Phospho-Switch: Regulation of the Activity of SAM-Dependent Methyltransferases Using H-Phosphinic SAM Analogue
Abstract
1. Introduction
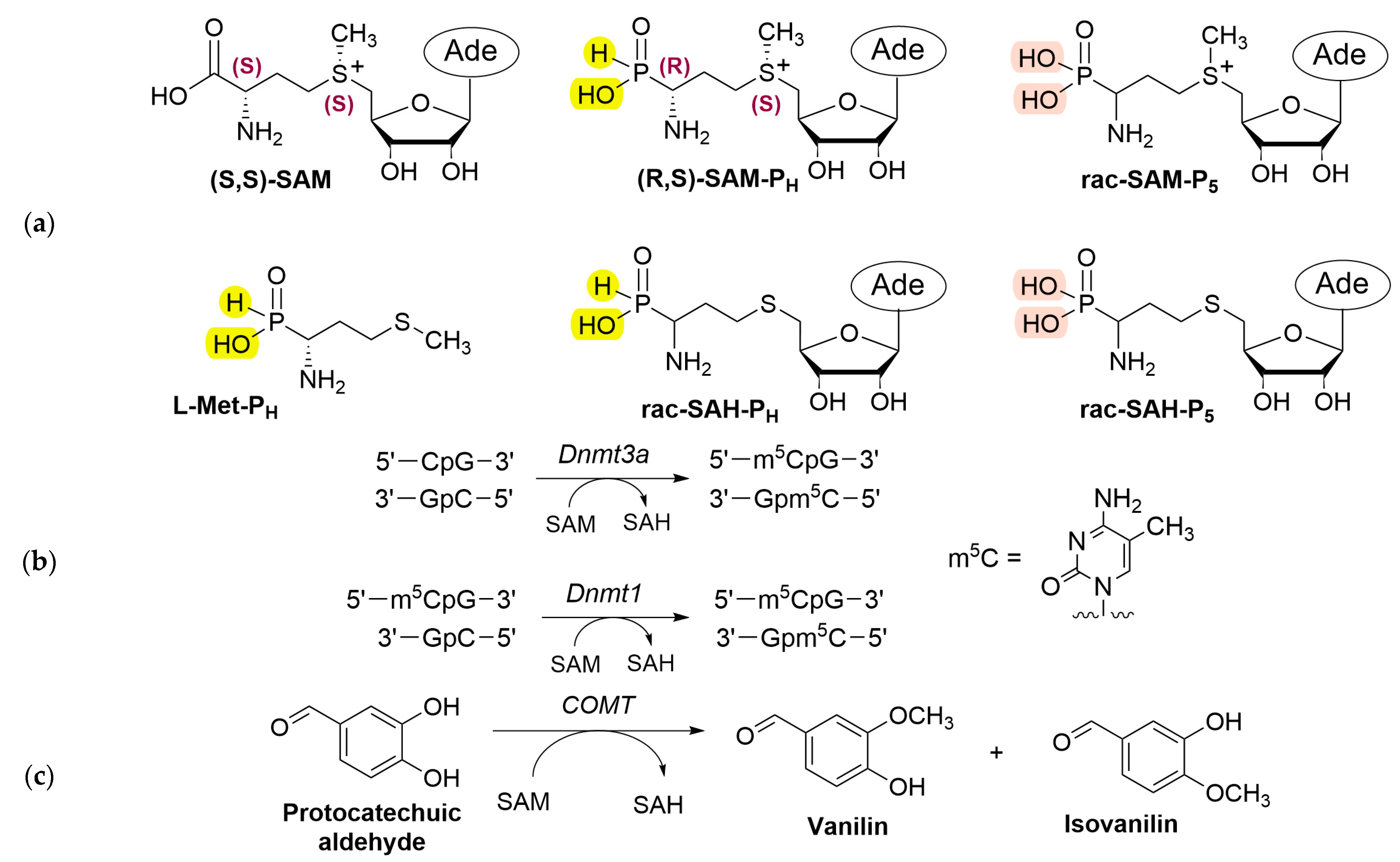
2. Results
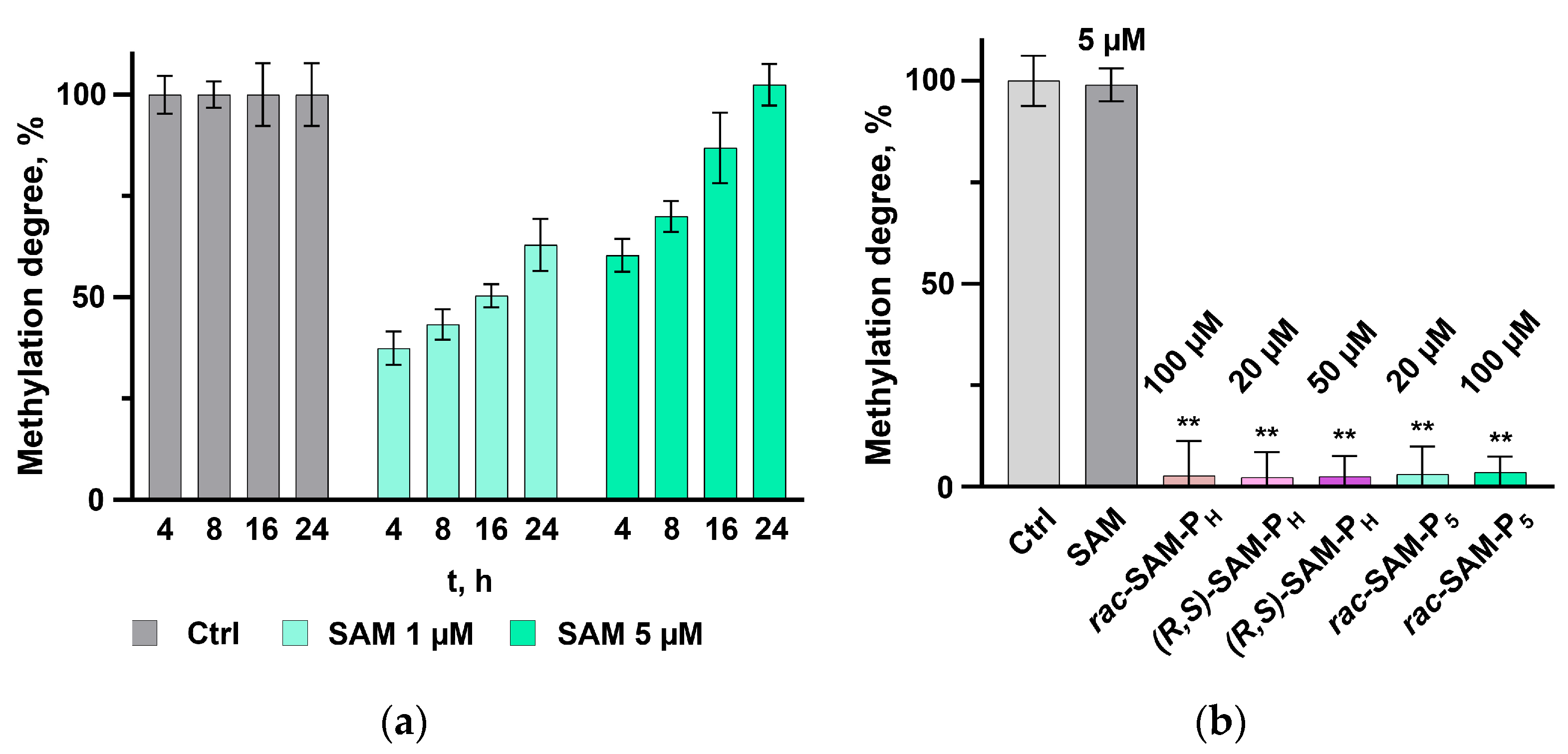
2.1. Phosphorus-Containing Analogues of SAM: Substrates for Dnmt3a and COMT, but Not Dnmt1
| Cofactor | Methylation Activity, % | ||
|---|---|---|---|
| Dnmt1 | Dnmt3a | COMT | |
| SAM | 100 | 100 | 100 |
| (R,S)-SAM-PH | Not a donor of methyl group | 59 [23] | 22 * |
| rac-SAM-P5 | Not a donor of methyl group | 51 [23] | Not a donor of methyl group |
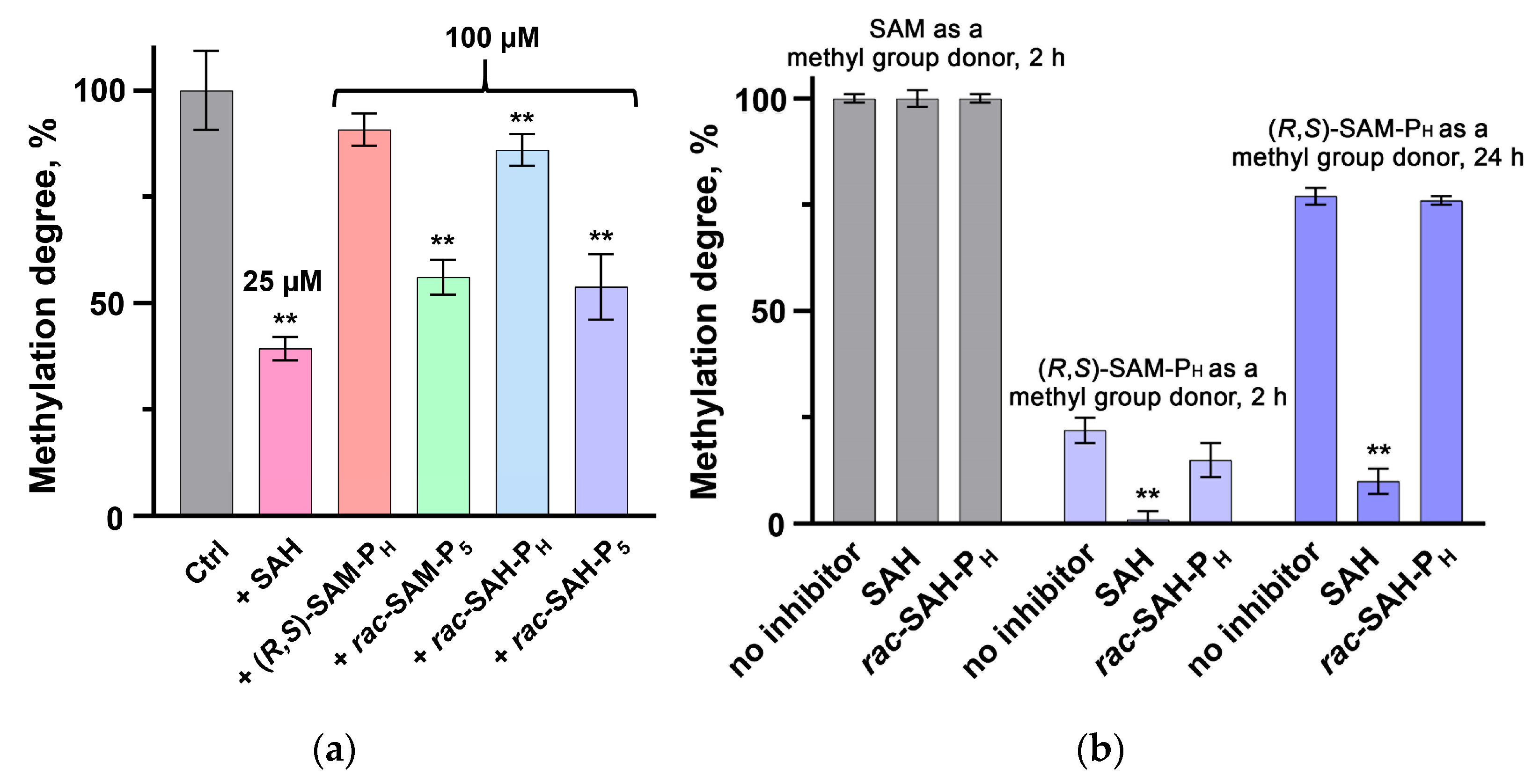
2.2. Inhibition of Dnmt1 by SAM and SAH Phosphorus-Containing Analogues
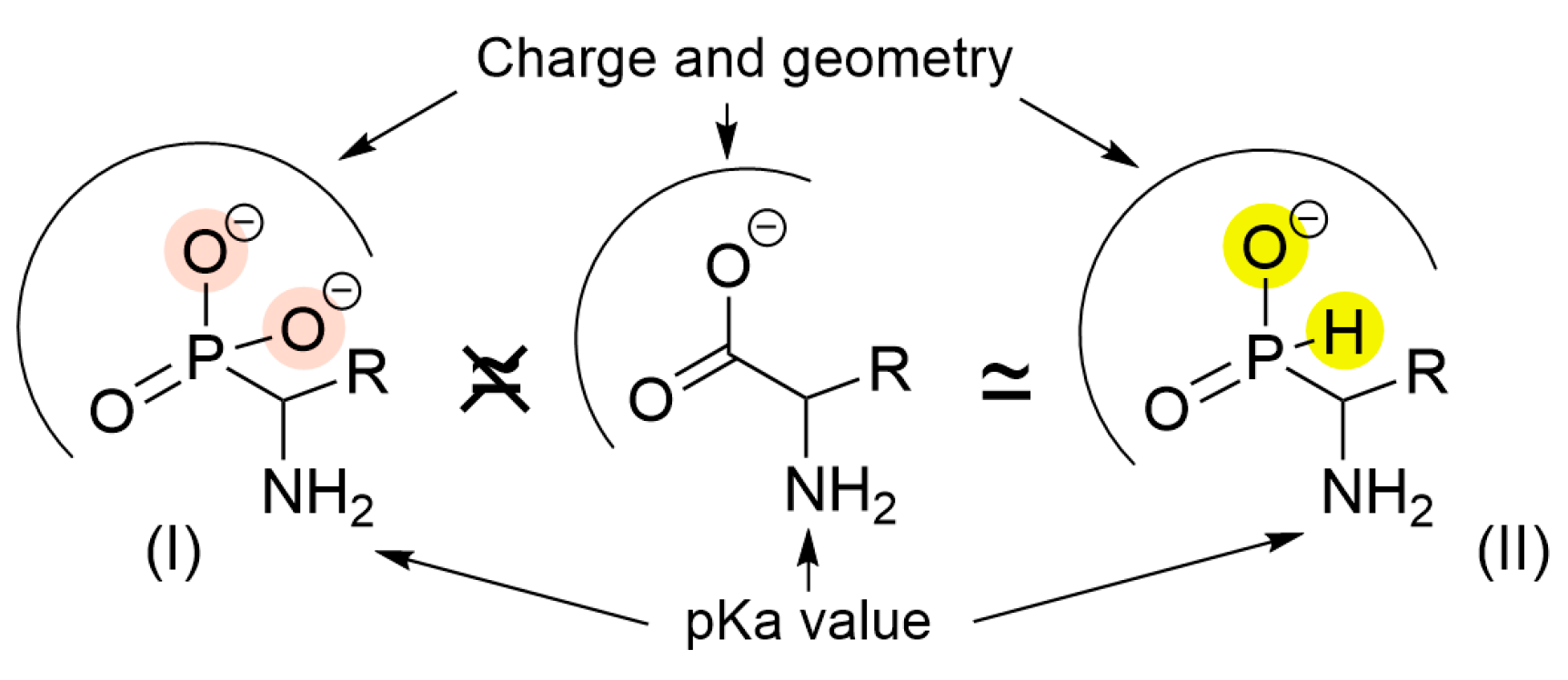
3. Discussion
4. Materials and Methods
4.1. Dnmt1 Activity Assay, Substrate Properties of Phosphorus-Containing Analogues of SAM and Inhibitory Activity of Phosphorus-Containing Analogues of SAH
4.2. COMT Activity Assay and Inhibitory Activity of Phosphorus-Containing Analogues of SAH
4.3. Statistical Analyzis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lee, Y.-H.; Ren, D.; Jeon, B.; Liu, H.-W. S-Adenosylmethionine: More than just a methyl donor. Nat. Prod. Rep. 2023, 40, 1521–1549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zheng, Y.G. SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases. ACS Chem. Biol. 2015, 11, 583–597. [Google Scholar] [CrossRef]
- Rudenko, A.Y.; Mariasina, S.S.; Sergiev, P.V.; Polshakov, V.I. Analogs of S-adenosyl-L-methionine in studies of methyltransferases. Mol. Biol. 2022, 56, 229–250. [Google Scholar] [CrossRef]
- Sun, Q.; Huang, M.; Wei, Y. Diversity of the reaction mechanisms of SAM-dependent enzymes. Acta Pharm. Sin. B 2021, 11, 632–650. [Google Scholar] [CrossRef]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining mysteries of molecular biology: The role of polyamines in the cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef]
