Unraveling Metabolic Dysfunction-Associated Steatotic Liver Disease Through the Use of Omics Technologies
Abstract
1. Introduction
1.1. The Evolution of Liver Disease Nomenclature: From NAFLD to MASLD
1.2. Prevalence, Race, and Ethnicity of the Disease
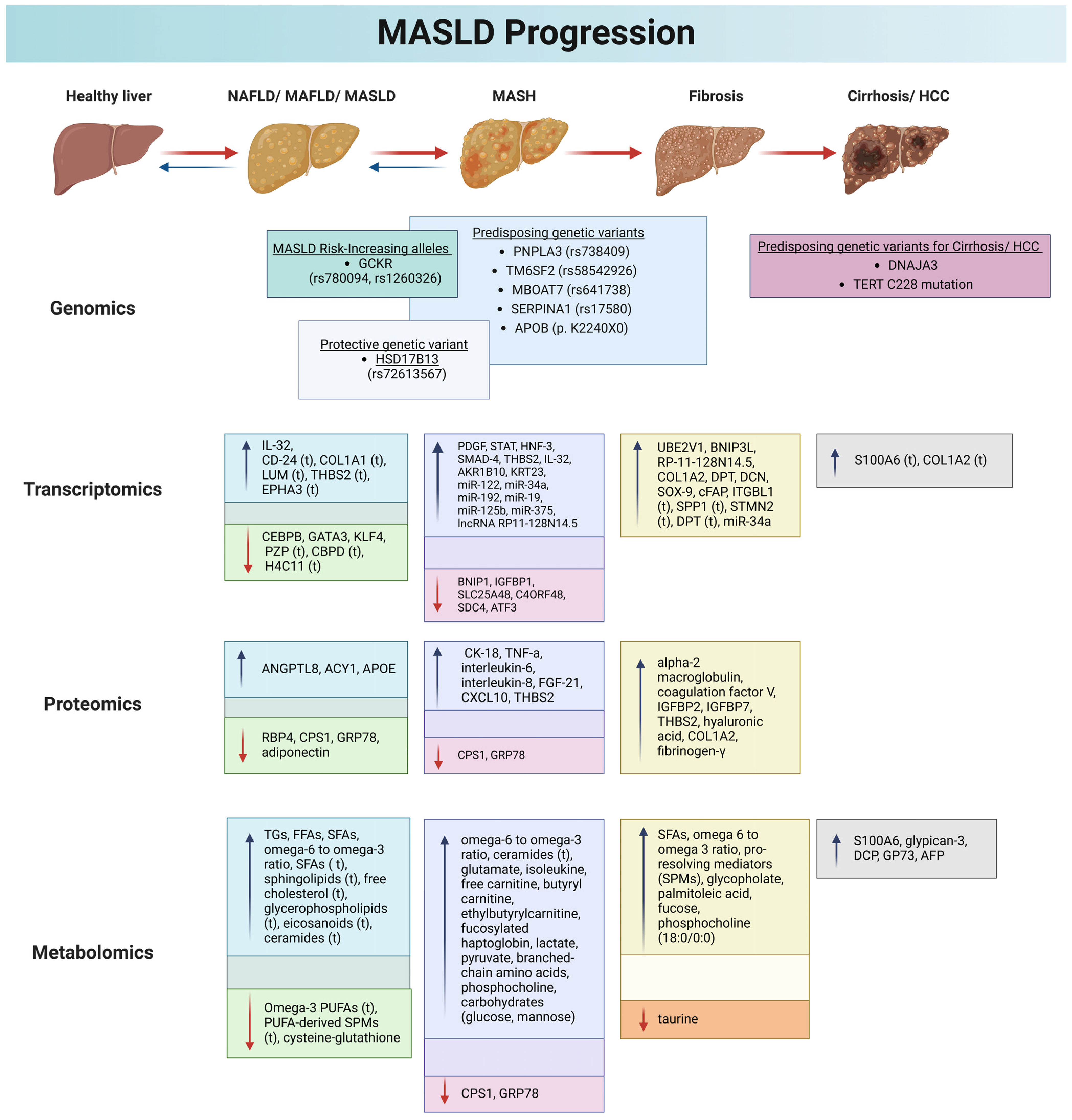
1.3. The Landscape of MASLD: Pathogenesis and Diagnosis
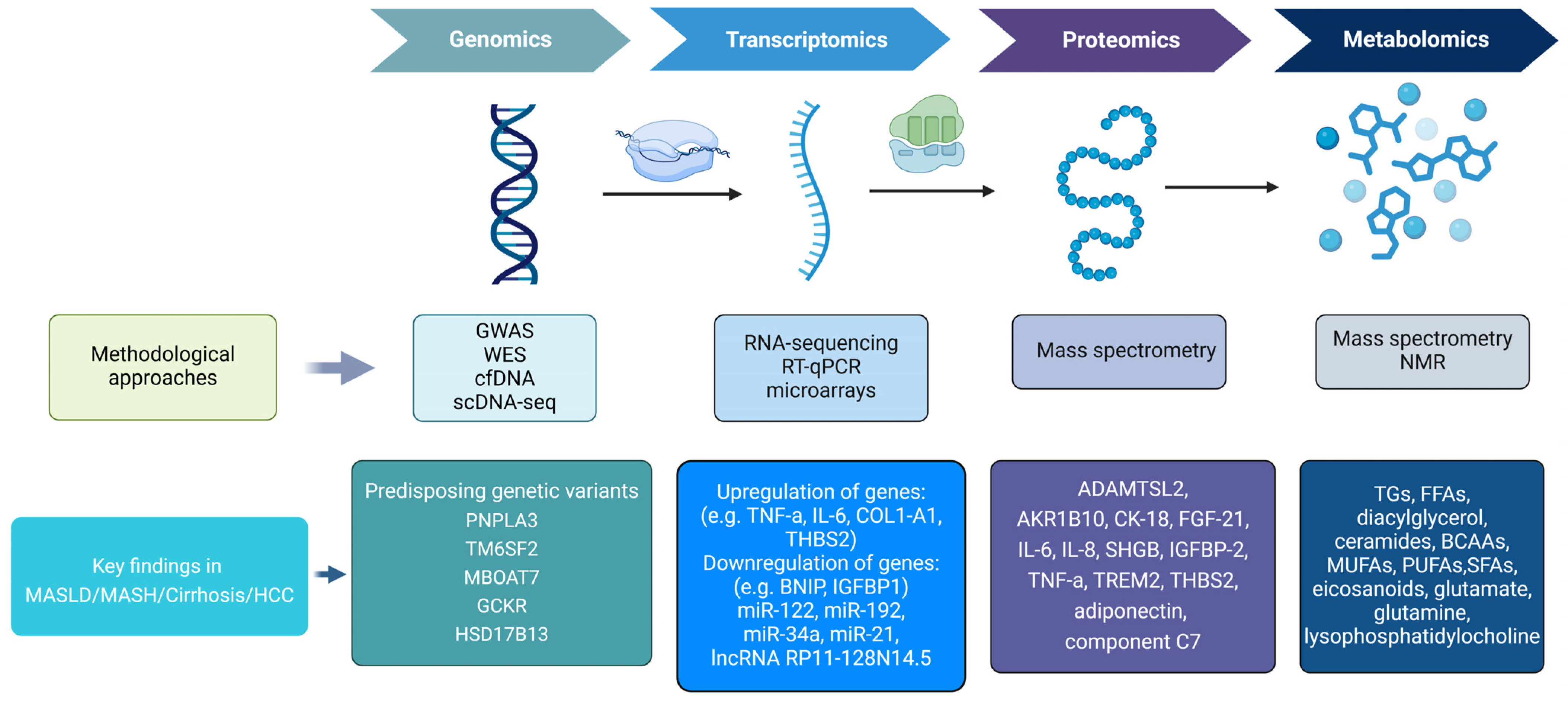
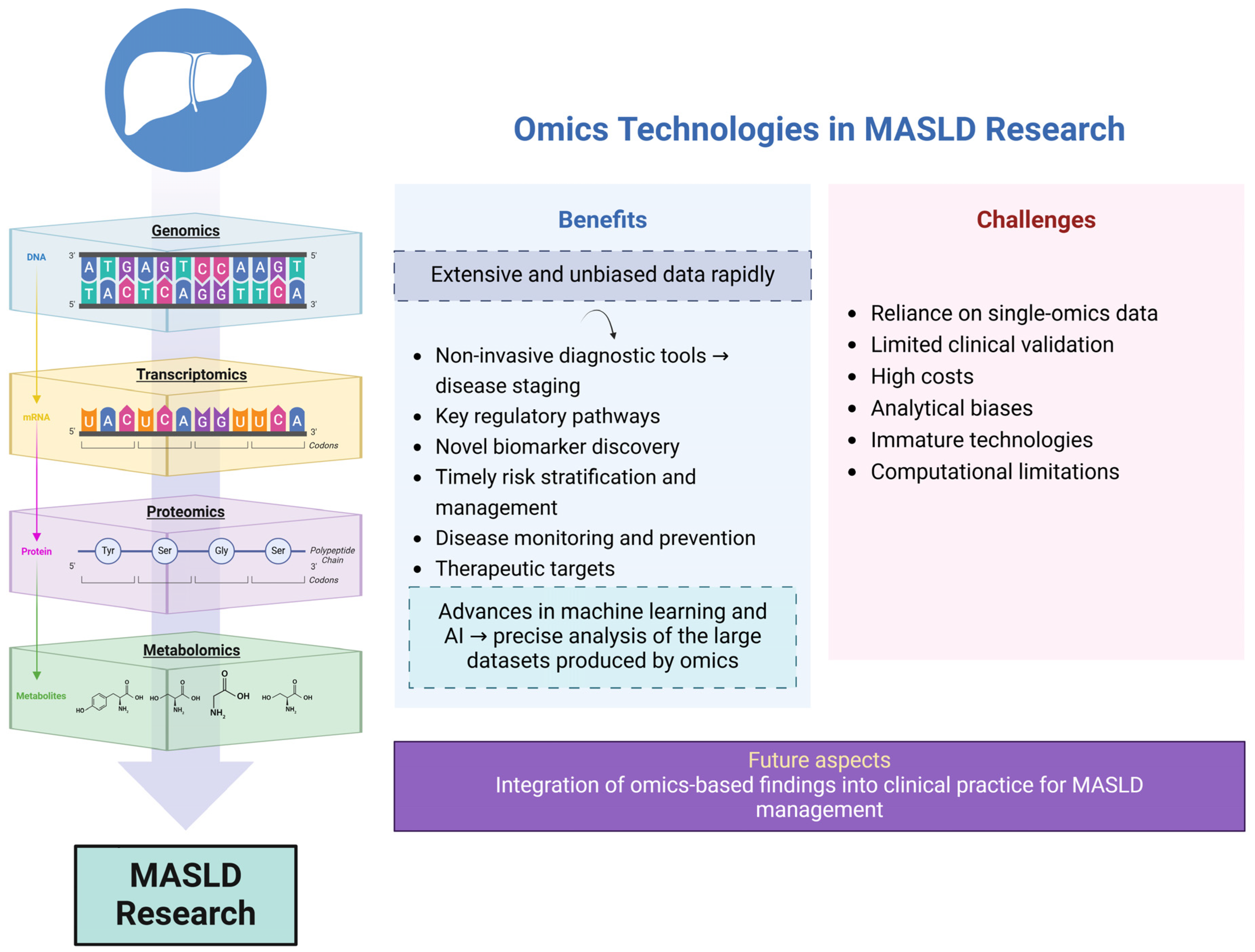
1.4. Omics Technologies
2. Genomics and MASLD
2.1. PNPLA3
2.2. TM6SF2
2.3. GCKR
2.4. MBOAT7
2.5. HSD17B13
2.6. Other Genes
3. Transcriptomics and MASLD
3.1. Micro-RNAs and Non-Coding RNAs
3.2. Long Non-Coding RNAs (lncRNAs)
3.3. Circular RNAs (circRNAs)
4. Proteomics and MASLD
5. Metabolomics and MASLD
5.1. Applied Metabolomics in MASLD
5.2. Applied Lipidomics in NAFLD
5.3. Applied Glycomics in NAFLD
6. Exposomics in NAFLD
7. Discussion
8. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Younossi, Z.M.; Golabi, P.; Paik, J.M.; Henry, A.; Van Dongen, C.; Henry, L. The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): A systematic review. Hepatology 2023, 77, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Riazi, K.; Azhari, H.; Charette, J.H.; E Underwood, F.; A King, J.; Afshar, E.E.; Swain, M.G.; E Congly, S.; Kaplan, G.G.; Shaheen, A.-A. The prevalence and incidence of NAFLD worldwide: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2022, 7, 851–861. [Google Scholar] [CrossRef]
- Karlsen, T.H.; Sheron, N.; Zelber-Sagi, S.; Carrieri, P.; Dusheiko, G.; Bugianesi, E.; Pryke, R.; Hutchinson, S.J.; Sangro, B.; Martin, N.K.; et al. The EASL–Lancet Liver Commission: Protecting the next generation of Europeans against liver disease complications and premature mortality. Lancet 2022, 399, 61–116. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 11–20. [Google Scholar] [CrossRef]
- Wong, V.W.-S.; Ekstedt, M.; Wong, G.L.-H.; Hagström, H. Changing epidemiology, global trends and implications for outcomes of NAFLD. J. Hepatol. 2023, 79, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Ye, L.; Liu, A.; Wen, S.W.; Deng, J.; Wu, X.; Lai, Z. Prevalence of nonalcoholic fatty liver disease in patients with type 2 diabetes mellitus. Medicine 2017, 96, e8179. [Google Scholar] [CrossRef]
- Rinella, M.E.; Rinella, M.E.; Lazarus, J.V.; Lazarus, J.V.; Ratziu, V.; Ratziu, V.; Francque, S.M.; Francque, S.M.; Sanyal, A.J.; Sanyal, A.J.; et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Ann. Hepatol. 2024, 29, 1542–1556. [Google Scholar] [CrossRef]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wong, V.W.-S.; Dufour, J.-F.; Schattenberg, J.M.; et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J. Hepatol. 2020, 73, 202–209. [Google Scholar] [CrossRef]
- Abdelhameed, F.; Kite, C.; Lagojda, L.; Dallaway, A.; Chatha, K.K.; Chaggar, S.S.; Dalamaga, M.; Kassi, E.; Kyrou, I.; Randeva, H.S. Non-invasive Scores and Serum Biomarkers for Fatty Liver in the Era of Metabolic Dysfunction-associated Steatotic Liver Disease (MASLD): A Comprehensive Review From NAFLD to MAFLD and MASLD. Curr. Obes. Rep. 2024, 13, 510–531. [Google Scholar] [CrossRef]
- Chen, L.; Tao, X.; Zeng, M.; Mi, Y.; Xu, L. Clinical and histological features under different nomenclatures of fatty liver disease: NAFLD, MAFLD, MASLD and MetALD. J. Hepatol. 2024, 80, e64–e66. [Google Scholar] [CrossRef]
- Lonardo, A.; Bril, F.; Caldwell, S.H.; Eslam, M.; Fan, J.-G.; Gish, R.G.; Gronbaek, H.; Sanal, M.G.; Stefan, N.; Suzuki, A.; et al. Researchers call for more flexible editorial conduct rather than abruptly adopting only the new MASLD nomenclature. J. Hepatol. 2024, 80, e192–e194. [Google Scholar] [CrossRef]
- Teng, M.L.; Ng, C.H.; Huang, D.Q.; Chan, K.E.; Tan, D.J.; Lim, W.H.; Yang, J.D.; Tan, E.; Muthiah, M.D. Global incidence and prevalence of nonalcoholic fatty liver disease. Clin. Mol. Hepatol. 2023, 29, S32–S42. [Google Scholar] [CrossRef]
- Riazi, K.; Swain, M.G.; Congly, S.E.; Kaplan, G.G.; Shaheen, A.-A. Race and Ethnicity in Non-Alcoholic Fatty Liver Disease (NAFLD): A Narrative Review. Nutrients 2022, 14, 4556. [Google Scholar] [CrossRef]
- Robinson, K.E.; Shah, V.H. Pathogenesis and pathways: Nonalcoholic fatty liver disease & alcoholic liver disease. Transl. Gastroenterol. Hepatol. 2020, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, S.K.; Tarzemani, S.; Aghajanzadeh, T.; Kasravi, M.; Hatami, B.; Zali, M.R.; Baghaei, K. Exploring the role of genetic variations in NAFLD: Implications for disease pathogenesis and precision medicine approaches. Eur. J. Med. Res. 2024, 29, 190. [Google Scholar] [CrossRef]
- Egede, L.E. Race, ethnicity, culture, and disparities in health care. J. Gen. Intern. Med. 2006, 21, 667–669. [Google Scholar] [CrossRef]
- Rich, N.E.; Oji, S.; Mufti, A.R.; Browning, J.D.; Parikh, N.D.; Odewole, M.; Mayo, H.; Singal, A.G. Racial and Ethnic Disparities in Nonalcoholic Fatty Liver Disease Prevalence, Severity, and Outcomes in the United States: A Systematic Review and Meta-analysis. Clin. Gastroenterol. Hepatol. 2018, 16, 198–210.e2. [Google Scholar] [CrossRef]
- Zhang, X.; Heredia, N.I.; Balakrishnan, M.; Thrift, A.P. Prevalence and factors associated with NAFLD detected by vibration controlled transient elastography among US adults: Results from NHANES 2017–2018. PLoS ONE 2021, 16, e0252164. [Google Scholar] [CrossRef]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef]
- Wagenknecht, L.E.; Palmer, N.D.; Bowden, D.W.; Rotter, J.I.; Norris, J.M.; Ziegler, J.; Chen, Y.-D.I.; Haffner, S.; Scherzinger, A.; Langefeld, C.D. Association of PNPLA3 with non-alcoholic fatty liver disease in a minority cohort: The Insulin Resistance Atherosclerosis Family Study. Liver Int. 2011, 31, 412–416. [Google Scholar] [CrossRef]
- Browning, J.D.; Szczepaniak, L.S.; Dobbins, R.; Nuremberg, P.; Horton, J.D.; Cohen, J.C.; Grundy, S.M.; Hobbs, H.H. Prevalence of hepatic steatosis in an urban population in the United States: Impact of ethnicity. Hepatology 2004, 40, 1387–1395. [Google Scholar] [CrossRef]
- Guerrero, R.; Vega, G.L.; Grundy, S.M.; Browning, J.D. Ethnic differences in hepatic steatosis. Hepatology 2009, 49, 791–801. [Google Scholar] [CrossRef]
- Sherif, Z.A.; Saeed, A.; Ghavimi, S.; Nouraie, S.-M.; Laiyemo, A.O.; Brim, H.; Ashktorab, H. Global Epidemiology of Nonalcoholic Fatty Liver Disease and Perspectives on US Minority Populations. Dig. Dis. Sci. 2016, 61, 1214–1225. [Google Scholar] [CrossRef]
- Rojas, Y.A.O.; Cuellar, C.L.V.; Barrón, K.M.A.; Arab, J.P.; Miranda, A.L. Non-alcoholic fatty liver disease prevalence in Latin America: A systematic review and meta-analysis. Ann. Hepatol. 2022, 27, 100706. [Google Scholar] [CrossRef]
- Le, M.H.; Yeo, Y.H.; Li, X.; Li, J.; Zou, B.; Wu, Y.; Ye, Q.; Huang, D.Q.; Zhao, C.; Zhang, J.; et al. 2019 global NAFLD prevalence—A systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2022, 20, 2809–2817.e28. [Google Scholar] [CrossRef]
- De Oliveira, C.P.M.S.; Cotrim, H.P.; Arrese, M. Nonalcoholic Fatty Liver Disease Risk Factors in Latin American Populations: Current Scenario and Perspectives. Clin. Liver Dis. 2019, 13, 39–42. [Google Scholar] [CrossRef]
- Patel, S.A.; Ali, M.K.; Alam, D.; Yan, L.L.; Levitt, N.S.; Bernabe-Ortiz, A.; Checkley, W.; Wu, Y.; Irazola, V.; Gutiérrez, L.; et al. Obesity and its Relation With Diabetes and Hypertension: A Cross-Sectional Study Across 4 Geographical Regions. Glob. Heart 2016, 11, 71–79.e4. [Google Scholar] [CrossRef]
- Pontoriero, A.C.; Trinks, J.; Hulaniuk, M.L.; Caputo, M.; Fortuny, L.; Pratx, L.B.; Frías, A.; Torres, O.; Nuñez, F.; Gadano, A.; et al. Influence of ethnicity on the distribution of genetic polymorphisms associated with risk of chronic liver disease in South American populations. BMC Genet. 2015, 16, 93. [Google Scholar] [CrossRef]
