, K.; et al. Neoadjuvant Immunotherapy in Locally Advanced Mismatch Repair-Deficient Colon Cancer. N. Engl. J. Med. 2024, 390, 1949–1958. [Google Scholar] [CrossRef]
- Chalabi, M.; Fanchi, L.F.; Dijkstra, K.K.; Van den Berg, J.G.; Aalbers, A.G.; Sikorska, K.; Lopez-Yurda, M.; Grootscholten, C.; Beets, G.L.; Snaebjornsson, P.; et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat. Med. 2020, 26, 566–576. [Google Scholar] [CrossRef]
- Sun, L.; Meng, C.; Zhang, X.; Gao, J.; Wei, P.; Zhang, J.; Zhang, Z. Management and prediction of immune-related adverse events for PD1/PDL-1 immunotherapy in colorectal cancer. Front. Pharmacol. 2023, 14, 1167670. [Google Scholar] [CrossRef]
- Locy, H.; de Mey, S.; de Mey, W.; De Ridder, M.; Thielemans, K.; Maenhout, S.K. Immunomodulation of the Tumor Microenvironment: Turn Foe into Friend. Front. Immunol. 2018, 9, 2909. [Google Scholar] [CrossRef]
- Takahashi, H.; Kojima, D.; Watanabe, M. Therapeutic potential of trained immunity for malignant disease. Immunol. Med. 2024, 48, 1–12. [Google Scholar] [CrossRef]
- Opzoomer, J.W.; Sosnowska, D.; Anstee, J.E.; Spicer, J.F.; Arnold, J.N. Cytotoxic Chemotherapy as an Immune Stimulus: A Molecular Perspective on Turning Up the Immunological Heat on Cancer. Front. Immunol. 2019, 10, 1654. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Al-Haidari, A.; Sun, J.; Kazi, J.U. T cell receptor (TCR) signaling in health and disease. Signal Transduct. Target. Ther. 2021, 6, 412. [Google Scholar] [CrossRef] [PubMed]
- Lythe, G.; Callard, R.E.; Hoare, R.L.; Molina-Paris, C. How many TCR clonotypes does a body maintain? J. Theor. Biol. 2016, 389, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Mazzotti, L.; Gaimari, A.; Bravaccini, S.; Maltoni, R.; Cerchione, C.; Juan, M.; Navarro, E.A.; Pasetto, A.; Nascimento Silva, D.; Ancarani, V.; et al. T-Cell Receptor Repertoire Sequencing and Its Applications: Focus on Infectious Diseases and Cancer. Int. J. Mol. Sci. 2022, 23, 8590. [Google Scholar] [CrossRef]
- Wang, C.Y.; Fang, Y.X.; Chen, G.H.; Jia, H.J.; Zeng, S.; He, X.B.; Feng, Y.; Li, S.J.; Jin, Q.W.; Cheng, W.Y.; et al. Analysis of the CDR3 length repertoire and the diversity of T cell receptor alpha and beta chains in swine CD4+ and CD8+ T lymphocytes. Mol. Med. Rep. 2017, 16, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Call, M.E.; Wucherpfennig, K.W. The T cell receptor: Critical role of the membrane environment in receptor assembly and function. Annu. Rev. Immunol. 2005, 23, 101–125. [Google Scholar] [CrossRef]
- Hodges, E.; Krishna, M.T.; Pickard, C.; Smith, J.L. Diagnostic role of tests for T cell receptor (TCR) genes. J. Clin. Pathol. 2003, 56, 1–11. [Google Scholar] [CrossRef]
- Kidman, J.; Principe, N.; Watson, M.; Lassmann, T.; Holt, R.A.; Nowak, A.K.; Lesterhuis, W.J.; Lake, R.A.; Chee, J. Characteristics of TCR Repertoire Associated with Successful Immune Checkpoint Therapy Responses. Front. Immunol. 2020, 11, 587014. [Google Scholar] [CrossRef]
- Sakimura, S.; Nagayama, S.; Fukunaga, M.; Hu, Q.; Kitagawa, A.; Kobayashi, Y.; Hasegawa, T.; Noda, M.; Kouyama, Y.; Shimizu, D.; et al. Impaired tumor immune response in metastatic tumors is a selective pressure for neutral evolution in CRC cases. PLoS Genet. 2021, 17, e1009113. [Google Scholar] [CrossRef]
