Abstract
Secondary peritonitis (SP) remains a major clinical challenge due to its high complication rates and it often results in sepsis and multi-organ dysfunction. This study investigated the association between four nitric oxide synthase (NOS) single-nucleotide polymorphisms (SNPs)—NOS3 c.-786T>C (rs2070744), NOS3 c.894G>T (rs1799983), NOS3 27 bp variable number tandem repeat (VNTR) (rs61722009), and NOS2 (rs2297518)—and sepsis-related complications in 202 patients with SP. Demographic and baseline clinical characteristics, Acute Physiology and Chronic Health Evaluation (APACHE) II scores, Mannheim Peritonitis Index, and complications (multiple organ dysfunction syndrome (MODS), multiple organ failure (MOF), acute respiratory distress syndrome (ARDS), and sepsis) were analyzed for associations with the NOS gene variants. Haplotype analysis was also performed. No SNP showed an association with in-hospital mortality. However, the NOS3 c.-786T>C TT genotype was significantly associated with an increased risk of MOF (p = 0.008), and remained independently associated after multivariate adjustment (pMOF = 0.006). The T4bG haplotype was significantly more frequent among patients with MODS (p = 0.026), MOF (p = 0.048), and sepsis (p = 0.018). These findings suggest that NOS gene variants, particularly NOS3 c.-786T>C and the T4bG haplotype, may potentially serve as biomarkers for risk stratification in critically ill patients.
1. Introduction
Secondary peritonitis arises from direct contamination of the peritoneum, typically resulting from a disruption of the integrity of the gastrointestinal or urogenital tract. It may occur due to perforation of hollow viscera or other pathological processes, such as intestinal ischemia, or intraperitoneal hemorrhage following trauma [1,2]. Despite significant advancements in diagnostic and treatment modalities, secondary peritonitis continues to be a major challenge in critical care settings globally, primarily due to its potentially life-threatening complications, such as intra-abdominal sepsis, and the high mortality rates associated with it [1]. Secondary peritonitis remains a leading cause of sepsis in intensive care unit (ICU) patients, underscoring the need for continued research and improved management strategies [2,3,4]. The Third International Consensus defines sepsis as a life-threatening organ dysfunction due to a dysregulated host response to infection [5], explaining the underlying pathophysiology of this complex condition. The response to infection may significantly differ between individuals and is believed to be influenced by several factors [6]. Research suggests that genetic predisposition plays a role in explaining the differences among hosts in terms of the clinical and pathophysiological aspects of sepsis, including the severity of complications and potential outcomes [7].
One of the key molecular players in sepsis pathophysiology is nitric oxide (NO), a potent vasodilator involved in regulating vascular tone, immune responses, and microcirculation. During sepsis, endothelial dysfunction and vascular dilation can lead to impaired blood flow and inadequate tissue oxygenation, which may contribute to organ dysfunction and septic shock [8,9,10]. While NO is essential for maintaining vascular tone by dilating blood vessels and lowering blood pressure under physiological conditions, excessive NO production in sepsis can worsen hypotension and exacerbate shock [11]. Genetic variations in NO synthase (NOS) genes, particularly NOS3 and NOS2, have been shown to influence the production and function of NO. Specific single-nucleotide polymorphisms (SNPs) in NOS3, such as c.-786T>C (rs2070744) and c.894G>T (p.Glu298Asp, rs1799983), affect endothelial NOS (eNOS) activity, which may exacerbate vascular dysfunction and worsen sepsis outcomes by impairing vascular tone and increasing the susceptibility to shock [8,9,12]. Similarly, NOS2, which encodes inducible NOS (iNOS), is upregulated during sepsis and contributes to excessive NO production. Specific polymorphisms in NOS2, such as c. 1823C>T (p.Ser608Leu, rs2297518), may enhance NOS2 gene expression, exacerbating endothelial dysfunction and contributing to critical septic complications such as hypotension and multiple organ failure (MOF) [12,13]. Identifying genetic variants, particularly SNPs, through genome-wide association studies (GWASs) could provide valuable insights for improved diagnosis, prediction of sepsis severity, and personalized treatment strategies [14,15,16]. The exploration of such biomarkers, alongside traditional inflammatory markers, holds promise for improving outcomes in patients with secondary peritonitis and sepsis [17,18].
In this study, we conducted a comprehensive analysis of four SNPs located within the NOS genes, NOS3 (including c.-786T>C (rs2070744), a 27 bp variable number tandem repeat (VNTR) in intron 4 (rs61722009), and c.894G>T, (p.Glu298Asp, rs1799983)) and NOS2 c.1823C>T (p.Ser608Leu, rs2297518), to investigate their potential association with sepsis onset and the development of severe complications and outcomes in patients with secondary peritonitis. The previously published findings on these NOS gene variants are summarized in Table 1.

Table 1.
Overview of the NOS gene polymorphisms analyzed in this study and their functional relevance.
