Spectral Region Optimization and Machine Learning-Based Nonlinear Spectral Analysis for Raman Detection of Cardiac Fibrosis Following Myocardial Infarction
Abstract
1. Introduction
2. Results
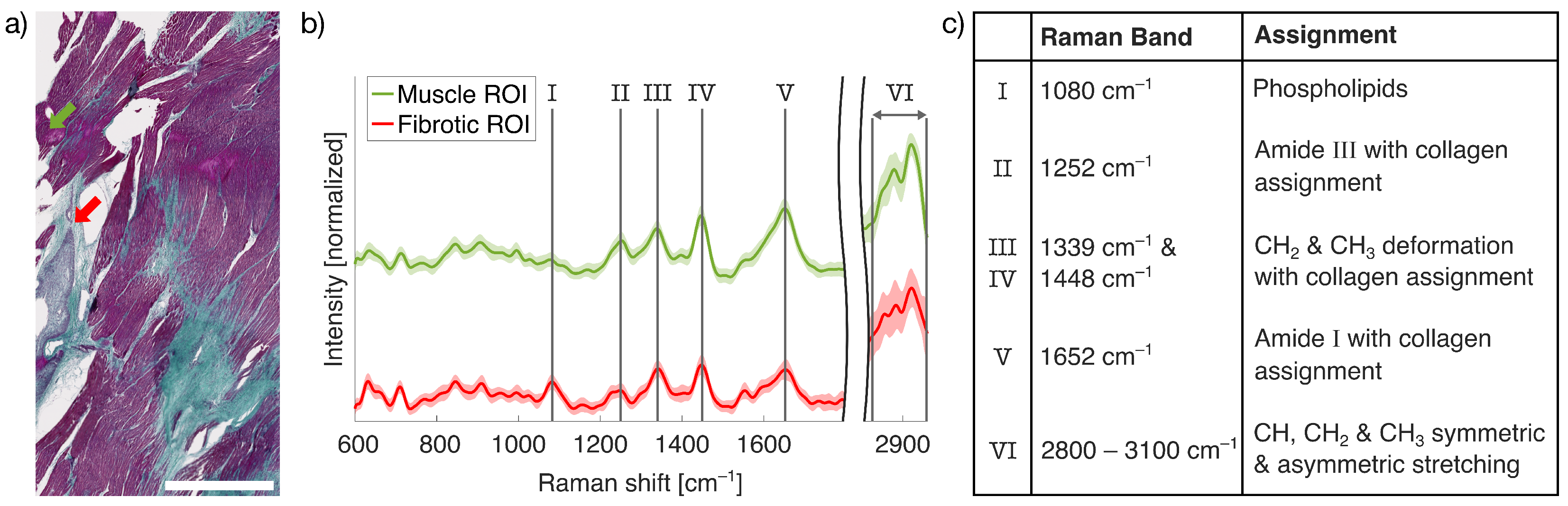
2.1. LSRM for Fibrotic Tissue Examination
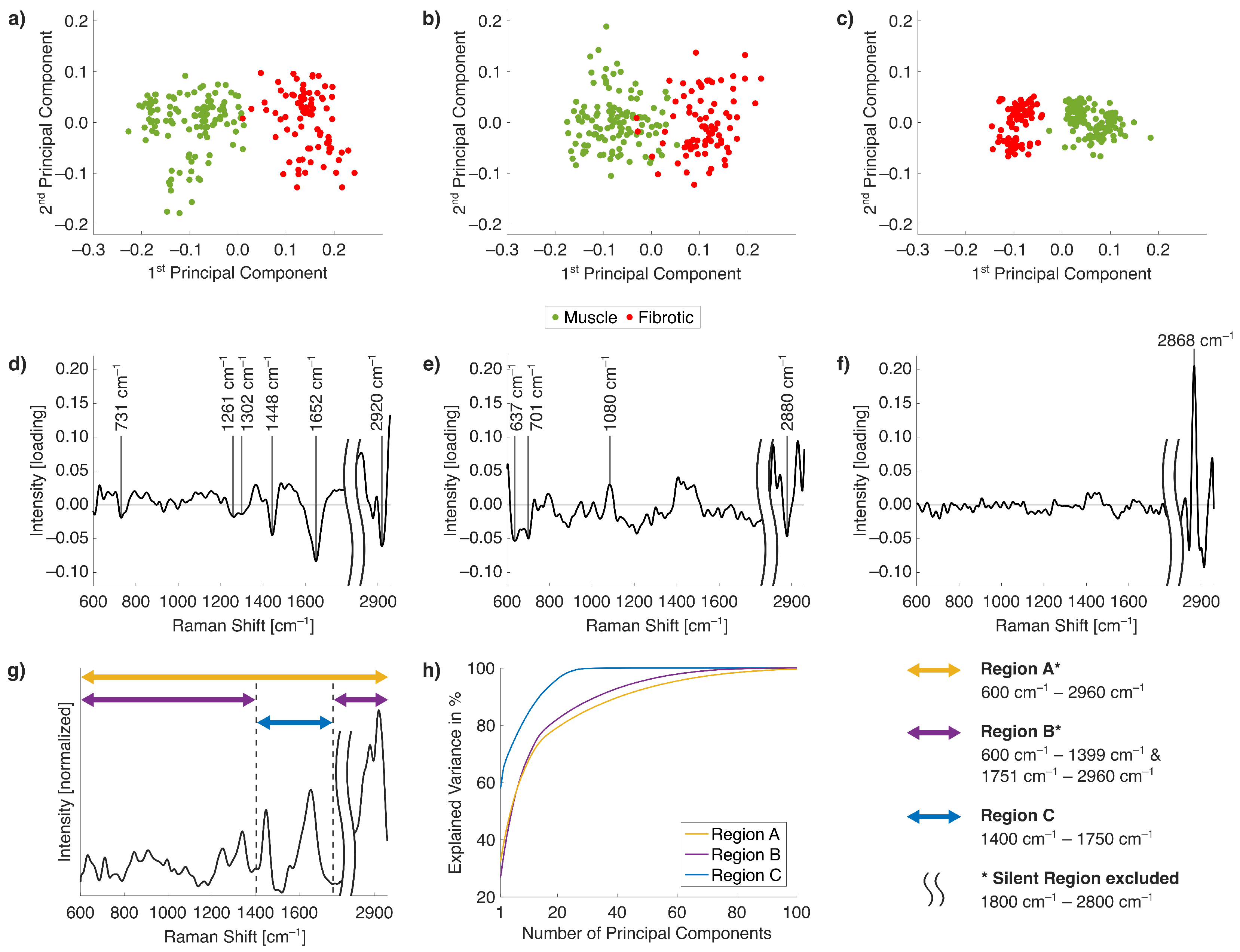
2.2. Raman Band Selection for Improved PCA
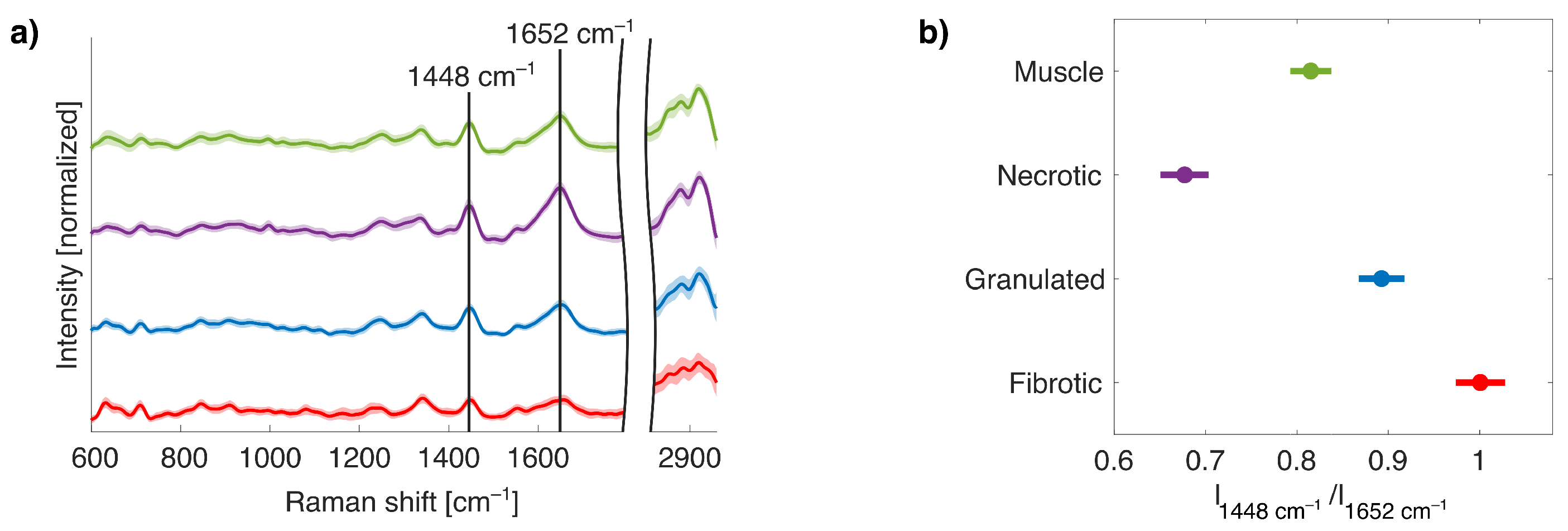
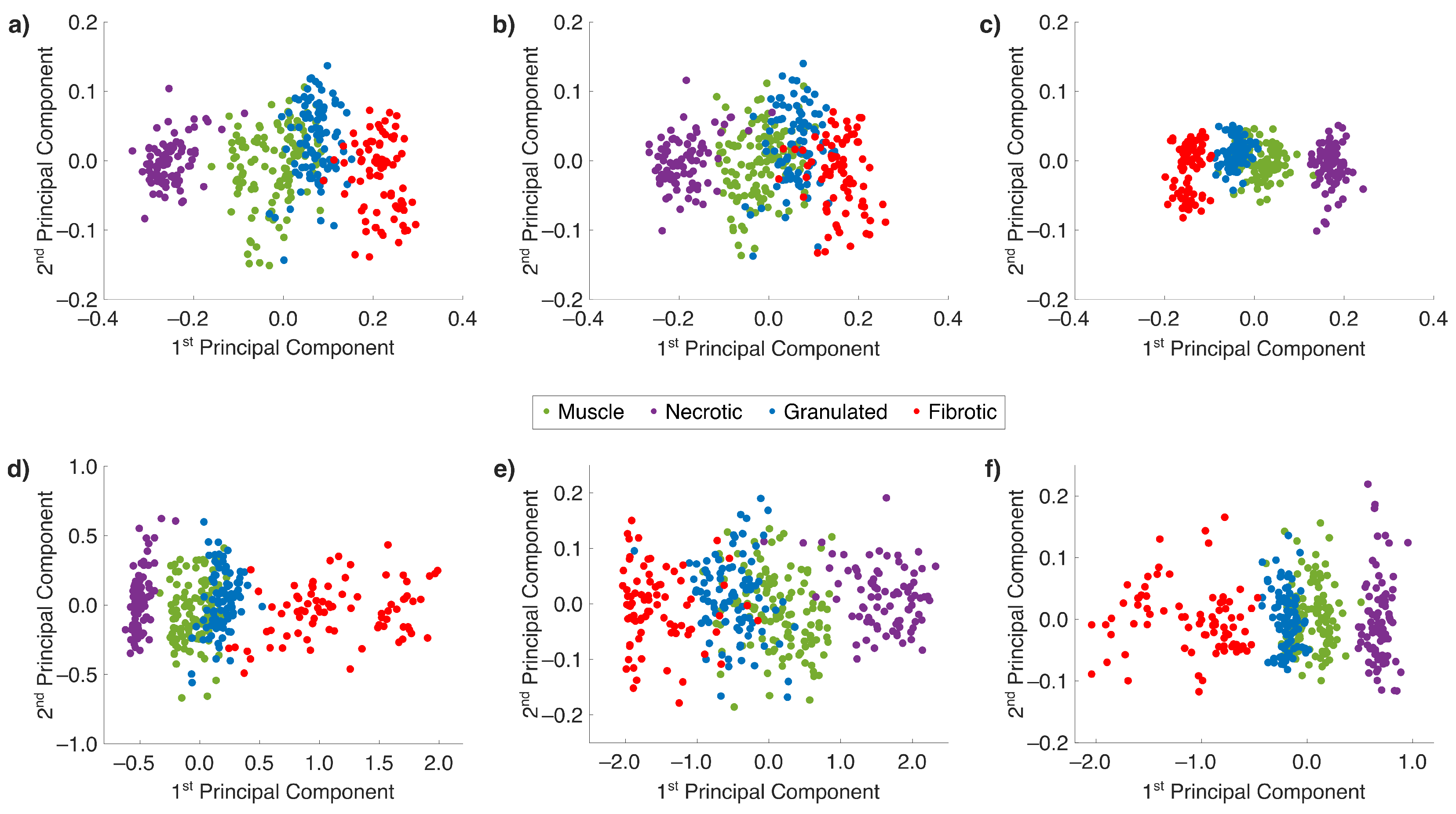
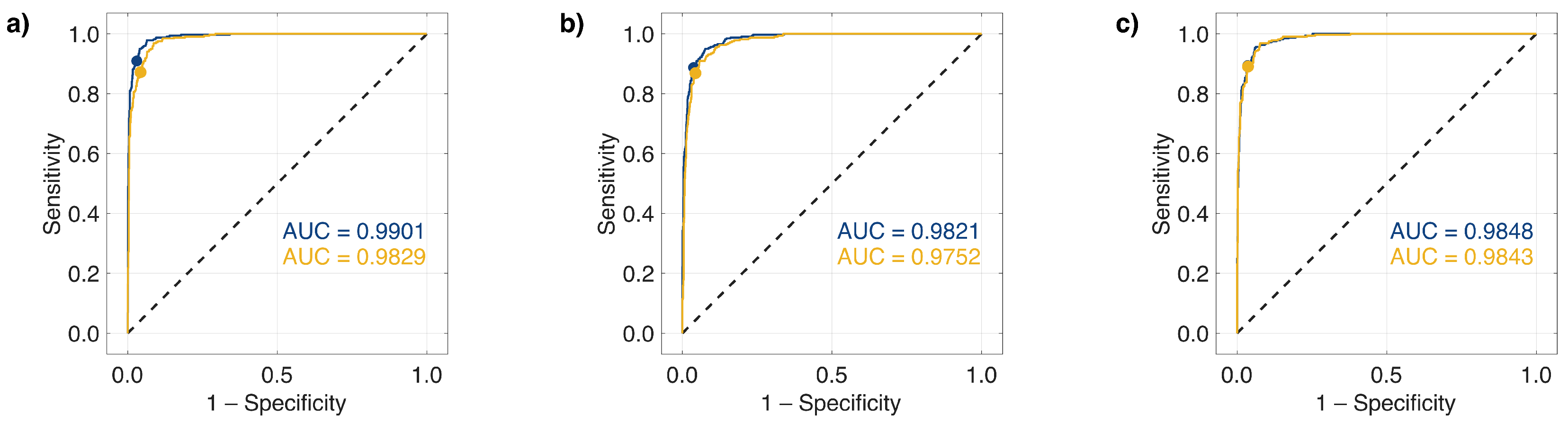
2.3. Nonlinear Dimensionality Reduction and ML with Collagen-Based Raman Band Selection
3. Discussion
4. Materials and Methods
4.1. Sample Preparation and Masson’s Trichrome Staining
4.2. Line Scan Microspectroscopy Setup
4.3. Spectral Processing and Classification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of Variance |
| AUC | Area under the ROC Curve |
| ECM | Extracellular Matrix |
| FOV | Field of View |
| LSRM | Line Scan Raman Microspectroscopy |
| MI | Myocardial Infarction |
| ML | Machine Learning |
| OCT | Optical Coherence Tomography |
| PBS | Phosphate-buffered Saline |
| PC | Principal Component |
| PCA | Principal Component Analysis |
| ROC | Receiver Operating Characteristic |
| ROI | Region of Interest |
| RS | Raman Spectroscopy |
| SCD | Sudden Cardiac Death |
| SRS | Stimulated Raman Scattering |
| SVM | Support Vector Machine |
| TS | Trichrome Staining |
| VF | Ventricular Fibrillation |
| VT | Ventricular Tachycardia |
References
- Wong, C.X.; Brown, A.; Lau, D.H.; Chugh, S.S.; Albert, C.M.; Kalman, J.M.; Sanders, P. Epidemiology of Sudden Cardiac Death: Global and Regional Perspectives. Hear. Lung Circ. 2019, 28, 6–14. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. The extracellular matrix in myocardial injury, repair, and remodeling. J. Clin. Investig. 2017, 127, 1600–1612. [Google Scholar] [CrossRef]
- Michaud, K.; Basso, C.; d’Amati, G.; Giordano, C.; Kholová, I.; Preston, S.D.; Rizzo, S.; Sabatasso, S.; Sheppard, M.N.; Vink, A.; et al. Diagnosis of myocardial infarction at autopsy: AECVP reappraisal in the light of the current clinical classification. Virchows Arch. 2020, 476, 179–194. [Google Scholar] [CrossRef]
- Richardson, W.J.; Clarke, S.A.; Quinn, T.A.; Holmes, J.W. Physiological implications of myocardial scar structure. Compr. Physiol. 2015, 5, 1877. [Google Scholar] [CrossRef]
