Detection of XPO1E571K Gene Mutation from Cell-Free DNA in Blood Circulation of Lymphoma Patients by FAST-COLD PCR
Abstract
1. Introduction
2. Results
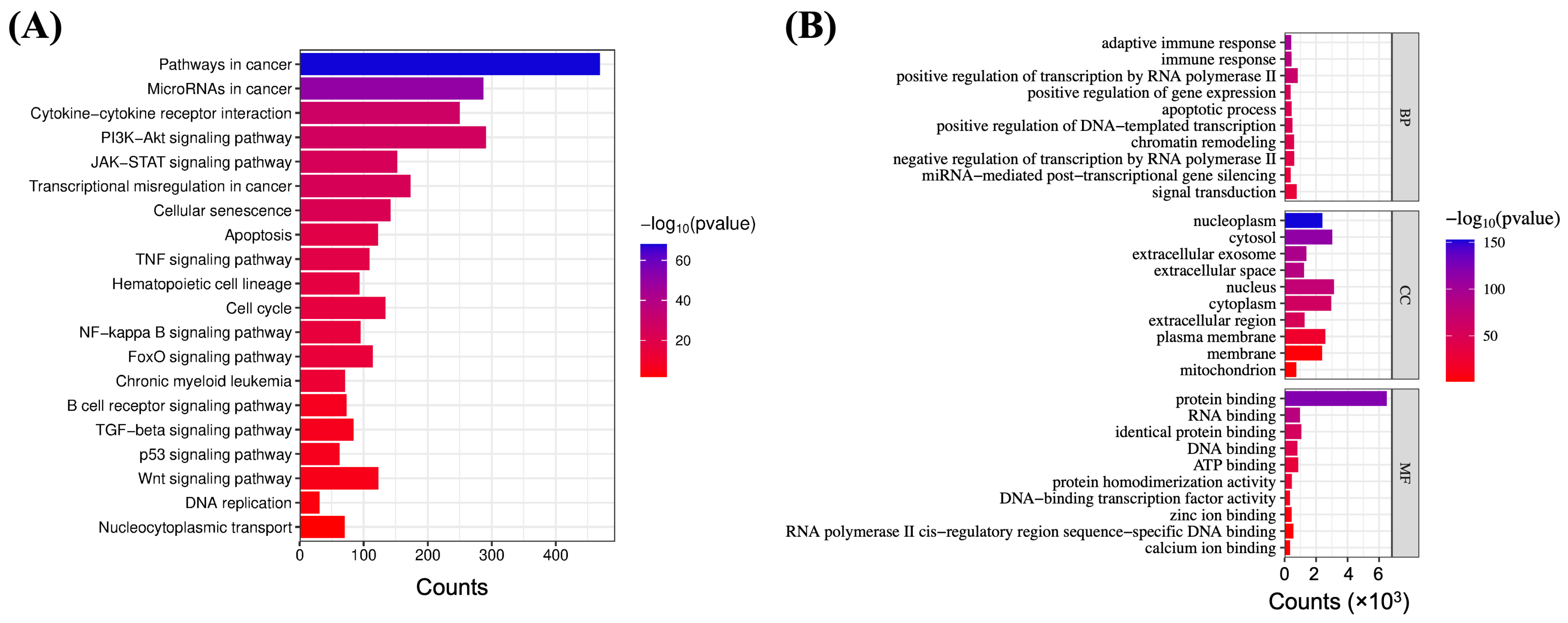
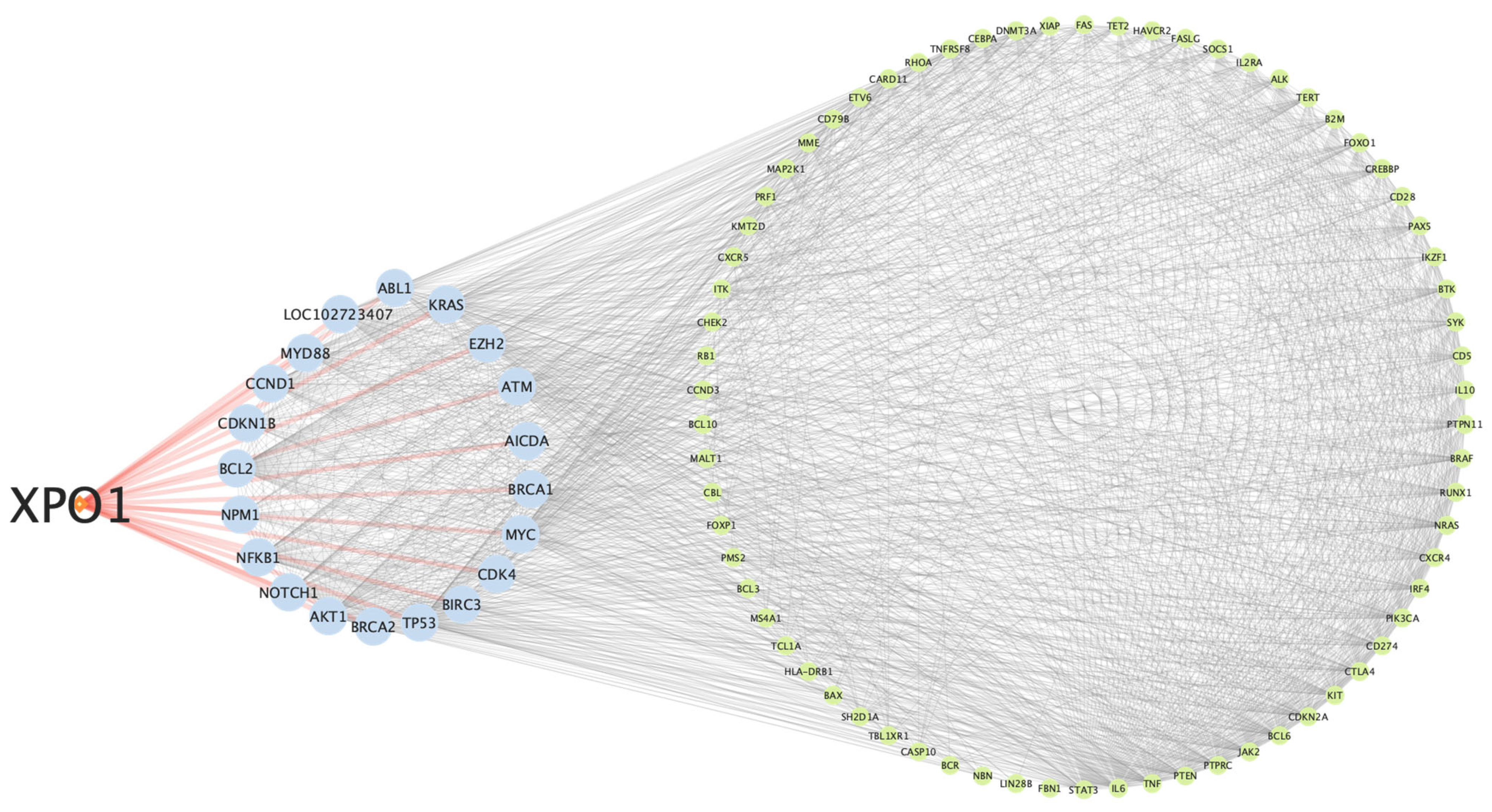
2.1. Network Analysis of XPO1 in Lymphoma
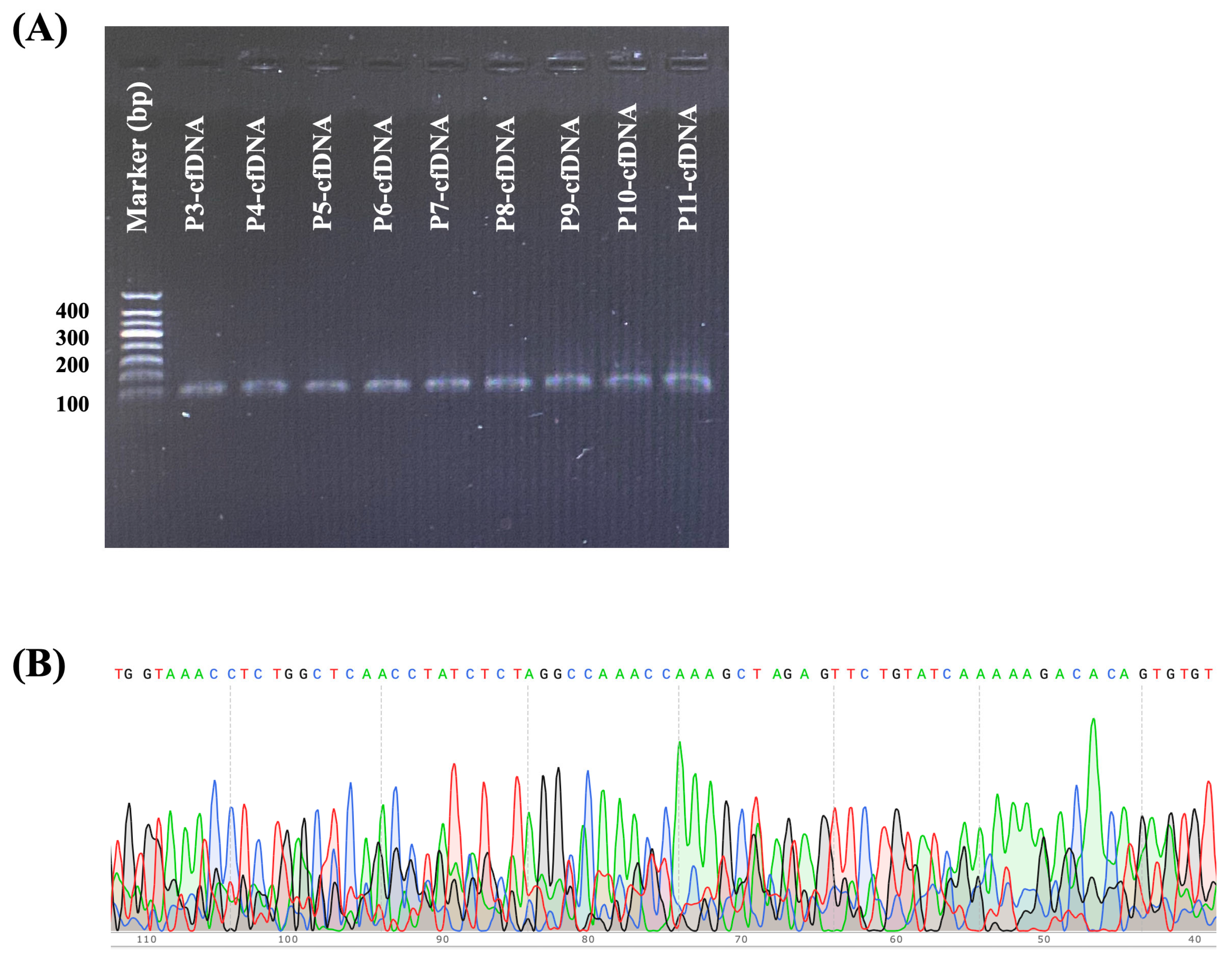
2.2. Assessment of Plasma and FFPE Tissue Samples for XPO1 Gene Amplification and Sequencing in Lymphoma Patients
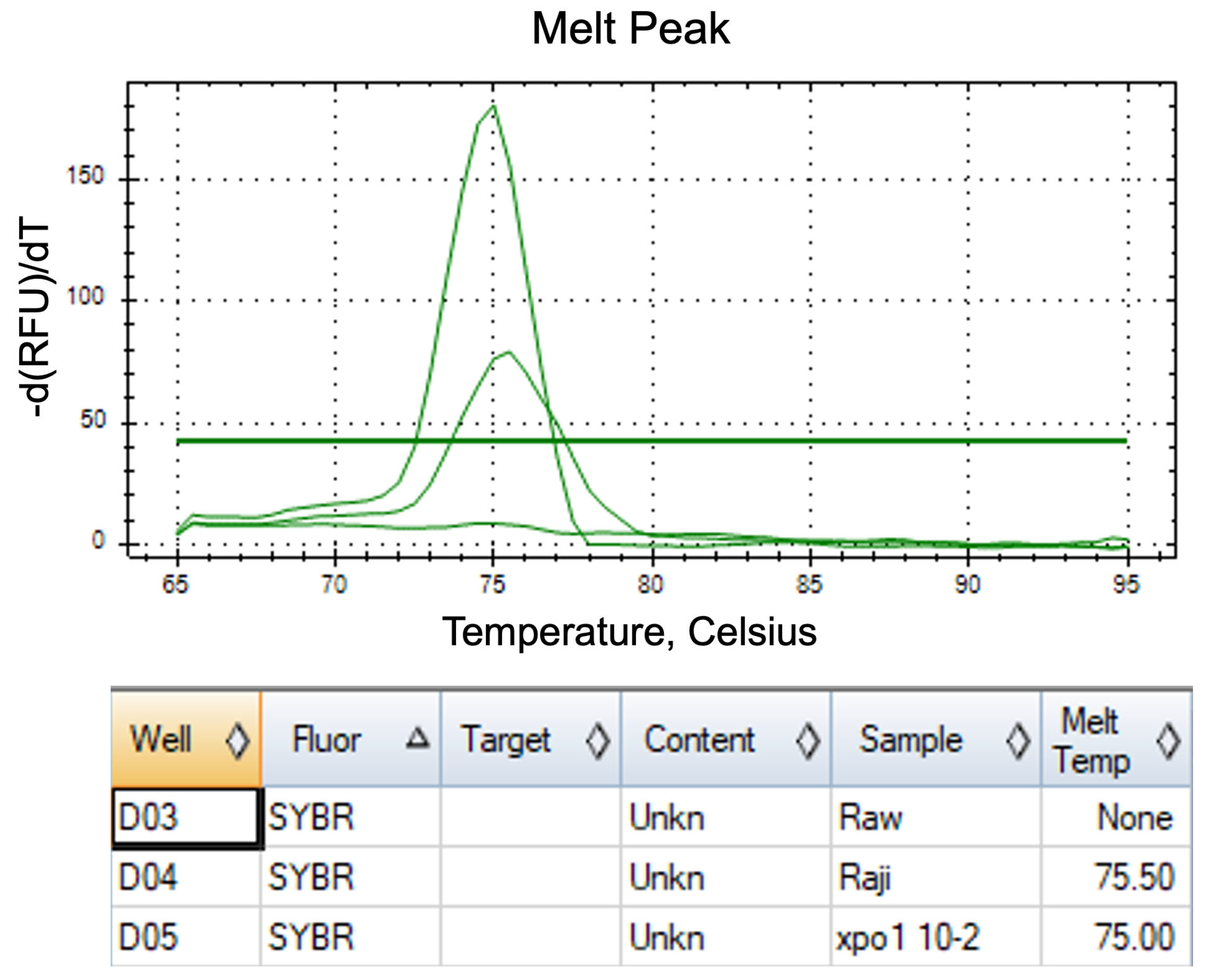
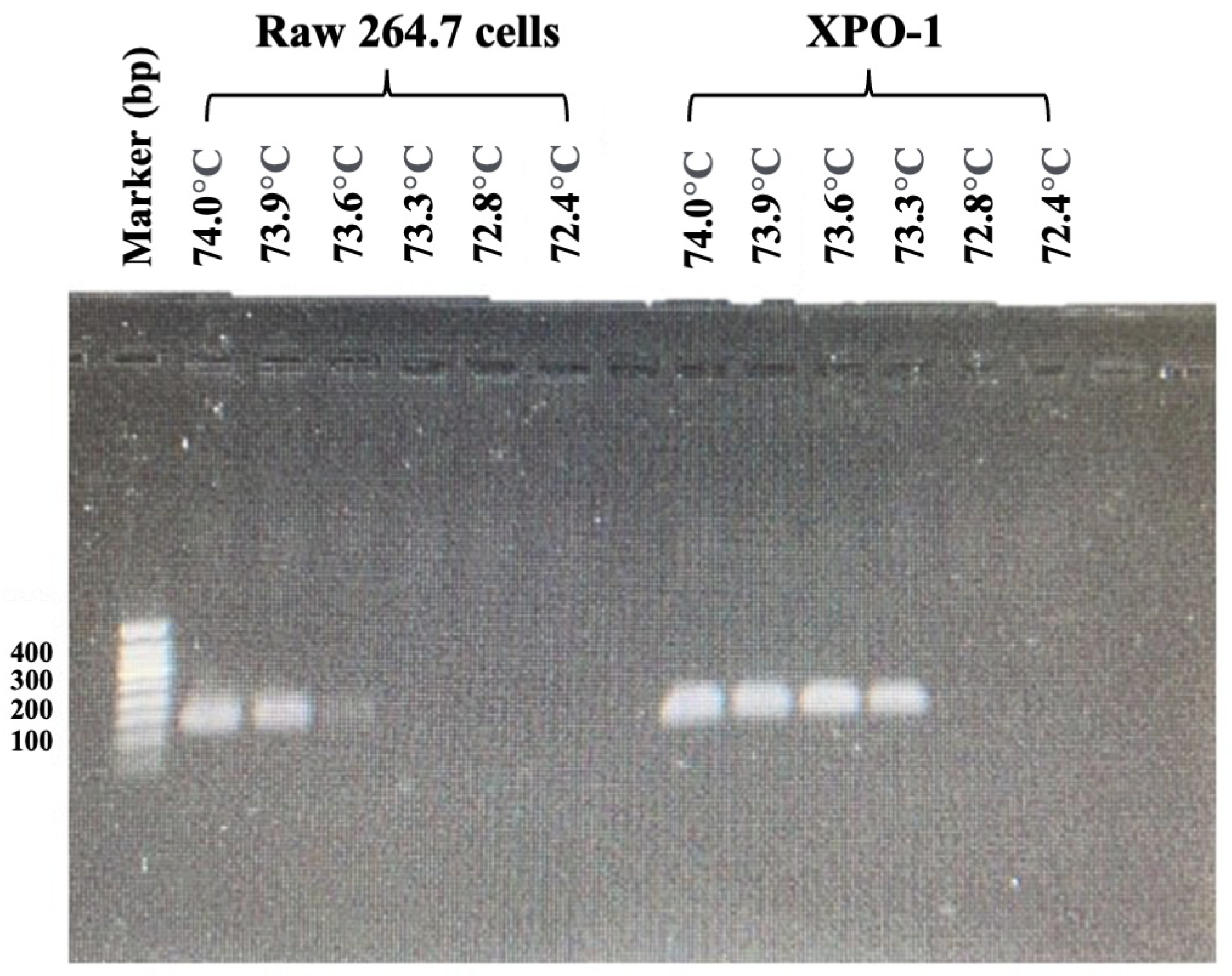
2.3. Optimal Critical Temperature (Tc) of FAST-COLD-PCR Assay and Verification of FAST-COLD-PCR Assay for XPO1E571K Detection
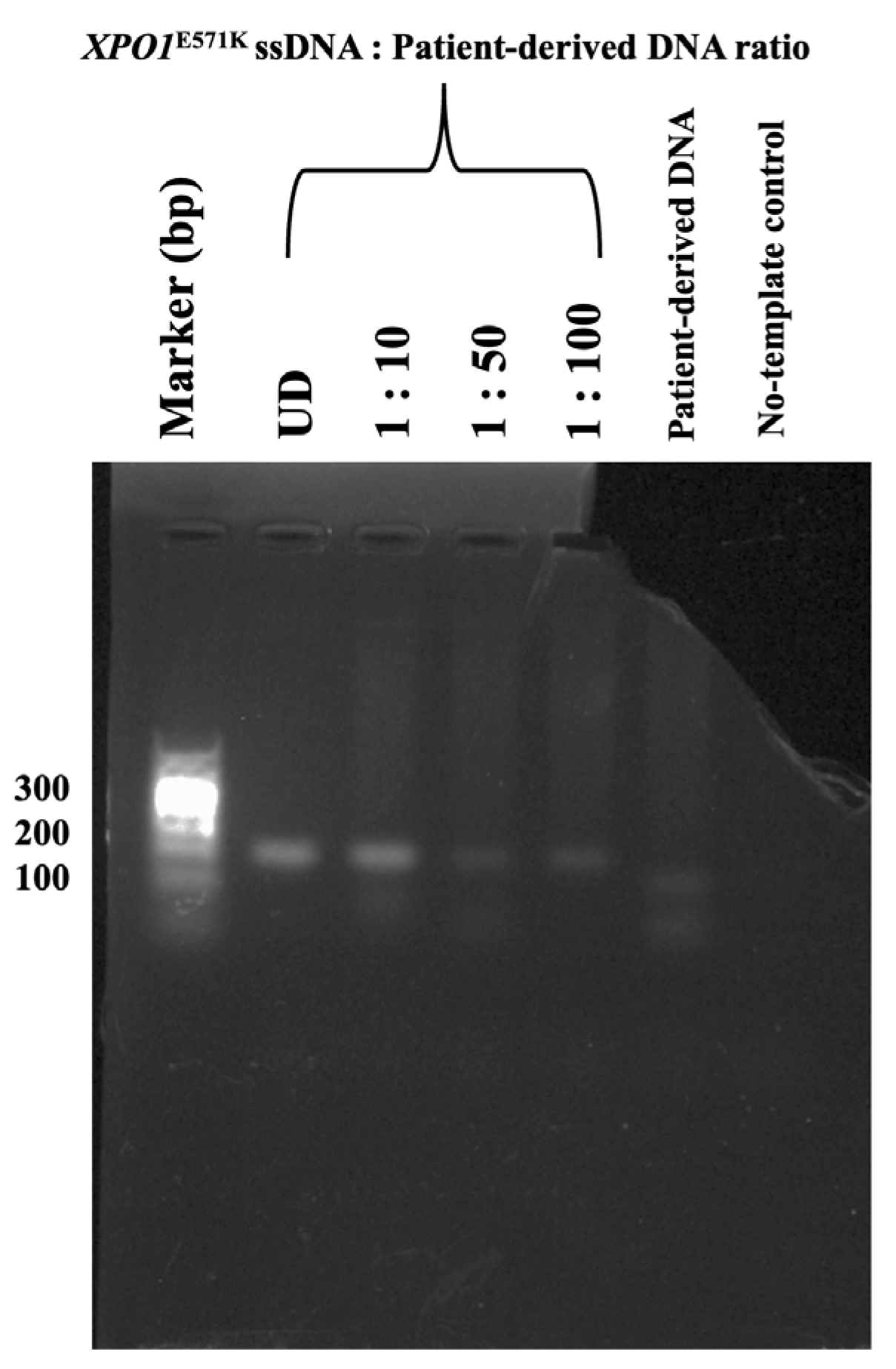
2.4. Limit of Detection and of FAST-COLD-PCR Assay for XPO1E571K Mutation Detection
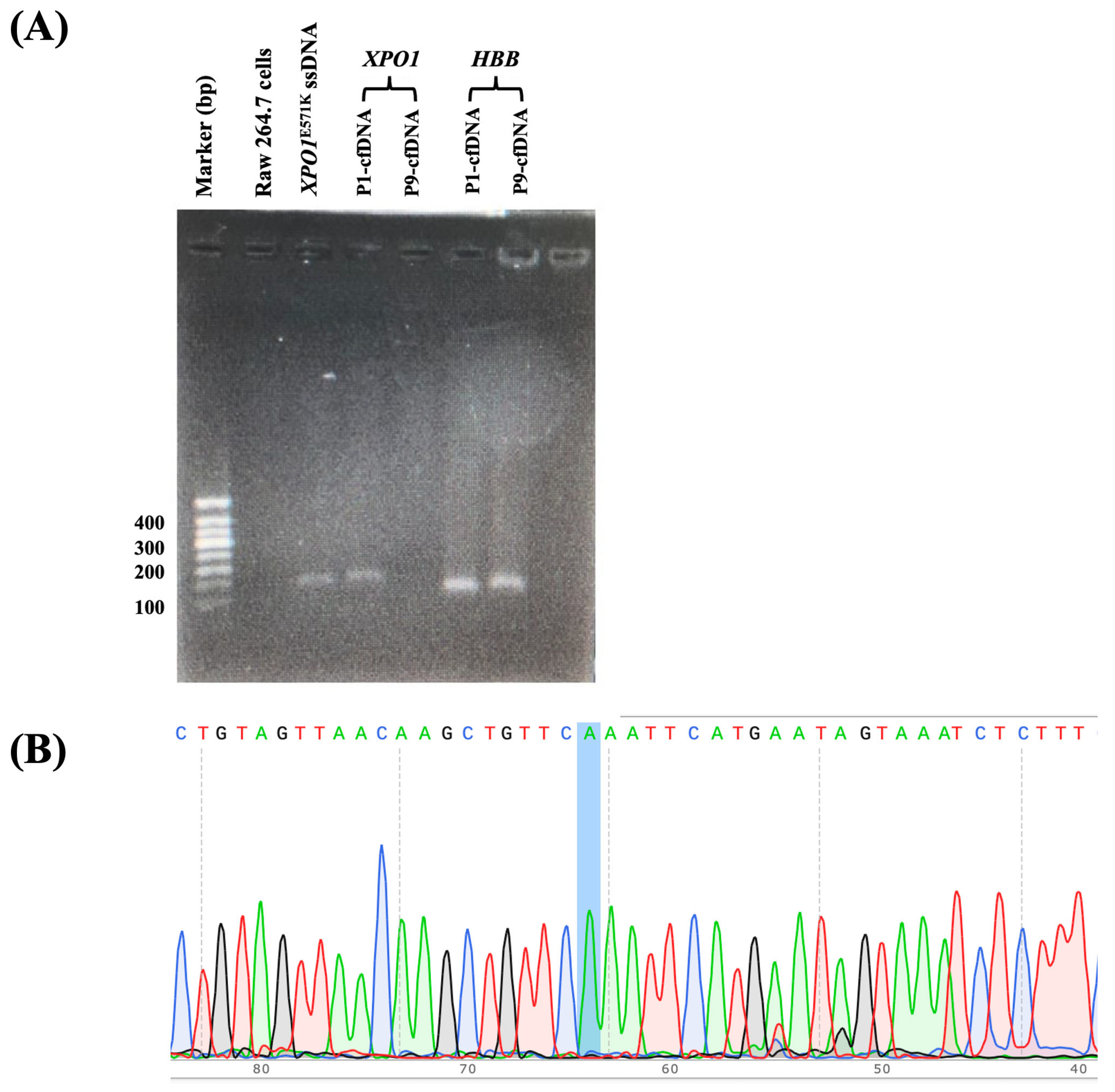
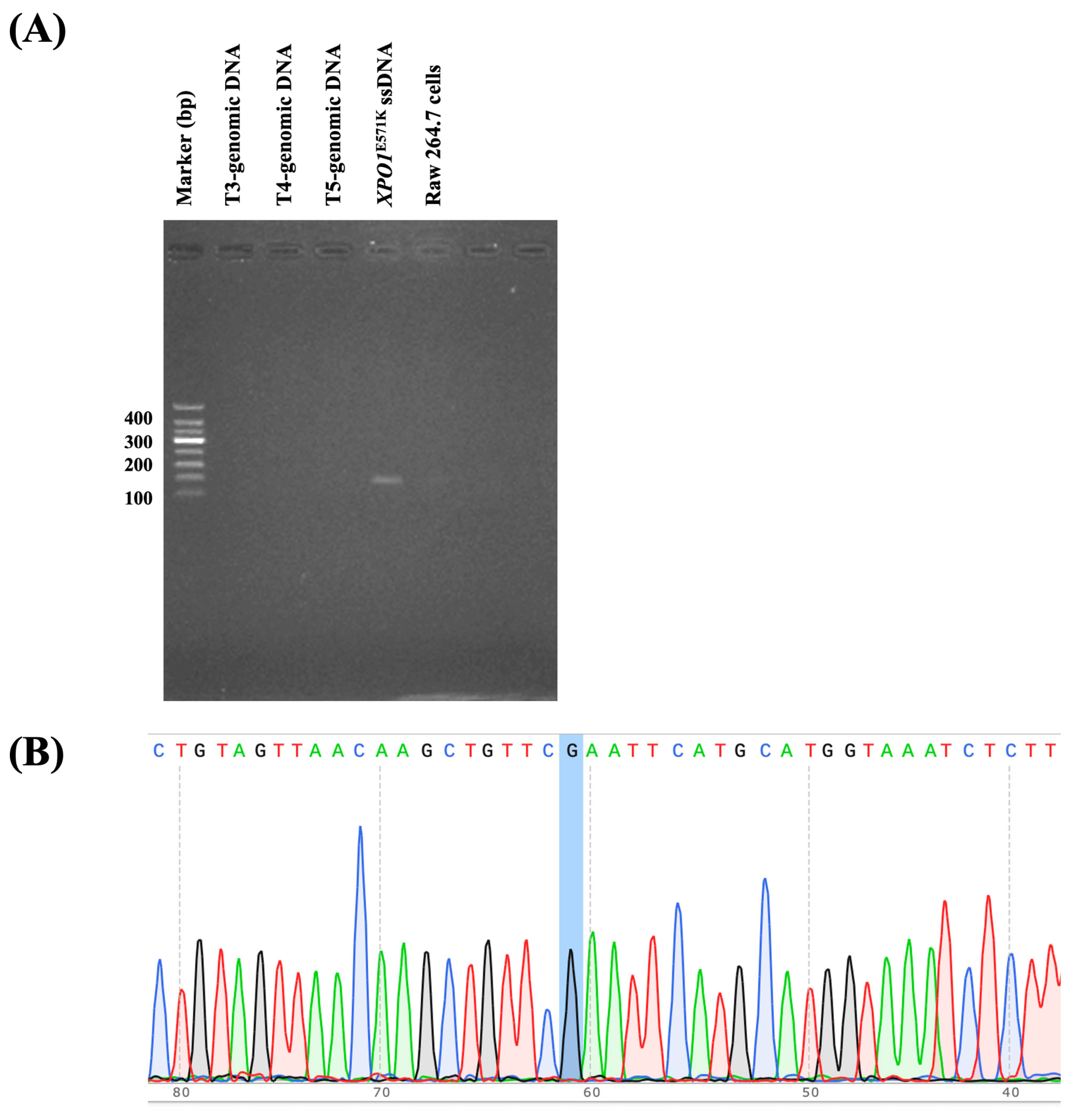
2.5. Detecting XPO1E571K Gene Mutation in Patient Samples and FFPE Tissue Samples Using the FAST-COLD-PCR Assay
3. Discussion
4. Materials and Methods
4.1. Bioinformatics Analysis
4.2. Patients and Sample Collections
4.3. Genomic DNA and cfDNA Preparations
4.4. Detection of XPO1E571K in Patient Samples and FFPE Tissue Samples Using Conventional PCR Assay
4.5. Development of FAST-COLD-PCR Assay
4.6. Assesment of the Limit of Detection of FAST-COLD-PCR Assay
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BCL | B-cell lymphoma |
| BL | Burkitt’s lymphoma |
| BP | Biological process |
| c-Myc | Cellular myelocytomatosis oncogene |
| CC | Cellular component |
| cfDNA | Cell-free circulating tumor DNA |
| cHL | Classical form of Hodgkin lymphoma |
| CRM1 | Chromosome region maintenance 1 |
| CMIR | Cell-mediated immune response |
| DLBCL | Diffuse large B-cell lymphoma |