- Frey, P.A.; Hegeman, A.D.; Ruzicka, F.J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 63–88. [Google Scholar] [CrossRef]
- Pey, A.L.; Majtan, T.; Sanchez-Ruiz, J.M.; Kraus, J.P. Human cystathionine β-synthase (CBS) contains two classes of binding sites for S-adenosylmethionine (SAM): Complex regulation of CBS activity and stability by SAM. Biochem. J. 2012, 449, 109–121. [Google Scholar] [CrossRef]
- Bhatia, M.; Thakur, J.; Suyal, S.; Oniel, R.; Chakraborty, R.; Pradhan, S.; Sharma, M.; Sengupta, S.; Laxman, S.; Masakapalli, S.K.; et al. Allosteric inhibition of MTHFR prevents futile SAM cycling and maintains nucleotide pools in one-carbon metabolism. J. Biol. Chem. 2020, 295, 16037–16057. [Google Scholar] [CrossRef]
- Pattyn, J.; Vaughan-Hirsch, J.; Van de Poel, B. The regulation of ethylene biosynthesis: A complex multilevel control circuitry. New Phytol. 2020, 229, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, A.Y.; Mariasina, S.S.; Ozhiganov, R.M.; Sergiev, P.V.; Polshakov, V.I. Enzymatic reactions of S-adenosyl-L-methionine: Synthesis and applications. Biochemistry 2025, 90, S105–S134. [Google Scholar] [CrossRef] [PubMed]
- Schluckebier, G.; Kozak, M.; Bleimling, N.; Weinhold, E.; Saenger, W. Differential binding of S-adenosylmethionine S-adenosylhomocysteine and sinefungin to the adenine-specific DNA methyltransferase M. TaqI. J. Mol. Biol. 1997, 265, 56–67. [Google Scholar] [CrossRef]
- Subramaniam, D.; Thombre, R.; Dhar, A.; Anant, S. DNA Methyltransferases: A novel target for prevention and therapy. Front. Oncol. 2014, 4, 80. [Google Scholar] [CrossRef]
- Stresemann, C.; Lyko, F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer 2008, 123, 8–13. [Google Scholar] [CrossRef]