- Sattler, S.M.; Skibsbye, L.; Linz, D.; Lubberding, A.F.; Tfelt-Hansen, J.; Jespersen, T. Ventricular Arrhythmias in First Acute Myocardial Infarction: Epidemiology, Mechanisms, and Interventions in Large Animal Models. Front. Cardiovasc. Med. 2019, 6, 158. [Google Scholar] [CrossRef]
- Huikuri, H.V.; Castellanos, A.; Myerburg, R.J. Sudden Death Due to Cardiac Arrhythmias. N. Engl. J. Med. 2001, 345, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Tfelt-Hansen, J.; Garcia, R.; Albert, C.; Merino, J.; Krahn, A.; Marijon, E.; Basso, C.; Wilde, A.A.M.; Haugaa, K.H. Risk stratification of sudden cardiac death: A review. EP Eur. 2023, 25, euad203. [Google Scholar] [CrossRef]
- Narayan, S.M.; John, R.M. Advanced Electroanatomic Mapping: Current and Emerging Approaches. Curr. Treat. Options Cardiovasc. Med. 2024, 26, 69–91. [Google Scholar] [CrossRef]
- Karamitsos, T.D.; Arvanitaki, A.; Karvounis, H.; Neubauer, S.; Ferreira, V.M. Myocardial Tissue Characterization and Fibrosis by Imaging. JACC Cardiovasc. Imaging 2020, 13, 1221–1234. [Google Scholar] [CrossRef]
- Anter, E.; Tschabrunn, C.M.; Buxton, A.E.; Josephson, M.E. High-Resolution Mapping of Postinfarction Reentrant Ventricular Tachycardia. Circulation 2016, 134, 314–327. [Google Scholar] [CrossRef]
- Leitgeb, R.A.; Baumann, B. Multimodal optical medical imaging concepts based on optical coherence tomography. Front. Phys. 2018, 6, 114. [Google Scholar] [CrossRef]
- Barkagan, M.; Leshem, E.; Shapira-Daniels, A.; Sroubek, J.; Buxton, A.E.; Saffitz, J.E.; Anter, E. Histopathological Characterization of Radiofrequency Ablation in Ventricular Scar Tissue. JACC Clin. Electrophysiol. 2019, 5, 920–931. [Google Scholar] [CrossRef]
- Cronin, E.M.; Bogun, F.M.; Maury, P.; Peichl, P.; Chen, M.; Namboodiri, N.; Aguinaga, L.; Leite, L.R.; Al-Khatib, S.M.; Anter, E.; et al. 2019 HRS/EHRA/APHRS/LAHRS expert consensus statement on catheter ablation of ventricular arrhythmias. EP Eur. 2019, 21, 1143–1144. [Google Scholar]
- Choo-Smith, L.P.; Edwards, H.G.M.; Endtz, H.P.; Kros, J.M.; Heule, F.; Barr, H.; Robinson, J.S., Jr.; Bruining, H.A.; Puppels, G.J. Medical applications of Raman spectroscopy: From proof of principle to clinical implementation. Biopolymers 2002, 67, 1–9. [Google Scholar] [CrossRef]