| DNA | Deoxyribonucleic acid. |
| E571K | A substitution of glutamate 571 to lysine |
| FFPE | Formalin-fixed, paraffin-embedded |
| GO | Gene Ontology |
| HBB | Hemoglobin Subunit Beta |
| HIR | Humoral immune response |
| HL | Hodgkin lymphoma |
| HRM | High-resolution melt |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LOD | Limit of detection |
| MF | Molecular function |
| NHL | Non-Hodgkin lymphoma |
| PCR | Polymerase chain reaction |
| PMBL | Primary mediastinal B-cell lymphoma |
| PPI | Protein–protein interaction |
| Tc | Critical temperature |
| Tm | Average melting temperature |
| UD | Undiluted |
| XPO1 | Exportin1 |
References
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125, S3–S23. [Google Scholar] [CrossRef]
- Intragumtornchai, T.; Bunworasate, U.; Wudhikarn, K.; Lekhakula, A.; Julamanee, J.; Chansung, K.; Sirijerachai, C.; Norasetthada, L.; Nawarawong, W.; Khuhapinant, A. Non-Hodgkin lymphoma in South East Asia: An analysis of the histopathology, clinical features, and survival from Thailand. Hematol. Oncol. 2018, 36, 28–36. [Google Scholar] [CrossRef]
- Armitage, J.O.; Gascoyne, R.D.; Lunning, M.A.; Cavalli, F. Non-hodgkin lymphoma. Lancet 2017, 390, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Granato, M.; Rizzello, C.; Romeo, M.A.; Yadav, S.; Santarelli, R.; D’Orazi, G.; Faggioni, A.; Cirone, M. Concomitant reduction of c-Myc expression and PI3K/AKT/mTOR signaling by quercetin induces a strong cytotoxic effect against Burkitt’s lymphoma. Int. J. Biochem. Cell Biol. 2016, 79, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Spender, L.C.; Inman, G.J. Developments in Burkitt’s lymphoma: Novel cooperations in oncogenic MYC signaling. Cancer Manag. Res. 2014, 6, 27–38. [Google Scholar] [PubMed]
