Biochemical Characterization of R-Loop Degradation by Chloroplast-Localized RNase H1 from Arabidopsis thaliana
Abstract
1. Introduction
2. Results
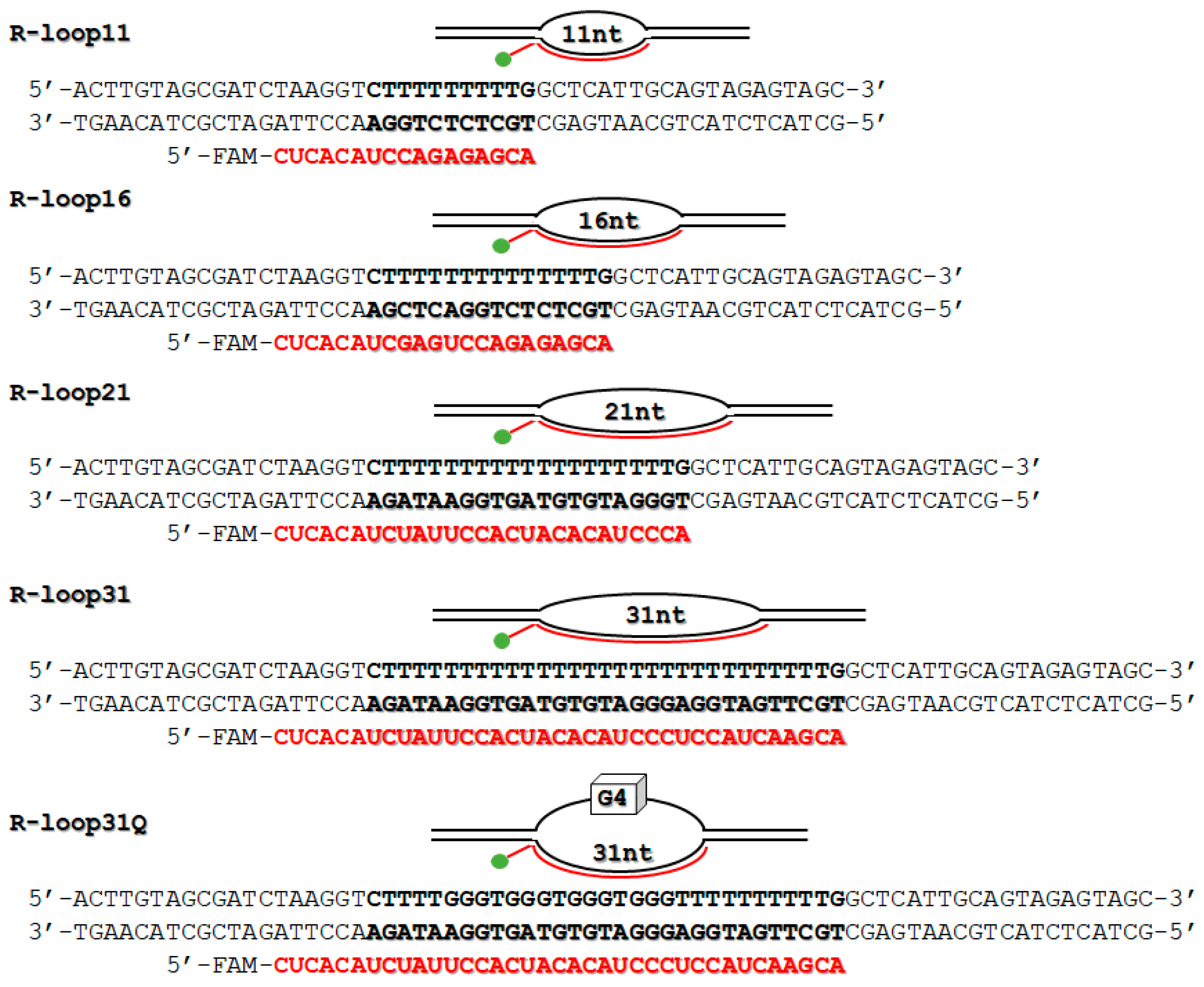
2.1. Design of R-Loops
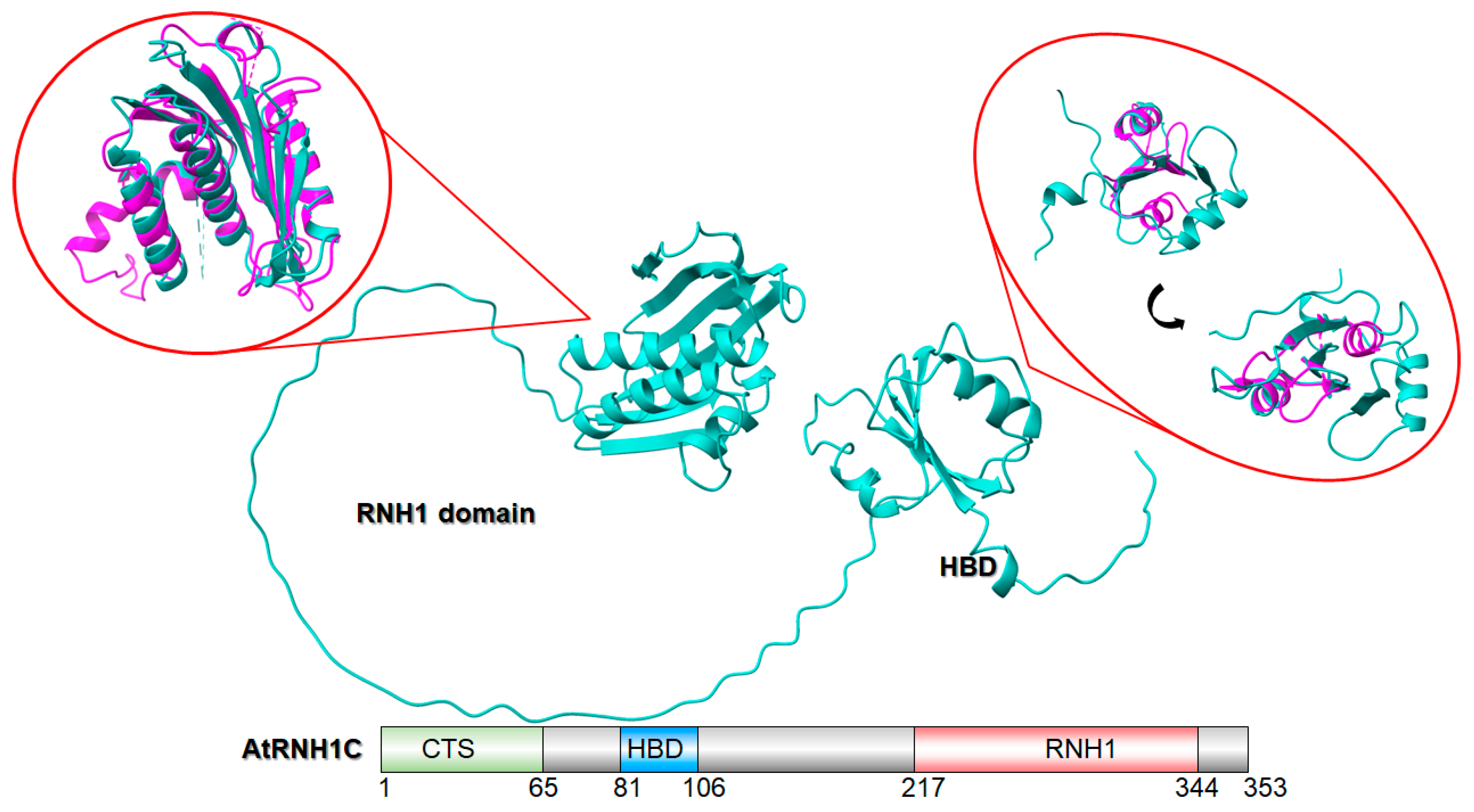
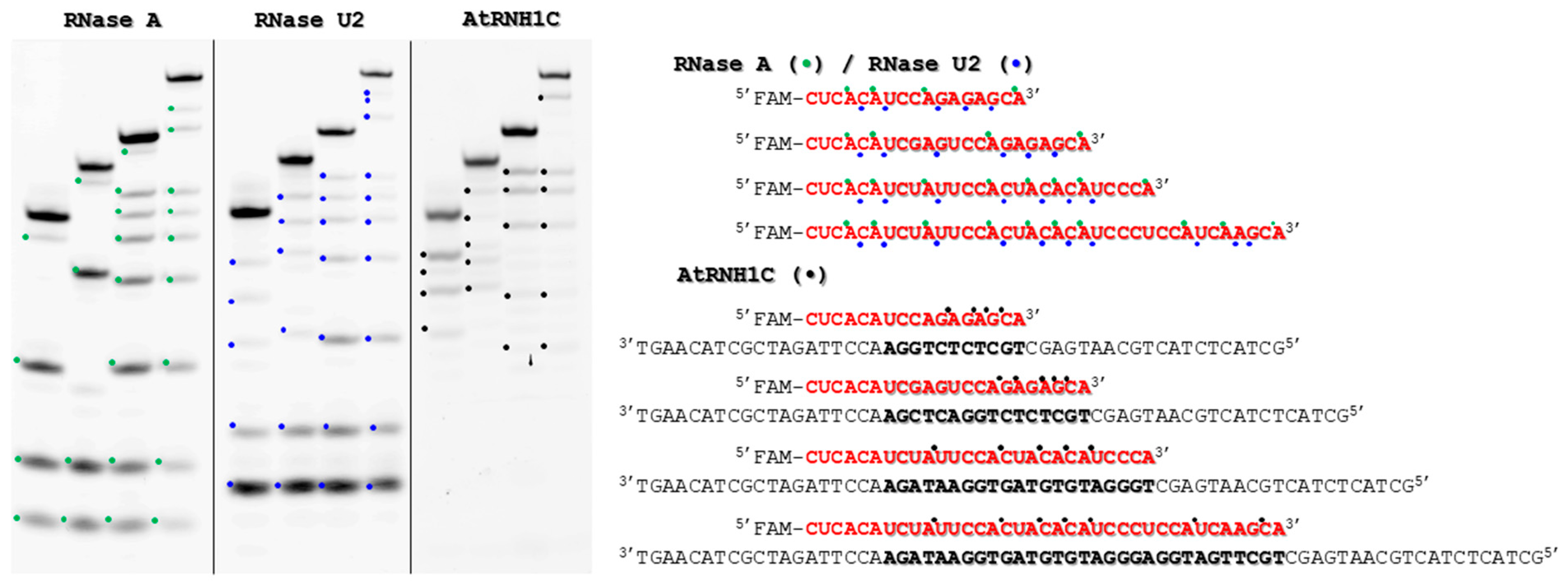
2.2. Sequence Preferences of Purified AtRNH1C
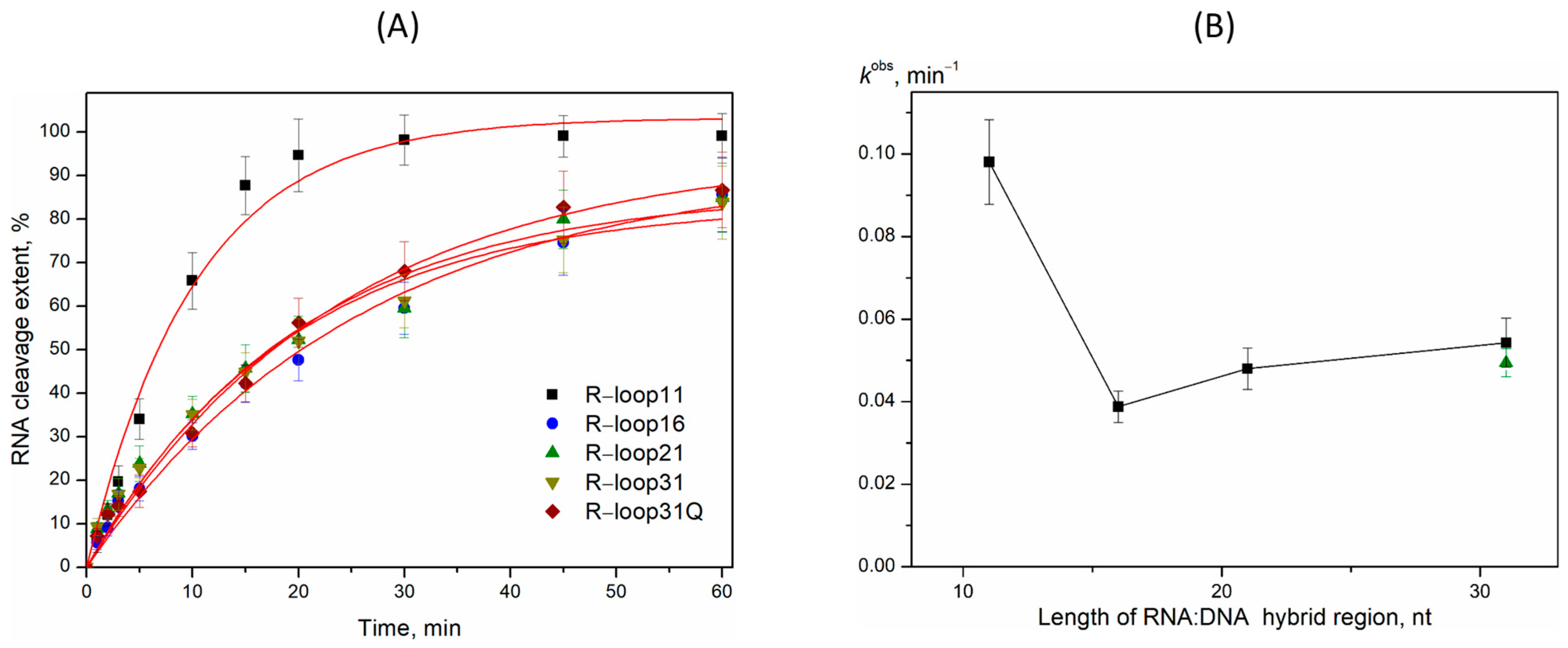
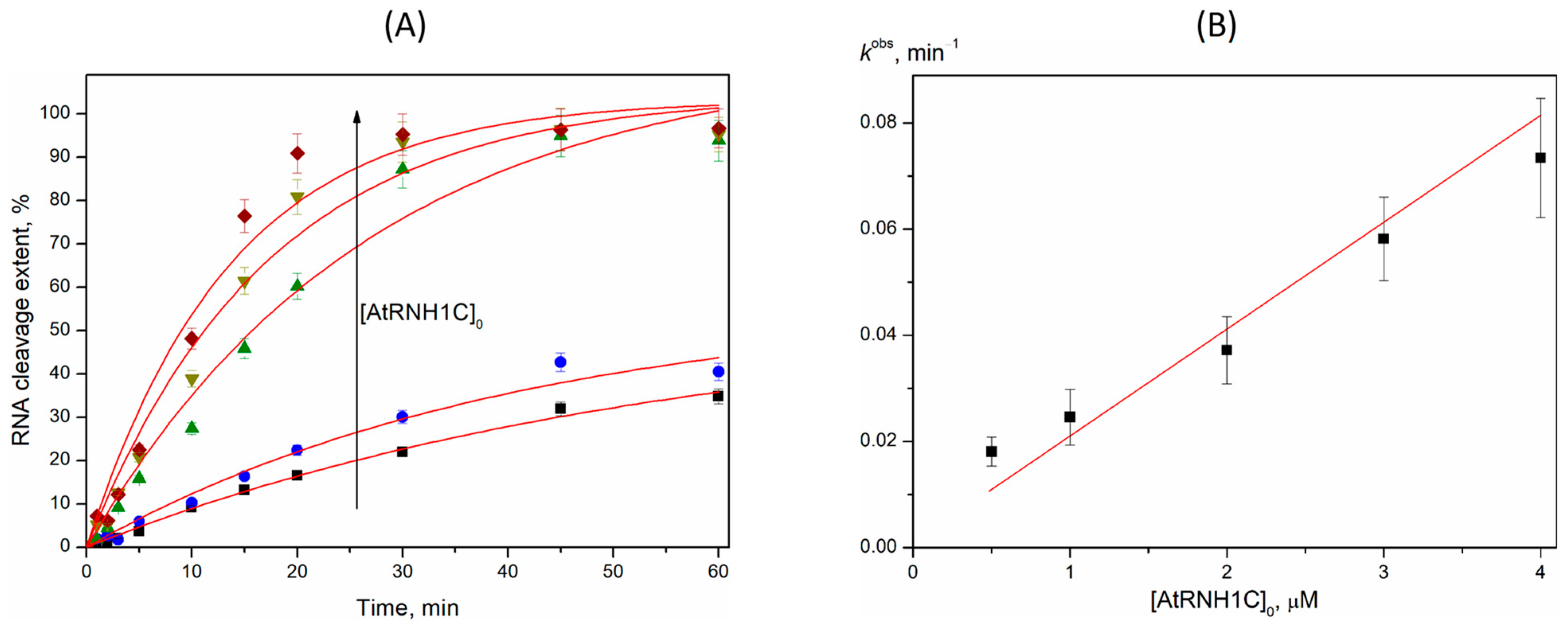
2.3. R-Loop Degradation Activity
3. Discussion
4. Materials and Methods
4.1. AtRNH1C Expression and Purification
4.2. Preparation of R-Loops
4.3. Cleavage Site Analysis
4.4. Activity Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, W.; Xu, H.; Li, K.; Fan, Y.; Liu, Y.; Yang, X.; Sun, Q. The R-Loop Is a Common Chromatin Feature of the Arabidopsis Genome. Nat. Plants 2017, 3, 704–714. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, W.; Sun, Q. R-loop: The New Genome Regulatory Element in Plants. J. Integr. Plant Biol. 2022, 64, 2275–2289. [Google Scholar] [CrossRef]
- Xu, Y.; Jiao, Y.; Liu, C.; Miao, R.; Liu, C.; Wang, Y.; Ma, C.; Liu, J. R-Loop and Diseases: The Cell Cycle Matters. Mol. Cancer 2024, 23, 84. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Y.; Sun, Y.; Liu, Q.; Gu, Y.; Huang, Y.; Zeng, Z.; Tang, F.; Ouyang, Y. Aberrant R-Loop–Mediated Immune Evasion, Cellular Communication, and Metabolic Reprogramming Affect Cancer Progression: A Single-Cell Analysis. Mol. Cancer 2024, 23, 11. [Google Scholar] [CrossRef]
- Brickner, J.R.; Garzon, J.L.; Cimprich, K.A. Walking a Tightrope: The Complex Balancing Act of R-Loops in Genome Stability. Mol. Cell 2022, 82, 2267–2297. [Google Scholar] [CrossRef]
- Khan, E.S.; Danckwardt, S. Pathophysiological Role and Diagnostic Potential of R-Loops in Cancer and Beyond. Genes 2022, 13, 2181. [Google Scholar] [CrossRef]
