Abstract
Wolfberry (Lycium barbarum L.) is a valued traditional medicinal plant and dietary supplement in China. The basic leucine zipper (bZIP) transcription factor (TF) family is a multifunctional group of regulatory proteins critical to plant biology, orchestrating processes such as growth and development, secondary metabolite biosynthesis, and stress responses to abiotic conditions. Despite its significance, limited information about this gene family in wolfberry is available. In this study, a total of 66 LbabZIP genes were identified, exhibiting a non-uniform distribution across all 12 chromosomes. Phylogenetic analysis divided these genes into 13 subgroups based on comparison with Arabidopsis bZIP proteins. Analysis of gene structures and conserved motifs revealed high similarities within individual subgroups. Gene duplication analysis indicated that dispersed duplication (DSD) and whole-genome duplication (WGD) events were the primary drivers of LbabZIP gene family expansion, with all duplicated genes subject to purifying selection. Cis-regulatory element (CRE) analysis of LbabZIP promoter regions identified numerous elements associated with plant growth and development, hormone signaling, and abiotic stress responses. Gene Ontology (GO) annotation further indicated that the LbabZIP genes are involved in transcriptional regulation, metabolism, and other biological processes. Transcriptome data and quantitative real-time PCR (qRT-PCR) analysis demonstrated tissue-specific expression patterns for several LbabZIP genes. Notably, LbaZIP21/40/49/65 showed significant involvement in wolfberry fruit development. Subcellular localization assays confirmed that these four proteins are nucleus-localized. This comprehensive analysis provides a theoretical foundation for future studies investigating the biological functions of LbabZIP genes, especially their role in wolfberry fruit development.
1. Introduction
Wolfberry (Lycium barbarum L.), a perennial shrub of the Solanaceae family, is widely cultivated in Northwest China’s arid and semi-arid regions. As an important medicinal and edible plant, the fruits of L. barbarum are rich in polysaccharides, carotenoids, amino acids, and flavonoids, and it has been reported to have immune-enhancing, anti-aging, cancer prevention, and anti-oxidative properties [1,2]. The development and maturation of wolfberry fruit represent crucial phases in its growth and overall development. The process of fruit development is highly regulated, with a set of genes playing key roles in the development of wolfberry fruit [3]. Moreover, with the report of the wolfberry genome sequence [4], several transcription factor (TF) families, such as R2R3-MYB, BBX, GATA, and PYL, have been noted [5,6,7,8]. However, this is just an iceberg of the complex transcriptional network in wolfberry.
The basic leucine zipper (bZIP) TF family is one of the largest and most diverse families of TF in plants and other eukaryotes. Structurally, members of the bZIP TFs are characterized by a highly conserved basic region at the N-terminus and a leucine zipper region at the C-terminus. The basic region is highly conserved, comprising ~16 amino acids with an invariant N-X7-R/K-X9 motif that recognizes ACGT-core DNA sequences, including A-box (TACGTA), C-box (GACGTC), and G-box (CACGTG) [9,10]. To date, the bZIP TF family has been identified and analyzed in several plants, including 78 in Arabidopsis [9], 86 in rice (Oryza sativa) [11], 69 in tomato (Solanum lycopersicum) [12], 92 in pear (Pyrus breschneideri) [13], 49 in potato (Solanum tuberosum) [14], 61 in wax gourd (Benincasa hispida) [15], and 45 in jujube (Ziziphus jujuba) [16]. However, basic information of the bZIP TFs in wolfberry is still uncovered.
Previous research reported that the roles of bZIP families are varied in regulating plant development and secondary metabolism [17,18]. For example, heterologous expression of FvbZIP11 in tomato significantly increased the fruit total soluble solid and soluble sugar content [19]. Overexpression of PgbZIP16 and PgbZIP34 from pomegranate plants in Arabidopsis promotes the accumulation of anthocyanin in Arabidopsis leaves [20]. In plum, PsbZIP1 and PsbZIP10 promoted anthocyanin synthesis by binding directly to the G-box of the PsUFGT promoter [21]. And citrus CsbZIP44 directly binds to the promoters of four carotenoid metabolism-related genes (CsDXR, CsGGPPs, CsBCH1, and CsNCED2) and activates their expression [22]. In addition, SlHY5 and SlPIF3 were involved in the biosynthesis of steroidal glycoalkaloids mediated by light signals by controlling the expression of SlGAME1, SlGAME4, and SlGAME17 in tomato leaves [23]. Therefore, the bZIP family of proteins is involved in and plays an important role in secondary metabolite synthesis during fruit development, such as for flavonoids, carotenoids, and alkaloids.
In this study, the potential role of LbabZIPs in regulating plant growth and development, particularly in wolfberry fruit development, was investigated. A total of 66 LbabZIP genes were identified and classified in the wolfberry genome using bioinformatics approaches. The gene structure, motif composition, and conserved domains were analyzed to identify the conservation and specificity of LbabZIPs. Furthermore, a comparative analysis of bZIP genes from wolfberry, tomato, pepper, Arabidopsis, and rice was performed to predict the functions of LbabZIP genes. The roles of specific LbabZIP genes in various biological processes were characterized through expression pattern analysis across different wolfberry tissues. More importantly, the expression of LbabZIP genes during fruit development comprehensively analyzed by RT-qPCR indicated that some LbabZIP genes were responsive to fruit development. This study provides valuable information for identifying candidate LbabZIP genes involved in regulating the growth and development of wolfberry, particularly fruit development.
2. Results
2.1. Identification and Characteristics of bZIP Genes in Wolfberry
To systematically identify bZIP TFs in the wolfberry genome, a homology-based BLASTP search was conducted using 78 experimentally validated Arabidopsis thaliana bZIP protein sequences as queries. The candidate sequences were confirmed to contain the bZIP domain using the Simple Modular Architecture Research Tool (SMART). A total of 66 LbabZIP genes were systematically identified in the Lycium barbarum genome and sequentially designated LbabZIP1 to LbabZIP66 according to their chromosomal locations (Supplementary Table S1). The amino acid lengths of the encoded LbabZIP proteins ranged from 129 amino acids (LbabZIP16) to 813 amino acids (LbabZIP31 and LbabZIP46), with molecular weight (MW) ranging from 14.98 kDa (LbabZIP16) to 92.17 kDa (LbabZIP31). The isoelectric points (pI) varied from 4.97 (LbabZIP37) and 9.93 (LbabZIP62). All LbabZIP proteins had negative grand average of hydropathicity (GRAVY) values, indicating their hydrophilic nature. Most of the LbabZIP proteins were unstable, except LbabZIP35 and LbabZIP31. Subcellular localization prediction revealed that the majority of LbabZIP proteins in Lycium barbarum are predominantly localized to the nucleus, consistent with their functional role as transcription factors regulating gene expression.
2.2. Phylogenetic Relationship and Classification of LbabZIP Proteins
To explore the evolutionary relationships and classification of the LbabZIP family, a phylogenetic tree was constructed using 353 bZIP protein sequences from L. barbarum (66), S. lycopersicum (69), C. annuum (54), O. sativa (86), and A. thaliana (78) (Figure 1). Based on their relationship with A. thaliana bZIP genes, the LbabZIP genes were divided into 13 subgroups: A, B, C, D, E, F, G, H, I, J, K, M, and S. Subgroup S contained the largest number of LbabZIP genes (14), followed by subgroup D (12) and subgroup A (11). Subgroups K and H contained only one gene each. Notably, no LbaZIP member was assigned to subgroup J.