- Camus, V.; Miloudi, H.; Taly, A.; Sola, B.; Jardin, F. XPO1 in B cell hematological malignancies: From recurrent somatic mutations to targeted therapy. J. Hematol. Oncol. 2017, 10, 47. [Google Scholar] [CrossRef]
- Zhang, W.; Ly, C.; Ishizawa, J.; Mu, H.; Ruvolo, V.; Shacham, S.; Daver, N.; Andreeff, M. Combinatorial targeting of XPO1 and FLT3 exerts synergistic anti-leukemia effects through induction of differentiation and apoptosis in FLT3-mutated acute myeloid leukemias: From concept to clinical trial. Haematologica 2018, 103, 1642. [Google Scholar] [CrossRef]
- Xu, Z.; Pan, B.; Miao, Y.; Li, Y.; Qin, S.; Liang, J.; Kong, Y.; Zhang, X.; Tang, J.; Xia, Y. Prognostic value and therapeutic targeting of XPO1 in chronic lymphocytic leukemia. Clin. Exp. Med. 2023, 23, 2651–2662. [Google Scholar] [CrossRef]
- Jardin, F.; Pujals, A.; Pelletier, L.; Bohers, E.; Camus, V.; Mareschal, S.; Dubois, S.; Sola, B.; Ochmann, M.; Lemonnier, F.; et al. Recurrent mutations of the exportin 1 gene (XPO1) and their impact on selective inhibitor of nuclear export compounds sensitivity in primary mediastinal B-cell lymphoma. Am. J. Hematol. 2016, 91, 923–930. [Google Scholar] [CrossRef]
- Camus, V.; Stamatoullas, A.; Mareschal, S.; Viailly, P.J.; Sarafan-Vasseur, N.; Bohers, E.; Dubois, S.; Picquenot, J.M.; Ruminy, P.; Maingonnat, C.; et al. Detection and prognostic value of recurrent exportin 1 mutations in tumor and cell-free circulating DNA of patients with classical Hodgkin lymphoma. Haematologica 2016, 101, 1094–1101. [Google Scholar] [CrossRef]