- Stone, M.L.; Chiappinelli, K.B.; Li, H.; Murphy, L.M.; Travers, M.E.; Topper, M.J.; Mathios, D.; Lim, M.; Shih, I.-M.; Wang, T.-L.; et al. Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden. Proc. Natl. Acad. Sci. USA 2017, 114, E10981–E10990. [Google Scholar] [CrossRef] [PubMed]
- Travers, M.; Brown, S.M.; Dunworth, M.; Holbert, C.E.; Wiehagen, K.R.; Bachman, K.E.; Foley, J.R.; Stone, M.L.; Baylin, S.B.; Casero, R.A., Jr.; et al. DFMO and 5-azacytidine increase M1 macrophages in the tumor microenvironment of murine ovarian cancer. Cancer Res. 2019, 79, 3445–3454. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.C.; Mato, J.M. S-adenosylmethionine in liver health, injury, and cancer. Physiol. Rev. 2012, 92, 1515–1542. [Google Scholar] [CrossRef]
- De Clercq, E. Another ten stories in antiviral drug discovery (part C): “Old” and “new” antivirals, strategies, and perspectives. Med. Res. Rev. 2009, 29, 611–645. [Google Scholar] [CrossRef]
- Loo, L.W.; Tiirikainen, M.; Cheng, I.; Lum-Jones, A.; Seifried, A.; Church, J.M.; Gryfe, R.; Weisenberger, D.J.; Lindor, N.M.; Gallinger, S.; et al. Integrated analysis of genome-wide copy number alterations and gene expression in microsatellite stable, CpG island methylator phenotype-negative colon cancer. Genes Chromosomes Cancer 2013, 52, 450–466. [Google Scholar] [CrossRef]
- Uchiyama, N.; Tanaka, Y.; Kawamoto, T. Aristeromycin and DZNeP cause growth inhibition of prostate cancer via induction of mir-26a. Eur. J. Pharmacol. 2017, 812, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Aury-Landas, J.; Girard, N.; Lhuissier, E.; Adouane, D.; Delépée, R.; Boumediene, K.; Baugé, C. The antitumoral effect of the S-adenosylhomocysteine hydrolase inhibitor, 3-deazaneplanocin A, is independent of EZH2 but is correlated with EGFR downregulation in chondrosarcomas. Cell. Physiol. Biochem. 2019, 53, 731–745. [Google Scholar] [CrossRef]