2. Results
2.1. Demographic and Clinical Characteristics of the Study Group
A total of 202 patients with secondary peritonitis were enrolled in the study. The demographic and clinical characteristics of the study population are presented in Table 2. The cohort had a mean age of 56.83 ± 17.7 years, with a slight male predominance (55.0%). The average Acute Physiology and Chronic Health Evaluation (APACHE) II score on admission was 10.63 ± 7.23, indicating moderate illness severity. The Mannheim Peritonitis Index (MPI) averaged 20.30 ± 7.46, reflecting the extent and severity of peritonitis in the cohort. Appendiceal perforation was the most frequent source of peritonitis, which was observed in 33.51% of the patients, followed by small bowel perforation (24.74%). Regarding the type of peritonitis, purulent peritonitis was the most prevalent, occurring in 80.73% of cases. The baseline laboratory findings are summarized in Table 2. Elevated white blood cell and neutrophil counts and increased C-reactive protein levels were consistent with systemic inflammatory response. The renal function parameters and bilirubin levels showed considerable variability, reflecting the heterogeneity in disease severity among the patients.

Table 2.
Demographic and baseline clinical characteristics of patients with secondary peritonitis.
2.2. Allele and Genotype Frequencies
The frequencies of the NOS gene polymorphisms are summarized in Table 3. For NOS3 c.-786T>C (rs2070744), the TT genotype was observed in 45.0% of the patients, and the T allele frequency was 0.69. The 4b/4b genotype of the NOS3 27 bp VNTR (rs61722009) was the most common (67.3%), with a 4b allele frequency of 0.83. The GG genotype of NOS3 c.894G>T (rs1799983) occurred in 52.0% of the patients, with a G allele frequency of 0.72. For NOS2 rs2297518 (alleles G/A based on genomic DNA; c.1823C>T based on HGVS (Human Genome Variation Society) coding nomenclature), the GG genotype was found in 58.9% of the patients, and the G allele frequency was 0.76. The genotype frequencies for all the analyzed SNPs were consistent with Hardy–Weinberg equilibrium (HWE). Overall, the allele frequencies closely resembled those reported in European reference populations.

Table 3.
Frequencies of genotypes and alleles of the NOS gene SNPs in patients with secondary peritonitis.
2.3. Associations of NOS Genotypes with In-Hospital Mortality and Severe Complications
The associations between the NOS genotypes and in-hospital mortality are shown in Table 4. None of the investigated SNPs demonstrated a statistically significant association with mortality.

Table 4.
Association between NOS gene polymorphisms and mortality in patients with secondary peritonitis.
The relationship between the NOS gene polymorphisms and severe clinical complications (ARDS, MODS, and MOF) is summarized in Table 5. No significant associations were observed between any of the analyzed SNPs and sepsis. Among the analyzed polymorphisms, only the NOS3 c.-786T>C (rs2070744) and NOS2 c.1823C>T (rs2297518) variants showed statistically significant associations with major complications. The individuals carrying the NOS3 c.-786T>C TT genotype exhibited an increased risk of both MODS (p = 0.017, OR = 2.67, 95% CI = 1.17–6.07) and MOF (p = 0.008, OR = 3.18, 95% CI = 1.31–7.69) compared to the individuals with at least one C allele (TC or CC). After applying the False Discovery Rate (FDR) correction (q = 0.05), the association between NOS3 c.-786T>C and MOF remained statistically significant (p = 0.032), whereas the association with MODS lost significance but showed a trend toward an increased risk (p = 0.068). On the other hand, the individuals carrying the NOS2 c.1823C>T GG genotype showed an increased risk of both ARDS (p = 0.046, OR = 2.59, 95% CI = 0.99–6.77) and MODS (p = 0.045, OR = 2.46, 95% CI = 0.99–6.07) compared to the individuals with at least one A allele (GA or AA). After applying the FDR correction for NOS2 c.1823C>T, neither ARDS (p = 0.503) nor MODS (p = 0.090) remained significant. Given these findings, both the NOS3 c.-786T>C TT and NOS2 c.1823C>T GG genotypes were further evaluated in a multivariate context to assess their independent contribution. After adjusting for sex, age, source of peritonitis, and APACHE II score in the logistic regression models, only the NOS3 c.-786T>C TT genotype remained significantly associated with a higher likelihood of developing MOF (p = 0.006, OR = 4.86, 95% CI = 1.56–15.13) and MODS (p = 0.047, OR = 3.67, 95% CI = 1.02–13.19). In the analyses of MOF and MODS outcomes, increasing age (pMODS = 0.009; pMOF = 0.030) and higher APACHE II scores (pMODS = 0.000; pMOF = 0.002) were consistently identified as significant predictors of adverse outcomes. In contrast, sex and source of peritonitis were not found to be significantly associated. The association of the NOS2 c.1823C>T GG genotype with an increased risk of developing ARDS or MODS did not reach statistical significance in the logistic regression models (pARDS = 0.211; pMODS = 0.146).

Table 5.
Association between NOS gene polymorphisms and severe complications in patients with secondary peritonitis.