- Baker, M.J.; Byrne, H.J.; Chalmers, J.; Gardner, P.; Goodacre, R.; Henderson, A.; Kazarian, S.G.; Martin, F.L.; Moger, J.; Stone, N.; et al. Clinical applications of infrared and Raman spectroscopy: State of play and future challenges. Analyst 2018, 143, 1735–1757. [Google Scholar] [CrossRef]
- Chaichi, A.; Prasad, A.; Gartia, M.R. Raman Spectroscopy and Microscopy Applications in Cardiovascular Diseases: From Molecules to Organs. Biosensors 2018, 8, 107. [Google Scholar] [CrossRef]
- Brauchle, E.; Knopf, A.; Bauer, H.; Shen, N.; Linder, S.; Monaghan, M.G.; Ellwanger, K.; Layland, S.L.; Brucker, S.Y.; Nsair, A.; et al. Non-invasive Chamber-Specific Identification of Cardiomyocytes in Differentiating Pluripotent Stem Cells. Stem Cell Rep. 2016, 6, 188–199. [Google Scholar] [CrossRef]
- Rog-Zielinska, E.A.; Norris, R.A.; Kohl, P.; Markwald, R. The Living Scar—Cardiac Fibroblasts and the Injured Heart. Trends Mol. Med. 2016, 22, 99–114. [Google Scholar] [CrossRef]
- Ogawa, M.; Harada, Y.; Yamaoka, Y.; Fujita, K.; Yaku, H.; Takamatsu, T. Label-free biochemical imaging of heart tissue with high-speed spontaneous Raman microscopy. Biochem. Biophys. Res. Commun. 2009, 382, 370–374. [Google Scholar] [CrossRef]
- Nishiki-Muranishi, N.; Harada, Y.; Minamikawa, T.; Yamaoka, Y.; Dai, P.; Yaku, H.; Takamatsu, T. Label-free evaluation of myocardial infarction and its repair by spontaneous Raman spectroscopy. Anal. Chem. 2014, 86, 6903–6910. [Google Scholar] [CrossRef]
- Yamamoto, T.; Minamikawa, T.; Harada, Y.; Yamaoka, Y.; Tanaka, H.; Yaku, H.; Takamatsu, T. Label-free evaluation of myocardial infarct in surgically excised ventricular myocardium by Raman spectroscopy. Sci. Rep. 2018, 8, 14671. [Google Scholar] [CrossRef]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Tech. Instrum. 2015, 2, 1–38. [Google Scholar] [CrossRef]
- Dong, D.; McAvoy, T. Nonlinear principal component analysis—Based on principal curves and neural networks. Comput. Chem. Eng. 1996, 20, 65–78. [Google Scholar] [CrossRef]
- Cleutjens, J.; Verluyten, M.; Smiths, J.; Daemen, M. Collagen remodeling after myocardial infarction in the rat heart. Am. J. Pathol. 1995, 147, 325–338. [Google Scholar]
- Dixon, I.M.; Ju, H.; Jassal, D.S.; Peterson, D.J. Effect of ramipril and losartan on collagen expression in right and left heart after myocardial infarction. Mol. Cell. Biochem. 1996, 165, 31–45. [Google Scholar] [CrossRef]