- Alt, F.W.; Oltz, E.M.; Young, F.; Gorman, J.; Taccioli, G.; Chen, J. VDJ recombination. Immunol. Today 1992, 13, 306–314. [Google Scholar] [CrossRef]
- Alarcon, B.; Gil, D.; Delgado, P.; Schamel, W.W. Initiation of TCR signaling: Regulation within CD3 dimers. Immunol. Rev. 2003, 191, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Notarangelo, L.D. Immunodeficiency and immune dysregulation associated with proximal defects of T cell receptor signaling. Curr. Opin. Immunol. 2014, 31, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Schrum, A.G.; Turka, L.A.; Palmer, E. Surface T-cell antigen receptor expression and availability for long-term antigenic signaling. Immunol. Rev. 2003, 196, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Aran, A.; Garrigos, L.; Curigliano, G.; Cortes, J.; Marti, M. Evaluation of the TCR Repertoire as a Predictive and Prognostic Biomarker in Cancer: Diversity or Clonality? Cancers 2022, 14, 1771. [Google Scholar] [CrossRef]
- Conroy, L.A.; Alexander, D.R. The role of intracellular signalling pathways regulating thymocyte and leukemic T cell apoptosis. Leukemia 1996, 10, 1422–1435. [Google Scholar]
- Takahashi, H.; Kuhtreiber, W.M.; Keefe, R.C.; Lee, A.H.; Aristarkhova, A.; Dias, H.F.; Ng, N.; Nelson, K.J.; Bien, S.; Scheffey, D.; et al. BCG vaccinations drive epigenetic changes to the human T cell receptor: Restored expression in type 1 diabetes. Sci. Adv. 2022, 8, eabq7240. [Google Scholar] [CrossRef]
- Xie, N.; Shen, G.; Gao, W.; Huang, Z.; Huang, C.; Fu, L. Neoantigens: Promising targets for cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 9. [Google Scholar] [CrossRef]
- Song, I.H.; Lee, S.B.; Jeong, B.K.; Park, J.; Kim, H.; Lee, G.; Cha, S.M.; Lee, H.; Gong, G.; Kwon, N.J.; et al. T cell receptor clonotype in tumor microenvironment contributes to intratumoral signaling network in patients with colorectal cancer. Immunol. Res. 2024, 72, 921–937. [Google Scholar] [CrossRef]
- Baker, A.M.; Nageswaran, G.; Nenclares, P.; Ronel, T.; Smith, K.; Kimberley, C.; Lacle, M.M.; Bhide, S.; Harrington, K.J.; Melcher, A.; et al. FUME-TCRseq Enables Sensitive and Accurate Sequencing of the T-Cell Receptor from Limited Input of Degraded RNA. Cancer Res. 2024, 84, 1560–1569. [Google Scholar] [CrossRef]
- Shen, Y.; Voigt, A.; Leng, X.; Rodriguez, A.A.; Nguyen, C.Q. A current and future perspective on T cell receptor repertoire profiling. Front. Genet. 2023, 14, 1159109. [Google Scholar] [CrossRef]
- Takahashi, H.; Yoshimatsu, G.; Faustman, D.L. The Roles of TNFR2 Signaling in Cancer Cells and the Tumor Microenvironment and the Potency of TNFR2 Targeted Therapy. Cells 2022, 11, 1952. [Google Scholar] [CrossRef] [PubMed]
