Abstract
Peptide antigens derived from tumors have been observed to elicit protective immune responses, categorized as either tumor-associated antigens (TAAs) or tumor-specific antigens (TSAs). Subunit cancer vaccines incorporating these antigens have shown promise in inducing protective immune responses, leading to cancer prevention or eradication. Over recent years, peptide-based cancer vaccines have gained popularity as a treatment modality and are often combined with other forms of cancer therapy. Several clinical trials have explored the safety and efficacy of peptide-based cancer vaccines, with promising outcomes. Advancements in techniques such as whole-exome sequencing, next-generation sequencing, and in silico methods have facilitated the identification of antigens, making it increasingly feasible. Furthermore, the development of novel delivery methods and a deeper understanding of tumor immune evasion mechanisms have heightened the interest in these vaccines among researchers. This article provides an overview of novel insights regarding advancements in the field of peptide-based vaccines as a promising therapeutic avenue for cancer treatment. It summarizes existing computational methods for tumor neoantigen prediction, ongoing clinical trials involving peptide-based cancer vaccines, and recent studies on human vaccination experiments.
1. Introduction
Cancer vaccines work by instructing the immune system to recognize tumor antigens as foreign [1]. They can be used prophylactically to stop or prevent tumor development or therapeutically to treat patients who have already been diagnosed with cancer [2]. FDA-approved cancer vaccines are presented in Table 1.

Table 1.
FDA-approved cancer vaccines.
Antigen selection strategy is the most import stage in the cancer vaccine development process. The ideal antigen should only be expressed by cancer cells and be highly immunogenic. The chosen antigen should also be present on all cancer cells which play a crucial role in cancer cell survival and protect against immune escape by mutations or loss of antigens in tumor cells. In order to induce an immune response, an antigen needs to be processed, presented to, and recognized by the immune cells [3]. Antigen processing and presentation refer to the processes that occur within a cell and result in fragmentation (proteolysis) of proteins, association of the fragments with MHC (major histocompatibility complex) molecules, and expression of the peptide–MHC molecules (p-MHC) at the cell surface where they can be recognized by the TCR (T-cell receptor) on a T cell (Figure 1) [4]. The TCR can recognize an antigen only in the form of a peptide bound to an MHC molecule on a human cell’s surface. The antigens recognized by T cells are peptides that arise from the breakdown of macromolecular structures, the unfolding of individual proteins, and their cleavage into short fragments through antigen processing. The peptides must be bound by an MHC molecule and presented at the cell surface. There are two classes of MHC molecules, MHC-I (MHC-Class I) and MHC-II (MHC-Class II). MHC-I molecules are specialized for the presentation of peptides derived from endogenous proteins (intracellular antigens) to the TCR of CD8+ T cells (CD8-expressing T cells) whilst MHC-II molecules are specialized for the presentation of extracellular antigens to the TCR of CD4+ T cells (CD4-expressing T cells) [5,6,7]. All T-cell epitopes can bind to MHC molecules; however, not all MHC binders are T-cell epitopes [8]. Feltkamp et al. showed that the binding affinity to MHC class I molecules is required but does not ensure T-cell immune responses [9]. Furthermore, factors other than MHC binding affinity are found to strongly influence T-cell immune responses, compared with the only moderate influence of MHC binding affinity [10]. Non-immunogenic epitopes may result from the following reasons: (a) p-MHC is truly unrecognized by TCR, (b) peptides are not presented by MHC, and (c) negative selection/clonal presentation is induced by excessive similarity to autologous peptides [11,12]. It has been estimated that only 1 in 200 potential peptide antigenic determinants will bind to a given MHC class I molecule with sufficient strength to elicit an immune response [13].

Figure 1.
Illustration of T-cell recognition. APC presents the antigen to the T cell in a complex with an MHC molecule. TCR of the T cell recognizes the complex. Other co-stimulatory signals also take part in the process. This figure was produced with the assistance of Servier Medical Art (https://smart.servier.com).