2.4. Haplotype Analysis
Haplotype analysis was also performed to evaluate the combined effects of three NOS3 polymorphisms (c.-786T>C (rs2070744), the 27 bp VNTR (rs61722009), and c.894G>T (rs1799983)) on clinical outcomes. The calculated linkage disequilibrium (LD) metrics (D′ and r2) are illustrated in Figure 1. No well-established haplotype block was identified within this cohort. Moreover, the frequencies of all the polymorphism haplotypes were consistent with the HWE. Analysis of the haplotype frequencies among the study participants revealed distinct haplotypes: T4bG, C4bT, T4bT, C4aG, C4bG, T4aG, and C4aT, with frequencies of 0.51, 0.13, 0.13, 0.11, 0.06, 0.05, and 0.02, respectively. Notably, the T4bG haplotype was significantly more frequent among the patients who developed severe complications. Specifically, its frequency was elevated in those with MODS (0.64 vs. 0.48; p = 0.026), MOF (0.61 vs. 0.48; p = 0.048), and sepsis (0.70 vs. 0.49; p = 0.018) compared to the patients without these complications. No other haplotypes demonstrated significant associations with these adverse clinical outcomes.
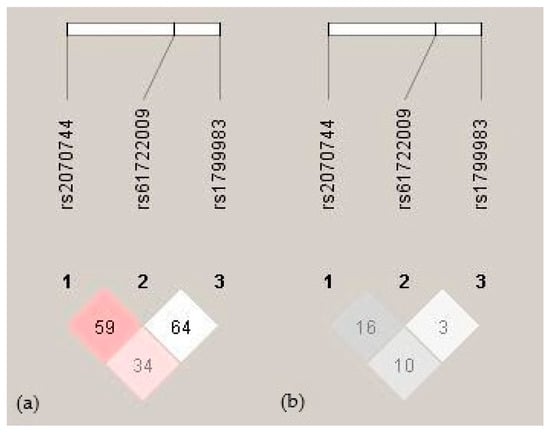
Figure 1.
Linkage disequilibrium between NOS3 gene polymorphisms. (a) Pairwise D′ values are indicated by red shading and numeric labels. Dark red indicates complete LD (D′ = 1) with an LOD > 2, while lighter shades represent lower values. (b) Pairwise r2 values are shown with gray shading and numeric values. Darker gray indicates a stronger correlation (higher r2) between variants.
3. Discussion
This study provides new insights into how genetic variability in NOS genes affects clinical outcomes in patients with secondary peritonitis, a population at high risk for systemic inflammation and organ failure. Although no significant associations were found between the studied NOS variants and mortality, our findings revealed that the NOS3 c.-786T>C (rs2070744) polymorphism, particularly the TT genotype, was linked to a significantly increased risk of MOF and showed a trend toward an increased risk of MODS. This association was significant for MOF and MODS after adjusting for potential confounders, suggesting that the NOS3 c.-786T>C polymorphism independently contributes to severe organ failure in this population. The haplotype analysis further supported this finding, revealing that the T4bG haplotype of NOS3 polymorphisms was significantly more frequent among the patients who developed MODS, MOF, and sepsis, indicating a potential combined effect of these variants on susceptibility to severe complications. Moreover, NOS2 c.1823C>T GG genotype showed a significant association with a higher risk of both ARDS and MODS, although this association did not persist after multivariate adjustment.
The association between NOS3 gene polymorphisms and sepsis has been the subject of several studies, which suggested a potential role of these variants in modulating susceptibility to and severity of the disease; however, the results remain inconclusive. Among these, the NOS3 c.-786T>C promoter variant is known to affect NOS3 gene expression, with the C allele typically linked to decreased transcriptional activity and, consequently, reduced NO production [19]. Interestingly, we found that the T allele, rather than the C allele, was associated with worse clinical outcomes, contradicting the expected protective role of higher NO bioavailability.
To our knowledge, our study is the first to assess the impact of selected variants on sepsis onset and the development of severe complications in patients with secondary peritonitis. Moreover, it is the first to report the T allele of NOS3 c.-786T>C as a potential genetic risk factor for severe sepsis-related complications in this population. However, similar research has been conducted. Ma et al. found no correlation between the NOS3 c.-786T>C variant and increased organ dysfunction or mortality in patients with severe sepsis [22], while Özkan et al. observed a higher frequency of the CC genotype and C allele in septic patients compared to healthy controls [10]. These discrepancies may reflect differences in study design, patient populations, or clinical outcomes, underscoring the complex role of NOS3 variants in inflammation and critical illness. However, the association of the T allele with adverse outcomes in our cohort suggests a context-dependent effect. Some evidence showing that NO acts as a double-edged mediator in sepsis supports this interpretation, as both its deficiency and excess can contribute to organ dysfunction depending on the specific physiological context [11,26]. An animal study highlighting the importance of the NOS3 gene demonstrated that NOS3-deficient mice experienced a greater degree of sepsis-associated MODS, with increased infiltration of mononuclear cells into tissues and heightened oxidative stress [27]. In secondary peritonitis, enhanced eNOS activity associated with the T allele may lead to excessive NO generation, promoting nitrosative stress through peroxynitrite (ONOO−) and other reactive nitrogen species (RNS), which can aggravate tissue injury and MOF [28,29,30,31]. This aligns with the studies underlying that modulation of the NO pathway should be finely balanced, as complete inhibition or uncontrolled upregulation of NO both worsen outcomes depending on when it occurs during the course of sepsis [26,32]. Consistently, clinical studies have shown that elevated circulating NO levels correlate with sepsis severity and adverse outcomes. Yu et al. reported that serum amyloid A and NO levels were positively correlated with APACHE II scores and mortality risk, reinforcing the idea that excessive production of NO contributes to disease progression [33]. Furthermore, recent insights summarized by Wu et al. highlight that the pathophysiology of sepsis involves a complex interplay of genetic predisposition, inflammatory signaling, and endothelial dysfunction, with eNOS-related pathways playing an important role [34].