- Cosson, A.; Chapiro, E.; Bougacha, N.; Lambert, J.; Herbi, L.; Cung, H.A.; Algrin, C.; Keren, B.; Damm, F.; Gabillaud, C.; et al. Gain in the short arm of chromosome 2 (2p+) induces gene overexpression and drug resistance in chronic lymphocytic leukemia: Analysis of the central role of XPO1. Leukemia 2017, 31, 1625–1629. [Google Scholar] [CrossRef]
- Aucamp, J.; Bronkhorst, A.J.; Badenhorst, C.P.; Pretorius, P.J. The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. 2018, 93, 1649–1683. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.-D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Zhivotovsky, B.; Samali, A.; Orrenius, S. Determination of apoptosis and necrosis. Curr. Protoc. Toxicol. 1999, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Newton, C.; Graham, A.; Heptinstall, L.; Powell, S.; Summers, C.; Kalsheker, N.; Smith, J.; Markham, A. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989, 17, 2503–2516. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Dong, L.; Wang, S.; Fu, B.; Wang, J. Evaluation of droplet digital PCR and next generation sequencing for characterizing DNA reference material for KRAS mutation detection. Sci. Rep. 2018, 8, 9650. [Google Scholar] [CrossRef]
- Coccaro, N.; Tota, G.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. Digital PCR: A reliable tool for analyzing and monitoring hematologic malignancies. Int. J. Mol. Sci. 2020, 21, 3141. [Google Scholar] [CrossRef]
- Chubarov, A.S.; Oscorbin, I.P.; Filipenko, M.L.; Lomzov, A.A.; Pyshnyi, D.V. Allele-specific PCR for KRAS mutation detection using phosphoryl guanidine modified primers. Diagnostics 2020, 10, 872. [Google Scholar] [CrossRef]
- Milbury, C.A.; Li, J.; Makrigiorgos, G.M. COLD-PCR–enhanced high-resolution melting enables rapid and selective identification of low-level unknown mutations. Clin. Chem. 2009, 55, 2130–2143. [Google Scholar] [CrossRef]