Several mechanisms alter the immune response against cancer cells, one of which is antigen spreading [14]. This phenomenon refers to the expansion of the immune responses from initially targeting one specific antigen (epitope) associated with the tumor to recognizing and attacking other antigens expressed by the same tumor or associated with it. As the immune system targets and eliminates tumor cells expressing the primary antigen, other tumor cells with different antigens may survive. As the tumor evolves, the immune system may begin to recognize these newly emerged antigens, leading to a secondary immune response. This secondary response involves the activation and expansion of immune cells specific to these new antigens. Epitope spreading can lead to a more robust and effective immune response against the tumor. By targeting multiple antigens, the immune system can overcome tumor heterogeneity and eliminate a broader range of cancer cells. The endogenous response of each individual patient against tumor antigens is considered a key factor in many current anticancer treatments [15]. These responses involve the recognition and elimination of cancerous or transformed cells by the immune system, particularly through the actions of cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells. However, tumor cells employ defense mechanisms against the immune response, including restricting antigen recognition, immune system inhibition, and inducing T-cell exhaustion [16]. Hence, understanding the mechanisms underlying these responses is essential for developing immunotherapeutic strategies to enhance anti-tumor immunity and improve outcomes for cancer patients.
2. Types of Tumor Antigens
Peptide cancer vaccines are based on the epitope peptides that can elicit humoral and cellular immune responses targeting tumor-associated antigens (TAAs) or tumor-specific antigens (TSAs) [17]. The main characteristics of TAAs and TSAs are presented in Table 2.

Table 2.
Characteristics of tumor antigens. WT1, Wilms tumor 1; p53, tumor protein 53; PSA, prostate-specific antigen; gp100, glycoprotein 100; NY-ESO-1, New York esophageal squamous cell carcinoma 1; MAGE-A3, melanoma antigen A3; HPV-E6/E7, human papillomavirus; HTLV-1, human T-cell lymphotropic virus type 1.
TAAs are derived from self-antigens, meaning they are present in both cancerous and non-cancerous cells, but they may be expressed at higher levels or have altered post-translational modifications in cancer cells. As a result, the immune system can recognize these antigens as potential targets for immune responses specifically directed against cancer cells. TAAs play a crucial role in the development of cancer vaccines. Peptides derived from TAAs can be used as antigens in cancer vaccines to stimulate an immune response specifically targeting tumor cells. By presenting TAA-derived peptides to immune cells, such as T cells, the vaccines aim to activate and mobilize the immune system to selectively attack and eliminate cancer cells bearing these antigens. A major disadvantage of TAAs is that since they are self-antigens in their nature, a potential break of immune self-tolerance may occur, thus leading to autoimmune reactions. There are three main groups of TAAs. Overexpressed antigens are a large and diverse group that includes any protein found at increased levels in tumors compared with normal healthy cells and tissues [18]. One such widely studied antigen is Wilms tumor 1 (WT1). It is a well-known TAA expressed in various types of cancers, including acute myeloid leukemia (AML). It has been targeted in T-cell receptor (TCR) gene therapy and shown to prevent AML relapse post-transplant [19]. Another example is MART-1 (Melan-A). That antigen is a TAA predominantly expressed in melanoma. It has been studied extensively as a target for T-cell-based immunotherapies in melanoma patients [20]. The second group of TAAs consists of differentiation antigens [20]. They are selectively expressed by the cell lineage from which malignant cells evolved. One representative example, the prostate-specific antigen, has highly restricted distribution and is expressed in normal epithelial cells of the prostate gland, as well as prostate carcinomas [21]. These two kinds of tumor antigens are currently proved not to fit well in the landscape of cancer vaccines, with high immunological tolerance and toxicity that threaten the efficacy and safety of cancer vaccines for patients [20]. In regard to this, the third group, cancer germline antigens (CGAs, also known as cancer/testis antigens (CTAs)), stand aside among other TAAs. Besides their nonspecific tumor expression, they have only been found to be expressed in immune-privileged tissues. Thus, their aberrant expression in tumors makes them highly immunogenic. In addition to segregation by tissue barriers, trophoblastic and male germ cells, which are a normal localization of CTAs, lack HLA class I molecules expression and therefore cannot present antigens to T cells [22,23,24]. NY-ESO-1 is a well-known CTA expressed in various tumor types, including melanoma, lung cancer, and ovarian cancer. It has been investigated as a potential target for cancer vaccines and adoptive T-cell therapies [25]. MAGE-A3 (melanoma antigen A3) is another CTA that has been extensively studied as a target for immunotherapeutic approaches in multiple cancer types, including non-small cell lung cancer and melanoma [26].