- Scheper, W.; Kelderman, S.; Fanchi, L.F.; Linnemann, C.; Bendle, G.; de Rooij, M.A.J.; Hirt, C.; Mezzadra, R.; Slagter, M.; Dijkstra, K.; et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med. 2019, 25, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.M.Q.; Nguyen, T.N.; Tran Nguyen, B.Q.; Diem Tran, T.P.; Diem Pham, N.M.; Phuc Nguyen, H.T.; Cuong Ho, T.K.; Linh Nguyen, D.V.; Nguyen, H.T.; Tran, D.H.; et al. The T cell receptor beta chain repertoire of tumor infiltrating lymphocytes improves neoantigen prediction and prioritization. eLife 2024, 13, RP94658. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ma, R.; Luo, R.; Sun, Y.; He, X.; Sun, W.; Tang, W.; Yao, X. Primary exploration of CDR3 spectratyping and molecular features of TCR beta chain in the peripheral blood and tissue of patients with colorectal carcinoma. Cancer Epidemiol. 2010, 34, 733–740. [Google Scholar] [CrossRef]
- Ochsenreither, S.; Fusi, A.; Wojtke, S.; Busse, A.; Nussler, N.C.; Thiel, E.; Keilholz, U.; Nagorsen, D. Comparison of T-cell receptor repertoire restriction in blood and tumor tissue of colorectal cancer patients. J. Transl. Med. 2010, 8, 35. [Google Scholar] [CrossRef]
- Nakamura, K.; Okuyama, R. Changes in the Immune Cell Repertoire for the Treatment of Malignant Melanoma. Int. J. Mol. Sci. 2022, 23, 12991. [Google Scholar] [CrossRef]
- Cui, J.H.; Lin, K.R.; Yuan, S.H.; Jin, Y.B.; Chen, X.P.; Su, X.K.; Jiang, J.; Pan, Y.M.; Mao, S.L.; Mao, X.F.; et al. TCR Repertoire as a Novel Indicator for Immune Monitoring and Prognosis Assessment of Patients with Cervical Cancer. Front. Immunol. 2018, 9, 2729. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Yang, Q.F.; Yang, J.S.; Cao, R.B.; Liang, J.Y.; Liu, Y.T.; Zeng, Y.L.; Chen, S.; Xia, X.F.; Zhang, K.; et al. Characteristics and prognostic significance of profiling the peripheral blood T-cell receptor repertoire in patients with advanced lung cancer. Int. J. Cancer 2019, 145, 1423–1431. [Google Scholar] [CrossRef]
- Britanova, O.V.; Putintseva, E.V.; Shugay, M.; Merzlyak, E.M.; Turchaninova, M.A.; Staroverov, D.B.; Bolotin, D.A.; Lukyanov, S.; Bogdanova, E.A.; Mamedov, I.Z.; et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 2014, 192, 2689–2698. [Google Scholar] [CrossRef]
- Tsai, Y.Y.; Nair, K.G.; Barot, S.V.; Xiang, S.; Kamath, S.; Melas, M.; Walker, C.P.; Srivastava, R.M.; Osborne, N.; Chan, T.A.; et al. Differences in tumor-associated T-cell receptor repertoires between early-onset and average-onset colorectal cancer. J. Natl. Cancer Inst. 2024, 116, 1645–1653. [Google Scholar] [CrossRef]
- Cai, Y.; Luo, M.; Yang, W.; Xu, C.; Wang, P.; Xue, G.; Jin, X.; Cheng, R.; Que, J.; Zhou, W.; et al. The Deep Learning Framework iCanTCR Enables Early Cancer Detection Using the T-Cell Receptor Repertoire in Peripheral Blood. Cancer Res. 2024, 84, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, J.; Hou, W.; Ding, Y.; Zhu, Y.; Zheng, J.; Huang, Q.; Cao, Z.; Xie, R.; Wei, Q.; et al. Colorectal cancer-associated T cell receptor repertoire abnormalities are linked to gut microbiome shifts and somatic cell mutations. Gut Microbes 2023, 15, 2263934. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cui, Y.; Zhang, Y.; Liu, Z.; Zhang, Q.; Wu, W.; Zheng, Z.; Li, S.; Zhang, Z.; Li, Y. A comprehensive study of immunology repertoires in both preoperative stage and postoperative stage in patients with colorectal cancer. Mol. Genet. Genom. Med. 2019, 7, e504. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Liao, W.J.; Ma, L.; Huang, Y.T.; Shi, M.; Wen, Q.; Wang, X.N. Dynamic monitoring the TCR CDR3 spectratypes in patients with metastatic CRC treated with a combination of bevacizumab, irinotecan, fluorouracil, and leucovorin. Cancer Immunol. Immunother. 2010, 59, 247–256. [Google Scholar] [CrossRef]