- Cholongitas, E.; Pavlopoulou, I.; Papatheodoridi, M.; Markakis, G.E.; Bouras, E.; Haidich, A.B.; Papatheodoridis, G. Epidemiology of nonalcoholic fatty liver disease in Europe: A systematic review and meta-analysis. Ann. Gastroenterol. 2021, 34, 404–414. [Google Scholar] [CrossRef]
- Nabi, O.; Lacombe, K.; Boursier, J.; Mathurin, P.; Zins, M.; Serfaty, L. Prevalence and Risk Factors of Nonalcoholic Fatty Liver Disease and Advanced Fibrosis in General Population: The French Nationwide NASH-CO Study. Gastroenterology 2020, 159, 791–793.e2. [Google Scholar] [CrossRef]
- Li, J.; Zou, B.; Yeo, Y.H.; Feng, Y.; Xie, X.; Lee, D.H.; Fujii, H.; Wu, Y.; Kam, L.Y.; Ji, F.; et al. Prevalence, incidence, and outcome of non-alcoholic fatty liver disease in Asia, 1999–2019: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2019, 4, 389–398. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver (EASL); European Association for the Study of Diabetes (EASD); European Association for the Study of Obesity (EASO). EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J. Hepatol. 2016, 64, 1388–1402. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Clin. Liver Dis. 2018, 11, 81. [Google Scholar] [CrossRef]
- Minich, A.; Arisar, F.A.Q.; Shaikh, N.-U.S.; Herman, L.; Azhie, A.; Orchanian-Cheff, A.; Patel, K.; Keshavarzi, S.; Bhat, M. Predictors of patient survival following liver transplant in non-alcoholic steatohepatitis: A systematic review and meta-analysis. eClinicalMedicine 2022, 50, 101534. [Google Scholar] [CrossRef]
- Bhat, M.; Mara, K.; Dierkhising, R.; Watt, K.D. Gender, Race and Disease Etiology Predict De Novo Malignancy Risk After Liver Transplantation: Insights for Future Individualized Cancer Screening Guidance. Transplantation 2019, 103, 91–100. [Google Scholar] [CrossRef]
- Lee, K.-C.; Wu, P.-S.; Lin, H.-C. Pathogenesis and treatment of non-alcoholic steatohepatitis and its fibrosis. Clin. Mol. Hepatol. 2023, 29, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Flessa, C.-M.; Kyrou, I.; Nasiri-Ansari, N.; Kaltsas, G.; Papavassiliou, A.G.; Kassi, E.; Randeva, H.S. Endoplasmic Reticulum Stress and Autophagy in the Pathogenesis of Non-alcoholic Fatty Liver Disease (NAFLD): Current Evidence and Perspectives. Curr. Obes. Rep. 2021, 10, 134–161. [Google Scholar] [CrossRef]
- Nasiri-Ansari, N.; Androutsakos, T.; Flessa, C.-M.; Kyrou, I.; Siasos, G.; Randeva, H.S.; Kassi, E.; Papavassiliou, A.G. Endothelial Cell Dysfunction and Nonalcoholic Fatty Liver Disease (NAFLD): A Concise Review. Cells 2022, 11, 2511. [Google Scholar] [CrossRef]
- Kassi, E.; Kyrou, I.; Randeva, H.S. Atherosclerotic and Cardio-Metabolic Diseases: From Molecular Basis to Therapeutic Advances. Int. J. Mol. Sci. 2023, 24, 9737. [Google Scholar] [CrossRef]
- Bassal, T.; Basheer, M.; Boulos, M.; Assy, N. Nonalcoholic Fatty Liver Disease—A Concise Review of Noninvasive Tests and Biomarkers. Metabolites 2022, 12, 1073. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Paik, J.M.; Stepanova, M.; Ong, J.; Alqahtani, S.; Henry, L. Clinical profiles and mortality rates are similar for metabolic dysfunction-associated steatotic liver disease and non-alcoholic fatty liver disease. J. Hepatol. 2024, 80, 694–701. [Google Scholar] [CrossRef]
- Thiele, M.; Villesen, I.F.; Niu, L.; Johansen, S.; Sulek, K.; Nishijima, S.; Van Espen, L.; Keller, M.; Israelsen, M.; Suvitaival, T.; et al. Opportunities and barriers in omics-based biomarker discovery for steatotic liver diseases. J. Hepatol. 2024, 81, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Baranova, A.; Liotta, L.; Petricoin, E.; Younossi, Z.M. The Role of Genomics and Proteomics: Technologies in Studying Non-alcoholic Fatty Liver Disease. Clin. Liver Dis. 2007, 11, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Wanichthanarak, K.; Fahrmann, J.F.; Grapov, D. Genomic, Proteomic, and Metabolomic Data Integration Strategies. Biomark. Insights 2015, 10, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Perakakis, N.; Stefanakis, K.; Mantzoros, C.S. The role of omics in the pathophysiology, diagnosis and treatment of non-alcoholic fatty liver disease. Metabolism 2020, 111, 154320. [Google Scholar] [CrossRef]
- Pelechá, M.; Villanueva-Bádenas, E.; Timor-López, E.; Donato, M.T.; Tolosa, L. Cell Models and Omics Techniques for the Study of Nonalcoholic Fatty Liver Disease: Focusing on Stem Cell-Derived Cell Models. Antioxidants 2021, 11, 86. [Google Scholar] [CrossRef]
- Masarone, M.; Motta, B.M.; Torre, P.; Aquino, M.; Belladonna, F.; Lombardi, M.; Troisi, J.; Persico, M. Evaluating cardiovascular risk in metabolic steatosis with precision medicine non-invasive approaches: Insights from a cohort study. Intern. Emerg. Med. 2024, 19, 2293–2307. [Google Scholar] [CrossRef] [PubMed]
- Kleinstein, S.E.; Rein, M.; Abdelmalek, M.F.; Guy, C.D.; Goldstein, D.B.; Diehl, A.M.; Moylan, C.A. Whole-Exome Sequencing Study of Extreme Phenotypes of NAFLD. Hepatol. Commun. 2018, 2, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Rakela, J.; Rule, J.; Ganger, D.; Lau, J.; Cunningham, J.; Dehankar, M.; Baheti, S.; Lee, W.M.; the Acute Liver Failure Study Group. Whole Exome Sequencing Among 26 Patients With Indeterminate Acute Liver Failure: A Pilot Study. Clin. Transl. Gastroenterol. 2019, 10, e00087. [Google Scholar] [CrossRef]
- Breher-Esch, S.; Sahini, N.; Trincone, A.; Wallstab, C.; Borlak, J. Genomics of lipid-laden human hepatocyte cultures enables drug target screening for the treatment of non-alcoholic fatty liver disease. BMC Med. Genom. 2018, 11, 111. [Google Scholar] [CrossRef] [PubMed]
- Alharthi, J.; Eslam, M. Biomarkers of Metabolic (Dysfunction)-associated Fatty Liver Disease: An Update. J. Clin. Transl. Hepatol. 2022, 10, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Trépo, E.; Valenti, L. Update on NAFLD genetics: From new variants to the clinic. J. Hepatol. 2020, 72, 1196–1209. [Google Scholar] [CrossRef]
- Singal, A.G.; Manjunath, H.; Yopp, A.C.; Beg, M.S.; A Marrero, J.; Gopal, P.; Waljee, A.K. The Effect of PNPLA3 on Fibrosis Progression and Development of Hepatocellular Carcinoma: A Meta-analysis. Am. J. Gastroenterol. 2014, 109, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S. PNPLA3, the triacylglycerol synthesis/hydrolysis/storage dilemma, and nonalcoholic fatty liver disease. World J. Gastroenterol. 2012, 18, 6018. [Google Scholar] [CrossRef]
- Kumari, M.; Schoiswohl, G.; Chitraju, C.; Paar, M.; Cornaciu, I.; Rangrez, A.Y.; Wongsiriroj, N.; Nagy, H.M.; Ivanova, P.T.; Scott, S.A.; et al. Adiponutrin Functions as a Nutritionally Regulated Lysophosphatidic Acid Acyltransferase. Cell Metab. 2012, 15, 691–702. [Google Scholar] [CrossRef]
- Luukkonen, P.K.; Nick, A.; Hölttä-Vuori, M.; Thiele, C.; Isokuortti, E.; Lallukka-Brück, S.; Zhou, Y.; Hakkarainen, A.; Lundbom, N.; Peltonen, M.; et al. Human PNPLA3-I148M variant increases hepatic retention of polyunsaturated fatty acids. J. Clin. Investig. 2019, 4, 127902. [Google Scholar] [CrossRef]
- Yuan, L.; Terrrault, N.A. PNPLA3 and nonalcoholic fatty liver disease: Towards personalized medicine for fatty liver. Hepatobiliary Surg. Nutr. 2020, 9, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.W.; Belbin, G.M.; Sorokin, E.P.; Van Vleck, T.; Wojcik, G.L.; Moscati, A.; Gignoux, C.R.; Cho, J.; Abul-Husn, N.S.; Nadkarni, G.; et al. A common variant in PNPLA3 is associated with age at diagnosis of NAFLD in patients from a multi-ethnic biobank. J. Hepatol. 2020, 72, 1070–1081. [Google Scholar] [CrossRef]
- Salari, N.; Darvishi, N.; Mansouri, K.; Ghasemi, H.; Hosseinian-Far, M.; Darvishi, F.; Mohammadi, M. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease: A systematic review and meta-analysis. BMC Endocr. Disord. 2021, 21, 125. [Google Scholar] [CrossRef] [PubMed]