- Shi, G.; Shen, X.; Ren, H.; Rao, Y.; Weng, S.; Tang, X. Kernel principal component analysis and differential non-linear feature extraction of pesticide residues on fruit surface based on surface-enhanced Raman spectroscopy. Front. Plant Sci. 2022, 13, 956778. [Google Scholar] [CrossRef]
- Sun, H.; Lv, G.; Mo, J.; Lv, X.; Du, G.; Liu, Y. Application of KPCA combined with SVM in Raman spectral discrimination. Optik 2019, 184, 214–219. [Google Scholar] [CrossRef]
- Yang, D.; Weigen, C.; Haiyang, S.; Fu, W.; Yongkuo, Z. Raman spectrum feature extraction and diagnosis of oil–paper insulation ageing based on kernel principal component analysis. High Volt. 2021, 6, 51–60. [Google Scholar] [CrossRef]
- Scholz, M. Validation of nonlinear PCA. Neural Process. Lett. 2012, 36, 21–30. [Google Scholar] [CrossRef]
- Sharma, V.; Green, A.; McLean, A.; Adegoke, J.; Gordon, C.; Starkey, G.; D’Costa, R.; James, F.; Afara, I.; Lal, S.; et al. Towards a point-of-care multimodal spectroscopy instrument for the evaluation of human cardiac tissue. Heart Vessel. 2023, 38, 1476–1485. [Google Scholar] [CrossRef]
- Bennett, K.P.; Campbell, C. Support vector machines: Hype or hallelujah? ACM SIGKDD Explor. Newsl. 2000, 2, 1–13. [Google Scholar] [CrossRef]
- Ouyang, J.; Guzman, M.; Desoto-Lapaix, F.; Pincus, M.R.; Wieczorek, R. Utility of desmin and a Masson’s trichrome method to detect early acute myocardial infarction in autopsy tissues. Int. J. Clin. Exp. Pathol. 2010, 3, 98. [Google Scholar]
- Movasaghi, Z.; Rehman, S.; Rehman, I. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Rachmawati, H.; Haryadi, B.; Anggadiredja, K.; Suendo, V. Intraoral Film Containing Insulin-Phospholipid Microemulsion: Formulation and In Vivo Hypoglycemic Activity Study. AAPS PharmSci 2014, 16, 692–703. [Google Scholar] [CrossRef]
- Pistritu, D.V.; Vasiliniuc, A.C.; Vasiliu, A.; Visinescu, E.F.; Visoiu, I.E.; Vizdei, S.; Martínez Anghel, P.; Tanca, A.; Bucur, O.; Liehn, E.A. Phospholipids, the Masters in the Shadows during Healing after Acute Myocardial Infarction. Int. J. Mol. Sci. 2023, 24, 8360. [Google Scholar] [CrossRef]