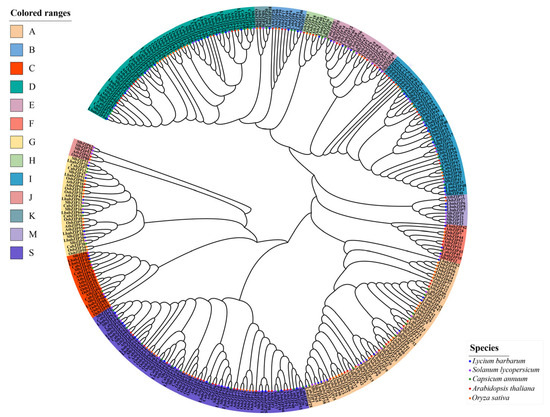
Figure 1.
Phylogenetic analysis of bZIP genes from wolfberry, tomato, pepper, Arabidopsis, and rice. Wolfberry bZIP proteins are divided into 13 distinct clades. Protein sequence alignment was performed using MUSCLE, and a neighbor-joining phylogenetic tree was reconstructed with 10,000 bootstrap replicates. Colored regions represent different subgroups. Wolfberry bZIP proteins (blue solid circles), tomato bZIP proteins (purple solid circles), pepper bZIP proteins (green solid circles), Arabidopsis bZIP proteins (red solid circles), and rice bZIP proteins (orange solid circles) are denoted.
2.3. Analysis of Gene Structure and Conserved Motifs in LbabZIP Genes
The gene structure of LbabZIP genes was analyzed by examining their exon–intron structure (Figure 2b). Gene structure analysis of the LbabZIP genes in Lycium barbarum revealed significant diversity in intron numbers, ranging from 0 to 11 introns per gene, with a mean of 3.9 introns. Subgroup-specific patterns were observed; for example, most genes in subgroup S slacked introns, while subgroup G had the largest number of introns.
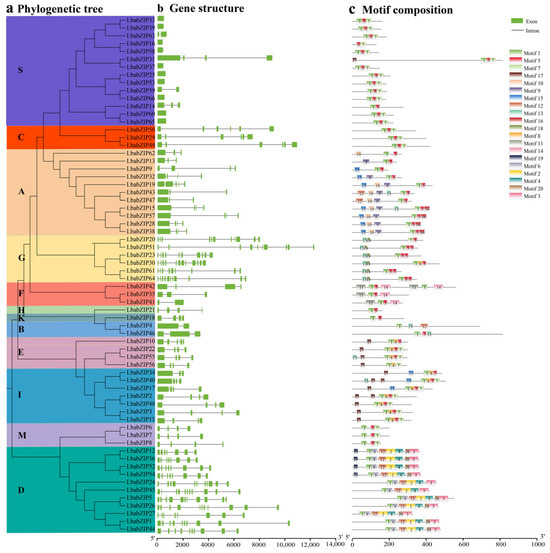
Figure 2.
Phylogenetic relationships, gene structure, and motif distributions of wolfberry bZIP genes. (a) The phylogenetic tree was generated from full-length sequences using MEGA 6.06. (b) The exon–intron structure of wolfberry bZIP genes, with yellow boxes indicating exons and gray lines indicating introns. (c) Conserved motif distributions in wolfberry proteins. Motifs 1–20 are represented by colored boxes. Motif sequence details are available in Supplementary Table S2. The scale bar indicates protein length.
The conserved motifs of LbabZIP proteins were identified using the MEME analysis, revealing 20 motifs (Figure 2c and Supplementary Table S2). Motif 1 was conserved in all LbabZIP proteins except LbabZIP20. Motifs 1, 5, and 7 were commonly detected across most subgroups, whereas subgroup-specific motifs included motif 13 and motif 18 (subgroup G), motif 10 (subgroup A), and motifs 2, 3, 4, and 6 (subgroup D). Motif 14 was unique to three members in subgroup F. These conserved motifs may reflect specific biological functions.
2.4. Chromosomal Distribution of bZIP Genes in Wolfberry
The 66 LbabZIP genes were unevenly distributed across 12 wolfberry chromosomes (Figure 3a). Chromosome 8 (Chr8) contained the largest number of LbabZIP genes (11, 16.6%), followed by Chr02 and 6, each with 10 genes (15.1%). Chromosomes 3 and 7 contained only one gene each (LbabZIP19 and LbabZIP36, respectively, Figure 3b). All chromosomes contained at least one bZIP gene. Interestingly, despite being the longest chromosome, chromosome 1 contained only eight bZIP genes, indicating no clear correlation between chromosome size and gene number (Figure 3c).
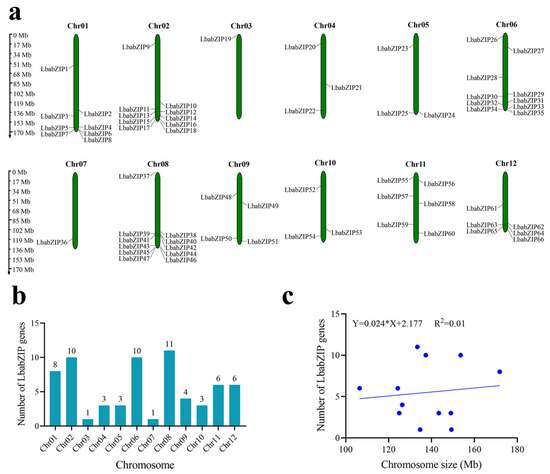
Figure 3.
The chromosomal localization of wolfberry bZIP genes. (a) The distribution of LbabZIP genes across 12 chromosomes, with chromosome numbers labeled at the top and scales in megabases (Mb). (b) The number of LbabZIP genes on each chromosome. (c) The correlation between chromosome size and the number of LbabZIP genes.
2.5. Synteny, Duplication Events, and Selective Pressure Analysis of LbabZIP Genes
Intra-species synteny analysis revealed 22 paralogous LbabZIP gene pairs, residing in 19 syntenic blocks. The smallest block contained 8 gene pairs and had 1 LbabZIP gene pair, while the largest block comprised 81 gene pairs and contained 1 LbabZIP gene pair. Comparative syntenic maps were plotted between L. barbarum and four plant species, including S. lycopersicum, C. annuum, A. thaliana, and O. sativa. These analyses revealed 53, 22, 55, and 27 homologous bZIP gene pairs, respectively (Figure 4b and Supplementary Table S3), suggesting a closer evolutionary relationship between L. barbarum and S. lycopersicum/A. thaliana. Further analysis revealed that 4 bZIP syntenic gene pairs were shared by all four plant species and 11 bZIP syntenic gene pairs were shared between two Solanaceae species (tomato and pepper) (Figure 4c).
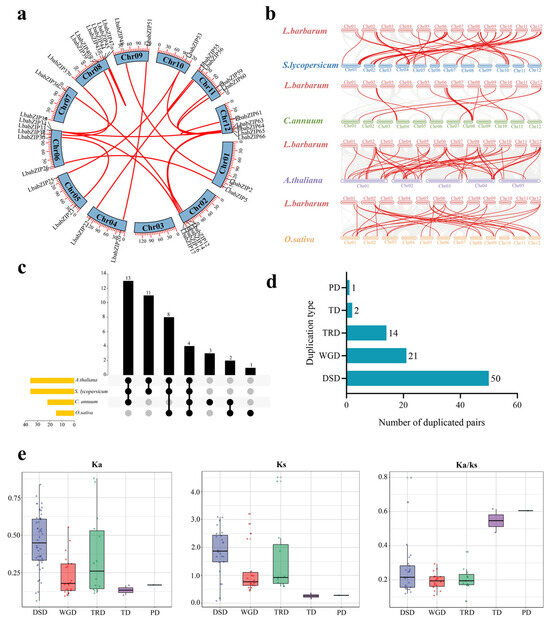
Figure 4.