- Mancini, I.; Santucci, C.; Sestini, R.; Simi, L.; Pratesi, N.; Cianchi, F.; Valanzano, R.; Pinzani, P.; Orlando, C. The use of COLD-PCR and high-resolution melting analysis improves the limit of detection of KRAS and BRAF mutations in colorectal cancer. J. Mol. Diagn. 2010, 12, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Rizaldos, E.; Liu, P.; Milbury, C.A.; Guha, M.; Brisci, A.; Cremonesi, L.; Ferrari, M.; Mamon, H.; Makrigiorgos, G.M. Temperature-tolerant COLD-PCR reduces temperature stringency and enables robust mutation enrichment. Clin. Chem. 2012, 58, 1130–1138. [Google Scholar] [CrossRef] [PubMed]
- Nelson, H.D.; O’meara, E.S.; Kerlikowske, K.; Balch, S.; Miglioretti, D. Factors associated with rates of false-positive and false-negative results from digital mammography screening: An analysis of registry data. Ann. Intern. Med. 2016, 164, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Team, N.L.S.T.R. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar]
- Aravanis, A.M.; Lee, M.; Klausner, R.D. Next-generation sequencing of circulating tumor DNA for early cancer detection. Cell 2017, 168, 571–574. [Google Scholar] [CrossRef]
- Azizian, N.G.; Li, Y. XPO1-dependent nuclear export as a target for cancer therapy. J. Hematol. Oncol. 2020, 13, 61. [Google Scholar] [CrossRef]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef]
- Glaviano, A.; Foo, A.S.; Lam, H.Y.; Yap, K.C.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef]
- Drakos, E.; Singh, R.R.; Rassidakis, G.; Schlette, E.; Li, J.; Claret, F.-X.; Ford, R.; Vega, F.; Medeiros, L.J. Activation of the p53 pathway by the MDM2 inhibitor nutlin-3a overcomes BCL2 overexpression in a preclinical model of diffuse large B-cell lymphoma associated with t (14; 18)(q32; q21). Leukemia 2011, 25, 856–867. [Google Scholar] [CrossRef]