- Rotman, Y.; Koh, C.; Zmuda, J.M.; Kleiner, D.E.; Liang, T.J.; Nash, C.R.N. The Association of Genetic Variability in Patatin-Like Phospholipase Domain-Containing Protein 3 (PNPLA3) with Histological Severity of Nonalcoholic Fatty Liver Disease. Hepatology 2010, 52, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Grimaudo, S.; Pipitone, R.M.; Pennisi, G.; Celsa, C.; Cammà, C.; Di Marco, V.; Barcellona, M.R.; Boemi, R.; Enea, M.; Giannetti, A.; et al. Association Between PNPLA3 rs738409 C>G Variant and Liver-Related Outcomes in Patients With Nonalcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2020, 18, 935–944.e3. [Google Scholar] [CrossRef]
- Kitamoto, T.; Kitamoto, A.; Yoneda, M.; Hyogo, H.; Ochi, H.; Nakamura, T.; Teranishi, H.; Mizusawa, S.; Ueno, T.; Chayama, K.; et al. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum. Genet. 2013, 132, 783–792. [Google Scholar] [CrossRef]
- Namjou, B.; Lingren, T.; Huang, Y.; Parameswaran, S.; Cobb1, B.L.; Stanaway, I.B.; Connolly, J.J.; Mentch, F.D.; Benoit, B.; Niu, X.; et al. GWAS and enrichment analyses of non-alcoholic fatty liver disease identify new trait-associated genes and pathways across eMERGE Network. BMC Med. 2019, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Flessa, C.-M.; Nasiri-Ansari, N.; Kyrou, I.; Leca, B.M.; Lianou, M.; Chatzigeorgiou, A.; Kaltsas, G.; Kassi, E.; Randeva, H.S. Genetic and Diet-Induced Animal Models for Non-Alcoholic Fatty Liver Disease (NAFLD) Research. Int. J. Mol. Sci. 2022, 23, 15791. [Google Scholar] [CrossRef]
- Santhekadur, P.K.; Kumar, D.P.; Sanyal, A.J. Preclinical models of non-alcoholic fatty liver disease. J. Hepatol. 2018, 68, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Mann, J.P.; Semple, R.K.; Armstrong, M.J. How Useful Are Monogenic Rodent Models for the Study of Human Non-Alcoholic Fatty Liver Disease? Front. Endocrinol. 2016, 7, 145. [Google Scholar] [CrossRef]
- Li, J.Z.; Huang, Y.; Karaman, R.; Ivanova, P.T.; Brown, H.A.; Roddy, T.; Castro-Perez, J.; Cohen, J.C.; Hobbs, H.H. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J. Clin. Investig. 2012, 122, 4130–4144. [Google Scholar] [CrossRef] [PubMed]
- Smagris, E.; BasuRay, S.; Li, J.; Huang, Y.; Ka-man, V.L.; Gromada, J.; Cohen, J.C.; Hobbs, H.H. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 2015, 61, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Kozlitina, J.; Smagris, E.; Stender, S.; Nordestgaard, B.G.; Zhou, H.H.; Tybjærg-Hansen, A.; Vogt, T.F.; Hobbs, H.H.; Cohen, J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014, 46, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Petta, S.; Maglio, C.; Fracanzani, A.L.; Pipitone, R.; Mozzi, E.; Motta, B.M.; Kaminska, D.; Rametta, R.; Grimaudo, S.; et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology 2015, 61, 506–514. [Google Scholar] [CrossRef]
- Luo, F.; Oldoni, F.; Das, A. TM6SF2: A Novel Genetic Player in Nonalcoholic Fatty Liver and Cardiovascular Disease. Hepatol. Commun. 2022, 6, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Wegermann, K.; Garrett, M.E.; Zheng, J.; Coviello, A.; Moylan, C.A.; Abdelmalek, M.F.; Chow, S.; Guy, C.D.; Diehl, A.M.; Ashley-Koch, A.; et al. Sex and Menopause Modify the Effect of Single Nucleotide Polymorphism Genotypes on Fibrosis in NAFLD. Hepatol. Commun. 2021, 5, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Carim-Todd, L.; Escarceller, M.; Estivill, X.; Sumoy, L. Cloning of the novel gene TM6SF1 reveals conservation of clusters of paralogous genes between human chromosomes 15q24→q26 and 19p13.3→p12. Cytogenet. Genome Res. 2000, 90, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Wang, F. APOC3rs2854116, PNPLA3rs738409, and TM6SF2rs58542926 polymorphisms might influence predisposition of NAFLD: A meta-analysis. IUBMB Life 2020, 72, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, S.; Gao, Y.; Ma, H.; Zhan, S.; Yang, Y.; Xin, Y.; Xuan, S. Association of TM6SF2 rs58542926 gene polymorphism with the risk of non-alcoholic fatty liver disease and colorectal adenoma in Chinese Han population. BMC Biochem. 2019, 20, 3. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef]
- Luukkonen, P.K.; Zhou, Y.; Haridas, P.N.; Dwivedi, O.P.; Hyötyläinen, T.; Ali, A.; Juuti, A.; Leivonen, M.; Tukiainen, T.; Ahonen, L.; et al. Impaired hepatic lipid synthesis from polyunsaturated fatty acids in TM6SF2 E167K variant carriers with NAFLD. J. Hepatol. 2017, 67, 128–136. [Google Scholar] [CrossRef]
- Krawczyk, M.; Rau, M.; Schattenberg, J.M.; Bantel, H.; Pathil, A.; Demir, M.; Kluwe, J.; Boettler, T.; Lammert, F.; Geier, A. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: A multicenter biopsy-based study. J. Lipid Res. 2017, 58, 247–255. [Google Scholar] [CrossRef]
- Liu, Y.-L.; Reeves, H.L.; Burt, A.D.; Tiniakos, D.; McPherson, S.; Leathart, J.B.S.; Allison, M.E.D.; Alexander, G.J.; Piguet, A.-C.; Anty, R.; et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 4309. [Google Scholar] [CrossRef]
- Sookoian, S.; Castaño, G.O.; Scian, R.; Mallardi, P.; Gianotti, T.F.; Burgueño, A.L.; Martino, J.S.; Pirola, C.J. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology 2015, 61, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Donati, B.; Dongiovanni, P.; Romeo, S.; Meroni, M.; McCain, M.; Miele, L.; Petta, S.; Maier, S.; Rosso, C.; De Luca, L.; et al. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci. Rep. 2017, 7, 4492. [Google Scholar] [CrossRef]
- Raimondo, A.; Rees, M.G.; Gloyn, A.L. Glucokinase regulatory protein. Curr. Opin. Lipidol. 2015, 26, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Zain, S.M.; Mohamed, Z.; Mohamed, R. A common variant in the glucokinase regulatory gene rs780094 and risk of nonalcoholic fatty liver disease: A meta-analysis. J. Gastroenterol. Hepatol. 2014, 30, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.F.; Vangipurapu, J.; Kuulasmaa, T.; Laakso, M. An intronic variant in the GCKR gene is associated with multiple lipids. Sci. Rep. 2019, 9, 10240. [Google Scholar] [CrossRef]
- Beer, N.L.; Tribble, N.D.; McCulloch, L.J.; Roos, C.; Johnson, P.R.; Orho-Melander, M.; Gloyn, A.L. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum. Mol. Genet. 2009, 18, 4081–4088. [Google Scholar] [CrossRef] [PubMed]
- Mancina, R.M.; Dongiovanni, P.; Petta, S.; Pingitore, P.; Meroni, M.; Rametta, R.; Borén, J.; Montalcini, T.; Pujia, A.; Wiklund, O.; et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 2016, 150, 1219–1230.e6. [Google Scholar] [CrossRef] [PubMed]
- Thangapandi, V.R.; Knittelfelder, O.; Brosch, M.; Patsenker, E.; Vvedenskaya, O.; Buch, S.; Hinz, S.; Hendricks, A.; Nati, M.; Herrmann, A.; et al. Loss of hepatic Mboat7 leads to liver fibrosis. Gut 2021, 70, 940–950. [Google Scholar] [CrossRef]
- Abul-Husn, N.S.; Cheng, X.; Li, A.H.; Xin, Y.; Schurmann, C.; Stevis, P.; Liu, Y.; Kozlitina, J.; Stender, S.; Wood, G.C.; et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N. Engl. J. Med. 2018, 378, 1096–1106. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Belyaeva, O.V.; Brown, P.M.; Fujita, K.; Valles, K.; Karki, S.; De Boer, Y.S.; Koh, C.; Chen, Y.; Du, X.; et al. 17-Beta Hydroxysteroid Dehydrogenase 13 Is a Hepatic Retinol Dehydrogenase Associated With Histological Features of Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 1504–1519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-B.; Su, W.; Xu, H.; Zhang, X.-Y.; Guan, Y.-F. HSD17B13: A Potential Therapeutic Target for NAFLD. Front. Mol. Biosci. 2022, 8, 824776. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.-P.; Ortiz-Pallardó, M.; Ko, Y.; Esch, C.; Zhou, H. Chronic liver disease in heterozygous α1-antitrypsin deficiency PiZ. J. Hepatol. 2000, 33, 883–892. [Google Scholar] [CrossRef]