Synteny, duplication events, and selective pressure analysis of wolfberry bZIP genes. (a) Synteny analysis of LbabZIP genes within wolfberry genome. Red lines highlight syntenic bZIP gene pairs. (b) Synteny analysis of bZIP genes between wolfberry and tomato, pepper, Arabidopsis, and rice genomes, with red lines indicating syntenic LbabZIP gene pairs. (c) Syntenic bZIP gene pairs across wolfberry and four other species are represented by vertically connected blocks and yellow columns. (d) Distribution of duplicated LbabZIP genes across five duplication types: transposed duplication (TRD), whole-genome duplication (WGD), dispersed duplication (DSD), tandem duplication (TD), and proximal duplication (PD). (e) Ka, Ks, and Ka/Ks values for duplicated LbabZIP gene pairs from five duplication events.
To investigate the expansion and evolution of the LbabZIP gene family, five duplication types (WGD—whole-genome duplication, DSD—dispersed duplication, TRD—transposed duplication, TD—tandem duplication, and PD—proximal duplication) were identified. A total of 86 duplication events were observed, with DSDs contributing the highest number (50 pairs), followed by WGD (21 pairs), TRD (14 pairs), TD (2 pairs), and PD (1 pair) (Figure 4d). These results suggest that dispersed duplication played a predominant role in the evolution of the LbabZIP gene family.
Selective pressure analysis showed that the Ka/Ks ratios for all duplicated gene pairs were less than 1, indicating that LbabZIP genes underwent purifying selection during evolution (Figure 4e and Supplementary Table S4).
2.6. Cis-Element Analysis of the LbabZIP Genes
A total of 896 cis-regulatory elements were identified and divided into three main subgroups: abiotic and biotic stress responses, plant growth and development, and phytohormone responsiveness (Figure 5 and Supplementary Table S5). In the abiotic and biotic stress subgroup, four types of cis-elements were identified, including MBS (involved in drought inducibility), G-Box (plant growth response to light), ARE (responsible for anaerobic induction), and LTR (low-temperature response). Among these, G-Boxes was the most prevalent cis-element, accounting for 43% of the total. In the plant growth and development subgroup, the identified cis-elements included Box 4, CAT-box, and O2-site, which are associated with light-induced growth, meristem-related processes, and zein metabolism, respectively. Box 4 accounted for the highest proportion (73%), suggesting a significant role for LbabZIP genes in light-regulated growth responses. In the phytohormone-responsive subgroup, five cis-elements were identified: ABRE (abscisic acid response), P-box, GARE-motif (gibberellin response), and CGTCA-motif and TGACG-motif (jasmonic acid methyl ester response). Notably, the LbabZIP genes contained 129 ABRE elements (40%), with the CGTCA-motif and TGACG-motif collectively accounting for 68%. These findings suggest that LbabZIP genes are involved in plant growth and development, hormone response, and stress adaptation.
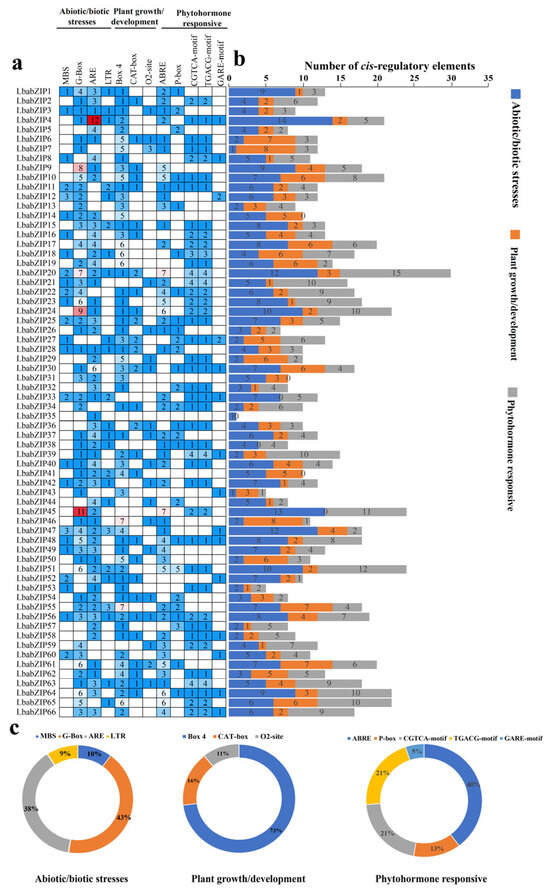
Figure 5.
Cis-regulatory elements analysis of LbabZIP gene promoters. (a) Number and type of cis-regulatory elements in LbabZIP genes, numbers, and colors indicating different elements. (b) Categorization of cis-regulatory elements of LbabZIP genes into three subgroups (biotic and abiotic stresses, plant hormones, and plant growth). (c) Pie charts showing the relative proportions of cis-regulatory elements in each subgroup.
2.7. Gene Enrichment Analysis of the LbabZIP Genes
To elucidate the functional classification of LbabZIP genes, the egg-NOG-mapper online tool was employed for Gene Ontology (GO) analysis. The predicted GO terms were categorized into three main groups: biological process, cellular component, and molecular function, distributed across 17 distinct pathways (Figure 6 and Supplementary Table S6). Among these pathways, nine were associated with biological processes, primarily signal transduction, nucleobase-containing compound metabolism, and cell communication. Molecular function-related pathways included DNA-binding transcription factor activity, transcription regulator activity, and DNA binding. The cellular component group had two pathways, reflecting the roles of LbabZIP proteins in specific cellular locations.
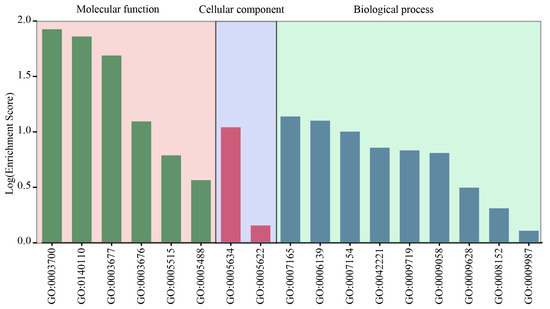
Figure 6.
GO enrichment analysis of wolfberry bZIP genes. Gene Ontology (GO) enrichment analysis categorized the LbabZIP genes into cellular components, biological processes, and molecular functions.
2.8. Expression Patterns of bZIP Genes in Different Tissues of Wolfberry
To explore roles of bZIP genes, we conducted a comprehensive expression analysis utilizing transcriptome sequencing data from multiple wolfberry tissues (stems, leaves, flowers, and fruits) and five distinct fruit developmental stages. Of the 66 LbabZIP genes, eight were not expressed in any tissue (Figure 7a and Supplementary Table S7), and nine showed no expression during fruit development (Figure 7b and Supplementary Table S8). Tissue-specific expression revealed diverse roles for LbabZIP genes. For example, LbabZIP4, LbabZIP12, LbabZIP40, and LbabZIP47 were highly expressed in stems, while LbabZIP18, LbabZIP54, and LbabZIP55 were highly expressed in flowers, indicating potential roles in flower development. Nine genes (LbabZIP3/9/17/19/21/24/33/50/65) exhibited high expression in fruits. Notably, the expression of LbabZIP17, LbabZIP21, and LbabZIP65 increased progressively during fruit development, whereas LbabZIP9, LbabZIP24, and LbabZIP47 showed a declining trend (Figure 7b). These results suggest that bZIP genes play crucial roles in fruit development.