- Taylor, J.; Sendino, M.; Gorelick, A.N.; Pastore, A.; Chang, M.T.; Penson, A.V.; Gavrila, E.I.; Stewart, C.; Melnik, E.M.; Herrejon Chavez, F. Altered nuclear export signal recognition as a driver of oncogenesis. Cancer Discov. 2019, 9, 1452–1467. [Google Scholar] [CrossRef]
- Caillot, M.; Miloudi, H.; Taly, A.; Profitós-Pelejà, N.; Santos, J.C.; Ribeiro, M.L.; Maitre, E.; Saule, S.; Roué, G.; Jardin, F. Exportin 1-mediated nuclear/cytoplasmic trafficking controls drug sensitivity of classical Hodgkin’s lymphoma. Mol. Oncol. 2023, 17, 2546–2564. [Google Scholar] [CrossRef] [PubMed]
- Durand, M.; Cabaud Gibouin, V.; Duplomb, L.; Salmi, L.; Caillot, M.; Sola, B.; Camus, V.; Jardin, F.; Garrido, C.; Jego, G. A first-in-class inhibitor of HSP110 to potentiate XPO1-targeted therapy in primary mediastinal B-cell lymphoma and classical Hodgkin lymphoma. J. Exp. Clin. Cancer Res. 2024, 43, 148. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
| Cycle Step | Temp | Time | Cycles |
|---|---|---|---|
| Initial denaturation | 94.0 °C | 2 min | 1 |
| Denaturation | 94.0 °C | 30 s | 10 |
| Annealing | 60.0 °C | 30 s | |
| Extension | 68.0 °C | 30 s | |
| Denaturation (Tc) | 73.3 °C | 30 s | 30 |
| Annealing | 60.0 °C | 30 s | |
| Extension | 68.0 °C | 30 s | |
| Hold | 4.0 °C | ∞ |
| Sample (Patient No.) | Sex | Age | Lymphoma Type | Patient Status | XPO1E571K Mutation |
|---|---|---|---|---|---|
| 1 | Female | 67 | DLBCL | New | + |
| 2 | Female | 54 | DLBCL | Refractory | + |
| 3 | Female | 64 | DLBCL | Refractory | + |
| 4 | Male | 70 | BCL | Refractory | + |
| 5 | Female | 71 | NHL | New | + |
| 6 | Female | 71 | NHL | Refractory | + |