Tumor-specific antigens (TSAs, also known as neoantigens) are a subset of antigens that are uniquely expressed by cancer cells but not found in normal cells. These antigens result from tumor-specific genetic alterations, such as somatic mutations, chromosomal rearrangements, or viral oncogene expression. They are highly specific to each individual. TSAs play a crucial role in cancer immunotherapy, particularly in the development of targeted cancer vaccines. Due to their tumor-specific expression, TSAs are attractive targets for therapeutic interventions aimed at stimulating an immune response against cancer cells while sparing healthy tissues, and due to their “non-self” features, they have a higher affinity to MHC molecules and T-cell receptors [27]. The specificity and selectivity of TSAs as targets for therapeutic interventions are their main advantages over TAAs. The field of neoantigen research is rapidly evolving, and new discoveries are continually being made. One of the most comprehensively studied neoantigens is KRAS. KRAS is a frequently mutated gene in various cancers, including pancreatic cancer, colorectal cancer, and lung adenocarcinoma. The G12V mutation in KRAS results in a constitutively active protein that drives oncogenic signaling pathways. This mutation generates a neoantigen that has been targeted in preclinical and clinical studies using personalized cancer vaccines [28] and adoptive T-cell therapies [29]. BRAF is another commonly mutated gene in cancers such as melanoma and colorectal cancer. The V600E mutation in BRAF leads to constitutive activation of the MAPK signaling pathway. This mutation generates a neoantigen that has been targeted using immune checkpoint inhibitors [30], adoptive T-cell therapies [31], and peptide-based vaccines [32], with promising results in clinical trials. EGFRvIII is a mutant variant of the epidermal growth factor receptor (EGFR) that is frequently observed in glioblastoma, a type of brain cancer. EGFRvIII arises from an in-frame deletion of exons 2-7, resulting in a truncated, constitutively active protein. This alteration creates a neoantigen that has been targeted in clinical trials using peptide-based vaccines [33] and adoptive T-cell therapies [34].
Also included in the group of TSAs are oncogenic viral antigens. In some cases, tumor cells may express antigens derived from viral proteins that are only present in infected cells. These antigens are attractive targets for immunotherapy because they are selectively expressed by tumor cells and can induce immune responses against both the virus and the tumor. Examples include the viral oncoproteins E6 and E7 derived from high-risk human papillomavirus (HPV) strains in HPV-associated cancers, such as cervical cancer [35].
3. Neoantigen Discovery and Selection for Peptide-Based Cancer Vaccines
Vaccination with TAAs has encountered limited efficacy, primarily attributed to their low tumor specificity, as these antigens are not exclusive to tumor cells [36]. Moreover, their utilization as cancer treatment candidates is hindered by suboptimal immune responses and the potential risk of autoimmune reactions. In contrast, TSAs, being exclusively expressed in tumor cells, offer a promising alternative. TSAs enable the development of personalized vaccines, providing an opportunity for selective tumor eradication while minimizing damage to healthy tissue.
The process of neoantigen discovery and selection for cancer treatment involves several steps to identify and prioritize potential neoantigens that can be targeted for immunotherapy [37] (Figure 2). The first step is to obtain tumor tissue or tumor-infiltrating lymphocytes (TILs) from the patient. The tumor DNA or RNA is then sequenced using techniques like whole-exome sequencing (WES) [38] or RNA sequencing (RNA-seq) [39] to identify genetic alterations and transcriptomic changes in the tumor cells. The sequencing data are analyzed to identify somatic mutations, including single-nucleotide variants (SNVs), insertions/deletions, and gene fusions, which result in the generation of potential neoantigens. These mutations are compared to the patient’s germline DNA to filter out non-tumor-specific alterations. In silico algorithms are used to predict and prioritize neoantigen candidates based on several factors [40]. These include binding affinity to human leukocyte antigen (HLA) molecules, processing and presentation capabilities, and potential immunogenicity.