- Minamikawa, T.; Harada, Y.; Koizumi, N.; Okihara, K.; Kamoi, K.; Yanagisawa, A.; Takamatsu, T. Label-free detection of peripheral nerve tissues against adjacent tissues by spontaneous Raman microspectroscopy. Histochem. Cell Biol. 2013, 139, 181–193. [Google Scholar] [CrossRef]
- Bro, R.; Acar, E.; Kolda, T.G. Resolving the Sign Ambiguity in the Singular Value Decomposition. J. Chemom. 2008, 22, 135–140. [Google Scholar] [CrossRef]
- Malthouse, E. Limitations of nonlinear PCA as performed with generic neural networks. IEEE Trans. Neural Netw. 1998, 9, 165–173. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Shlens, J. A tutorial on principal component analysis. arXiv 2014, arXiv:1404.1100. [Google Scholar]
- Kjeldahl, K.; Bro, R. Some common misunderstandings in chemometrics. J. Chemom. 2010, 24, 558–564. [Google Scholar] [CrossRef]
- Ruiz, A.; López-de Teruel, P.E. Nonlinear kernel-based statistical pattern analysis. IEEE Trans. Neural Netw. 2001, 12, 16–32. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. Cardiac fibrosis: Cell biological mechanisms, molecular pathways and therapeutic opportunities. Mol. Asp. Med. 2019, 65, 70–99. [Google Scholar] [CrossRef]
- Pioppi, L.; Parvan, R.; Samrend, A.; Silva, G.; Paolantoni, M.; Sassi, P.; Cataliotti, A. Vibrational spectroscopy identifies myocardial chemical modifications in heart failure with preserved ejection fraction. J. Transl. Med. 2023, 21, 617. [Google Scholar] [CrossRef]
- de Campos Vidal, B.; Mello, M.L.S. Collagen type I amide I band infrared spectroscopy. Micron 2011, 42, 283–289. [Google Scholar] [CrossRef]
- Cleutjens, J.P.; Kandala, J.C.; Guarda, E.; Guntaka, R.V.; Weber, K.T. Regulation of collagen degradation in the rat myocardium after infarction. J. Mol. Cell. Cardiol. 1995, 27, 1281–1292. [Google Scholar] [CrossRef]
- Becker, L.; Lu, C.E.; Montes-Mojarro, I.A.; Layland, S.L.; Khalil, S.; Nsair, A.; Duffy, G.P.; Fend, F.; Marzi, J.; Schenke-Layland, K. Raman microspectroscopy identifies fibrotic tissues in collagen-related disorders via deconvoluted collagen type I spectra. Acta Biomater. 2023, 162, 278–291. [Google Scholar] [CrossRef]
- Hinderer, S.; Schenke-Layland, K. Cardiac fibrosis – A short review of causes and therapeutic strategies. Adv. Drug Deliv. Rev. 2019, 146, 77–82. [Google Scholar] [CrossRef]
- Cárcamo-Vega, J.; Aliaga, A.; Clavijo, E.; Manuel, B.; Vallette, M. Raman and surface-enhanced Raman scattering in the study of human rotator cuff tissues after shock wave treatment. J. Raman Spectrosc. 2012, 43, 248–254. [Google Scholar] [CrossRef]