- Wu, G.; Wang, N.; Luo, Y.; Zhang, Y.; Wang, P.; Zhu, Z.; Gao, Y.; Du, Z.; Yang, B. Metabolic perturbation of epigenome by inhibiting S-adenosylhomocysteine hydrolase elicits senescence through DNA damage response in hepatoma cells. Tumor Biol. 2017, 39, 1010428317699117. [Google Scholar] [CrossRef]
- Rudenko, A.Y.; Mariasina, S.S.; Bolikhova, A.K.; Nikulin, M.V.; Ozhiganov, R.M.; Vasil’eV, V.G.; Ikhalaynen, Y.A.; Khandazhinskaya, A.L.; Khomutov, M.A.; Sergiev, P.V.; et al. Organophosphorus S-adenosyl-L-methionine mimetics: Synthesis, stability, and substrate properties. Front. Chem. 2024, 12, 1448747. [Google Scholar] [CrossRef]
- Filonov, V.L.; Khomutov, M.A.; Sergeev, A.V.; Khandazhinskaya, A.L.; Kochetkov, S.N.; Gromova, E.S.; Khomutov, A.R. Interaction of DNA methyltransferase Dnmt3a with phosphorus analogs of S-adenosylmethionine and S-adenosylhomocysteine. Mol. Biol. 2023, 57, 747–754. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Pradhan, S.; Bacolla, A.; Wells, R.D.; Roberts, R.J. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999, 274, 33002–33010. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.J.; McKelvie, J.C.; Maynard-Smith, M.D.; Roach, P.L. A real-time assay for CpG-specific cytosine-C5 methyltransferase activity. Nucleic Acids Res. 2010, 38, e107. [Google Scholar] [CrossRef] [PubMed]
- Huber, T.D.; Wang, F.; Singh, S.; Johnson, B.R.; Zhang, J.; Sunkara, M.; Van Lanen, S.G.; Morris, A.J.; Phillips, G.N.; Thorson, J.S. Functional AdoMet isosteres resistant to classical AdoMet degradation pathways. ACS Chem. Biol. 2016, 11, 2484–2491. [Google Scholar] [CrossRef]
- Halby, L.; Menon, Y.; Rilova, E.; Pechalrieu, D.; Masson, V.; Faux, C.; Bouhlel, M.A.; David-Cordonnier, M.-H.; Novosad, N.; Aussagues, Y.; et al. Rational design of bisubstrate-type analogues as inhibitors of DNA methyltransferases in cancer cells. J. Med. Chem. 2017, 60, 4665–4679. [Google Scholar] [CrossRef]