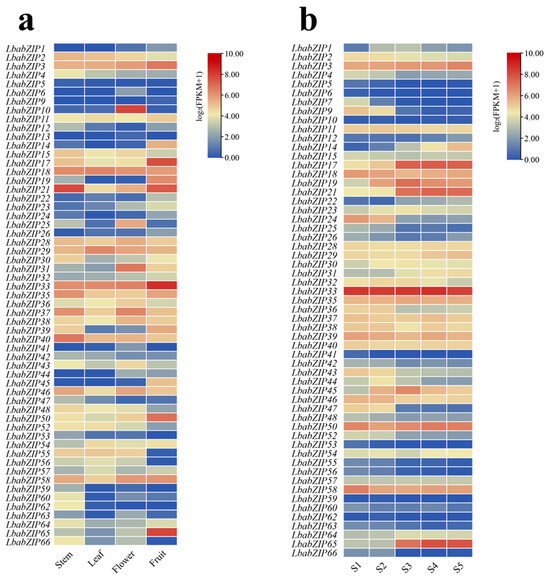
Figure 7.
Expression profiles of LbabZIP genes in wolfberry tissues and fruits. (a) Heatmap representing LbabZIP gene expression in four wolfberry tissues. (b) Heatmap of LbabZIP gene expression during five fruit developmental stages of Lycium barbarum var. auranticarpum. S1, S2, S3, S4, and S5 periods represent 12, 19, 25, 30, and 37 days after full bloom (DAF), respectively. FPKM values were Log2-transformed (value + 1), with red and blue indicating high and low expression levels, respectively.
2.9. Expression Validation by qRT-PCR
To validate transcriptome results, the expression of 15 representative LbabZIP genes was analyzed at five stages of wolfberry fruit development using qRT-PCR (Figure 8 and Supplementary Table S9). Consistent with transcriptome data, the expression levels of LbabZIP17, LbabZIP21, LbabZIP22, and LbabZIP65 gradually increased and peaked at the S5 stage. In contrast, LbabZIP9, LbabZIP24, LbabZIP47, and LbabZIP52 exhibited decreasing expression patterns. This consistency suggests that LbabZIP genes play a regulatory role in fruit development.
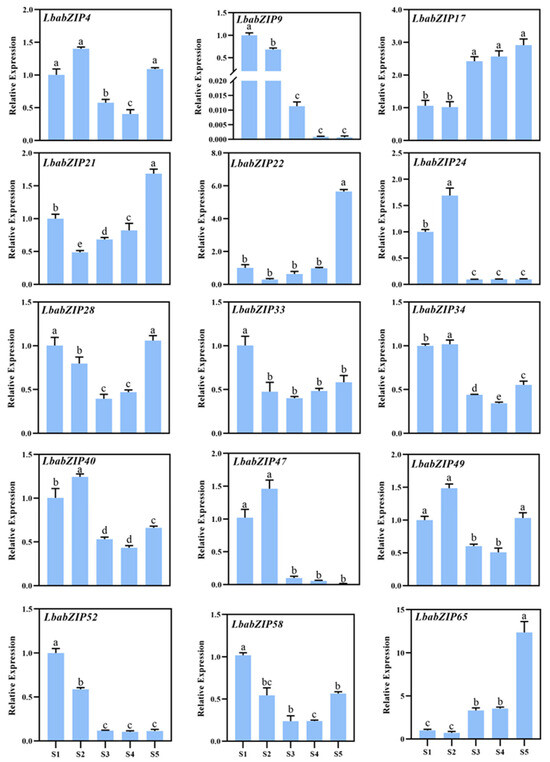
Figure 8.
Expression patterns of 15 LbabZIP gene determined by qRT-PCR. Expression levels were normalized to the wolfberry LbaActin gene and analyzed using the 2−ΔΔCt method. S1, S2, S3, S4, and S5 period represent 12, 19, 25, 30, and 37 days after full bloom (DAF), respectively. Data are presented as the mean ± standard deviation (SD). Different letters indicate significant differences (p < 0.05, n = 3).
2.10. Subcellular Localization of LbabZIP Proteins
To determine the subcellular localization of LbabZIP21/40/49/65 proteins, GFP-tagged fusion proteins were transiently expressed in Nicotiana benthamiana leaf epidermal cells. Fluorescence microscopy revealed that GFP signals for the control were ubiquitously distributed throughout the cells, while the fusion proteins (LbabZIP21/40/49/65) were confined in the nucleus (Figure 9). These results confirm that LbabZIP21, LbabZIP40, LbabZIP49, and LbabZIP65 encode nuclear-localized proteins.
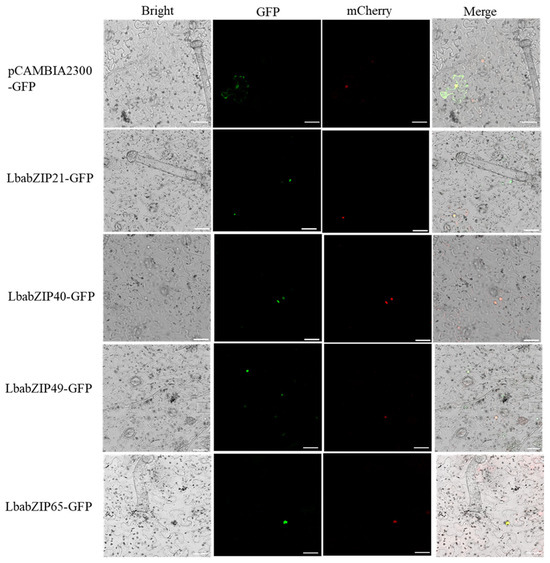
Figure 9.
Subcellular localization of LbabZIPs proteins. LbabZIPs-GFP fusion proteins were transiently expressed in N. benthamiana leaves and visualized via fluorescence microscopy 48 h post-infiltration. The 35S-GFP was used as positive control. From left to right: bright field; green fluorescent protein (GFP); mCherry is a red fluorescent protein (RFP) fused with the nucleus marker; Merge indicates merger between GFP and mCherry fluorescence images. Scale bars = 20 μm.
To evaluate the transcriptional activity of four LbabZIP TFs (LbabZIP21, LbabZIP40, LbabZIP49, and LbabZIP65) in vivo, a yeast two-hybrid (Y2H) assay was conducted using the pGBKT7-LbabZIP constructs expressed in Saccharomyces cerevisiae strain AH109. Yeast cells harboring the pGBKT7-LbabZIP40 and pGBKT7-LbabZIP49 constructs and the positive control (pGBKT7-53 + PGADT7-RecT) grew successfully on the selective medium SD/-Ade/-His/-Leu/-Trp. In contrast, yeast cells containing pGBKT7-LbabZIP21 and pGBKT7-LbabZIP65 and the negative control (empty vector pGBKT7) failed to grow in the same medium (Figure 10). These findings demonstrate that LbabZIP40 and LbabZIP49 have transactivation activities in the yeast assay system, whereas LbabZIP21 and LbabZIP65 do not exhibit such activity under these experimental conditions.
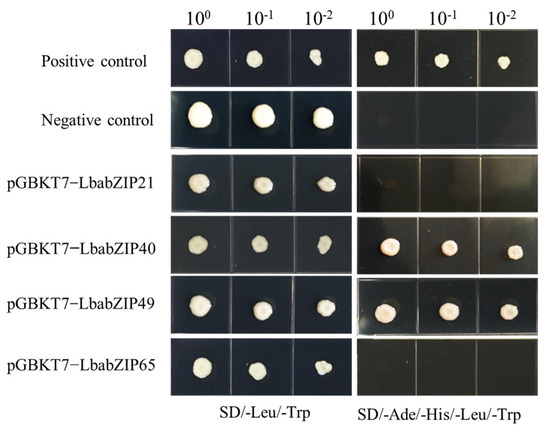
Figure 10.