- Schulze, H.G.; Rangan, S.; Vardaki, M.Z.; Blades, M.W.; Turner, R.F.B.; Piret, J.M. Critical Evaluation of Spectral Resolution Enhancement Methods for Raman Hyperspectra. Appl. Spectrosc. 2022, 76, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Santangeli, P.; Frankel, D.S.; Marchlinski, F.E. End points for ablation of scar-related ventricular tachycardia. Circ. Arrhythm. Electrophysiol. 2014, 7, 949–960. [Google Scholar] [CrossRef]
- Laflamme, M.A.; Murry, C.E. Regenerating the heart. Nat. Biotechnol. 2005, 23, 845–856. [Google Scholar] [CrossRef]
- Guadagnoli, E.; Velicer, W.F. Relation of sample size to the stability of component patterns. Psychol. Bull. 1988, 103, 265. [Google Scholar] [CrossRef]
- Garde, A.; Voss, A.; Caminal, P.; Benito, S.; Giraldo, B.F. SVM-based feature selection to optimize sensitivity–specificity balance applied to weaning. Comput. Biol. Med. 2013, 43, 533–540. [Google Scholar] [CrossRef]
- Kuo, P.H.; Chang, C.W.; Chang, C.C.; Yau, H.T. Synthetic minority oversampling and iterative fluorescence-suppression integrated algorithm for Raman spectrum pesticide detection system. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2025, 326, 125162. [Google Scholar] [CrossRef]
- Hsu, C.W.; Lin, C.J. A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Netw. 2002, 13, 415–425. [Google Scholar] [CrossRef]
- Han, H.; Jiang, X. Overcome support vector machine diagnosis overfitting. Cancer Inform. 2014, 13, CIN–S13875. [Google Scholar] [CrossRef]
- Ghosh, S.; Dasgupta, A.; Swetapadma, A. A Study on Support Vector Machine based Linear and Non-Linear Pattern Classification. In Proceedings of the 2019 International Conference on Intelligent Sustainable Systems (ICISS), Palladam, India, 21–22 February 2019; pp. 24–28. [Google Scholar] [CrossRef]
- Rangel-Castilla, L.; Russin, J.J.; Britz, G.W.; Spetzler, R.F. Update on transient cardiac standstill in cerebrovascular surgery. Neurosurg. Rev. 2015, 38, 595–602. [Google Scholar] [CrossRef]
- Intarakhao, P.; Thiarawat, P.; Tewaritrueangsri, A.; Pojanasupawun, S. Low-dose adenosine-induced transient asystole during intracranial aneurysm surgery. Surg. Neurol. Int. 2020, 11, 235. [Google Scholar] [CrossRef]