| 7 | Male | 61 | DLBCL | Refractory | + |
| 8 | Female | 69 | DLBCL | Refractory | + |
| 9 | Female | 20 | NHL | Refractory | − |
| 10 | Female | NA | N/A | N/A | − |
| 11 | Male | 70 | NHL | N/A | − |
| 12 | Female | N/A | N/A | N/A | − |
| 13 | Male | N/A | N/A | N/A | − |
| 14 | Male | N/A | N/A | N/A | − |
| 15 | Female | 20 | NHL | N/A | − |
| 16 | Male | 73 | N/A | N/A | − |
| 17 | Male | 29 | NHL | N/A | − |
| 18 | Male | 58 | N/A | N/A | − |
| 19 | Male | 28 | HL | Refractory | − |
| 20 | Female | N/A | BCL | N/A | − |
| 21 | Female | 51 | BCL | N/A | − |
| 22 | Male | 60 | HL | N/A | − |
| 23 | Female | 53 | NHL | N/A | − |
| 24 | Female | 32 | NHL | N/A | − |
| 25 | Male | 40 | N/A | N/A | − |
| 26 | Female | 21 | HL | N/A | − |
| 27 | Male | N/A | NHL | N/A | − |
| 28 | Female | 64 | DLBCL | New | − |
| 29 | Female | N/A | N/A | N/A | − |
| 30 | Female | 55 | N/A | N/A | − |
| Sample (Patient No.) | Sex | Age | Lymphoma Type | Patient Status | XPO1E571K Mutation |
|---|---|---|---|---|---|
| 1 | Male | 44 | DLBCL | Refractory | − |
| 2 | Female | 54 | DLBCL | Refractory | − |
| 3 | Male | 53 | DLBCL | New | − |
| 4 | Male | 56 | DLBCL | New | − |
| 5 | Male | 21 | DLBCL | New | − |
| 6 | Male | 59 | DLBCL | New | − |
| 7 | Female | 44 | DLBCL | New | − |
| 8 | Female | 55 | DLBCL | New | − |
| 9 | Female | 64 | DLBCL | New | − |
| 10 | Male | 73 | DLBCL | New | − |
| 11 | Male | 75 | DLBCL | Refractory | − |
| 12 | Female | 66 | DLBCL | Refractory | − |
| 13 | Male | 74 | DLBCL | New | − |
| 14 | Female | 68 | DLBCL | New | − |
| 15 | Male | 68 | DLBCL | New | − |
| 16 | Female | 64 | NHL | Relapsed | − |
| 17 | Male | 66 | DLBCL | Relapsed | − |
| 18 | Female | 68 | DLBCL | Relapsed | − |