- Krawczyk, M.; Liebe, R.; Lammert, F. Toward Genetic Prediction of Nonalcoholic Fatty Liver Disease Trajectories: PNPLA3 and Beyond. Gastroenterology 2020, 158, 1865–1880.e1. [Google Scholar] [CrossRef] [PubMed]
- Luukkonen, P.K.; Salomaa, V.; Åberg, F. The Pi∗MZ Allele in Alpha-1 Antitrypsin Increases Liver-Related Outcomes in a Population-Based Study. Gastroenterology 2021, 160, 1874–1875. [Google Scholar] [CrossRef]
- Strnad, P.; Buch, S.; Hamesch, K.; Fischer, J.; Rosendahl, J.; Schmelz, R.; Brueckner, S.; Brosch, M.; Heimes, C.V.; Woditsch, V.; et al. Heterozygous carriage of the alpha1-antitrypsin Pi*Z variant increases the risk to develop liver cirrhosis. Gut 2019, 68, 1099–1107. [Google Scholar] [CrossRef]
- Semmler, G.; Balcar, L.; Oberkofler, H.; Zandanell, S.; Strasser, M.; Niederseer, D.; Feldman, A.; Stickel, F.; Strnad, P.; Datz, C.; et al. PNPLA3 and SERPINA1 Variants Are Associated with Severity of Fatty Liver Disease at First Referral to a Tertiary Center. J. Pers. Med. 2021, 11, 165. [Google Scholar] [CrossRef]
- Lu, W.; Mei, J.; Yang, J.; Wu, Z.; Liu, J.; Miao, P.; Chen, Y.; Wen, Z.; Zhao, Z.; Kong, H.; et al. ApoE deficiency promotes non-alcoholic fatty liver disease in mice via impeding AMPK/mTOR mediated autophagy. Life Sci. 2020, 252, 117601. [Google Scholar] [CrossRef] [PubMed]
- Berg, E.H.v.D.; Corsetti, J.P.; Bakker, S.J.L.; Dullaart, R.P.F. Plasma ApoE elevations are associated with NAFLD: The PREVEND Study. PLoS ONE 2019, 14, e0220659. [Google Scholar] [CrossRef]
- Xu, F.; Liu, C.; Zhou, D.; Zhang, L. TGF-β/SMAD Pathway and Its Regulation in Hepatic Fibrosis. J. Histochem. Cytochem. 2016, 64, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Emdin, C.A.; Haas, M.; Ajmera, V.; Simon, T.G.; Homburger, J.; Neben, C.; Jiang, L.; Wei, W.-Q.; Feng, Q.; Zhou, A.; et al. Association of Genetic Variation With Cirrhosis: A Multi-Trait Genome-Wide Association and Gene–Environment Interaction Study. Gastroenterology 2021, 160, 1620–1633.e13. [Google Scholar] [CrossRef] [PubMed]
- Cefalù, A.B.; Pirruccello, J.P.; Noto, D.; Gabriel, S.; Valenti, V.; Gupta, N.; Spina, R.; Tarugi, P.; Kathiresan, S.; Averna, M.R. A Novel APOB Mutation Identified by Exome Sequencing Cosegregates With Steatosis, Liver Cancer, and Hypocholesterolemia. Arter. Thromb. Vasc. Biol. 2013, 33, 2021–2025. [Google Scholar] [CrossRef] [PubMed]
- Baselli, G.A.; Jamialahmadi, O.; Pelusi, S.; Ciociola, E.; Malvestiti, F.; Saracino, M.; Santoro, L.; Cherubini, A.; Dongiovanni, P.; Maggioni, M.; et al. Rare ATG7 genetic variants predispose patients to severe fatty liver disease. J. Hepatol. 2022, 77, 596–606. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, W.; Huang, S.; Xu, L.; Miao, M.; Wu, C.; Yu, C.; Li, Y.; Xu, C. Serum apoB levels independently predict the development of non-alcoholic fatty liver disease: A 7-year prospective study. Liver Int. 2017, 37, 1202–1208. [Google Scholar] [CrossRef] [PubMed]
- Ueno, M.; Takeda, H.; Takai, A.; Seno, H. Risk factors and diagnostic biomarkers for nonalcoholic fatty liver disease-associated hepatocellular carcinoma: Current evidence and future perspectives. World J. Gastroenterol. 2022, 28, 3410–3421. [Google Scholar] [CrossRef]
- Akuta, N.; Kawamura, Y.; Arase, Y.; Saitoh, S.; Fujiyama, S.; Sezaki, H.; Hosaka, T.; Kobayashi, M.; Suzuki, Y.; Suzuki, F.; et al. TERT Promoter Mutation in Serum Cell-Free DNA Is a Diagnostic Marker of Primary Hepatocellular Carcinoma in Patients with Nonalcoholic Fatty Liver Disease. Oncology 2021, 99, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Casamassimi, A.; Federico, A.; Rienzo, M.; Esposito, S.; Ciccodicola, A. Transcriptome Profiling in Human Diseases: New Advances and Perspectives. Int. J. Mol. Sci. 2017, 18, 1652. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Baranova, A.; Ziegler, K.; Del Giacco, L.; Schlauch, K.; Born, T.L.; Elariny, H.; Gorreta, F.; VanMeter, A.; Younoszai, A.; et al. A Genomic and Proteomic Study of the Spectrum of Nonalcoholic Fatty Liver Disease *. Hepatology 2005, 42, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Ryaboshapkina, M.; Hammar, M. Human hepatic gene expression signature of non-alcoholic fatty liver disease progression, a meta-analysis. Sci. Rep. 2017, 7, 12361. [Google Scholar] [CrossRef] [PubMed]
- Teufel, A.; Itzel, T.; Erhart, W.; Brosch, M.; Wang, X.Y.; Kim, Y.O.; Von Schonfels, W.; Herrmann, A.; Brückner, S.; Stickel, F.; et al. Comparison of Gene Expression Patterns Between Mouse Models of Nonalcoholic Fatty Liver Disease and Liver Tissues From Patients. Gastroenterology 2016, 151, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Sun, C.; Hou, Y.; Tang, Y.; Zhu, Z.; Zhang, Z.; Zhang, Y.; Wang, L.; Zhao, Q.; Chen, M.-G.; et al. A comprehensive bioinformatics analysis on multiple Gene Expression Omnibus datasets of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Sci. Rep. 2018, 8, 7630. [Google Scholar] [CrossRef] [PubMed]
- Suppli, M.P.; Rigbolt, K.T.G.; Veidal, S.S.; Heebøll, S.; Eriksen, P.L.; Demant, M.; Bagger, J.I.; Nielsen, J.; Oró, D.; Thrane, S.W.; et al. Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am. J. Physiol. Liver Physiol. 2019, 316, G462–G472. [Google Scholar] [CrossRef]
- Starmann, J.; Fälth, M.; Spindelböck, W.; Lanz, K.-L.; Lackner, C.; Zatloukal, K.; Trauner, M.; Sültmann, H. Gene Expression Profiling Unravels Cancer-Related Hepatic Molecular Signatures in Steatohepatitis but Not in Steatosis. PLoS ONE 2012, 7, e46584. [Google Scholar] [CrossRef]
- Moylan, C.A.; Pang, H.; Dellinger, A.; Suzuki, A.; Garrett, M.E.; Guy, C.D.; Murphy, S.K.; Ashley-Koch, A.E.; Choi, S.S.; Michelotti, G.A.; et al. Hepatic Gene Expression Profiles Differentiate Presymptomatic Patients With Mild Versus Severe Nonalcoholic Fatty Liver Disease. Hepatology 2014, 59, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Yan, X.; Jing, S.; Wu, Y.; Shan, Y.; Guo, W.; Gu, J.; Li, Y.; Zhang, H.; Li, H. Single-cell and spatially resolved transcriptomics for liver biology. Hepatology 2024, 80, 698–720. [Google Scholar] [CrossRef] [PubMed]
- Govaere, O.; Cockell, S.; Tiniakos, D.; Queen, R.; Younes, R.; Vacca, M.; Alexander, L.; Ravaioli, F.; Palmer, J.; Petta, S.; et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci. Transl. Med. 2020, 12, eaba4448. [Google Scholar] [CrossRef] [PubMed]
- Govaere, O.; Hasoon, M.; Alexander, L.; Cockell, S.; Tiniakos, D.; Ekstedt, M.; Schattenberg, J.M.; Boursier, J.; Bugianesi, E.; Ratziu, V.; et al. A proteo-transcriptomic map of non-alcoholic fatty liver disease signatures. Nat. Metab. 2023, 5, 572–578. [Google Scholar] [CrossRef]
- Chen, P.; Zhu, Z.; Geng, H.; Cui, X.; Han, Y.; Wang, L.; Zhang, Y.; Lu, H.; Wang, X.; Zhang, Y.; et al. Integrated spatial metabolomics and transcriptomics decipher the hepatoprotection mechanisms of wedelolactone and demethylwedelolactone on non-alcoholic fatty liver disease. J. Pharm. Anal. 2024, 14, 100910. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; He, J.; Shi, L.; Zhang, M.; Zheng, J.; Fan, Y. Identification of biomarkers in nonalcoholic fatty liver disease: A machine learning method and experimental study. Front. Genet. 2022, 13, 1020899. [Google Scholar] [CrossRef] [PubMed]
- Baselli, G.A.; Dongiovanni, P.; Rametta, R.; Meroni, M.; Pelusi, S.; Maggioni, M.; Badiali, S.; Pingitore, P.; Maurotti, S.; Montalcini, T.; et al. Liver transcriptomics highlights interleukin-32 as novel NAFLD-related cytokine and candidate biomarker. Gut 2020, 69, 1855–1866. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, M.; Endo, H.; Mawatari, H.; Nozaki, Y.; Fujita, K.; Akiyama, T.; Higurashi, T.; Uchiyama, T.; Yoneda, K.; Takahashi, H.; et al. Gene expression profiling of non-alcoholic steatohepatitis using gene set enrichment analysis. Hepatol. Res. 2008, 38, 1204–1212. [Google Scholar] [CrossRef] [PubMed]