- Marabitti, V.; Valenzisi, P.; Lillo, G.; Malacaria, E.; Palermo, V.; Pichierri, P.; Franchitto, A. R-Loop-Associated Genomic Instability and Implication of WRN and WRNIP1. Int. J. Mol. Sci. 2022, 23, 1547. [Google Scholar] [CrossRef]
- San Martin-Alonso, M.; Soler-Oliva, M.E.; García-Rubio, M.; García-Muse, T.; Aguilera, A. Harmful R-Loops Are Prevented via Different Cell Cycle-Specific Mechanisms. Nat. Commun. 2021, 12, 4451. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, M.; Shinriki, S.; Matsui, H. R-Loops in Normal and Malignant Hematopoiesis. Front. Hematol. 2023, 2, 1297657. [Google Scholar] [CrossRef]
- Skourti-Stathaki, K.; Proudfoot, N.J. A Double-Edged Sword: R Loops as Threats to Genome Integrity and Powerful Regulators of Gene Expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, D.; Zhao, G.; Wang, D. Unraveling R-Loops: The Hidden Drivers of Inflammation and Immune Dysregulation. Medicine 2025, 104, e42833. [Google Scholar] [CrossRef]
- Castillo-Guzman, D.; Chédin, F. Defining R-Loop Classes and Their Contributions to Genome Instability. DNA Repair 2021, 106, 103182. [Google Scholar] [CrossRef]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zafar, A.; Luo, L.; Denning, A.M.; Gu, J.; Bennett, A.; Yuan, F.; Zhang, Y. R-Loops in Genome Instability and Cancer. Cancers 2023, 15, 4986. [Google Scholar] [CrossRef] [PubMed]
- Westover, K.R.; Jin, P.; Yao, B. Bridging the Gap: R-Loop Mediated Genomic Instability and Its Implications in Neurological Diseases. Epigenomics 2024, 16, 589–608. [Google Scholar] [CrossRef] [PubMed]
- Hegazy, Y.A.; Fernando, C.M.; Tran, E.J. The Balancing Act of R-Loop Biology: The Good, the Bad, and the Ugly. J. Biol. Chem. 2020, 295, 905–913. [Google Scholar] [CrossRef]