- Chen, Y.T.; Hsu, H.C.; Lee, Y.S.; Liu, H.; Tan, B.C.; Chin, C.Y.; Chang, I.Y.; Yang, C.Y. Longitudinal High-Throughput Sequencing of the T-Cell Receptor Repertoire Reveals Dynamic Change and Prognostic Significance of Peripheral Blood TCR Diversity in Metastatic Colorectal Cancer During Chemotherapy. Front. Immunol. 2021, 12, 743448. [Google Scholar] [CrossRef]
- Luo, W.; Liao, W.J.; Huang, Y.T.; Shi, M.; Zhang, Y.; Wen, Q.; Zhou, M.Q.; Ma, L. Cancer of the gastrointestinal tract results in a restricted T-cell repertoire dependent upon tumor differentiation. Cell. Immunol. 2011, 270, 47–52. [Google Scholar] [CrossRef]
- Matsutani, T.; Shiiba, K.; Yoshioka, T.; Tsuruta, Y.; Suzuki, R.; Ochi, T.; Itoh, T.; Musha, H.; Mizoi, T.; Sasaki, I. Evidence for existence of oligoclonal tumor-infiltrating lymphocytes and predominant production of T helper 1/T cytotoxic 1 type cytokines in gastric and colorectal tumors. Int. J. Oncol. 2004, 25, 133–141. [Google Scholar] [CrossRef]
- Nakanishi, K.; Kukita, Y.; Segawa, H.; Inoue, N.; Ohue, M.; Kato, K. Characterization of the T-cell receptor beta chain repertoire in tumor-infiltrating lymphocytes. Cancer Med. 2016, 5, 2513–2521. [Google Scholar] [CrossRef]
- Tamura, K.; Hazama, S.; Yamaguchi, R.; Imoto, S.; Takenouchi, H.; Inoue, Y.; Kanekiyo, S.; Shindo, Y.; Miyano, S.; Nakamura, Y.; et al. Characterization of the T cell repertoire by deep T cell receptor sequencing in tissues and blood from patients with advanced colorectal cancer. Oncol. Lett. 2016, 11, 3643–3649. [Google Scholar] [CrossRef]
- Ye, X.; Waite, J.C.; Dhanik, A.; Gupta, N.; Zhong, M.; Adler, C.; Malahias, E.; Ni, M.; Wei, Y.; Gurer, C.; et al. Endogenous retroviral proteins provide an immunodominant but not requisite antigen in a murine immunotherapy tumor model. Oncoimmunology 2020, 9, 1758602. [Google Scholar] [CrossRef]
- Sukegawa, K.; Shitaoka, K.; Hamana, H.; Kobayashi, E.; Miyahara, Y.; Fujii, K.; Tsuda, K.; Saeki, S.; Nagata, T.; Ozawa, T.; et al. Relationship between T cell receptor clonotype and PD-1 expression of tumor-infiltrating lymphocytes in colorectal cancer. Eur. J. Immunol. 2020, 50, 1580–1590. [Google Scholar] [CrossRef]
- Saha, S.; Ghosh, S.; Ghosh, S.; Nandi, S.; Nayak, A. Unraveling the complexities of colorectal cancer and its promising therapies—An updated review. Int. Immunopharmacol. 2024, 143, 113325. [Google Scholar] [CrossRef] [PubMed]
- Barnd, D.L.; Kerr, L.A.; Metzgar, R.S.; Finn, O.J. Human tumor-specific cytotoxic T cell lines generated from tumor-draining lymph node infiltrate. Transplant. Proc. 1988, 20, 339–341. [Google Scholar] [PubMed]