- Chen, S.; Wang, Y.; Zhou, W.; Li, S.; Peng, J.; Shi, Z.; Hu, J.; Liu, Y.-C.; Ding, H.; Lin, Y.; et al. Identifying novel selective non-nucleoside DNA methyltransferase 1 inhibitors through docking-based virtual screening. J. Med. Chem. 2014, 57, 9028–9041. [Google Scholar] [CrossRef] [PubMed]
- Kafarski, P.; Lejczak, B. Aminophosphonic acids of potential medical importance. Curr. Med. Chem. Agents 2001, 1, 301–312. [Google Scholar] [CrossRef]
- Badet, B.; Inagaki, K.; Soda, K.; Walsh, C.T. Time-dependent inhibition of Bacillus stearothermophilus alanine racemase by (1-aminoethyl)phosphonate isomers by isomerization to noncovalent slowly dissociating enzyme-(1-aminoethyl)phosphonate complexes. Biochemistry 1986, 25, 3275–3282. [Google Scholar] [CrossRef]
- A Steere, J.; Sampson, P.B.; Honek, J.F. Synthesis of an α-aminophosphonate nucleoside as an inhibitor of S-adenosyl-L-homocysteine hydrolase. Bioorganic Med. Chem. Lett. 2002, 12, 457–460. [Google Scholar] [CrossRef] [PubMed]
- De Biase, D.; Cappadocio, F.; Pennacchietti, E.; Giovannercole, F.; Coluccia, A.; Vepsäläinen, J.; Khomutov, A. Enzymatic kinetic resolution of desmethylphosphinothricin indicates that phosphinic group is a bioisostere of carboxyl group. Commun. Chem. 2020, 3, 121. [Google Scholar] [CrossRef]
- Baylis, E.K.; Campbell, C.D.; Dingwall, J.G. 1-Aminoalkylphosphonous acids. Part Isosteres of the protein amino acids. J. Chem. Soc. Perkin Trans. 1 1984, 2845–2853. [Google Scholar] [CrossRef]
- Laber, B.; Amrhein, N. Metabolism of 1-aminoethylphosphinate generates acetylphosphinate, a potent inhibitor of pyruvate dehydrogenase. Biochem. J. 1987, 248, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Khomutov, R.M.; Khurs, E.N.; Dzhavakhiya, V.G.; Voinova, T.M.; Ermolinskii, B.S. 1-Aminoethylphosphonic acid, a new inhibitor of the polyketide pathway for the biosynthesis of natural compounds. Bioorg. Khim. 1987, 13, 1422–1424. [Google Scholar]