Transactivation activity of LbabZIP genes. LbabZIP21, LbabZIP40, LbabZIP49, and LbabZIP65 coding sequences were cloned into the pGBKT7 vector and transformed into yeast strain AH109. Yeast cells were dotted at 10−1 dilution on SD/-Leu/-Trp and SD/-Ade/-His/-Leu/-Trp media. Positive control: pGBKT7-53 + PGADT7-RecT; negative control: pBGKT7 vector alone.
3. Discussion
bZIP TFs is one of the largest transcription factor families that widely exist in eukaryotes and they have been highly conserved in evolution [9]. bZIP TFs play crucial roles in regulating plant growth and development, secondary metabolism, and responses to abiotic stress [17,18,24]. The bZIP gene family has been extensively studied in many plants, with 78 members identified in Arabidopsis [9], 86 in Oryza sativa [11], 69 in Solanum lycopersicum [12], 52 in Capsicum annuum [25], 62 in Pyrus bretschneideri [13], 59 in Castanea mollissima [26], 92 in Hordeum vulgare [27], and 45 in Liriodendron chinensis [28]. However, until now, although the entire wolfberry genome has been sequenced [4], studies of bZIP gene family in wolfberry are limited. In this study, we performed a genome-wide analysis of wolfberry and identified a total of 66 LbabZIP genes. It is similar to the number of bZIP gene family members in Solanum lycopersicum (66) [12], Pyrus bretschneideri (62) [13], and of other species. The bZIP genes in wolfberry were classified into 13 subgroups by constructing phylogenetic trees of the bZIP gene family in Lycium barbarum, Solanum lycopersicum, Capsicum annuum, Oryza sativa, and Arabidopsis, which is similar to the grouping of the Arabidopsis bZIP gene family [9] (Figure 2). In addition to the absence of bZIP gene family members of L. barbarum in group J, the bZIP gene numbers of five species were distributed in other subgroups. The suggests that the number of bZIP genes is more conserved during evolution. Due to the highly conserved nature of bZIP sequences, genes within the same bZIP group exhibit identical or similar functions. This conservation provides a valuable reference for investigating the functional roles of this gene family [29].
The analysis of gene structure and conserved motifs can facilitate the prediction of gene functions [15]. The results of structural analysis of wolfberry bZIP genes revealed that the bZIP gene structure of wolfberry is relatively simple, with the number of introns ranging from 0 to 11. It can be divided into two categories according to the presence or absence of introns. A total of 10 bZIP genes without introns was found in the genome of wolfberry. All 10 bZIP genes were classified in group S. A similar situation exists in species such as Arabidopsis, Nelumbo, and Suaeda australis [9,30,31]. The bZIP genes categorized into various subgroups may serve distinct functions in plant growth and development. For example, most of members of group A are involved in abiotic stress responses, such as drought, salinity, and cold stress. Genes located in group C are associated with seed maturation and storage protein synthesis [32]. Genes located in group S are often linked to sugar signaling and metabolism [33]. Genes located in group G are implicated in light signaling and photomorphogenesis [34]. The number of conserved motifs present in each bZIP protein also exhibits significant variability. Some bZIP proteins contain only two conserved motifs; most proteins have three conserved motifs (Figure 2). In addition to the specific bZIP_1 (PF00170) and bZIP_2 (PF07716) domains, plant bZIP proteins also possess additional functionally conserved domains that are involved in various biological processes. In wolfberry, the identified bZIP members contain three distinct conserved domains: BRLZ, bZIP_C, and bZIP_2. Among them, the BRLZ conserved domain is widely present in the bZIP gene family of wolfberry. bZIP_C is found in two members (LbabZIP29 and LbabZIP49), while bZIP_2 is only present in LbabZIP31 (Figure 2).
Previous studies have found that the expansion and contraction of gene families are mainly driven by gene duplication, such as whole-genome duplication, dispersed duplication, and tandem duplication [35,36]. For example, the expansion of BBX and LBD gene families was primarily driven by whole-genome duplication and dispersed duplication [35,37]. In this study, gene duplication analysis revealed that dispersed duplication and whole-genome duplication events drove the expansion of the LbabZIP gene family. Fifty LbabZIP genes (58.13%) were grouped into the DSD type, and 21 genes (24.41%) belonged to the WGD type, which might be due to the high ratio of self-incompatibility and the domestication process of wolfberry [4]. Following gene duplication, duplicated gene pairs can experience three main different types of selection pressures, including positive selection, negative selection (also known as purifying selection), and neutral selection [37]. Based on the estimated results of Ka/Ks, our analysis revealed that the evolution of the LbabZIP gene family was mainly driven by purifying selection.
Gene transcription is significantly impacted by the cis-acting element of the promoter, which can directly alter gene expression and function [38]. The function of the gene can be predicted or inferred based on the types of cis-acting element present in its promoter region. Among the promoters of the 66 LbabZIP gene family members in wolfberry, most of cis-elements were related to stresses, plant growth, and development and phytohormones. Previous studies have been verified that show that the bZIP genes are regulated by hormones such as ABA and IAA in the regulation of plant growth and development [39,40,41]. For instance, OsABF2 in rice binds to ABREs to regulate abiotic stress-responsive genes via an ABA-dependent pathway [42], and PpABF3 targets the G-box element in the PpWRKY44 promoter, enhancing salinity-induced malate accumulation [43]. In apples, MdbZIP44 enhances anthocyanin accumulation in response to ABA by facilitating MdMYB1 binding to target gene promoters [41]. Many hormone-responsive elements were also detected in the LbabZIP promoter region: ABRE, P-box, CGTA-motif, TGACG-motif, and GARE-motif. Therefore, the LbabZIP gene family plays a crucial role in plant growth, development, and stress resistance in wolfberry by mediating plant hormone regulation.
Increasing evidence suggests that bZIP genes have varied functions during plant development processes. Generally, patterns of gene expression can offer significant insights for predicting gene function [37]. To investigate LbabZIP gene expression patters, transcriptome sequencing and qRT-PCR expression profiling were performed across four tissues and five fruit developmental stages. The bZIP gene family showed diverse expression patterns in four distinct tissues. This result showed that LbabZIP genes could be involved in a range of biological processes. Forty-seven genes are expressed throughout all tissues, indicating their extensive role in wolfberry growth and development. In addition, 12.12% of LbaZIPs were observed to have high transcriptional levels in wolfberry fruit, suggesting these genes may have important roles in development. Furthermore, qRT-PCR was used to confirm the expression levels of 15 randomly selected members of the LbabZIP gene family. Interestingly, we found that the S subgroup gene LbabZIP65 was highly expressed in mature fruit, and is orthologous with AtbZIP6 (AT2G22850) and OsbZIP16 (Os02g0191600). Therefore, we speculated that the LbabZIP65 gene might be involved in the regulation of fruit ripening and related color changes. However, the function of bZIP genes in fruits is still unknown and needs to be investigated further in future studies. Future research should employ techniques such as CRISPR/Cas9-mediated gene editing and RNA interference (RNAi) to directly manipulate LbabZIP gene expression in wolfberry. Investigation at the protein level, including protein–protein interaction studies and post-translational modification analyses, is needed to fully understand the regulatory mechanisms of bZIP TFs in wolfberry.
In this study, the LbabZIP gene family was identified, and its expression during fruit development was analyzed. Genes associated with fruit development, such as LbabZIP65, were excavated, which provided opportunities for manipulating fruit quality traits. Breeding strategies can be designed to regulate the expression of these genes, potentially leading to improvements in the fruit ripening process, the enhancement of coloration, and an increase in nutritional content. This provides important targets for the genetic improvement of wolfberry and holds significant importance for its breeding and production.