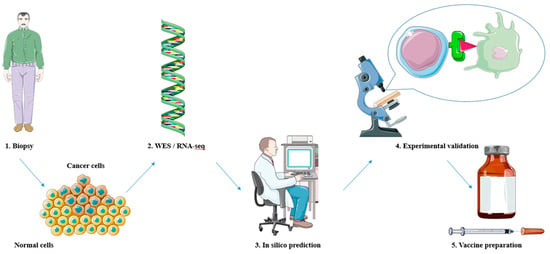
Figure 2.
Neoantigen discovery and selection for peptide-based cancer vaccines. 1. Obtaining tumor tissue via biopsy. 2. Whole-exome sequencing (WES) or RNA sequencing (RNA-seq) on tumor DNA or RNA to identify mutations. 3. Neoantigen prediction and prioritization through in silico methods. 4. Experimental validation of predicted neoantigens. 5. The most promising neoantigens are included in vaccine preparations and carried on to preclinical and clinical trials. The figure was produced with the assistance of Servier Medical Art (https://smart.servier.com).
Tumors typically harbor an average of over 100,000 mutations, emphasizing the vast number of potential neoantigens that may arise [41]. However, evaluating all these neoantigens experimentally is neither feasible nor practical. In response, in silico prediction algorithms play a crucial role by significantly reducing the pool of neoantigens for experimental validation. These algorithms utilize various information sources, such as amino acid sequences binding to the HLA, sequences recognized by the TCR, or the significance of specific amino acids in stabilizing the HLA–peptide–TCR complex [42]. They rely on principles of sequence alignment [43], machine learning [44], and artificial intelligence [45] to predict neoantigens accurately and efficiently. The latest advancements in neoantigen prediction primarily focus on supervised learning techniques, leveraging known data of both neoantigens and non-neoantigens. These approaches involve building predictive models capable of distinguishing between these two classes. Through supervised learning, these models are trained on labeled datasets and subsequently evaluated using validation datasets to measure their predictive accuracy, sensitivity, specificity, and other performance metrics [46]. Once trained and validated, these models are then deployed to predict candidate neoantigens efficiently and reliably.
Various computational tools (Table 3) are commonly used for neoantigen prediction. HLA typing of the patient is performed to determine the specific HLA alleles expressed by their immune cells [47]. The process is performed by in silico prediction tools [48]. This information is crucial for predicting neoantigen presentation and the subsequent immune response. The predicted neoantigens are further validated through experimental methods, such as peptide–HLA binding assays [49] or mass spectrometry [50], to confirm their binding affinity to HLA molecules and their presentation on the tumor cell surface. These assays help refine the list of potential neoantigens and prioritize those with high immunogenicity and HLA binding. Selected neoantigens can be further evaluated through functional assays to assess their ability to elicit immune responses. This may involve in vitro studies [51] using T cells from the patient or healthy donors, assessing T-cell reactivity against the neoantigen-expressing tumor cells. In vivo studies [52] using animal models or patient-derived xenograft (PDX) models can also provide insights into neoantigen-specific immune responses and their impact on tumor growth. Finally, the most promising neoantigens are considered for clinical translation, such as the development of personalized cancer vaccines or adoptive cell therapies. These therapies involve the administration of neoantigen-specific vaccines [53] or the genetic modification of patient-derived T cells to express neoantigen-specific TCRs [54] or chimeric antigen receptors (CARs) [55].

Table 3.
Summary of existing computational methods for tumor neoantigen prediction.
Personalized peptide-based cancer vaccines are still in the early stages of development and are primarily being tested in clinical trials (Table 4). Numerous challenges persist in the advancement and extensive application of this type of immunotherapy [68]. The protracted process encompassing cancer genome sequencing, neoantigen detection, and validation of immunogenicity contributes to the substantial financial burden associated with personalized vaccines, impeding their feasibility for broad implementation in cancer patients. The emergence of enhanced bioinformatics tools tailored for neoantigen characterization, deeper insights into tumor immunology, and advancements in vaccine formulation and administration techniques are imperative for the realization and utilization of innovative neoantigen-based cancer vaccines.

Table 4.
Peptide-based cancer vaccines currently in active, not recruiting clinical trials (participants are receiving an intervention or being examined, but new participants are not currently being recruited or enrolled). Variants of HPV and HBV vaccines are not included.