| 19 | Female | 80 | DLBCL | New | − |
| 20 | Male | 75 | DLBCL | New | − |
| 21 | Female | 71 | DLBCL | New | − |
| 22 | Female | 68 | DLBCL | New | − |
| 23 | Male | 66 | DLBCL | New | − |
| 24 | Female | 64 | DLBCL | New | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duangmano, S.; Viriyaadhammaa, N.; Khamphikham, P.; Intasai, N.; Tantiworawit, A.; Daroontum, T.; Chiampanichayakul, S.; Anuchapreeda, S. Detection of XPO1E571K Gene Mutation from Cell-Free DNA in Blood Circulation of Lymphoma Patients by FAST-COLD PCR. Int. J. Mol. Sci. 2025, 26, 7324. https://doi.org/10.3390/ijms26157324
Duangmano S, Viriyaadhammaa N, Khamphikham P, Intasai N, Tantiworawit A, Daroontum T, Chiampanichayakul S, Anuchapreeda S. Detection of XPO1E571K Gene Mutation from Cell-Free DNA in Blood Circulation of Lymphoma Patients by FAST-COLD PCR. International Journal of Molecular Sciences. 2025; 26(15):7324. https://doi.org/10.3390/ijms26157324
Chicago/Turabian StyleDuangmano, Suwit, Natsima Viriyaadhammaa, Pinyaphat Khamphikham, Nutjeera Intasai, Adisak Tantiworawit, Teerada Daroontum, Sawitree Chiampanichayakul, and Songyot Anuchapreeda. 2025. "Detection of XPO1E571K Gene Mutation from Cell-Free DNA in Blood Circulation of Lymphoma Patients by FAST-COLD PCR" International Journal of Molecular Sciences 26, no. 15: 7324. https://doi.org/10.3390/ijms26157324
APA StyleDuangmano, S., Viriyaadhammaa, N., Khamphikham, P., Intasai, N., Tantiworawit, A., Daroontum, T., Chiampanichayakul, S., & Anuchapreeda, S. (2025). Detection of XPO1E571K Gene Mutation from Cell-Free DNA in Blood Circulation of Lymphoma Patients by FAST-COLD PCR. International Journal of Molecular Sciences, 26(15), 7324. https://doi.org/10.3390/ijms26157324