In contrast to the significant associations observed for the NOS3 c.-786T>C polymorphism, the 27 bp VNTR polymorphism in intron 4 (rs61722009) did not show a significant link with mortality or major complications in our cohort. Although the 4a allele has been previously associated with reduced plasma NO levels and decreased eNOS activity [20], as well as increased susceptibility to osteomyelitis [21], our study suggests that this variant has a limited impact on outcomes in secondary peritonitis. Regarding the NOS3 c.894G>T (rs1799983) polymorphism, which causes a p.Glu298Asp substitution and affects eNOS protein stability and NO bioavailability [19], we did not observe a significant correlation with mortality or severe complications. This is consistent with the results of Özkan et al. [10] but contrasts with the findings of Ma et al. [22] and Martin et al. [9], who reported associations between the T allele and increased organ dysfunction or sepsis susceptibility. Additionally, Huttunen et al. [23] found that the T allele is linked to hypotension in E. coli bacteremia, suggesting that this variant may affect vascular function during infection.
Haplotype analysis provided additional insights into the combined effect of NOS3 variants on clinical outcomes. In a study on healthy volunteers, Metzger et al. identified the C-4b-Glu haplotype (–786C, 4b, p.Glu298) as being associated with lower vascular NO production, thereby illustrating the functional relevance of specific variant combinations [35]. In contrast, Özkan et al. investigated haplotypes in septic patients and identified the C-4b-G haplotype (comprising-786C, 27 bp 4b, and 894G) as more frequent among those with sepsis, suggesting a role of this combination in increasing disease susceptibility [10]. Collectively, these findings underscore that NOS3 haplotypes can modulate NO bioavailability and impact clinical outcomes in inflammatory conditions, though the specific combinations associated with an increased risk of adverse outcomes may differ across populations and disease contexts. Our results extend these observations to secondary peritonitis, emphasizing the potential contribution of specific NOS3 haplotypes to organ dysfunction and severe complications. Further functional studies are warranted to elucidate how these combined polymorphisms affect eNOS expression and NO production in acute inflammatory states.
In the present study, the GG genotype of NOS2 rs2297518 polymorphism (p.Ser608Leu), located in exon 16, initially showed a significant association with an increased risk of developing ARDS and MODS in patients with secondary peritonitis. In contrast, previous research identified the A allele as the one associated with an increased risk of sepsis and septic shock [12]. However, in our study, the association of the GG genotype of the rs2297518 polymorphism did not reach statistical significance in the multivariable logistic regression models, suggesting that the observed effect may be influenced by confounding factors such as sex, age, source of peritonitis, or APACHE II score. Unlike the other NOS isoforms, NOS2 is not constitutively expressed and is upregulated in response to cytokines, bacterial lipopolysaccharides (LPSs), and other inflammatory mediators or pathogen-associated molecular patterns (PAMPs), leading to substantial NO production. This mechanism has been implicated in the pathophysiology of septic shock, particularly through profound vasodilatation and subsequent hypotension [30,36,37]. Previous studies have suggested a functional role for rs2297518 in sepsis. Wang et al. reported an increased risk of septic shock among carriers of the GA and AA genotypes in two independent Chinese cohorts [12] while Martin et al. identified a higher prevalence of another NOS2 variant (rs374198) in septic patients compared to uninfected controls [9]. However, no direct correlation between genotype and circulating NOx levels was observed, promoting the idea that genotype alone may be insufficient to predict clinical outcomes.
Although NOS2 polymorphisms have also been linked to chronic inflammatory diseases such as inflammatory bowel disease (IBD) [24,25], the role of rs2297518 may differ in acute settings. In very-early-onset IBD, iNOS overexpression contributes directly to chronic mucosal damage, whereas in acute conditions like peritonitis, its role may be more limited and influenced by multiple overlapping inflammatory pathways. Taken together, our data suggest that rs2297518 does not substantially influence clinical outcomes in secondary peritonitis, possibly due to the predominance of other clinical predictors such as age and APACHE II scores. Further studies across diverse populations and clinical settings are needed to clarify the broader relevance of NOS2 in infectious disease susceptibility and progression.