- Lynch, P.G.; Das, A.; Alam, S.; Rich, C.C.; Frontiera, R.R. Mastering femtosecond stimulated Raman spectroscopy: A practical guide. Acs Phys. Chem. Au 2023, 4, 1–18. [Google Scholar] [CrossRef]
- Lombardini, A.; Mytskaniuk, V.; Sivankutty, S.; Andresen, E.R.; Chen, X.; Wenger, J.; Fabert, M.; Joly, N.; Louradour, F.; Kudlinski, A.; et al. High-resolution multimodal flexible coherent Raman endoscope. Light. Sci. Appl. 2018, 7, 10. [Google Scholar] [CrossRef]
- Kim, J.; Kim, S.; Song, J.W.; Kim, H.J.; Lee, M.W.; Han, J.; Kim, J.W.; Yoo, H. Flexible endoscopic micro-optical coherence tomography for three-dimensional imaging of the arterial microstructure. Sci. Rep. 2020, 10, 9248. [Google Scholar] [CrossRef]
- Dib, N.; Diethrich, E.B.; Campbell, A.; Gahremanpour, A.; McGarry, M.; Opie, S.R. A percutaneous swine model of myocardial infarction. J. Pharmacol. Toxicol. Methods 2006, 53, 256–263. [Google Scholar] [CrossRef]
- Pallares-Lupon, N.; Bayer, J.D.; Guillot, B.; Caluori, G.; Ramlugun, G.S.; Kulkarni, K.; Loyer, V.; Bloquet, S.; El Hamrani, D.; Naulin, J.; et al. Tissue preparation techniques for contrast-enhanced micro computed tomography imaging of large mammalian cardiac models with chronic disease. JoVE (J. Vis. Exp.) 2022, 180, e62909. [Google Scholar] [CrossRef]
- Ramlugun, G.S.; Kulkarni, K.; Pallares-Lupon, N.; Boukens, B.J.; Efimov, I.R.; Bernus, O.; Walton, R.D. A comprehensive framework for evaluation of high pacing frequency and arrhythmic optical mapping signals. Front. Physiol. 2023, 14, 734356. [Google Scholar] [CrossRef]
- Giardina, G.; Micko, A.; Bovenkamp, D.; Krause, A.; Placzek, F.; Papp, L.; Krajnc, D.; Spielvogel, C.P.; Winklehner, M.; Höftberger, R.; et al. Morpho-Molecular Metabolic Analysis and Classification of Human Pituitary Gland and Adenoma Biopsies Based on Multimodal Optical Imaging. Cancers 2021, 13, 3234. [Google Scholar] [CrossRef]
- Bovenkamp, D.; Micko, A.; Püls, J.; Placzek, F.; Höftberger, R.; Vila, G.; Leitgeb, R.; Drexler, W.; Andreana, M.; Wolfsberger, S.; et al. Line Scan Raman Microspectroscopy for Label-Free Diagnosis of Human Pituitary Biopsies. Molecules 2019, 24, 3577. [Google Scholar] [CrossRef]
- McCreery Group. Standard Spectra. Available online: https://www.chem.ualberta.ca/~mccreery/ramanmaterials.html (accessed on 22 September 2022).