4. Materials and Methods
4.1. Plant Materials
Lycium barbarum var. auranticarpum plants were cultivated at the National Wolfberry Engineering Research Center, Yinchuan, Ningxia, China (37°53′ N, 105°72′ E). The fruit samples were collected on days 12 (S1), 19 (S2), 25 (S3), 30 (S4), and 37 (S5) after full-bloom (DAF), respectively. The fruit is yellow in color and nearly round in nature. The samples were immediately frozen in liquid nitrogen, and stored at −80 °C until further analysis.
4.2. Identification of bZIP Genes in the Wolfberry Genome
The completed L. barbarum genome sequence (Accession: GCA_019175385.2) was retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/datasets/genome/?taxon=112863, accessed on 15 January 2024) [4]. Two complementary methods were employed to identify bZIP genes in the wolfberry genome, as described in previous studies [6]. Initially, protein sequences of all Arabidopsis bZIP proteins, obtained from the TAIR database (https://www.arabidopsis.org/, accessed on 8 February 2024), were utilized as queries to perform a BLASTP search against the wolfberry genome database (E-value < 1 × 10−5). Secondly, the Hidden Markov Model (HMM) profile for the bZIP domain (PF00170) was downloaded from the Pfam database (http://pfam-legacy.xfam.org/, accessed on 15 January 2024) and used to screen the wolfberry protein database, with a threshold of an E-value < 1 × 10−5. Subsequently, candidate LbabZIP genes were validated by confirming the presence of the bZIP domain using SMART (http://smart.embl.de/, accessed on 15 January 2024) and the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/, accessed on 15 January 2024). Protein properties, including molecular weight (Mw), isoelectric point (pI), and grand average of hydropathicity (GRAVY), were calculated using the Expasy online tool (https://web.expasy.org/compute_pi/, accessed on 16 January 2024). Subcellular localization predictions for LbabZIP proteins were performed using WoLF PSORT (https://wolfpsort.hgc.jp/, accessed on 16 January 2024) [44].
4.3. Phylogenetic Analysis of LbabZIP Proteins
To explore the evolutionary relationships of the wolfberry bZIP gene family, protein sequences of Arabidopsis bZIP genes (AtbZIP) were obtained from the TAIR database (https://www.arabidopsis.org/, accessed on 25 January 2024). Sequences of bZIP genes from two Solanaceae species (Solanum lycopersicum and Capsicum annuum) were retrieved from the SGN database (https://solgenomics.sgn.cornell.edu/, accessed on 25 January 2024). O. sativa bZIP genes were identified from previous studies [11]. All sequences were aligned using Muscle v5 [45], and a phylogenetic tree was constructed using the neighbor-joining method in MEGA6.06 [46] 1000 bootstrap replicates. Classification of the LbabZIP family was based on Arabidopsis bZIP classification criteria [9]. The phylogenetic tree was visualized using the iTOL platform (https://itol.embl.de/, accessed on 25 January 2024) [47].
4.4. Gene Structure and Conserved Motif Analysis of LbabZIP Genes
Exon–intron structures of LbabZIP genes were visualized using the Gene Structure Display Server 2.0 (GSDS) (https://gsds.gao-lab.org/Gsds_help.php, accessed on 20 January 2024) [48]. Conserved motifs in LbabZIP proteins were identified using the MEME suite (https://meme-suite.org/meme/, accessed on 20 January 2024), with the maximum number of motifs set to 20 [49].
4.5. Chromosomal Localization of LbabZIP Genes
Physical mapping of LbabZIPs genes was mapped to wolfberry chromosomes using MG2C v2.1 [50] based on genome location information.
4.6. Synteny, Duplication Events, and Selective Pressure Analysis of LbabZIP Gene Family
Collinearity analysis of LbabZIP genes was performed using TBtools-II [51]. Homology analysis between L. bararum and four other plant species (S. lycopersicum, C. annuum, A. thaliana, and O. sativa) was conducted using the dual synteny plotter in TBtools-II. Gene duplication events were identified using the Gen_finder tool (https://github.com/qiao-xin/DupGen_finder, accessed on 20 January 2024). The Ka, Ks, and Ka/Ks ratios were calculated using the Simple Ka/Ks Calculator (NG) model in TBtools v2.225 [51].
4.7. Cis-Element Analysis for LbabZIP Genes
To investigate regulatory elements associated with LbabZIP gene expression, promoter sequences (2000 bp sequence upstream of the start codon) were extracted using TBtools-II [52]. Cis-elements were identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/PlantCARE/html/, accessed on 25 March 2024), and the results were visualized with TBtools v2.225 [51].
4.8. GO Analysis of the LbaZIP Genes
Gene Ontology (GO) term prediction for the LbabZIP genes was performed using the EggNOG-mapper tool (http://eggnog-mapper.embl.de/, accessed on 25 June 2024). The identified GO terms were categorized into three major categories: cellular components, biological processes, and molecular functions.
4.9. Expression Profiles of LbabZIP Genes Determined from RNA-Seq Datasets
Transcriptome sequencing (RNA-seq) data from five distinct developmental stages and four tissues of Lycium barbarum var. auranticarpum were retrieved from the NCBI database (accession number: PRJNA845109). The FPKM (fragments per kilobase of transcript per million mapped reads) values of LbabZIP genes were normalized through K-means clustering analysis using R software (version 3.2.2) [53]. Subsequently, the expression patterns were visualized using TBtools v2.225 software [51].
4.10. Total RNA Extraction and Expression Analysis of LbabZIP Genes
Primers specific to the LbabZIP genes were designed using Primer Premier 5 (Supplemental Table S9). Total RNA was extracted from wolfberry fruit of different stages using the E.Z.N.A.®Plant RNA Kit (OMEGA, Guangzhou, China). The first-strand cDNA was synthesized using the EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen, Beijing, China). qRT-PCR was performed using the PerfextStartTM Green qPCR Super Mix (TransGen, Beijing, China) following the manufacturer’s instructions. The wolfberry LbaActin gene was used as the internal control for the normalization of gene expression. Relative expression levels were calculated using the 2−ΔΔCt method [53]. Three independent biological replicates were used in the analysis. Statistical analyses were conducted using one-way analysis of variance (ANOVA) followed by Turkey’s honestly significant difference (HSD) test. Differences were considered significant at p < 0.05, with different letters indicating significant differences.
4.11. Subcellular Localization
Full-length coding sequences of LbabZIP15, LbabZIP21, LbabZIP31, LbabZIP34, LbabZIP49, and LbabZIP65 (excluding stop codons) were cloned into the pCAMBIA2300-GFP vector. Recombinant constructs (pCAMBIA2300-LbabZIP15/21/31/34/49/65-GFP) were introduced into A. tumefaciens strain GV3101 via the heat shock method. Four-week-old N. benthamiana seedlings were used for transient expression via agrobacterium-mediated infiltration. Infiltrated leaves were kept in the dark for 12 h, and fluorescence signals were observed 72 h post-injection using a laser confocal microscope (Zeiss, Oberkochen, Germany). The pCAMBIA2300-GFP empty vector served as a negative control. Primer sequences are listed in Supplementary Table S10.
4.12. Transactivation Assays
The coding sequences (CDS) of LbabZIP21/40/49/65 were cloned into the pGBKT7 vector to generate recombinant plastids (pGBKT7-LbabZIP21/40/49/65). Recombinant plasmids, along with the positive control (pGBKT7-53 + pGADT7-RecT) and the negative control (empty vector pGBKT7), were transformed into yeast strain AH109. Transformed yeast cells were grown on SD/-Leu/-Trp medium and transferred to SD/-Ade/-His/-Leu/-Trp medium to assess transactivation activity. Plates were incubated at 28 °C for three days, and transactivation activity was evaluated based on yeast cell growth. Primer sequences are listed in Supplemental Table S11.