A primary limitation of our study is the absence of functional validation of the identified genetic associations. While we discussed potential mechanisms involving NO production and nitrosative stress, they remain speculative without direct measurements such as NO levels or enzyme activity. Additionally, since this was a single-center study, the generalizability of our findings to other populations may be limited. Although the allele frequencies in our cohort align with European reference data, validation in diverse, multi-center cohorts is needed.
To advance our understanding of NOS gene variants in secondary peritonitis and sepsis, future research should integrate larger, multi-center cohorts; include functional analyses of variant-specific effects on NO production; and explore gene–environment and gene–pathogen interactions. Integrating genetic data with transcriptomic and proteomic profiling may help uncover the biological mechanisms linking NOS polymorphisms to clinical outcomes in acute inflammatory states.
In conclusion, genetic predisposition may influence the host inflammatory response, potentially offering a path toward risk stratification or targeted therapies in this critically ill population. Our findings suggest that NOS3 gene variants, both individually and in haplotypic combinations, may potentially serve as biomarkers or therapeutic targets for secondary peritonitis. However, further functional studies and analyses across diverse populations are needed to translate these genetic insights into clinical applications.
4. Materials and Methods
4.1. Study Population
This cohort study was conducted at the Clinic for Emergency Surgery at the University Clinical Center of Serbia and the Institute of Human Genetics in Belgrade, Serbia. A total of 202 patients aged 18 years and older who were diagnosed with secondary peritonitis were enrolled after being treated at the Clinic for Emergency Surgery. We excluded patients in the terminal phase of malignant or chronic systemic diseases, those receiving steroid or immunosuppressive therapy, immunocompromised patients, and pregnant individuals to reduce potential confounding factors. The study was approved by the Ethics Committee of the Clinical Center of Serbia (No. 5030/2) and the Ethics Committee of the Faculty of Medicine, University of Belgrade (No. 1322/IX-71). The study included all patients who had undergone surgery due to peritonitis. Postoperative treatment followed standard protocols for abdominal infections, beginning with empirical broad-spectrum antibiotic therapy based on clinical suspicion of the cause of the infection. Antibiotic regimens were later adjusted according to antibiogram results obtained from microbiological analyses of intraoperatively collected peritoneal fluid, postoperative blood cultures, and swabs.
The APACHE II score was calculated for each patient using parameters collected within the first 24 h of admission to assess illness severity and the risk of complications or mortality [38]. Severe complications monitored included ARDS, MODS, MOF, and sepsis. The diagnosis of sepsis in patients treated before 2016 was based on the criteria established by the 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference [39]. For patients treated from 2016 onwards, the diagnosis followed the criteria outlined in The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) [5]. Additionally, a specific MPI score was used to evaluate the prognosis of peritonitis for each patient [40]. Patient monitoring included clinical evaluations, laboratory tests (e.g., complete blood count, inflammatory markers, coagulation profile, and biochemical parameters), and imaging studies, including ultrasound, X-ray, and multi-slice computed tomography (MSCT). These procedures were carried out during hospitalization considering the length of stay in the ICU as well as the morbidity and mortality rates.
4.2. DNA Extraction and Genotyping
Molecular genetic testing was performed at the Institute of Human Genetics, Faculty of Medicine, University of Belgrade. Total genomic DNA was extracted from 5 mL of whole blood using the salting-out method [41]. The concentration and quality of the isolated DNA were assessed using a fluorimeter (Qubit 3.0, ThermoFisher Scientific, Waltham, MA, USA). Standard quality control procedures were applied for all SNP genotyping assays. To assess reproducibility, a randomly selected 10% subset of samples was re-genotyped in duplicate, resulting in a concordance rate exceeding 99%. The overall genotype call rate was 98.5%. Negative controls (no DNA template) were included in each PCR run to monitor for potential contamination. Additionally, samples with ambiguous genotype results were re-analyzed to ensure accurate genotype assignment.
4.2.1. NOS3 Genotyping
Genotypes for the NOS3 27 bp VNTR in intron 4 were identified using the polymerase chain reaction (PCR) method. Additionally, genotyping for the NOS3 c.-786 T>C (rs2070744) and NOS3 c.894G>T (rs1799983) SNPs was performed using PCR analysis followed by restriction fragment length polymorphism (RFLP) analysis. Table 6 provides detailed information on the primer sequences, reaction conditions, restriction enzymes utilized, fragment sizes, and corresponding genotypes.

Table 6.
Polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) conditions.
4.2.2. NOS2 Genotyping
The NOS2 c.1823C>T (p.Ser608Leu, rs2297518) SNP located in exon 16 was genotyped using the TaqMan SNP Genotyping Assay (assay ID: C_11889257_10, ThermoFisher Scientific, Waltham, MA, USA). The analysis was conducted in a 96-well plate using a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The genotypes for rs2297518 are reported according to the genomic DNA strand, where G represents the reference allele and A represents the variant allele. The corresponding HGVS nomenclature is c.1823C>T based on the coding DNA sequence of NOS2. Each reaction was carried out in a final volume of 12.5 μL and included 2x Probe qPCR Mix EURx (EURx, Gdańsk, Poland), 1x specific TaqMan assay, and 5–50 ng of genomic DNA; the thermal cycling conditions recommended by the manufacturer were used.