- Matsuda, T.; Miyauchi, E.; Hsu, Y.W.; Nagayama, S.; Kiyotani, K.; Zewde, M.; Park, J.H.; Kato, T.; Harada, M.; Matsui, S.; et al. TCR sequencing analysis of cancer tissues and tumor draining lymph nodes in colorectal cancer patients. Oncoimmunology 2019, 8, e1588085. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Wang, H.; Jiang, R.; Wang, F.; Chen, C.; Xu, Z.; Xiao, R. Characterization of the T-cell receptor repertoire associated with lymph node metastasis in colorectal cancer. Front. Oncol. 2024, 14, 1354533. [Google Scholar] [CrossRef]
- Haraguchi, M.; Kiyotani, K.; Tate, T.; Sakata, S.; Sagawa, R.; Takagi, S.; Nagayama, S.; Takeuchi, K.; Takahashi, K.; Katayama, R. Spatiotemporal commonality of the TCR repertoire in a T-cell memory murine model and in metastatic human colorectal cancer. Cancer Immunol. Immunother. 2023, 72, 2971–2989. [Google Scholar] [CrossRef]
- Campana, L.G.; Mansoor, W.; Hill, J.; Macutkiewicz, C.; Curran, F.; Donnelly, D.; Hornung, B.; Charleston, P.; Bristow, R.; Lord, G.M.; et al. T-Cell Infiltration and Clonality May Identify Distinct Survival Groups in Colorectal Cancer: Development and Validation of a Prognostic Model Based on The Cancer Genome Atlas (TCGA) and Clinical Proteomic Tumor Analysis Consortium (CPTAC). Cancers 2022, 14, 5883. [Google Scholar] [CrossRef]
- Sugiyarto, G.; Prossor, D.; Dadas, O.; Arcia-Anaya, E.D.; Elliott, T.; James, E. Protective low-avidity anti-tumour CD8+ T cells are selectively attenuated by regulatory T cells. Immunother. Adv. 2021, 1, ltaa001. [Google Scholar] [CrossRef]
- Shen, L.S.; Wang, J.; Shen, D.F.; Yuan, X.L.; Dong, P.; Li, M.X.; Xue, J.; Zhang, F.M.; Ge, H.L.; Xu, D. CD4+CD25+CD127low/− regulatory T cells express Foxp3 and suppress effector T cell proliferation and contribute to gastric cancers progression. Clin. Immunol. 2009, 131, 109–118. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, X.; Zheng, L.; Zhang, Y.; Li, Y.; Fang, Q.; Gao, R.; Kang, B.; Zhang, Q.; Huang, J.Y.; et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018, 564, 268–272. [Google Scholar] [CrossRef]
- Hui, Z.; Zhang, J.; Zheng, Y.; Yang, L.; Yu, W.; An, Y.; Wei, F.; Ren, X. Single-Cell Sequencing Reveals the Transcriptome and TCR Characteristics of pTregs and In Vitro Expanded iTregs. Front. Immunol. 2021, 12, 619932. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Kondou, R.; Iizuka, A.; Miyata, H.; Maeda, C.; Kanematsu, A.; Ashizawa, T.; Nagashima, T.; Urakami, K.; Shimoda, Y.; et al. Characterization of the Immunological Status of Hypermutated Solid Tumors in the Cancer Genome Analysis Project HOPE. Anticancer Res. 2022, 42, 3537–3549. [Google Scholar] [CrossRef] [PubMed]
- Borras, D.M.; Verbandt, S.; Ausserhofer, M.; Sturm, G.; Lim, J.; Verge, G.A.; Vanmeerbeek, I.; Laureano, R.S.; Govaerts, J.; Sprooten, J.; et al. Single cell dynamics of tumor specificity vs bystander activity in CD8+ T cells define the diverse immune landscapes in colorectal cancer. Cell Discov. 2023, 9, 114. [Google Scholar] [CrossRef]
- Inamori, K.; Togashi, Y.; Fukuoka, S.; Akagi, K.; Ogasawara, K.; Irie, T.; Motooka, D.; Kobayashi, Y.; Sugiyama, D.; Kojima, M.; et al. Importance of lymph node immune responses in MSI-H/dMMR colorectal cancer. JCI Insight 2021, 6, e137365. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Roh, W.; Chen, P.L.; Reuben, A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Gopalakrishnan, V.; Wang, F.; Cooper, Z.A.; Reddy, S.M.; et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 2017, 9, eaah3560. [Google Scholar] [CrossRef]