5. Conclusions
This study provides the first genome-wide analysis of the bZIP gene family in wolfberry. A total of sixty-six wolfberry bZIP genes were identified, cis-acting element prediction was performed, and analyses of phylogenetic relationship, gene structure, motif composition, chromosomal distribution, collinearity, and expression patterns were performed. Phylogenetic analysis of bZIP genes from wolfberry, tomato, pepper, Arabidopsis, and rice revealed both conserved and divergent evolutionary patterns, providing instructive information for further investigation on the function of LbaZIP genes. Furthermore, study of expression patterns in diverse tissues revealed that LbabZIP played an important role in wolfberry growth and development. In addition, we analyzed the expression of LbabZIP genes at different fruit development stages and identified some candidate genes involved in the growth and development of wolfberry fruits. Our findings significantly advance understanding of bZIP-mediated regulation in wolfberry, providing both a framework for future functional studies of individual LbabZIP genes and valuable targets for molecular breeding of improved fruit quality traits.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms26104665/s1.
Author Contributions
H.G. designed the project and drafted the manuscript. X.C. performed gene expression. Y.Y. and Y.M. collected the genome data and performed the bioinformatics analysis. X.Q. and X.Z. contributed to collect materials. X.B. and A.X. contributed to collect samples. Y.Y. and R.Z. contributed to review drafts of the paper and approved the final draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key Research and Development projects of Ningxia Hui Autonomous Region (No. 2022BBF02008), Central Guidance Fund for Local Science and Technology Development (2024FRD05009), Natural Science Foundation of Ningxia (2020AAC03284), and the National Natural Science Foundation of China (31560418).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The wolfberry genome datasets used during the current study are available in NCBI database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA640228).The RNA-seq data used in this study were downloaded from the NCBI Database (PRJNA845109).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wenli, S.; Shahrajabian, M.H.; Qi, C. Health benefits of wolfberry (Gou Qi Zi, Fructus barbarum L.) on the basis of ancient Chineseherbalism and Western modern medicine. Avicenna J. Phytomed. 2021, 11, 109–119. [Google Scholar] [PubMed]
- Vidović, B.B.; Milinčić, D.D.; Marčetić, M.D.; Djuriš, J.D.; Ilić, T.D.; Kostić, A.; Pešić, M.B. Health Benefits and Applications of Goji Berries in Functional Food Products Development: A Review. Antioxidants 2022, 11, 248. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Luo, Y.; Zhang, C.; An, W.; Zhou, J.; Jin, C.; Zhang, Y.; Zhao, J. Integrated Metabolome and Transcriptome during Fruit Development Reveal Metabolic Differences and Molecular Basis between Lycium barbarum and Lycium ruthenicum. Metabolites 2023, 13, 680. [Google Scholar] [CrossRef]
- Cao, Y.-L.; Li, Y.-l.; Fan, Y.-F.; Li, Z.; Yoshida, K.; Wang, J.-Y.; Ma, X.-K.; Wang, N.; Mitsuda, N.; Kotake, T.; et al. Wolfberry genomes and the evolution of Lycium (Solanaceae). Commun. Biol. 2021, 4, 671. [Google Scholar] [CrossRef]
- Yin, Y.; Guo, C.; Shi, H.; Zhao, J.; Ma, F.; An, W.; He, X.; Luo, Q.; Cao, Y.; Zhan, X. Genome-Wide Comparative Analysis of the R2R3-MYB Gene Family in Five Solanaceae Species and Identification of Members Regulating Carotenoid Biosynthesis in Wolfberry. Int. J. Mol. Sci. 2022, 23, 2259. [Google Scholar] [CrossRef]
- Yin, Y.; Shi, H.; Mi, J.; Qin, X.; Zhao, J.; Zhang, D.; Guo, C.; He, X.; An, W.; Cao, Y.; et al. Genome-Wide Identification and Analysis of the BBX Gene Family and Its Role in Carotenoid Biosynthesis in Wolfberry (Lycium barbarum L.). Int. J. Mol. Sci. 2022, 23, 8440. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, Y.; Shi, X.; Wang, X.; Yin, Y. Comparative Analysis of the GATA Transcription Factors in Five Solanaceae Species and Their Responses to Salt Stress in Wolfberry (Lycium barbarum L.). Genes 2023, 14, 1943. [Google Scholar] [CrossRef]
- Li, Z.; Liu, J.; Chen, Y.; Liang, A.; He, W.; Qin, X.; Qin, K.; Mu, Z. Genome-Wide Identification of PYL/RCAR ABA Receptors and Functional Analysis of LbPYL10 in Heat Tolerance in Goji (Lycium barbarum). Plants 2024, 13, 887. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Guo, Z.; Dzinyela, R.; Yang, L.; Hwarari, D. bZIP Transcription Factors: Structure, Modification, Abiotic Stress Responses and Application in Plant Improvement. Plants 2024, 13, 2058. [Google Scholar] [CrossRef]
- Zg, E.; Zhang, Y.P.; Zhou, J.H.; Wang, L. Mini review roles of the bZIP gene family in rice. Genet. Mol. Res. GMR 2014, 13, 3025–3036. [Google Scholar] [CrossRef]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 771. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, M.A.; Manzoor, M.M.; Li, G.; Abdullah, M.; Han, W.; Wenlong, H.; Shakoor, A.; Riaz, M.W.; Rehman, S.; Cai, Y. Genome-wide identification and characterization of bZIP transcription factors and their expression profile under abiotic stresses in Chinese pear (Pyrus bretschneideri). BMC Plant Biol. 2021, 21, 413. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, C.; Li, Z.; Sun, J.; Wang, D.; Xu, L.; Li, X.; Guo, Y. Identification and Analysis of bZIP Family Genes in Potato and Their Potential Roles in Stress Responses. Front. Plant Sci. 2021, 12, 637343. [Google Scholar] [CrossRef]
- Liu, W.; Wang, M.; Zhong, M.; Luo, C.; Shi, S.; Qian, Y.; Kang, Y.; Jiang, B. Genome-wide identification of bZIP gene family and expression analysis of BhbZIP58 under heat stress in wax gourd. BMC Plant Biol. 2023, 23, 598. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, W.; Li, H.; Wang, Y.; Li, D.; Xue, C.; Liu, Z.; Liu, M.; Zhao, J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2020, 21, 483. [Google Scholar] [CrossRef]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Han, H.; Wang, C.; Yang, X.; Wang, L.; Ye, J.; Xu, F.; Liao, Y.; Zhang, W. Role of bZIP transcription factors in the regulation of plant secondary metabolism. Planta 2023, 258, 13. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Chen, Y.; Liu, Y.; Lin, Y.; Li, M.; Wang, Y.; He, W.; Chen, Q.; Zhang, Y. Heterologous overexpression of strawberry bZIP11 induces sugar accumulation and inhibits plant growth of tomato. Sci. Hortic. 2022, 292, 110634. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, X.; Li, B.; Zhao, X.; Shen, Y.; Yuan, Z. Genome-wide identification and characterization of bZIP gene family and cloning of candidate genes for anthocyanin biosynthesis in pomegranate (Punica granatum). BMC Plant Biol. 2022, 22, 170. [Google Scholar] [CrossRef]