4.3. Statistical Analysis
During the planning phase of the study, the minimum sample size was determined using G*Power software (version 3.1.9.7). Assuming an effect size of 0.3, a Type I error (α) rate of 0.05, and a statistical power of 0.95 with 2 degrees of freedom (df), the necessary sample size was 172 participants.
Genotype and allele frequency differences were analyzed using the Chi-square test or Fisher’s exact test depending on the sample size and assumptions about the expected frequencies. Deviations from HWE were tested using the simple Pearson’s χ2 test. Statistical analyses were conducted with SPSS software, version 16.0 (SPSS Inc., Chicago, IL, USA). For genetic association analyses, correction for multiple testing was implemented using the Benjamini–Hochberg step-up procedure for FDR with cut-off values q = 0.05. FDR correction was applied using the False Discovery Rate Online Calculator [42]. Haplotype analysis of the NOS3 gene was performed using Haploview software 4.2 (Broad Institute, Cambridge, MA, USA) [43]. A p value of 0.05 or lower was considered statistically significant.
Author Contributions
Conceptualization: N.M. and K.D.V.; methodology: N.M., K.D.V. and D.P.; writing—original draft preparation: M.R.; writing—review and editing: N.M., K.D.V., I.N. and P.R.; data collection and database formatting: M.R., M.S. and M.P.; laboratory analysis: M.R., M.D.P., M.G., A.D.U. and N.S.; statistical analysis: N.M., M.G., T.D. and M.D.P.; funding acquisition: I.N. and T.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Ministry of Science, Technological Development, and Innovation of the Republic of Serbia [Grant No. 451-03-66/2024-03/200110].
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Clinical Center of Serbia (Approval Code: 5030/2; Approval Date: 13 December 2012) and the Ethics Committee of the Faculty of Medicine, University of Belgrade (Approval Code: 1322/IX-71; Approval Date: 30 September 2021).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ross, J.T.; Matthay, M.A.; Harris, H.W. Secondary Peritonitis: Principles of Diagnosis and Intervention. BMJ 2018, 361, k1407. [Google Scholar] [CrossRef] [PubMed]
- Dumitrascu, C.O.; Gherghe, M.; Costache, M.; Cretu, B.; Cirstoiu, C. The Role of Serum and Peritoneal Biomarkers in Predicting Sepsis and Septic Multiorgan Failure in Patients with Secondary Peritonitis. Cureus 2023, 15, e41724. [Google Scholar] [CrossRef] [PubMed]
- Clements, T.W.; Tolonen, M.; Ball, C.G.; Kirkpatrick, A.W. Secondary Peritonitis and Intra-Abdominal Sepsis: An Increasingly Global Disease in Search of Better Systemic Therapies. Scand. J. Surg. 2021, 110, 139–149. [Google Scholar] [CrossRef]
- La Via, L.; Sangiorgio, G.; Stefani, S.; Marino, A.; Nunnari, G.; Cocuzza, S.; La Mantia, I.; Cacopardo, B.; Stracquadanio, S.; Spampinato, S.; et al. The Global Burden of Sepsis and Septic Shock. Epidemiologia 2024, 5, 456–478. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA—J. Am. Med. Assoc. 2016, 315, 801–810. [Google Scholar] [CrossRef]
- David, V.L.; Ercisli, M.F.; Rogobete, A.F.; Boia, E.S.; Horhat, R.; Nitu, R.; Diaconu, M.M.; Pirtea, L.; Ciuca, I.; Horhat, D.; et al. Early Prediction of Sepsis Incidence in Critically Ill Patients Using Specific Genetic Polymorphisms. Biochem. Genet. 2017, 55, 193–203. [Google Scholar] [CrossRef]
- Villar, J.; Maca-Meyer, N.; Pérez-Méndez, L.; Flores, C. Bench-to-Bedside Review: Understanding Genetic Predisposition to Sepsis. Crit. Care 2004, 8, 180–189. [Google Scholar] [CrossRef][Green Version]
- Winkler, M.S.; Kluge, S.; Holzmann, M.; Moritz, E.; Robbe, L.; Bauer, A.; Zahrte, C.; Priefler, M.; Schwedhelm, E.; Böger, R.H.; et al. Markers of Nitric Oxide Are Associated with Sepsis Severity: An Observational Study. Crit. Care 2017, 21, 189. [Google Scholar] [CrossRef]