- Arakawa, A.; Vollmer, S.; Tietze, J.; Galinski, A.; Heppt, M.V.; Burdek, M.; Berking, C.; Prinz, J.C. Clonality of CD4+ Blood T Cells Predicts Longer Survival with CTLA4 or PD-1 Checkpoint Inhibition in Advanced Melanoma. Front. Immunol. 2019, 10, 1336. [Google Scholar] [CrossRef]
- Inoue, H.; Park, J.H.; Kiyotani, K.; Zewde, M.; Miyashita, A.; Jinnin, M.; Kiniwa, Y.; Okuyama, R.; Tanaka, R.; Fujisawa, Y.; et al. Intratumoral expression levels of PD-L1, GZMA, and HLA-A along with oligoclonal T cell expansion associate with response to nivolumab in metastatic melanoma. Oncoimmunology 2016, 5, e1204507. [Google Scholar] [CrossRef]
- Zhao, L.; Pang, Y.; Zhou, Y.; Chen, J.; Fu, H.; Guo, W.; Xu, W.; Xue, X.; Su, G.; Sun, L.; et al. Antitumor efficacy and potential mechanism of FAP-targeted radioligand therapy combined with immune checkpoint blockade. Signal Transduct. Target. Ther. 2024, 9, 142. [Google Scholar] [CrossRef]
- Monjazeb, A.M.; Giobbie-Hurder, A.; Lako, A.; Thrash, E.M.; Brennick, R.C.; Kao, K.Z.; Manuszak, C.; Gentzler, R.D.; Tesfaye, A.; Jabbour, S.K.; et al. A Randomized Trial of Combined PD-L1 and CTLA-4 Inhibition with Targeted Low-Dose or Hypofractionated Radiation for Patients with Metastatic Colorectal Cancer. Clin. Cancer Res. 2021, 27, 2470–2480. [Google Scholar] [CrossRef]
- Zhao, W.; Lei, J.; Ke, S.; Chen, Y.; Xiao, J.; Tang, Z.; Wang, L.; Ren, Y.; Alnaggar, M.; Qiu, H.; et al. Fecal microbiota transplantation plus tislelizumab and fruquintinib in refractory microsatellite stable metastatic colorectal cancer: An open-label, single-arm, phase II trial (RENMIN-215). eClinicalMedicine 2023, 66, 102315. [Google Scholar] [CrossRef] [PubMed]
- Philip, H.; Snir, T.; Gordin, M.; Shugay, M.; Zilberberg, A.; Efroni, S. A T cell repertoire timestamp is at the core of responsiveness to CTLA-4 blockade. iScience 2021, 24, 102100. [Google Scholar] [CrossRef]
- Ree, A.H.; Hoye, E.; Esbensen, Y.; Beitnes, A.R.; Negard, A.; Bernklev, L.; Tetlie, L.K.; Fretland, A.A.; Hamre, H.M.; Kersten, C.; et al. Complete response of metastatic microsatellite-stable BRAF V600E colorectal cancer to first-line oxaliplatin-based chemotherapy and immune checkpoint blockade. Oncoimmunology 2024, 13, 2372886. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Pamplona, R.; Melas, M.; Maoz, A.; Schmit, S.L.; Rennert, H.; Lejbkowicz, F.; Greenson, J.K.; Sanjuan, X.; Lopez-Zambrano, M.; Alonso, M.H.; et al. Lymphocytic infiltration in stage II microsatellite stable colorectal tumors: A retrospective prognosis biomarker analysis. PLoS Med. 2020, 17, e1003292. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Tosolini, M.; Charoentong, P.; Kirilovsky, A.; Bindea, G.; Berger, A.; Camus, M.; Gillard, M.; Bruneval, P.; Fridman, W.H.; et al. Biomolecular network reconstruction identifies T-cell homing factors associated with survival in colorectal cancer. Gastroenterology 2010, 138, 1429–1440. [Google Scholar] [CrossRef]