- Aguilera, P.; Aguilera, A. R-Loop Homeostasis in Genome Dynamics, Gene Expression and Development. Curr. Opin. Genet. Dev. 2025, 92, 102325. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, X.; Zhao, H.; Wang, Z. Update on R-Loops in Genomic Integrity: Formation, Functions, and Implications for Human Diseases. Genes Dis. 2025, 12, 101401. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kwak, M.J.; Kim, J.J. R-Loops: A Key Driver of Inflammatory Responses in Cancer. Exp. Mol. Med. 2025, 57, 1455–1466. [Google Scholar] [CrossRef]
- Niehrs, C.; Luke, B. Regulatory R-Loops as Facilitators of Gene Expression and Genome Stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Cao, Y.; Lv, Y.; Lei, K.; Tu, Z.; Gong, C.; Wang, H.; Liu, F.; Huang, K. Mechanisms Underlining R-Loop Biology and Implications for Human Disease. Front. Cell Dev. Biol. 2025, 13, 1537731. [Google Scholar] [CrossRef]
- Gong, Y.; Liu, Y. R-Loops at Chromosome Ends: From Formation, Regulation, and Cellular Consequence. Cancers 2023, 15, 2178. [Google Scholar] [CrossRef]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and Mechanism of Helicases and Nucleic Acid Translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef]
- Cristini, A.; Groh, M.; Kristiansen, M.S.; Gromak, N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep. 2018, 23, 1891–1905. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Hotz-Wagenblatt, A.; Voit, R.; Grummt, I. SIRT7 and the DEAD-Box Helicase DDX21 Cooperate to Resolve Genomic R Loops and Safeguard Genome Stability. Genes Dev. 2017, 31, 1370–1381. [Google Scholar] [CrossRef]