- Martin, G.; Asensi, V.; Montes, A.H.; Collazos, J.; Alvarez, V.; Pérez-Is, L.; Carton, J.A.; Taboada, F.; Valle-Garay, E. Endothelial (NOS3 E298D) and Inducible (NOS2 Exon 22) Nitric Oxide Synthase Polymorphisms, as Well as Plasma NOx, Influence Sepsis Development. Nitric Oxide—Biol. Chem. 2014, 42, 79–86. [Google Scholar] [CrossRef]
- Özkan, M.; Günay, N.; Sener, E.F.; Karcıoglu, Ö.; Tahtasakal, R.; Dal, F.; Günay, N.E.; Demiryürek, A.T. Variants in TNF and NOS3 (ENOS) Genes Associated with Sepsis in Adult Patients. J. Gene Med. 2021, 23, e3323. [Google Scholar] [CrossRef]
- Lambden, S. Bench to Bedside Review: Therapeutic Modulation of Nitric Oxide in Sepsis—An Update. Intensive Care Med. Exp. 2019, 7, 64. [Google Scholar] [CrossRef]
- Wang, Z.; Feng, K.; Yue, M.; Lu, X.; Zheng, Q.; Zhang, H.; Zhai, Y.; Li, P.; Yu, L.; Cai, M.; et al. A Non-Synonymous SNP in the NOS2 Associated with Septic Shock in Patients with Sepsis in Chinese Populations. Hum. Genet. 2013, 132, 337–346. [Google Scholar] [CrossRef]
- Raia, L.; Zafrani, L. Endothelial Activation and Microcirculatory Disorders in Sepsis. Front. Med. 2022, 9, 907992. [Google Scholar] [CrossRef]
- Vera, S.; Martinez, R.; Gormaz, J.G.; Gajardo, A.; Galleguillos, F.; Rodrigo, R. Novel Relationships between Oxidative Stress and Angiogenesis-Related Factors in Sepsis: New Biomarkers and Therapies. Ann. Med. 2015, 47, 289–300. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the Missing Heritability of Complex Diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Agnello, L.; Ciaccio, M. Biomarkers of Sepsis. Diagnostics 2023, 13, 435. [Google Scholar] [CrossRef] [PubMed]
- Pierrakos, C.; Vincent, J.L. Sepsis Biomarkers: A Review. Crit. Care 2010, 14, R15. [Google Scholar] [CrossRef] [PubMed]
- Póvoa, P.; Coelho, L.; Dal-Pizzol, F.; Ferrer, R.; Huttner, A.; Conway Morris, A.; Nobre, V.; Ramirez, P.; Rouze, A.; Salluh, J.; et al. How to Use Biomarkers of Infection or Sepsis at the Bedside: Guide to Clinicians. Intensive Care Med. 2023, 49, 142–153. [Google Scholar] [CrossRef]
- Oliveira-Paula, G.H.; Lacchini, R.; Tanus-Santos, J.E. Endothelial Nitric Oxide Synthase: From Biochemistry and Gene Structure to Clinical Implications of NOS3 Polymorphisms. Gene 2016, 575, 584–599. [Google Scholar] [CrossRef]
- Tsukada, T.; Yokoyama, K.; Arai, T.; Takemoto, F.; Hara, S.; Yamada, A.; Kawaguchi, Y.; Hosoya, T.; Igari, J. Evidence of Association of the EcNOS Gene Polymorphism with Plasma NO Metabolite Levels in Humans. Biochem. Biophys. Res. Commun. 1998, 245, 190–193. [Google Scholar] [CrossRef]
- Asensi, V.; Montes, A.H.; Valle, E.; Ocaña, M.G.; Astudillo, A.; Alvarez, V.; López-Anglada, E.; Solís, A.; Coto, E.; Meana, A.; et al. The NOS3 (27-Bp Repeat, Intron 4) Polymorphism Is Associated with Susceptibility to Osteomyelitis. Nitric Oxide—Biol. Chem. 2007, 16, 44–53. [Google Scholar] [CrossRef]
- Ma, P.; Zhu, Y.; Qiu, H.; Liu, J.; Wang, Y.; Zeng, L. Endothelial Nitric Oxide Synthase 894G→T but Not -786T→C Gene Polymorphism Is Associated with Organ Dysfunction and Increased Mortality in Patients with Severe Sepsis. J. Trauma—Inj. Infect. Crit. Care 2011, 71, 872–877. [Google Scholar] [CrossRef]
- Huttunen, R.; Hurme, M.; Laine, J.; Eklund, C.; Vuento, R.; Aittoniemi, J.; Huhtala, H.; Syrjänen, J. Endothelial Nitric Oxide Synthase G894T (GLU298ASP) Polymorphism Is Associated with Hypotension in Patients with E. Coli Bacteremia but Not in Bacteremia Caused by a Gram-Positive Organism. Shock 2009, 31, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.C.; Martinez, A.; Mendoza, J.L.; Taxonera, C.; Díaz-Rubio, M.; Fernández-Arquero, M.; De La Concha, E.G.; Urcelay, E. Influence of the Inducible Nitric Oxide Synthase Gene (NOS2A) on Inflammatory Bowel Disease Susceptibility. Immunogenetics 2007, 59, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S.S.; Mastropaolo, L.A.; Murchie, R.; Griffiths, C.; Thöni, C.; Elkadri, A.; Xu, W.; Mack, A.; Walters, T.; Guo, C.; et al. Higher Activity of the Inducible Nitric Oxide Synthase Contributes to Very Early Onset Inflammatory Bowel Disease. Clin. Transl. Gastroenterol. 2014, 5, e46. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Lee, Y.; Kellum, J.A. A New Perspective on NO Pathway in Sepsis and ADMA Lowering as a Potential Therapeutic Approach. Crit. Care 2022, 26, 246. [Google Scholar] [CrossRef]