- Bocklitz, T.; Walter, A.; Hartmann, K.; Rösch, P.; Popp, J. How to pre-process Raman spectra for reliable and stable models? Anal. Chim. Acta 2011, 704, 47–56. [Google Scholar] [CrossRef]
- Green, P.; Silverman, B. Nonparametric Regression and Generalized Linear Models: A Roughness Penalty Approach; Chapman & Hall/CRC Monographs on Statistics & Applied Probability; Taylor & Francis: New York, NY, USA, 1993. [Google Scholar]
- Afseth, N.K.; Kohler, A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemom. Intell. Lab. Syst. 2012, 117, 92–99. [Google Scholar] [CrossRef]
- Lieber, C.A.; Mahadevan-Jansen, A. Automated Method for Subtraction of Fluorescence from Biological Raman Spectra. Appl. Spectrosc. 2003, 57, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Guo, S.; Popp, J.; Bocklitz, T. Chemometric analysis in Raman spectroscopy from experimental design to machine learning–based modeling. Nat. Protoc. 2021, 16, 5426–5459. [Google Scholar] [CrossRef]
- Landgrebe, J.; Wurst, W.; Welzl, G. Permutation-validated principal components analysis of microarray data. Genome Biol. 2002, 3, research0019-1. [Google Scholar] [CrossRef]
| Principal Component | Spectral Region A | Spectral Region B | Spectral Region C |
|---|---|---|---|
| First | |||
| Second | |||
| Third | |||
| Sum |
| Principal | Spectral Region A | Spectral Region B | Spectral Region C | |||
|---|---|---|---|---|---|---|
| Component |
Linear
PCA |
Nonlinear
PCA |
Linear
PCA |
Nonlinear
PCA |
Linear
PCA |
Nonlinear
PCA |
| First | ||||||
| Second | ||||||
| Sum | ||||||
| Spectral Region A | Spectral Region B | Spectral Region C | ||||
|---|---|---|---|---|---|---|
|
Linear
SVM |
Quadratic
SVM |
Linear
SVM |
Quadratic
SVM |
Linear
SVM |
Quadratic
SVM | |
| Number of PCs | 15 | 15 | 3 | |||
| Explained Variance | ||||||
| RMSE | ||||||
| Total Sensitivity | ||||||
| Total Specificity | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krause, A.; Andreana, M.; Walton, R.D.; Marchant, J.; Pallares-Lupon, N.; Kulkarni, K.; Drexler, W.; Unterhuber, A. Spectral Region Optimization and Machine Learning-Based Nonlinear Spectral Analysis for Raman Detection of Cardiac Fibrosis Following Myocardial Infarction. Int. J. Mol. Sci. 2025, 26, 7240. https://doi.org/10.3390/ijms26157240
Krause A, Andreana M, Walton RD, Marchant J, Pallares-Lupon N, Kulkarni K, Drexler W, Unterhuber A. Spectral Region Optimization and Machine Learning-Based Nonlinear Spectral Analysis for Raman Detection of Cardiac Fibrosis Following Myocardial Infarction. International Journal of Molecular Sciences. 2025; 26(15):7240. https://doi.org/10.3390/ijms26157240
Chicago/Turabian StyleKrause, Arno, Marco Andreana, Richard D. Walton, James Marchant, Nestor Pallares-Lupon, Kanchan Kulkarni, Wolfgang Drexler, and Angelika Unterhuber. 2025. "Spectral Region Optimization and Machine Learning-Based Nonlinear Spectral Analysis for Raman Detection of Cardiac Fibrosis Following Myocardial Infarction" International Journal of Molecular Sciences 26, no. 15: 7240. https://doi.org/10.3390/ijms26157240
APA StyleKrause, A., Andreana, M., Walton, R. D., Marchant, J., Pallares-Lupon, N., Kulkarni, K., Drexler, W., & Unterhuber, A. (2025). Spectral Region Optimization and Machine Learning-Based Nonlinear Spectral Analysis for Raman Detection of Cardiac Fibrosis Following Myocardial Infarction. International Journal of Molecular Sciences, 26(15), 7240. https://doi.org/10.3390/ijms26157240