- Faleev, N.G.; Zhukov, Y.N.; Khurs, E.N.; Gogoleva, O.I.; Barbolina, M.V.; Bazhulina, N.P.; Belikov, V.M.; Demidkina, T.V.; Khomutov, R.M. Interaction of tyrosine phenol-lyase with phosphoroorganic analogues of substrate amino acids. Eur. J. Biochem. 2000, 267, 6897–6902. [Google Scholar] [CrossRef]
- Faleev, N.G.; Alferov, K.V.; Tsvetikova, M.A.; Morozova, E.A.; Revtovich, S.V.; Khurs, E.N.; Vorob’ev, M.M.; Phillips, R.S.; Demidkina, T.V.; Khomutov, R.M. Methionine gamma-lyase: Mechanistic deductions from the kinetic pH-effects. The role of the ionic state of a substrate in the enzymatic activity. Biochim. Biophys. Acta 2009, 1794, 1414–1420. [Google Scholar] [CrossRef]
- Filonov, V.L.; Khomutov, M.A.; Tkachev, Y.V.; Udod, A.V.; Yanvarev, D.V.; Giovannercole, F.; Khurs, E.N.; Kochetkov, S.N.; De Biase, D.; Khomutov, A.R. Enzymatic synthesis of biologically active H-phosphinic analogue of α-ketoglutarate. Biomolecules 2024, 14, 1574. [Google Scholar] [CrossRef]
- Wijayasinghe, Y.S.; Blumenthal, R.M.; Viola, R.E. Producing proficient methyl donors from alternative substrates of S-adenosylmethionine synthetase. Biochemistry 2014, 53, 1521–1526. [Google Scholar] [CrossRef]
- Takeshita, K.; Suetake, I.; Yamashita, E.; Suga, M.; Narita, H.; Nakagawa, A.; Tajima, S. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc. Natl. Acad. Sci. USA 2011, 108, 9055–9059. [Google Scholar] [CrossRef]
- Yu, J.; Xie, T.; Wang, Z.; Wang, X.; Zeng, S.; Kang, Y.; Hou, T. DNA methyltransferases: Emerging targets for the discovery of inhibitors as potent anticancer drugs. Drug Discov. Today 2019, 24, 2323–2331. [Google Scholar] [CrossRef]