- Coletta, C.; Módis, K.; Oláh, G.; Brunyánszki, A.; Herzig, D.S.; Sherwood, E.R.; Ungvári, Z.; Szabo, C. Endothelial Dysfunction Is a Potential Contributor to Multiple Organ Failure and Mortality in Aged Mice Subjected to Septic Shock: Preclinical Studies in a Murine Model of Cecal Ligation and Puncture. Crit. Care 2014, 18, 511. [Google Scholar] [CrossRef]
- Ni, J.; McLoughlin, R.M.; Brodovitch, A.; Moulin, P.; Brouckaert, P.; Casadei, B.; Feron, O.; Topley, N.; Balligand, J.L.; Devuyst, O. Nitric Oxide Synthase Isoforms Play Distinct Roles during Acute Peritonitis. Nephrol. Dial. Transplant. 2010, 25, 86–96. [Google Scholar] [CrossRef]
- Pacher, P.; Beckman, J.S.; Liaudet, L. Nitric Oxide and Peroxynitrite in Health and Disease. Physiol. Rev. 2007, 87, 315–424. [Google Scholar] [CrossRef]
- Spiller, F.; Oliveira Formiga, R.; Fernandes da Silva Coimbra, J.; Alves-Filho, J.C.; Cunha, T.M.; Cunha, F.Q. Targeting Nitric Oxide as a Key Modulator of Sepsis, Arthritis and Pain. Nitric Oxide—Biol. Chem. 2019, 89, 32–40. [Google Scholar] [CrossRef]
- Srdić, T.; Đurašević, S.; Lakić, I.; Ružičić, A.; Vujović, P.; Jevđović, T.; Dakić, T.; Đorđević, J.; Tosti, T.; Glumac, S.; et al. From Molecular Mechanisms to Clinical Therapy: Understanding Sepsis-Induced Multiple Organ Dysfunction. Int. J. Mol. Sci. 2024, 25, 7770. [Google Scholar] [CrossRef]
- Hollenberg, S.M.; Cinel, I. Bench-to-Bedside Review: Nitric Oxide in Critical Illness—Update 2008. Crit. Care 2009, 13, 218. [Google Scholar] [CrossRef]
- Yu, M.H.; Chen, M.H.; Han, F.; Li, Q.; Sun, R.H.; Tu, Y.X. Prognostic Value of the Biomarkers Serum Amyloid A and Nitric Oxide in Patients with Sepsis. Int. Immunopharmacol. 2018, 62, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Li, Y.; Zhao, Y.; Wu, M.; Mi, B.; Liu, L.; Ma, H.; Jiang, C. Genetic Polymorphisms, Biomarkers and Signaling Pathways Associated with Septic Shock: From Diagnosis to Therapeutic Targets. Burn. Trauma 2024, 12, tkae006. [Google Scholar] [CrossRef] [PubMed]
- Metzger, I.F.; Sertório, J.T.C.; Tanus-Santos, J.E. Modulation of Nitric Oxide Formation by Endothelial Nitric Oxide Synthase Gene Haplotypes. Free Radic. Biol. Med. 2007, 43, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, D.; Assreuy, J. Nitric Oxide and Vascular Reactivity in Sepsis. Shock 2008, 30, 10–13. [Google Scholar] [CrossRef]
- Förstermann, U.; Sessa, W.C. Nitric Oxide Synthases: Regulation and Function. Eur. Heart J. 2012, 33, 829–837. [Google Scholar] [CrossRef]
- Koperna, T. Risk Stratification in Emergency Surgical Patients. Arch. Surg. 2001, 136, 55. [Google Scholar] [CrossRef]
- Levy, M.M.; Fink, M.P.; Marshall, J.C.; Abraham, E.; Angus, D.; Cook, D.; Cohen, J.; Opal, S.M.; Vincent, J.L.; Ramsay, G. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Intensive Care Med. 2003, 29, 530–538. [Google Scholar] [CrossRef]
- Salamone, G.; Licari, L.; Falco, N.; Augello, G.; Tutino, R.; Campanella, S.; Guercio, G.; Gulotta, G. Mannheim Peritonitis Index (MPI) and Elderly Population: Prognostic Evaluation in Acute Secondary Peritonitis. G. Chir.—J. Surg. 2016, 27, 243. [Google Scholar] [CrossRef]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A Simple Salting out Procedure for Extracting DNA from Human Nucleated Cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Carbocation Corporation False Discovery Rate Online Calculator. Available online: https://tools.carbocation.com/FDR (accessed on 16 October 2025).
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).