- Gerhard, G.S.; Legendre, C.; Still, C.D.; Chu, X.; Petrick, A.; DiStefano, J.K. Transcriptomic Profiling of Obesity-Related Nonalcoholic Steatohepatitis Reveals a Core Set of Fibrosis-Specific Genes. J. Endocr. Soc. 2018, 2, 710–726. [Google Scholar] [CrossRef] [PubMed]
- Kozumi, K.; Kodama, T.; Murai, H.; Sakane, S.; Govaere, O.; Cockell, S.; Motooka, D.; Kakita, N.; Yamada, Y.; Kondo, Y.; et al. Transcriptomics Identify Thrombospondin-2 as a Biomarker for NASH and Advanced Liver Fibrosis. Hepatology 2021, 74, 2452–2466. [Google Scholar] [CrossRef] [PubMed]
- Di Mauro, S.; Scamporrino, A.; Petta, S.; Urbano, F.; Filippello, A.; Ragusa, M.; Di Martino, M.T.; Scionti, F.; Grimaudo, S.; Pipitone, R.M.; et al. Serum coding and non-coding RNAs as biomarkers of NAFLD and fibrosis severity. Liver Int. 2019, 39, 1742–1754. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yuan, S.-L.; Yin, J.-Y.; Yang, K.; Zhou, X.-G.; Xie, W.; Wang, Q. Differences of core genes in liver fibrosis and hepatocellular carcinoma: Evidence from integrated bioinformatics and immunohistochemical analysis. World J. Gastrointest. Oncol. 2022, 14, 1265–1280. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gong, Q.; Zhang, J.; Chen, L.; Zhang, Z.; Lu, L.; Yu, D.; Han, Y.; Zhang, D.; Chen, P.; et al. Characterization of gene expression profiles in HBV-related liver fibrosis patients and identification of ITGBL1 as a key regulator of fibrogenesis. Sci. Rep. 2017, 7, 43446. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.; de Ribeiro, A.V.; Prakoso, E.; Veillard, A.; Shackel, N.; Bu, Y.; Brooks, B.; Cavanagh, E.; Raleigh, J.; McLennan, S.; et al. Lower serum fibroblast activation protein shows promise in the exclusion of clinically significant liver fibrosis due to non-alcoholic fatty liver disease in diabetes and obesity. Diabetes Res. Clin. Pract. 2015, 108, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Syn, W.-K.; Choi, S.S.; Liaskou, E.; Karaca, G.F.; Agboola, K.M.; Oo, Y.H.; Mi, Z.; Pereira, T.A.; Zdanowicz, M.; Malladi, P.; et al. Osteopontin is induced by hedgehog pathway activation and promotes fibrosis progression in nonalcoholic steatohepatitis. Hepatology 2011, 53, 106–115. [Google Scholar] [CrossRef]
- Paradis, V.; Dargere, D.; Bieche, Y.; Asselah, T.; Marcellin, P.; Vidaud, M.; Bedossa, P. SCG10 Expression on activation of hepatic stellate cells promotes cell motility through interference with microtubules. Am. J. Pathol. 2010, 177, 1791–1797. [Google Scholar] [CrossRef]
- Lefebvre, P.; Lalloyer, F.; Baugé, E.; Pawlak, M.; Gheeraert, C.; Dehondt, H.; Vanhoutte, J.; Woitrain, E.; Hennuyer, N.; Mazuy, C.; et al. Interspecies NASH disease activity whole-genome profiling identifies a fibrogenic role of PPARα-regulated dermatopontin. J. Clin. Investig. 2017, 2, e92264. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, M.; Ling, B.; Liu, S.; Zheng, Y.; Nie, C.; Yuan, Z.; Zhou, L.; Guo, G.; Tong, A.; et al. Increased expression of S100A6 promotes cell proliferation and migration in human hepatocellular carcinoma. J. Mol. Med. 2014, 92, 291–303. [Google Scholar] [CrossRef]
- Sun, J.; Shi, R.; Wu, Y.; Lou, Y.; Nie, L.; Zhang, C.; Cao, Y.; Yan, Q.; Ye, L.; Zhang, S.; et al. Integration of transcriptomic analysis and multiple machine learning approaches identifies NAFLD progression-specific hub genes to reveal distinct genomic patterns and actionable targets. J. Big Data 2024, 11, 40. [Google Scholar] [CrossRef]
- Gharanei, S.; Shabir, K.; Brown, J.E.; Weickert, M.O.; Barber, T.M.; Kyrou, I.; Randeva, H.S. Regulatory microRNAs in Brown, Brite and White Adipose Tissue. Cells 2020, 9, 2489. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xiong, Y.; Sheng, Q.; Zhao, S.; Wattacheril, J.; Flynn, C.R. A micro-RNA expression signature for human NAFLD progression. J. Gastroenterol. 2016, 51, 1022–1030. [Google Scholar] [CrossRef]
- Cheung, O.; Puri, P.; Eicken, C.; Contos, M.J.; Mirshahi, F.; Maher, J.W.; Kellum, J.M.; Min, H.; Luketic, V.A.; Sanyal, A.J. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 2008, 48, 1810–1820. [Google Scholar] [CrossRef]
- Huang, R.; Duan, X.; Fan, J.; Li, G.; Wang, B. Role of Noncoding RNA in Development of Nonalcoholic Fatty Liver Disease. BioMed Res. Int. 2019, 2019, 8690592. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Meroni, M.; Longo, M.; Fargion, S.; Fracanzani, A.L. miRNA Signature in NAFLD: A Turning Point for a Non-Invasive Diagnosis. Int. J. Mol. Sci. 2018, 19, 3966. [Google Scholar] [CrossRef] [PubMed]
- Tobaruela-Resola, A.L.; Milagro, F.I.; Elorz, M.; Benito-Boillos, A.; Herrero, J.I.; Mogna-Peláez, P.; Tur, J.A.; Martínez, J.A.; Abete, I.; Zulet, M.Á. Circulating miR-122-5p, miR-151a-3p, miR-126-5p and miR-21-5p as potential predictive biomarkers for Metabolic Dysfunction-Associated Steatotic Liver Disease assessment. J. Physiol. Biochem. 2024, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hochreuter, M.Y.; Dall, M.; Treebak, J.T.; Barrès, R. MicroRNAs in non-alcoholic fatty liver disease: Progress and perspectives. Mol. Metab. 2022, 65, 101581. [Google Scholar] [CrossRef] [PubMed]
- Pirola, C.J.; Fernandez Gianotti, T.; Castano, G.O.; Mallardi, P.; San Martino, J.; Mora Gonzalez Lopez Ledesma, M.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015, 64, 800–812. [Google Scholar] [CrossRef]
- Miyaaki, H.; Ichikawa, T.; Kamo, Y.; Taura, N.; Honda, T.; Shibata, H.; Milazzo, M.; Fornari, F.; Gramantieri, L.; Bolondi, L.; et al. Significance of serum and hepatic microRNA-122 levels in patients with non-alcoholic fatty liver disease. Liver Int. 2014, 34, e302–e307. [Google Scholar] [CrossRef] [PubMed]
- Latorre, J.; Moreno-Navarrete, J.M.; Mercader, J.M.; Sabater, M.; Rovira, Ò.; Gironès, J.; Ricart, W.; Fernández-Real, J.M.; Ortega, F.J. Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int. J. Obes. 2017, 41, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.D.; Sharapova, T.; Lake, A.D.; Blomme, E.; Maher, J.; Cherrington, N.J. Circulating microRNA 122 in the methionine and choline-deficient mouse model of non-alcoholic steatohepatitis. J. Appl. Toxicol. 2013, 34, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.M.; Xu, Z.; Shek, F.H.; Wong, K.-F.; Lee, N.P.; Poon, R.T.; Chen, J.; Luk, J.M. miR-122 Targets Pyruvate Kinase M2 and Affects Metabolism of Hepatocellular Carcinoma. PLoS ONE 2014, 9, e86872. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, S.; Pfeffer, S.; Baumert, T.F.; Zeisel, M.B. miR-122—A key factor and therapeutic target in liver disease. J. Hepatol. 2015, 62, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Al Ageeli, E. Dual Roles of microRNA-122 in Hepatocellular Carcinoma and Breast Cancer Progression and Metastasis: A Comprehensive Review. Curr. Issues Mol. Biol. 2024, 46, 11975–11992. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-H.; Ampuero, J.; Gil-Gómez, A.; Montero-Vallejo, R.; Rojas, Á.; Muñoz-Hernández, R.; Gallego-Durán, R.; Romero-Gómez, M. miRNAs in patients with non-alcoholic fatty liver disease: A systematic review and meta-analysis. J. Hepatol. 2018, 69, 1335–1348. [Google Scholar] [CrossRef]
- Matias-Garcia, P.R.; Wilson, R.; Mussack, V.; Reischl, E.; Waldenberger, M.; Gieger, C.; Anton, G.; Peters, A.; Kuehn-Steven, A. Impact of long-term storage and freeze-thawing on eight circulating microRNAs in plasma samples. PLoS ONE 2020, 15, e0227648. [Google Scholar] [CrossRef] [PubMed]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating MicroRNAs in Patients with Chronic Hepatitis C and Non-Alcoholic Fatty Liver Disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.P.; Rau, M.; Schmitt, J.; Malsch, C.; Hammer, C.; Bantel, H.; Müllhaupt, B.; Geier, A. Performance of Serum microRNAs -122, -192 and -21 as Biomarkers in Patients with Non-Alcoholic Steatohepatitis. PLoS ONE 2015, 10, e0142661. [Google Scholar] [CrossRef]