- Weinreb, J.T.; Ghazale, N.; Pradhan, K.; Gupta, V.; Potts, K.S.; Tricomi, B.; Daniels, N.J.; Padgett, R.A.; De Oliveira, S.; Verma, A.; et al. Excessive R-Loops Trigger an Inflammatory Cascade Leading to Increased HSPC Production. Dev. Cell 2021, 56, 627–640.e5. [Google Scholar] [CrossRef]
- Mersaoui, S.Y.; Yu, Z.; Coulombe, Y.; Karam, M.; Busatto, F.F.; Masson, J.; Richard, S. Arginine Methylation of the DDX5 Helicase RGG/RG Motif by PRMT5 Regulates Resolution of RNA:DNA Hybrids. EMBO J. 2019, 38, e100986. [Google Scholar] [CrossRef]
- Yang, Z.; Li, M.; Sun, Q. RHON1 Co-Transcriptionally Resolves R-Loops for Arabidopsis Chloroplast Genome Maintenance. Cell Rep. 2020, 30, 243–256.e5. [Google Scholar] [CrossRef] [PubMed]
- Manzo, S.G.; Hartono, S.R.; Sanz, L.A.; Marinello, J.; De Biasi, S.; Cossarizza, A.; Capranico, G.; Chedin, F. DNA Topoisomerase I Differentially Modulates R-Loops across the Human Genome. Genome Biol. 2018, 19, 100. [Google Scholar] [CrossRef]
- Uruci, S.; Lo, C.S.Y.; Wheeler, D.; Taneja, N. R-Loops and Its Chro-Mates: The Strange Case of Dr. Jekyll and Mr. Hyde. Int. J. Mol. Sci. 2021, 22, 8850. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of Action and Regulation of ATP-Dependent Chromatin-Remodelling Complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Bhadouriya, S.L.; Mehrotra, S.; Basantani, M.K.; Loake, G.J.; Mehrotra, R. Role of Chromatin Architecture in Plant Stress Responses: An Update. Front. Plant Sci. 2021, 11, 603380. [Google Scholar] [CrossRef]
- Banerjee, S.; Roy, S. An Insight into Understanding the Coupling between Homologous Recombination Mediated DNA Repair and Chromatin Remodeling Mechanisms in Plant Genome: An Update. Cell Cycle 2021, 20, 1760–1784. [Google Scholar] [CrossRef] [PubMed]
- Hyjek, M.; Figiel, M.; Nowotny, M. RNases H: Structure and Mechanism. DNA Repair 2019, 84, 102672. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Crouch, R.J. Ribonuclease H: The Enzymes in Eukaryotes. FEBS J. 2009, 276, 1494–1505. [Google Scholar] [CrossRef]