- Kumar, S.; Cheng, X.; Klimasauskas, S.; Mi, S.; Posfai, J.; Roberts, R.J.; Wilson, G.G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994, 22, 1–10. [Google Scholar] [CrossRef]
- Song, J.; Rechkoblit, O.; Bestor, T.H.; Patel, D.J. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 2011, 331, 1036–1040. [Google Scholar] [CrossRef]
- Jeltsch, A.; Jurkowska, R.Z. Allosteric control of mammalian DNA methyltransferases—A new regulatory paradigm. Nucleic Acids Res. 2016, 44, 8556–8575. [Google Scholar] [CrossRef] [PubMed]
- Gowher, H.; Jeltsch, A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 2002, 277, 20409–20414. [Google Scholar] [CrossRef]
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and function of mammalian DNA methyltransferases. Chembiochem 2011, 12, 206–222. [Google Scholar] [CrossRef]
- Yoo, J.; Choi, S.; Medina-Franco, J.L.; Velasco, G. Molecular modeling studies of the novel inhibitors of DNA methyltransferases SGI-1027 and CBC12: Implications for the mechanism of inhibition of DNMTs. PLoS ONE 2013, 8, e62152. [Google Scholar] [CrossRef]
- Xie, T.; Yu, J.; Fu, W.; Wang, Z.; Xu, L.; Chang, S.; Wang, E.; Zhu, F.; Zeng, S.; Kang, Y.; et al. Insight into the selective binding mechanism of DNMT1 and DNMT3A inhibitors: A molecular simulation study. Phys. Chem. Chem. Phys. 2019, 21, 12931–12947. [Google Scholar] [CrossRef] [PubMed]
- Tajima, S.; Suetake, I.; Takeshita, K.; Nakagawa, A.; Kimura, H. Domain structure of the Dnmt1, Dnmt3a, and Dnmt3b DNA methyltransferases. Adv. Exp. Med. Boil. 2016, 945, 63–86. [Google Scholar] [CrossRef]
- Giovannercole, F.; Gonçalves, L.G.; Armengaud, J.; Coelho, A.V.; Khomutov, A.; De Biase, D. Integrated multi-omics unveil the impact of H-phosphinic analogs of glutamate and α-ketoglutarate on Escherichia coli metabolism. J. Biol. Chem. 2024, 300, 107803. [Google Scholar] [CrossRef] [PubMed]