- Okamoto, K.; Koda, M.; Okamoto, T.; Onoyama, T.; Miyoshi, K.; Kishina, M.; Matono, T.; Kato, J.; Tokunaga, S.; Sugihara, T.; et al. Serum miR-379 expression is related to the development and progression of hypercholesterolemia in non-alcoholic fatty liver disease. PLoS ONE 2020, 15, e0219412. [Google Scholar] [CrossRef]
- Fang, Z.; Dou, G.; Wang, L. MicroRNAs in the Pathogenesis of Nonalcoholic Fatty Liver Disease. Int. J. Biol. Sci. 2021, 17, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.A.; Ratziu, V.; Boursier, J.; Francque, S.; Bedossa, P.; Majd, Z.; Cordonnier, G.; Ben Sudrik, F.; Darteil, R.; Liebe, R.; et al. A blood-based biomarker panel (NIS4) for non-invasive diagnosis of non-alcoholic steatohepatitis and liver fibrosis: A prospective derivation and global validation study. Lancet Gastroenterol. Hepatol. 2020, 5, 970–985. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118, Correction in Nat. Rev. Mol. Cell Biol. 2021, 22, 159. [Google Scholar] [CrossRef]
- Sun, C.; Liu, X.; Yi, Z.; Xiao, X.; Yang, M.; Hu, G.; Liu, H.; Liao, L.; Huang, F. Genome-wide analysis of long noncoding RNA expression profiles in patients with non-alcoholic fatty liver disease. IUBMB Life 2015, 67, 847–852. [Google Scholar] [CrossRef]
- Atanasovska, B.; Rensen, S.S.; Van Der Sijde, M.R.; Marsman, G.; Kumar, V.; Jonkers, I.; Withoff, S.; Shiri-Sverdlov, R.; Greve, J.W.M.; Faber, K.N.; et al. A liver-specific long noncoding RNA with a role in cell viability is elevated in human nonalcoholic steatohepatitis. Hepatology 2017, 34, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhang, Y.; Ji, X.; Li, B.; Wang, Y.; Huang, Y.; Zhang, X.; Yu, J.; Zou, R.; Qin, D.; et al. Long Noncoding RNA lncRHPL Regulates Hepatic VLDL Secretion by Modulating hnRNPU/BMAL1/MTTP Axis. Diabetes 2022, 71, 1915–1928. [Google Scholar] [CrossRef]
- Jin, S.-S.; Lin, C.-J.; Lin, X.-F.; Zheng, J.-Z.; Guan, H.-Q. Silencing lncRNA NEAT1 reduces nonalcoholic fatty liver fat deposition by regulating the miR-139-5p/c-Jun/SREBP-1c pathway. Ann. Hepatol. 2022, 27, 100584. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, J.; Tang, X.; Li, Y.; Xia, P.; Gao, X. Berberine ameliorates nonalcoholic fatty liver disease by a global modulation of hepatic mRNA and lncRNA expression profiles. J. Transl. Med. 2015, 13, 24. [Google Scholar] [CrossRef]
- Shao, T.; Pan, Y.-H.; Xiong, X.-D. Circular RNA: An important player with multiple facets to regulate its parental gene expression. Mol. Ther.-Nucleic Acids 2021, 23, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Liu, C.-H.; Ampuero, J.; Wu, D.; Jiang, W.; Zhou, L.; Li, H.; Bai, L.; Romero-Gómez, M.; Tang, H. Circular RNAs in non-alcoholic fatty liver disease: Functions and clinical significance. RNA Biol. 2024, 21, 65–79. [Google Scholar] [CrossRef]
- Li, P.; Shan, K.; Liu, Y.; Zhang, Y.; Xu, L.; Xu, L. CircScd1 Promotes Fatty Liver Disease via the Janus Kinase 2/Signal Transducer and Activator of Transcription 5 Pathway. Dig. Dis. Sci. 2019, 64, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-Y.; He, C.-X.; Wang, Y.-Q.; Sun, C.; Li, G.-M.; Su, Q.; Pan, Q.; Fan, J.-G. Circular RNA Profiling and Bioinformatic Modeling Identify Its Regulatory Role in Hepatic Steatosis. BioMed. Res. Int. 2017, 2017, 5936171. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tan, Q.-Q.; Tan, X.-R.; Li, S.-J.; Zhang, X.-X. Circ_0057558 promotes nonalcoholic fatty liver disease by regulating ROCK1/AMPK signaling through targeting miR-206. Cell Death Dis. 2021, 12, 809. [Google Scholar] [CrossRef]
- Lim, M.S.; Elenitoba-Johnson, K.S.J. Proteomics in pathology research. Mod. Pathol. 2004, 84, 1227–1244. [Google Scholar] [CrossRef]
- Cyranoski, D. China takes centre stage for liver proteome. Nature 2003, 425, 441. [Google Scholar] [CrossRef][Green Version]
- He, F. Human Liver Proteome Project. Mol. Cell. Proteom. 2005, 4, 1841–1848. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Aebersold, R.; Baker, M.S.; Bian, X.; Bo, X.; Chan, D.W.; Chang, C.; Chen, L.; Chen, X.; Chen, Y.-J.; et al. π-HuB: The proteomic navigator of the human body. Nature 2024, 636, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Bell, L.N.; Theodorakis, J.L.; Vuppalanchi, R.; Saxena, R.; Bemis, K.G.; Wang, M.; Chalasani, N. Serum Proteomics and Biomarker Discovery Across the Spectrum of Nonalcoholic Fatty Liver Disease. Hepatology 2010, 51, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhuang, X.; Zhang, J.; Li, M.; Du, S.; Tian, J.; Yuan, Y.; Ji, G.; Hu, C. Clinical characterization and proteomic profiling of lean nonalcoholic fatty liver disease. Front. Endocrinol. 2023, 14, 1171397. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, A.J.; Williams, S.A.; Lavine, J.E.; Neuschwander-Tetri, B.A.; Alexander, L.; Ostroff, R.; Biegel, H.; Kowdley, K.V.; Chalasani, N.; Dasarathy, S.; et al. Defining the serum proteomic signature of hepatic steatosis, inflammation, ballooning and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2023, 78, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Altomare, A.A.; Aiello, G.; Garcia, J.L.; Garrone, G.; Zoanni, B.; Carini, M.; Aldini, G.; D’amato, A. Protein Profiling of a Cellular Model of NAFLD by Advanced Bioanalytical Approaches. Int. J. Mol. Sci. 2022, 23, 9025. [Google Scholar] [CrossRef]
- Rodríguez-Suárez, E.; Duce, A.M.; Caballería, J.; Arrieta, F.M.; Fernández, E.; Gómara, C.; Alkorta, N.; Ariz, U.; Martínez-Chantar, M.L.; Lu, S.C.; et al. Non-alcoholic fatty liver disease proteomics. Proteom.—Clin. Appl. 2010, 4, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wadhawan, S.; Greenfield, A.; Decato, B.E.; Oseini, A.M.; Collen, R.; Shevell, D.E.; Thompson, J.; Jarai, G.; Charles, E.D.; et al. SOMAscan Proteomics Identifies Serum Biomarkers Associated With Liver Fibrosis in Patients With NASH. Hepatol. Commun. 2021, 5, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Sveinbjornsson, G.; Ulfarsson, M.O.; Thorolfsdottir, R.B.; Jonsson, B.A.; Einarsson, E.; Gunnlaugsson, G.; Rognvaldsson, S.; Arnar, D.O.; Baldvinsson, M.; Bjarnason, R.G.; et al. Multiomics study of nonalcoholic fatty liver disease. Nat. Genet. 2022, 54, 1652–1663. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Chen, Q.; Shen, T.; Wang, X.; Fang, W.; Wu, X.; Yuan, Z.; Chen, G.; Ling, W.; Chen, Y. Association of sex hormone-binding globulin with nonalcoholic fatty liver disease in Chinese adults. Nutr. Metab. 2018, 15, 79. [Google Scholar] [CrossRef]
- Hua, M.-C.; Su, H.-M.; Yao, T.-C.; Kuo, M.-L.; Lai, M.-W.; Tsai, M.-H.; Huang, J.-L. Alternation of plasma fatty acids composition and desaturase activities in children with liver steatosis. PLoS ONE 2017, 12, e0182277. [Google Scholar] [CrossRef]
- Dong, J.; Liu, C.; Lu, J.; Wang, L.; Xie, S.; Ji, L.; Lu, B. The relationship between sex hormone-binding protein and non-alcoholic fatty liver disease using Mendelian randomisation. Eur. J. Clin. Investig. 2024, 54, e14082. [Google Scholar] [CrossRef] [PubMed]
- Castera, L.; Friedrich-Rust, M.; Loomba, R. Noninvasive Assessment of Liver Disease in Patients with Nonalcoholic Fatty Liver Disease. Gastroenterology 2019, 156, 1264–1281.e4. [Google Scholar] [CrossRef]
- Grigorescu, M.; Crisan, D.; Radu, C.; Grigorescu, M.D.; Sparchez, Z.; Serban, A. A novel pathophysiological-based panel of biomarkers for the diagnosis of nonalcoholic steatohepatitis. J. Physiol. Pharmacol. 2012, 63, 347–353. [Google Scholar] [PubMed]
- Kyrou, I.; Randeva, H.S.; Tsigos, C.; Kaltsas, G.; Weickert, M.O. Clinical Problems Caused by Obesity. 2000. Available online: https://www.ncbi.nlm.nih.gov/pubmed/25905207 (accessed on 1 October 2024).