- Mach, J. Thrown for a Loop: How RNase H1 and DNA Gyrases Limit R-Loops and Maintain Genome Stability in Chloroplasts. Plant Cell 2017, 29, 2311–2312. [Google Scholar] [CrossRef] [PubMed]
- Kuciński, J.; Chamera, S.; Kmera, A.; Rowley, M.J.; Fujii, S.; Khurana, P.; Nowotny, M.; Wierzbicki, A.T. Evolutionary History and Activity of RNase H1-Like Proteins in Arabidopsis Thaliana. Plant Cell Physiol. 2020, 61, 1107–1119. [Google Scholar] [CrossRef]
- Jarvis, P.; López-Juez, E. Biogenesis and Homeostasis of Chloroplasts and Other Plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802, Erratum in Nat. Rev. Mol. Cell Biol. 2014, 15, 147. https://doi.org/10.1038/nrm3744. [Google Scholar] [CrossRef]
- Powikrowska, M.; Oetke, S.; Jensen, P.E.; Krupinska, K. Dynamic Composition, Shaping and Organization of Plastid Nucleoids. Front. Plant Sci. 2014, 5, 424. [Google Scholar] [CrossRef]
- Oldenburg, D.J.; Bendich, A.J. DNA Maintenance in Plastids and Mitochondria of Plants. Front. Plant Sci. 2015, 6, 883. [Google Scholar] [CrossRef]
- Raven, J.A. Implications of Mutation of Organelle Genomes for Organelle Function and Evolution. J. Exp. Bot. 2015, 66, 5639–5650. [Google Scholar] [CrossRef][Green Version]
- Katayanagi, K.; Miyagawa, M.; Matsushima, M.; Ishikawa, M.; Kanaya, S.; Ikehara, M.; Matsuzaki, T.; Morikawa, K. Three-Dimensional Structure of Ribonuclease H from E. Coli. Nature 1990, 347, 306–309. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; Frolova, E.G.; Feng, C.; Grinberg, A.; Love, P.E.; Crouch, R.J. Failure to Produce Mitochondrial DNA Results in Embryonic Lethality in Rnaseh1 Null Mice. Mol. Cell 2003, 11, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Cerritelli, S.M.; Crouch, R.J. Cloning, Expression, and Mapping of Ribonucleases H of Human and Mouse Related to Bacterial RNase HI. Genomics 1998, 53, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lima, W.F.; Crooke, S.T. Investigating the Structure of Human RNase H1 by Site-Directed Mutagenesis. J. Biol. Chem. 2001, 276, 23547–23553. [Google Scholar] [CrossRef]
- Gaidamakov, S.A. Eukaryotic RNases H1 Act Processively by Interactions through the Duplex RNA-Binding Domain. Nucleic Acids Res. 2005, 33, 2166–2175. [Google Scholar] [CrossRef] [PubMed]
- Nowotny, M.; Cerritelli, S.M.; Ghirlando, R.; Gaidamakov, S.A.; Crouch, R.J.; Yang, W. Specific Recognition of RNA/DNA Hybrid and Enhancement of Human RNase H1 Activity by HBD. EMBO J. 2008, 27, 1172–1181. [Google Scholar] [CrossRef]
- Yang, Z.; Hou, Q.; Cheng, L.; Xu, W.; Hong, Y.; Li, S.; Sun, Q. RNase H1 Cooperates with DNA Gyrases to Restrict R-Loops and Maintain Genome Integrity in Arabidopsis Chloroplasts. Plant Cell 2017, 29, 2478–2497. [Google Scholar] [CrossRef]
- Nudler, E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009, 78, 335–361. [Google Scholar] [CrossRef]
- Yadav, V.; Hemansi; Kim, N.; Tuteja, N.; Yadav, P. G Quadruplex in Plants: A Ubiquitous Regulatory Element and Its Biological Relevance. Front. Plant Sci. 2017, 8, 1163. [Google Scholar] [CrossRef]
- Liu, H.; Chu, Z.; Yang, X. A Key Molecular Regulator, RNA G-Quadruplex and Its Function in Plants. Front. Plant Sci. 2022, 13, 926953. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-Quadruplexes and Their Regulatory Roles in Biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Kosarev, I.A.; Timofeyeva, N.A.; Novopashina, D.S.; Kuznetsov, N.A. Kinetic Features of Degradation of R-Loops by RNase H1 from Escherichia Coli. Int. J. Mol. Sci. 2024, 25, 12263. [Google Scholar] [CrossRef]