- Porciello, N.; Franzese, O.; D’Ambrosio, L.; Palermo, B.; Nistico, P. T-cell repertoire diversity: Friend or foe for protective antitumor response? J. Exp. Clin. Cancer Res. 2022, 41, 356. [Google Scholar] [CrossRef]
- Kuang, M.; Cheng, J.; Zhang, C.; Feng, L.; Xu, X.; Zhang, Y.; Zu, M.; Cui, J.; Yu, H.; Zhang, K.; et al. A novel signature for stratifying the molecular heterogeneity of the tissue-infiltrating T-cell receptor repertoire reflects gastric cancer prognosis. Sci. Rep. 2017, 7, 7762. [Google Scholar] [CrossRef]
- Hu, W.; Chen, J.; Qi, L.; Ge, W.; Zheng, S.; Yang, Y. Hypermutated tumours across 11 cancer types show three distinct immune subtypes. Eur. J. Cancer 2021, 148, 230–238. [Google Scholar] [CrossRef]
- Spiotto, M.; Fu, Y.X.; Weichselbaum, R.R. The intersection of radiotherapy and immunotherapy: Mechanisms and clinical implications. Sci. Immunol. 2016, 1, eaag1266. [Google Scholar] [CrossRef]
- Xu, M.; Tsunedomi, R.; Kiyotani, K.; Tomochika, S.; Furuya, K.; Nakajima, M.; Matsui, H.; Tokumitsu, Y.; Shindo, Y.; Yoshida, S.; et al. Anti-VEGF and Anti-EGFR Antibody Therapy on T-Cell Infiltration and TCR Variation in Metastatic Colorectal Cancer. Anticancer Res. 2023, 43, 613–620. [Google Scholar] [CrossRef]
- Akiyoshi, T.; Gotoh, O.; Tanaka, N.; Kiyotani, K.; Yamamoto, N.; Ueno, M.; Fukunaga, Y.; Mori, S. T-cell complexity and density are associated with sensitivity to neoadjuvant chemoradiotherapy in patients with rectal cancer. Cancer Immunol. Immunother. 2021, 70, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Hoye, E.; Dagenborg, V.J.; Torgunrud, A.; Lund-Andersen, C.; Fretland, A.A.; Lorenz, S.; Edwin, B.; Hovig, E.; Fromm, B.; Inderberg, E.M.; et al. T cell receptor repertoire sequencing reveals chemotherapy-driven clonal expansion in colorectal liver metastases. Gigascience 2022, 12, giad032. [Google Scholar] [CrossRef] [PubMed]
- Donahue, R.E.; Srinivasula, S.; Uchida, N.; Kim, I.; St Claire, A.; Duralde, G.; DeGrange, P.; St Claire, M.; Reba, R.C.; Bonifacino, A.C.; et al. Discordance in lymphoid tissue recovery following stem cell transplantation in rhesus macaques: An in vivo imaging study. Blood 2015, 126, 2632–2641. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, H.; Hanaoka, K.; Wada, H.; Kojima, D.; Watanabe, M. The Current Status of T Cell Receptor (TCR) Repertoire Analysis in Colorectal Cancer. Int. J. Mol. Sci. 2025, 26, 2698. https://doi.org/10.3390/ijms26062698
Takahashi H, Hanaoka K, Wada H, Kojima D, Watanabe M. The Current Status of T Cell Receptor (TCR) Repertoire Analysis in Colorectal Cancer. International Journal of Molecular Sciences. 2025; 26(6):2698. https://doi.org/10.3390/ijms26062698
Chicago/Turabian StyleTakahashi, Hiroyuki, Katsuzo Hanaoka, Hideo Wada, Daibo Kojima, and Masato Watanabe. 2025. "The Current Status of T Cell Receptor (TCR) Repertoire Analysis in Colorectal Cancer" International Journal of Molecular Sciences 26, no. 6: 2698. https://doi.org/10.3390/ijms26062698
APA StyleTakahashi, H., Hanaoka, K., Wada, H., Kojima, D., & Watanabe, M. (2025). The Current Status of T Cell Receptor (TCR) Repertoire Analysis in Colorectal Cancer. International Journal of Molecular Sciences, 26(6), 2698. https://doi.org/10.3390/ijms26062698