- Zhukov, Y.N.; Vavilova, N.A.; Osipova, T.I.; Voinova, T.M.; Khurs, E.N.; Dzhavakhia, V.G.; Khomutov, R.M. Fungicidal activity of phosphinic analogues of amino acids involved in methionine metabolism. Dokl. Biochem. Biophys. 2004, 397, 210–212. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filonov, V.L.; Khomutov, M.A.; Rudenko, A.Y.; Mariasina, S.S.; Ozhiganov, R.M.; Sergeev, A.V.; Kochetkov, S.N.; Polshakov, V.I.; Gromova, E.S.; Khandazhinskaya, A.L.; et al. Phospho-Switch: Regulation of the Activity of SAM-Dependent Methyltransferases Using H-Phosphinic SAM Analogue. Int. J. Mol. Sci. 2025, 26, 8590. https://doi.org/10.3390/ijms26178590
Filonov VL, Khomutov MA, Rudenko AY, Mariasina SS, Ozhiganov RM, Sergeev AV, Kochetkov SN, Polshakov VI, Gromova ES, Khandazhinskaya AL, et al. Phospho-Switch: Regulation of the Activity of SAM-Dependent Methyltransferases Using H-Phosphinic SAM Analogue. International Journal of Molecular Sciences. 2025; 26(17):8590. https://doi.org/10.3390/ijms26178590
Chicago/Turabian StyleFilonov, Vsevolod L., Maxim A. Khomutov, Alexander Yu. Rudenko, Sofia S. Mariasina, Ratislav M. Ozhiganov, Alexander V. Sergeev, Sergei N. Kochetkov, Vladimir I. Polshakov, Elizaveta S. Gromova, Anastasia L. Khandazhinskaya, and et al. 2025. "Phospho-Switch: Regulation of the Activity of SAM-Dependent Methyltransferases Using H-Phosphinic SAM Analogue" International Journal of Molecular Sciences 26, no. 17: 8590. https://doi.org/10.3390/ijms26178590
APA StyleFilonov, V. L., Khomutov, M. A., Rudenko, A. Y., Mariasina, S. S., Ozhiganov, R. M., Sergeev, A. V., Kochetkov, S. N., Polshakov, V. I., Gromova, E. S., Khandazhinskaya, A. L., & Khomutov, A. R. (2025). Phospho-Switch: Regulation of the Activity of SAM-Dependent Methyltransferases Using H-Phosphinic SAM Analogue. International Journal of Molecular Sciences, 26(17), 8590. https://doi.org/10.3390/ijms26178590