- Kim, Y.-S.; Lee, S.-H.; Park, S.G.; Won, B.Y.; Chun, H.; Cho, D.-Y.; Kim, M.-J.; Lee, J.E.; Haam, J.-H.; Han, K. Low levels of total and high-molecular-weight adiponectin may predict non-alcoholic fatty liver in Korean adults. Metabolism 2020, 103, 154026. [Google Scholar] [CrossRef] [PubMed]
- Mavilia, M.G.; Wu, G.Y. Liver and serum adiponectin levels in non-alcoholic fatty liver disease. J. Dig. Dis. 2022, 22, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Polyzos, S.A.; Toulis, K.A.; Goulis, D.G.; Zavos, C.; Kountouras, J. Serum total adiponectin in nonalcoholic fatty liver disease: A systematic review and meta-analysis. Metabolism 2011, 60, 313–326. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, J.; Man, K.; Chu, E.S.; Yau, T.O.; Go, M.Y.; Deng, J.; Lu, L.; Wong, V.W.; Sung, J.J.; et al. CXCL10 plays a key role as an inflammatory mediator and a non-invasive biomarker of non-alcoholic steatohepatitis. J. Hepatol. 2014, 61, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Xu, D.; Li, Q.; Xie, N.; Xia, J.; Huo, Q.; Li, P.; Chen, Q.; Huang, S. Metabonomics screening of serum identifies pyroglutamate as a diagnostic biomarker for nonalcoholic steatohepatitis. Clin. Chim. Acta 2017, 473, 89–95. [Google Scholar] [CrossRef]
- Shen, J.; Chan, H.L.; Wong, G.L.; Choi, P.C.; Chan, A.W.; Chan, H.Y.; Chim, A.M.; Yeung, D.K.; Chan, F.K.; Woo, J.; et al. Non-invasive diagnosis of non-alcoholic steatohepatitis by combined serum biomarkers. J. Hepatol. 2012, 56, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Baranova, A.; Stepanova, M.; Page, S.; Calvert, V.S.; Afendy, A.; Goodman, Z.; Chandhoke, V.; Liotta, L.; Petricoin, E. Phosphoproteomic Biomarkers Predicting Histologic Nonalcoholic Steatohepatitis and Fibrosis. J. Proteome Res. 2010, 9, 3218–3224. [Google Scholar] [CrossRef] [PubMed]
- Trak-Smayra, V.; Dargere, D.; Noun, R.; Albuquerque, M.; Yaghi, C.; Gannage-Yared, M.-H.; Bedossa, P.; Paradis, V. Serum proteomic profiling of obese patients: Correlation with liver pathology and evolution after bariatric surgery. Gut 2009, 58, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Sun, Y.; Cheng, Q.; Hu, K.; Ye, J.; Zhao, Y.; Wu, J.; Shao, X.; Fang, L.; Ding, Y.; et al. Proteomic analysis to identify differentially expressed proteins between subjects with metabolic healthy obesity and non-alcoholic fatty liver disease. J. Proteom. 2022, 221, 103683. [Google Scholar] [CrossRef]
- Wood, G.C.; Chu, X.; Argyropoulos, G.; Benotti, P.; Rolston, D.; Mirshahi, T.; Petrick, A.; Gabrielson, J.; Carey, D.J.; DiStefano, J.K.; et al. A multi-component classifier for nonalcoholic fatty liver disease (NAFLD) based on genomic, proteomic, and phenomic data domains. Sci. Rep. 2017, 7, 43238. [Google Scholar] [CrossRef] [PubMed]
- Lockman, K.A.; Htun, V.; Sinha, R.; Treskes, P.; Nelson, L.J.; Martin, S.F.; Rogers, S.M.; Le Bihan, T.; Hayes, P.C.; Plevris, J.N. Proteomic profiling of cellular steatosis with concomitant oxidative stress in vitro. Lipids Health Dis. 2016, 15, 114. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Xu, Q.; Li, Z.; Ren, Y.; Jiao, Q.; Wang, L.; Wang, Y. Role of the angiopoietin-like protein family in the progression of NAFLD. Heliyon 2024, 10, e27739. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Lee, S.-G.; Lee, C.J.; Kim, S.H.; Song, Y.-M.; Yoon, M.R.; Jeon, B.H.; Lee, J.H.; Lee, B.-W.; Kang, E.S.; et al. Association between betatrophin/ANGPTL8 and non-alcoholic fatty liver disease: Animal and human studies. Sci. Rep. 2016, 6, 24013. [Google Scholar] [CrossRef]
- Yang, L.-Y.; Zhao, D.; Ma, Y.; Wang, Y.; Zhang, Q.; Lang, J.-N. Serum ANGPTL2 and ANGPTL3 as potential biomarkers for diagnosis of non-alcoholic fatty liver disease. Environ. Dis. 2020, 5, 29. [Google Scholar] [CrossRef]
- Altun, Ö.; Dikker, O.; Arman, Y.; Ugurlukisi, B.; Kutlu, O.; Cil, E.O.; Yoldemir, S.A.; Akarsu, M.; Ozcan, M.; Kalyon, S.; et al. Serum Angiopoietin-like peptide 4 levels in patients with hepatic steatosis. Cytokine 2018, 111, 496–499. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Liu, S.; Zhang, Z.; Hu, J. Circulating angiopoietin-like proteins in metabolic-associated fatty liver disease: A systematic review and meta-analysis. Lipids Health Dis. 2022, 20, 55. [Google Scholar] [CrossRef] [PubMed]
- Marrero, J.A.; Feng, Z.; Wang, Y.; Nguyen, M.H.; Befeler, A.S.; Roberts, L.R.; Reddy, K.R.; Harnois, D.; Llovet, J.M.; Normolle, D.; et al. α-Fetoprotein, Des-γ Carboxyprothrombin, and Lectin-Bound α-Fetoprotein in Early Hepatocellular Carcinoma. Gastroenterology 2009, 137, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Parikh, N.D.; Mehta, A.S.; Singal, A.G.; Block, T.; Marrero, J.A.; Lok, A.S. Biomarkers for the Early Detection of Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2495–2503. [Google Scholar] [CrossRef]
- Xu, Q.; Feng, M.; Ren, Y.; Liu, X.; Gao, H.; Li, Z.; Su, X.; Wang, Q.; Wang, Y. From NAFLD to HCC: Advances in noninvasive diagnosis. Biomed. Pharmacother. 2023, 165, 115028. [Google Scholar] [CrossRef] [PubMed]
- Best, J.; Bechmann, L.P.; Sowa, J.P.; Sydor, S.; Dechêne, A.; Pflanz, K.; Bedreli, S.; Schotten, C.; Geier, A.; Berg, T.; et al. GALAD Score Detects Early Hepatocellular Carcinoma in an International Cohort of Patients With Nonalcoholic Steatohepatitis. Clin. Gastroenterol. Hepatol. 2020, 18, 728–735. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, M.-H.; Liu, D.; Lin, Y.; Song, S.J.; Chu, E.S.-H.; Liu, D.; Singh, S.; Berman, M.; Lau, H.C.-H.; et al. A blood-based biomarker panel for non-invasive diagnosis of metabolic dysfunction-associated steatohepatitis. Cell Metab. 2024, 37, 59–68.e3. [Google Scholar] [CrossRef]
- Giera, M.; Ivanisevic, J. Introduction. In A Practical Guide to Metabolomics Applications in Health and Disease; Learning Materials in Biosciences; Springer: New York, NY, USA, 2023; pp. 3–30. [Google Scholar] [CrossRef]
- Qiu, S.; Cai, Y.; Yao, H.; Lin, C.; Xie, Y.; Tang, S.; Zhang, A. Small molecule metabolites: Discovery of biomarkers and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 132. [Google Scholar] [CrossRef]
- Masoodi, M.; Gastaldelli, A.; Hyötyläinen, T.; Arretxe, E.; Alonso, C.; Gaggini, M.; Brosnan, J.; Anstee, Q.M.; Millet, O.; Ortiz, P.; et al. Metabolomics and lipidomics in NAFLD: Biomarkers and non-invasive diagnostic tests. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 835–856. [Google Scholar] [CrossRef] [PubMed]
- Puri, P.; Wiest, M.M.; Cheung, O.; Mirshahi, F.; Sargeant, C.; Min, H.; Contos, M.J.; Sterling, R.K.; Fuchs, M.; Zhou, H.; et al. The plasma lipidomic signature of nonalcoholic steatohepatitis†‡. Hepatology 2009, 50, 1827–1838. [Google Scholar] [CrossRef]
- Gorden, D.; Myers, D.S.; Ivanova, P.T.; Fahy, E.; Maurya, M.R.; Gupta, S.; Min, J.; Spann, N.J.; McDonald, J.G.; Kelly, S.L.; et al. Biomarkers of NAFLD progression: A lipidomics approach to an epidemic. J. Lipid Res. 2015, 56, 722–736. [Google Scholar] [CrossRef]
- Chiappini, F.; Desterke, C.; Bertrand-Michel, J.; Guettier, C.; Le Naour, F. Hepatic and serum lipid signatures specific to nonalcoholic steatohepatitis in murine models. Sci. Rep. 2016, 6, 31587. [Google Scholar] [CrossRef] [PubMed]
- Chiappini, F.; Coilly, A.; Kadar, H.; Gual, P.; Tran, A.; Desterke, C.; Samuel, D.; Duclos-Vallée, J.-C.; Touboul, D.; Bertrand-Michel, J.; et al. Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients. Sci. Rep. 2017, 7, 46658. [Google Scholar] [CrossRef] [PubMed]
- Musso, G.; Cassader, M.; Paschetta, E.; Gambino, R. Bioactive Lipid Species and Metabolic Pathways in Progression and Resolution of Nonalcoholic Steatohepatitis. Gastroenterology 2018, 155, 282–302.e8. [Google Scholar] [CrossRef] [PubMed]
- Gormaz, J.G.; Rodrigo, R.; Videla, L.A.; Beems, M. Biosynthesis and bioavailability of long-chain polyunsaturated fatty acids in non-alcoholic fatty liver disease. Prog. Lipid Res. 2010, 49, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Choudhury, J.; Cheung, O.; Sargeant, C.; Contos, M.J.; Sanyal, A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef]
- Horrillo, R.; González-Périz, A.; Martínez-Clemente, M.; López-Parra, M.; Ferré, N.; Titos, E.; Morán-Salvador, E.; Deulofeu, R.; Arroyo, V.; Clària, J. 5-Lipoxygenase Activating Protein Signals Adipose Tissue Inflammation and Lipid Dysfunction in Experimental Obesity. J. Immunol. 2010, 184, 3978–3987. [Google Scholar] [CrossRef] [PubMed]
- Rius, B.; Duran-Güell, M.; Flores-Costa, R.; López-Vicario, C.; Lopategi, A.; Alcaraz-Quiles, J.; Casulleras, M.; Lozano, J.J.; Titos, E.; Clària, J. The specialized proresolving lipid mediator maresin 1 protects hepatocytes from lipotoxic and hypoxia-induced endoplasmic reticulum stress. FASEB J. 2017, 31, 5384–5398. [Google Scholar] [CrossRef] [PubMed]
- Perakakis, N.; Polyzos, S.A.; Yazdani, A.; Sala-Vila, A.; Kountouras, J.; Anastasilakis, A.D.; Mantzoros, C.S. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: A proof of concept study. Metabolism 2019, 101, 154005. [Google Scholar] [CrossRef]
- Zhou, Y.; Orešič, M.; Leivonen, M.; Gopalacharyulu, P.; Hyysalo, J.; Arola, J.; Verrijken, A.; Francque, S.; Van Gaal, L.; Hyötyläinen, T.; et al. Noninvasive Detection of Nonalcoholic Steatohepatitis Using Clinical Markers and Circulating Levels of Lipids and Metabolites. Clin. Gastroenterol. Hepatol. 2016, 14, 1463–1472.e6. [Google Scholar] [CrossRef]
- Caussy, C.; Ajmera, V.H.; Puri, P.; Hsu, C.L.-S.; Bassirian, S.; Mgdsyan, M.; Singh, S.; Faulkner, C.; A Valasek, M.; Rizo, E.; et al. Serum metabolites detect the presence of advanced fibrosis in derivation and validation cohorts of patients with non-alcoholic fatty liver disease. Gut 2019, 68, 1884–1892. [Google Scholar] [CrossRef]
- Robinson, E.J.; Taddeo, M.C.; Chu, X.; Shi, W.; Wood, C.; Still, C.; Rovnyak, V.G.; Rovnyak, D. Aqueous Metabolite Trends for the Progression of Nonalcoholic Fatty Liver Disease in Female Bariatric Surgery Patients by Targeted 1H-NMR Metabolomics. Metabolites 2021, 11, 737. [Google Scholar] [CrossRef]
- Li, H.; Wang, L.; Yan, X.; Liu, Q.; Yu, C.; Wei, H.; Li, Y.; Zhang, X.; He, F.; Jiang, Y. A Proton Nuclear Magnetic Resonance Metabonomics Approach for Biomarker Discovery in Nonalcoholic Fatty Liver Disease. J. Proteome Res. 2011, 10, 2797–2806. [Google Scholar] [CrossRef]
- Kalhan, S.C.; Guo, L.; Edmison, J.; Dasarathy, S.; McCullough, A.J.; Hanson, R.W.; Milburn, M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metab. Clin. Exp. 2011, 60, 404–413. [Google Scholar] [CrossRef]
- Kozyra, M.; Johansson, I.; Nordling, Å.; Ullah, S.; Lauschke, V.M.; Ingelman-Sundberg, M. Human hepatic 3D spheroids as a model for steatosis and insulin resistance. Sci. Rep. 2018, 8, 14297. [Google Scholar] [CrossRef] [PubMed]
- Salvador, A.F.; Shyu, C.-R.; Parks, E.J. Measurement of lipid flux to advance translational research: Evolution of classic methods to the future of precision health. Exp. Mol. Med. 2022, 54, 1348–1353. [Google Scholar] [CrossRef] [PubMed]