- Kiełpiński, Ł.J.; Hagedorn, P.H.; Lindow, M.; Vinther, J. RNase H Sequence Preferences Influence Antisense Oligonucleotide Efficiency. Nucleic Acids Res. 2017, 45, 12932–12944. [Google Scholar] [CrossRef]
- Chakraborty, P. New Insight into the Biology of R-Loops. Mutat. Res. Mol. Mech. Mutagen. 2020, 821, 111711. [Google Scholar] [CrossRef] [PubMed]
- Palmer, B.; Lee, C.-Y.; Yang, L.; Paul, T.; Myong, S. High-Frequency Transcription Leads to Rapid R-Loop Formation. J. Biol. Chem. 2025, 301, 108514. [Google Scholar] [CrossRef] [PubMed]
- Lim, G.; Hwang, S.; Yu, K.; Kang, J.Y.; Kang, C.; Hohng, S. Translocating RNA Polymerase Generates R-Loops at DNA Double-Strand Breaks without Any Additional Factors. Nucleic Acids Res. 2023, 51, 9838–9848. [Google Scholar] [CrossRef]
- Belotserkovskii, B.P.; Tornaletti, S.; D’Souza, A.D.; Hanawalt, P.C. R-Loop Generation during Transcription: Formation, Processing and Cellular Outcomes. DNA Repair 2018, 71, 69–81. [Google Scholar] [CrossRef]
- Nowotny, M.; Gaidamakov, S.A.; Ghirlando, R.; Cerritelli, S.M.; Crouch, R.J.; Yang, W. Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription. Mol. Cell 2007, 28, 264–276. [Google Scholar] [CrossRef]
- Kuznetsova, A.A.; Novopashina, D.S.; Fedorova, O.S.; Kuznetsov, N.A. Effect of the Substrate Structure and Metal Ions on the Hydrolysis of Undamaged RNA by Human AP Endonuclease APE1. Acta Naturae 2020, 12, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, A.A.; Gavrilova, A.A.; Novopashina, D.S.; Fedorova, O.S.; Kuznetsov, N.A. Mutational and Kinetic Analysis of APE1 Endoribonuclease Activity. Mol. Biol. 2021, 55, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, A.A.; Akhmetgalieva, A.A.; Ulyanova, V.V.; Ilinskaya, O.N.; Fedorova, O.S.; Kuznetsov, N.A. Efficiency of RNA Hydrolysis by Binase from Bacillus Pumilus: The Impact of Substrate Structure, Metal Ions, and Low Molecular Weight Nucleotide Compounds. Mol. Biol. 2020, 54, 769–776. [Google Scholar] [CrossRef]
- Johnson, K.A. Kinetic Analysis for the New Enzymology, 1st ed.; KinTek Corporation: Austin, TX, USA, 2019; ISBN 978-1-7339982-0-8. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gavrilova, A.A.; Kuznetsova, A.A.; Novopashina, D.S.; Zheng, C.; Sun, Q.; Kuznetsov, N.A. Biochemical Characterization of R-Loop Degradation by Chloroplast-Localized RNase H1 from Arabidopsis thaliana. Int. J. Mol. Sci. 2025, 26, 11125. https://doi.org/10.3390/ijms262211125
Gavrilova AA, Kuznetsova AA, Novopashina DS, Zheng C, Sun Q, Kuznetsov NA. Biochemical Characterization of R-Loop Degradation by Chloroplast-Localized RNase H1 from Arabidopsis thaliana. International Journal of Molecular Sciences. 2025; 26(22):11125. https://doi.org/10.3390/ijms262211125
Chicago/Turabian StyleGavrilova, Anastasia A., Aleksandra A. Kuznetsova, Darya S. Novopashina, Chengxia Zheng, Qianwen Sun, and Nikita A. Kuznetsov. 2025. "Biochemical Characterization of R-Loop Degradation by Chloroplast-Localized RNase H1 from Arabidopsis thaliana" International Journal of Molecular Sciences 26, no. 22: 11125. https://doi.org/10.3390/ijms262211125
APA StyleGavrilova, A. A., Kuznetsova, A. A., Novopashina, D. S., Zheng, C., Sun, Q., & Kuznetsov, N. A. (2025). Biochemical Characterization of R-Loop Degradation by Chloroplast-Localized RNase H1 from Arabidopsis thaliana. International Journal of Molecular Sciences, 26(22), 11125. https://doi.org/10.3390/ijms262211125