- Shen, S.; Hu, X.; Cheng, J.; Lou, L.; Huan, C.; Zheng, X. PsbZIP1 and PsbZIP10 induce anthocyanin synthesis in plums (Prunus salicina cv. Taoxingli) via PsUFGT by methyl salicylate treatment during postharvest. Postharvest Biol. Technol. 2023, 203, 112396. [Google Scholar] [CrossRef]
- Sun, Q.; He, Z.; Wei, R.; Zhang, Y.; Ye, J.; Chai, L.; Xie, Z.; Guo, W.; Xu, J.; Cheng, Y.; et al. The transcriptional regulatory module CsHB5-CsbZIP44 positively regulates abscisic acid-mediated carotenoid biosynthesis in citrus (Citrus spp.). Plant Biotechnol. J. 2024, 22, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-C.; Meng, L.-H.; Gao, Y.; Grierson, D.; Fu, D.-Q. Manipulation of Light Signal Transduction Factors as a Means of Modifying Steroidal Glycoalkaloids Accumulation in Tomato Leaves. Front. Plant Sci. 2018, 9, 437. [Google Scholar] [CrossRef]
- Li, Z.; Fu, D.; Wang, X.; Zeng, R.; Zhang, X.; Tian, J.; Zhang, S.; Yang, X.; Tian, F.; Lai, J.; et al. The transcription factor bZIP68 negatively regulates cold tolerance in maize. Plant Cell 2022, 34, 2833–2851. [Google Scholar] [CrossRef]
- Gai, W.X.; Ma, X.; Qiao, Y.M.; Shi, B.H.; Ul Haq, S.; Li, Q.H.; Wei, A.M.; Liu, K.K.; Gong, Z.H. Characterization of the bZIP Transcription Factor Family in Pepper (Capsicum annuum L.): CabZIP25 Positively Modulates the Salt Tolerance. Front. Plant Sci. 2020, 11, 139. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, J.; Jia, N.; Wang, M.; Lu, Y.; Wang, D.; Zhang, J.; Zhang, H.; Wang, X. Genome-wide identification and characterization of the bZIP gene family and their function in starch accumulation in Chinese chestnut (Castanea mollissima Blume). Front. Plant Sci. 2023, 14, 1166717. [Google Scholar] [CrossRef]
- Zhong, X.; Feng, X.; Li, Y.; Guzmán, C.; Lin, N.; Xu, Q.; Zhang, Y.; Tang, H.; Qi, P.; Deng, M.; et al. Genome-wide identification of bZIP transcription factor genes related to starch synthesis in barley (Hordeum vulgare L.). Genome 2021, 64, 1067–1080. [Google Scholar] [CrossRef]
- Liang, J.; Fang, Y.; An, C.; Yao, Y.; Wang, X.; Zhang, W.; Liu, R.; Wang, L.; Aslam, M.; Cheng, Y.; et al. Genome-wide identification and expression analysis of the bHLH gene family in passion fruit (Passiflora edulis) and its response to abiotic stress. Int. J. Biol. Macromol. 2022, 225, 389–403. [Google Scholar] [CrossRef]
- Feng, X.; Wang, C.; Jia, S.; Wang, J.; Zhou, L.; Song, Y.; Guo, Q.; Zhang, C. Genome-Wide Analysis of bZIP Transcription Factors and Expression Patterns in Response to Salt and Drought Stress in Vaccinium corymbosum. Int. J. Mol. Sci. 2025, 26, 843. [Google Scholar] [CrossRef]
- Zhou, P.; Li, J.; Jiang, H.; Jin, Q.; Wang, Y.; Xu, Y. Analysis of bZIP gene family in lotus (Nelumbo) and functional study of NnbZIP36 in regulating anthocyanin synthesis. BMC Plant Biol. 2023, 23, 429. [Google Scholar] [CrossRef]
- Qu, Y.; Wang, J.; Gao, T.; Qu, C.; Mo, X.; Zhang, X. Systematic analysis of bZIP gene family in Suaeda australis reveal their roles under salt stress. BMC Plant Biol. 2024, 24, 816. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Nie, K.; Zhou, H.; Yan, X.; Zhan, Q.; Zheng, Y.; Song, C.P. ABI5 modulates seed germination via feedback regulation of the expression of the PYR/PYL/RCAR ABA receptor genes. New Phytol. 2020, 228, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Viana, A.J.C.; Matiolli, C.C.; Newman, D.W.; Vieira, J.G.P.; Duarte, G.T.; Martins, M.C.M.; Gilbault, E.; Hotta, C.T.; Caldana, C.; Vincentz, M. The sugar-responsive circadian clock regulator bZIP63 modulates plant growth. New Phytol. 2021, 231, 1875–1889. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shi, Y.; Li, M.; Fu, D.; Wu, S.; Li, J.; Gong, Z.; Liu, H.; Yang, S. The CRY2-COP1-HY5-BBX7/8 module regulates blue light-dependent cold acclimation in Arabidopsis. Plant Cell 2021, 33, 3555–3573. [Google Scholar] [CrossRef]
- Song, B.; Tang, Z.; Li, X.; Li, J.; Zhang, M.; Zhao, K.; Liu, H.; Zhang, S.; Wu, J. Mining and evolution analysis of lateral organ boundaries domain (LBD) genes in Chinese white pear (Pyrus bretschneideri). BMC Genom. 2020, 21, 644. [Google Scholar] [CrossRef]
- Liu, C.; Qiao, X.; Li, Q.; Zeng, W.; Wei, S.; Wang, X.; Chen, Y.; Wu, X.; Wu, J.; Yin, H.; et al. Genome-wide comparative analysis of the BAHD superfamily in seven Rosaceae species and expression analysis in pear (Pyrus bretschneideri). BMC Plant Biol. 2020, 20, 14. [Google Scholar] [CrossRef]
- Zeng, W.; Qiao, X.; Li, Q.; Liu, C.; Wu, J.; Yin, H.; Zhang, S. Genome-wide identification and comparative analysis of the ADH gene family in Chinese white pear (Pyrus bretschneideri) and other Rosaceae species. Genomics 2020, 112, 3484–3496. [Google Scholar] [CrossRef]
- Xin, H.; Liu, X.; Chai, S.; Yang, X.; Li, H.; Wang, B.; Xu, Y.; Lin, S.; Zhong, X.; Liu, B.; et al. Identification and functional characterization of conserved cis-regulatory elements responsible for early fruit development in cucurbit crops. Plant Cell 2024, 36, 2272–2288. [Google Scholar] [CrossRef]
- Yao, L.; Hao, X.; Cao, H.; Ding, C.; Yang, Y.; Wang, L.; Wang, X. ABA-dependent bZIP transcription factor, CsbZIP18, from Camellia sinensis negatively regulates freezing tolerance in Arabidopsis. Plant Cell Rep. 2020, 39, 553–565. [Google Scholar] [CrossRef]
- An, J.-P.; Yao, J.-F.; Xu, R.-R.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018, 41, 2678–2692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, X.; Cao, P.; Xiao, Z.A.; Zhan, C.; Liu, M.; Nvsvrot, T.; Wang, N. The bZIP53–IAA4 module inhibits adventitious root development in Populus. J. Exp. Bot. 2020, 71, 3485–3498. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Cho, J.I.; Han, M.; Ahn, C.H.; Jeon, J.S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Alabd, A.; Cheng, H.; Ahmad, M.; Wu, X.; Peng, L.; Wang, L.; Yang, S.; Bai, S.; Ni, J.; Teng, Y. ABRE-BINDING FACTOR3-WRKY DNA-BINDING PROTEIN44 module promotes salinity-induced malate accumulation in pear. Plant Physiol. 2023, 192, 1982–1996. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chao, J.; Li, Z.; Sun, Y.; Aluko, O.O.; Wu, X.; Wang, Q.; Liu, G. MG2C: A user-friendly online tool for drawing genetic maps. Mol. Hortic. 2021, 1, 16. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Kang, Z.; Xu, Z.; Liu, Q. Robust deep k-means: An effective and simple method for data clustering. Pattern Recognit. 2021, 117, 107996. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).