- Deng, K.-Q.; Huang, X.; Lei, F.; Zhang, X.-J.; Zhang, P.; She, Z.-G.; Cai, J.; Ji, Y.-X.; Li, H. Role of hepatic lipid species in the progression of nonalcoholic fatty liver disease. Am. J. Physiol. Physiol. 2022, 323, C630–C639. [Google Scholar] [CrossRef] [PubMed]
- Ooi, G.J.; Meikle, P.J.; Huynh, K.; Earnest, A.; Roberts, S.K.; Kemp, W.; Parker, B.L.; Brown, W.; Burton, P.; Watt, M.J. Hepatic lipidomic remodeling in severe obesity manifests with steatosis and does not evolve with non-alcoholic steatohepatitis. J. Hepatol. 2021, 75, 524–535. [Google Scholar] [CrossRef]
- Mayo, R.; Crespo, J.; Martínez-Arranz, I.; Banales, J.M.; Arias, M.; Mincholé, I.; de la Fuente, R.A.; Jimenez-Agüero, R.; Alonso, C.; de Luis, D.A.; et al. Metabolomic-based noninvasive serum test to diagnose nonalcoholic steatohepatitis: Results from discovery and validation cohorts. Hepatol. Commun. 2018, 2, 807–820. [Google Scholar] [CrossRef]
- Perez-Diaz-Del-Campo, N.; Riezu-Boj, J.I.; Marin-Alejandre, B.A.; Monreal, J.I.; Elorz, M.; Herrero, J.I.; Benito-Boillos, A.; Milagro, F.I.; Tur, J.A.; Abete, I.; et al. Three Different Genetic Risk Scores Based on Fatty Liver Index, Magnetic Resonance Imaging and Lipidomic for a Nutrigenetic Personalized Management of NAFLD: The Fatty Liver in Obesity Study. Diagnostics 2021, 11, 1083. [Google Scholar] [CrossRef]
- Ogawa, Y.; Kobayashi, T.; Honda, Y.; Kessoku, T.; Tomeno, W.; Imajo, K.; Nakahara, T.; Oeda, S.; Nagaoki, Y.; Amano, Y.; et al. Metabolomic/lipidomic-based analysis of plasma to diagnose hepatocellular ballooning in patients with non-alcoholic fatty liver disease: A multicenter study. Hepatol. Res. 2020, 50, 955–965. [Google Scholar] [CrossRef]
- Wang, Z.-H.; I Zheng, K.; Wang, X.-D.; Qiao, J.; Li, Y.-Y.; Zhang, L.; Zheng, M.-H.; Wu, J. LC-MS-based lipidomic analysis in distinguishing patients with nonalcoholic steatohepatitis from nonalcoholic fatty liver. Hepatobiliary Pancreat. Dis. Int. 2021, 20, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Jambulingam, N.; Forlano, R.; Preston, B.; Mullish, B.H.; Portone, G.; Baheer, Y.; Yee, M.; Goldin, R.D.; Thursz, M.R.; Manousou, P. Metabolic Profile Reflects Stages of Fibrosis in Patients with Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2023, 24, 3563. [Google Scholar] [CrossRef]
- Lovric, A.; Granér, M.; Bjornson, E.; Arif, M.; Benfeitas, R.; Nyman, K.; Ståhlman, M.; Pentikäinen, M.O.; Lundbom, J.; Hakkarainen, A.; et al. Characterization of different fat depots in NAFLD using inflammation-associated proteome, lipidome and metabolome. Sci. Rep. 2018, 8, 14200. [Google Scholar] [CrossRef] [PubMed]
- Preuss, C.; Jelenik, T.; Bódis, K.; Müssig, K.; Burkart, V.; Szendroedi, J.; Roden, M.; Markgraf, D.F. A New Targeted Lipidomics Approach Reveals Lipid Droplets in Liver, Muscle and Heart as a Repository for Diacylglycerol and Ceramide Species in Non-Alcoholic Fatty Liver. Cells 2019, 8, 277. [Google Scholar] [CrossRef]
- Okada, L.S.d.R.R.; Oliveira, C.P.; Stefano, J.T.; Nogueira, M.A.; da Silva, I.D.C.G.; Cordeiro, F.B.; Alves, V.A.F.; Torrinhas, R.S.; Carrilho, F.J.; Puri, P.; et al. Omega-3 PUFA modulate lipogenesis, ER stress, and mitochondrial dysfunction markers in NASH—Proteomic and lipidomic insight. Clin. Nutr. 2018, 37, 1474–1484. [Google Scholar] [CrossRef] [PubMed]
- Cantero, I.; Abete, I.; del Bas, J.M.; Caimari, A.; Arola, L.; Zulet, M.A.; Martinez, J.A. Changes in lysophospholipids and liver status after weight loss: The RESMENA study. Nutr. Metab. 2018, 15, 51. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, J. Chromatographic and Mass Spectrometric Techniques. In Handbook of Glycomics; Elsevier: Amsterdam, The Netherlands, 2010; pp. 161–176. [Google Scholar] [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Kannagi, R.; Toole, B.; Stanley, P. Glycosylation Changes in Cancer. 2015. Available online: https://www.ncbi.nlm.nih.gov/pubmed/2579340 (accessed on 29 August 2024).
- Rosso, N.; Stephenson, A.M.; Giraudi, P.J.; Tiribelli, C. Diagnostic management of nonalcoholic fatty liver disease: A transformational period in the development of diagnostic and predictive tools—A narrative review. Ann. Transl. Med. 2021, 9, 727. [Google Scholar] [CrossRef]
- He, J.; Mao, W.; Zhang, J.; Jin, X. Association between Serum Sialic Acid Levels and Nonalcoholic Fatty Liver Disease: A Cross-Sectional Study. Ann. Nutr. Metab. 2015, 67, 69–75. [Google Scholar] [CrossRef]
- Li, J.; Hsu, H.-C.; Mountz, J.D.; Allen, J.G. Unmasking Fucosylation: From Cell Adhesion to Immune System Regulation and Diseases. Cell Chem. Biol. 2018, 25, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Herrera, H.; Block, T. GlycosylaTion and Liver Cancer; Elsevier: Amsterdam, The Netherlands, 2015; pp. 257–279. [Google Scholar] [CrossRef]
- Kamada, Y.; Ono, M.; Hyogo, H.; Fujii, H.; Sumida, Y.; Mori, K.; Tanaka, S.; Yamada, M.; Akita, M.; Mizutani, K.; et al. A novel noninvasive diagnostic method for nonalcoholic steatohepatitis using two glycobiomarkers. Hepatology 2015, 62, 1433–1443. [Google Scholar] [CrossRef]
- Zhou, Y.; Fukuda, T.; Hang, Q.; Hou, S.; Isaji, T.; Kameyama, A.; Gu, J. Inhibition of fucosylation by 2-fluorofucose suppresses human liver cancer HepG2 cell proliferation and migration as well as tumor formation. Sci. Rep. 2017, 7, 11563. [Google Scholar] [CrossRef] [PubMed]
- Barouki, R.; Samson, M.; Blanc, E.B.; Colombo, M.; Zucman-Rossi, J.; Lazaridis, K.N.; Miller, G.W.; Coumoul, X. The exposome and liver disease—How environmental factors affect liver health. J. Hepatol. 2023, 79, 492–505. [Google Scholar] [CrossRef]
- Price, E.J.; Vitale, C.M.; Miller, G.W.; David, A.; Barouki, R.; Audouze, K.; Walker, D.I.; Antignac, J.-P.; Coumoul, X.; Bessonneau, V.; et al. Merging the exposome into an integrated framework for “omics” sciences. iScience 2022, 25, 103976. [Google Scholar] [CrossRef] [PubMed]
- Siroux, V.; Agier, L.; Slama, R. The exposome concept: A challenge and a potential driver for environmental health research. Eur. Respir. Rev. 2016, 25, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Merra, G.; Gualtieri, P.; La Placa, G.; Frank, G.; Della Morte, D.; De Lorenzo, A.; Di Renzo, L. The Relationship between Exposome and Microbiome. Microorganisms 2024, 12, 1386. [Google Scholar] [CrossRef] [PubMed]
- Vilar-Gomez, E.; Calzadilla-Bertot, L.; Wong, V.W.-S.; Castellanos, M.; La Fuente, R.A.-D.; Metwally, M.; Eslam, M.; Gonzalez-Fabian, L.; Sanz, M.A.-Q.; Conde-Martín, A.F.; et al. Fibrosis Severity as a Determinant of Cause-Specific Mortality in Patients With Advanced Nonalcoholic Fatty Liver Disease: A Multi-National Cohort Study. Gastroenterology 2018, 155, 443–457.e17. [Google Scholar] [CrossRef] [PubMed]
- Roth, K.; Yang, Z.; Agarwal, M.; Liu, W.; Peng, Z.; Long, Z.; Birbeck, J.; Westrick, J.; Liu, W.; Petriello, M.C. Exposure to a mixture of legacy, alternative, and replacement per- and polyfluoroalkyl substances (PFAS) results in sex-dependent modulation of cholesterol metabolism and liver injury. Environ. Int. 2021, 157, 106843. [Google Scholar] [CrossRef]
- Laparra, J.M.; Fernández-Musoles, R.; Tejedor, A.G. Immunonutritional contribution of gut microbiota to fatty liver disease. Nutr. Hosp. 2019, 37, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, R. Transforming Clinical Research: The Power of High-Throughput Omics Integration. Proteomes 2024, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Jung, E.-S.; Chae, S.-W.; Jung, S.-J.; Daily, J.W.; Park, S. Biomarkers for Health Functional Foods in Metabolic Dysfunction-Associated Steatotic Liver Disorder (MASLD) Prevention: An Integrative Analysis of Network Pharmacology, Gut Microbiota, and Multi-Omics. Nutrients 2024, 16, 3061. [Google Scholar] [CrossRef] [PubMed]
- Dar, M.A.; Arafah, A.; Bhat, K.A.; Khan, A.; Khan, M.S.; Ali, A.; Ahmad, S.M.; Rashid, S.M.; Rehman, M.U. Multiomics technologies: Role in disease biomarker discoveries and therapeutics. Briefings Funct. Genom. 2023, 22, 76–96. [Google Scholar] [CrossRef]

| Omics | Key Contributions of Omics in MASLD Research | References |
|---|---|---|
| Genomics | • Identification of genetic risk factors | [46,48,49,53] |
| • Understanding disease mechanisms | [51] | |
| • Personalized treatment and risk stratification | [46,52] | |
| • Therapeutic target identification | [51] | |
| • Improved monitoring and management | [46] | |
| • Addressing population-specific challenges | [52] | |
| Transcriptomics | • Insight into disease mechanisms | [106] |
| • Gene expression regulation | [46,106] | |
| • Identification of disease pathways | [46,107,108,109,110,111,112] | |
| • Stratification of disease stages | [115] | |
| • Identification of biomarkers for diagnosis | [116,129,138] | |
| Proteomics | • Identification of biomarkers and improved biomarker combinations | [169,179,180] |
| • Non-invasive diagnostic tools | [119,171] | |
| • Disease stage differentiation | [169,173,185,186] | |
| • Uncover therapeutic targets | [168,172] | |
| • Personalized medicine potential | [168] | |
| Metabolomics | • Comprehensive insight into disease mechanisms | [205] |
| • Biomarker discovery—non-invasive diagnostic | [202,205] | |
| • Precision medicine applications (Algorithm development) | [215,216] | |
| • Real-time physiological status | [204,205] | |
| • Advancing therapeutics | [205] | |
| Exposomics | • Holistic risk assessment | [243,247] |
| • Insight into pathological mechanisms | [243,247,248] | |
| • Targeted prevention and intervention | [243,247,249] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bourganou, M.V.; Chondrogianni, M.E.; Kyrou, I.; Flessa, C.-M.; Chatzigeorgiou, A.; Oikonomou, E.; Lambadiari, V.; Randeva, H.S.; Kassi, E. Unraveling Metabolic Dysfunction-Associated Steatotic Liver Disease Through the Use of Omics Technologies. Int. J. Mol. Sci. 2025, 26, 1589. https://doi.org/10.3390/ijms26041589
Bourganou MV, Chondrogianni ME, Kyrou I, Flessa C-M, Chatzigeorgiou A, Oikonomou E, Lambadiari V, Randeva HS, Kassi E. Unraveling Metabolic Dysfunction-Associated Steatotic Liver Disease Through the Use of Omics Technologies. International Journal of Molecular Sciences. 2025; 26(4):1589. https://doi.org/10.3390/ijms26041589
Chicago/Turabian StyleBourganou, Maria V., Maria Eleni Chondrogianni, Ioannis Kyrou, Christina-Maria Flessa, Antonios Chatzigeorgiou, Evangelos Oikonomou, Vaia Lambadiari, Harpal S. Randeva, and Eva Kassi. 2025. "Unraveling Metabolic Dysfunction-Associated Steatotic Liver Disease Through the Use of Omics Technologies" International Journal of Molecular Sciences 26, no. 4: 1589. https://doi.org/10.3390/ijms26041589
APA StyleBourganou, M. V., Chondrogianni, M. E., Kyrou, I., Flessa, C.-M., Chatzigeorgiou, A., Oikonomou, E., Lambadiari, V., Randeva, H. S., & Kassi, E. (2025). Unraveling Metabolic Dysfunction-Associated Steatotic Liver Disease Through the Use of Omics Technologies. International Journal of Molecular Sciences, 26(4), 1589. https://doi.org/10.3390/ijms26041589