4. Studies with Tumor Antigens
To date, there are few confirmed neoantigens in humans. While there are abundant data from lab tests and animal studies, applying these findings to human cancer patients is complex due to differences in our immune systems. Yet, recent human vaccination experiments bring hope that soon there will be more comprehensive studies. The studies on cancer epitopes that have been validated in vivo, along with other studies providing valuable insights into this field of research, are presented in Table 5.

Table 5.
Studies regarding advancements made in the research field of cancer peptide vaccination.
Most studies focus on finding patient-specific neoantigens and checking the immune response against them in real patients. Melanoma is a primary focus due to its high mutation rate [91], followed by lung cancer [92]. However, pancreatic cancer patients with longer survival times tend to have stronger T-cell activity and fewer immunogenic mutations, highlighting the importance of neoantigen quality in therapy [93]. While these three types of cancer are the primary focus of human immunization experiments, other cancers investigated in these studies typically have lower tumor mutational burdens, and neoantigen vaccinations against them are mostly reported as single-case studies.
4.1. Melanoma Studies
Khong and colleagues [69] investigated a vaccination approach in 37 patients with metastatic melanoma. They used epitopes from the NY-ESO-1 antigen presented by HLA-A0201 and/or HLA-DPbeta104 molecules. Interestingly, the vaccination not only targeted NY-ESO-1 but also triggered responses against other melanoma antigens, suggesting a broader anti-tumor immune reaction. This phenomenon, known as antigen spreading, was further explored by Corbiere et al. [70], who studied the mechanisms behind melanoma metastasis regression post-vaccination. They found that antigen-specific T cells could recognize various tumor antigens beyond the original targets.
Carreno et al. [71] demonstrated an increase in natural neoantigen-specific immunity and identified new HLA class I-restricted neoantigens in advanced melanoma patients after receiving a dendritic cell vaccine loaded with neoantigens. They confirmed the presentation of these neoantigens in human melanoma using mass spectrometry, revealing previously unrecognized targets.
Cohen et al. [72] developed a method to isolate neoantigen-specific T cells from both tumor and peripheral blood. They identified nine mutated epitopes from five out of eight patients using whole-exome sequencing and peptide–MHC tetramers. They successfully isolated mutation-reactive T cells from peripheral blood, showing reactivity towards eight out of the nine identified epitopes.
Linette et al. [73] explored how intratumoral heterogeneity (ITH) influences T-cell immunity in melanoma. They found clonal and subclonal neoantigens in multiple metastases from four melanoma patients. Although CD8+ T-cell responses against these neoantigens were limited to a subset with a restricted TCR repertoire, mature dendritic cell vaccination using tumor-encoded amino acid-substituted (AAS) peptides revealed diverse CD8+ T-cell responses specific to neoantigens. These findings highlight the importance of therapeutic vaccination because many T-cell clones that can recognize melanoma neoantigens are in a naive state and may not be easily identified.
Hu et al. [74] studied personalized neoantigen vaccines in eight patients with surgically resected stage IIIB/C or IVM1a/b melanoma. They observed tumor infiltration by neoantigen-specific T-cell clones post-vaccination, along with epitope spreading.
Sahin et al. [75] explored personalized RNA-based vaccines targeting distinct tumor mutations in 13 patients. By administering tumor-specific neoantigens, they aimed to induce a comprehensive immune response against cancer cells. These personalized RNA mutanome vaccines hold promise for revolutionizing cancer treatments through their tailored approach.
4.2. Pancreatic Carcinoma Studies
The study by Abou-Alfa et al. [76] aimed to test a vaccine targeting KRAS codon 12 mutations, often seen in pancreatic adenocarcinoma, as potential tumor-specific neoantigens. They vaccinated 24 patients who had their pancreatic cancer removed and showed KRAS mutations with a 21-mer peptide vaccine containing their tumor’s KRAS mutation. Results showed the vaccine was safe, with no severe side effects, but it did not trigger strong immune responses against the KRAS mutation. Median recurrence-free survival was 8.6 months, and median overall survival was 20.3 months, indicating limited effectiveness.
Dillard et al. [77] explored TCR redirected T cells as a therapy for solid tumors, focusing on KRAS mutations. They identified and studied four specific KRAS-targeting T cells from a pancreatic cancer patient who had received a vaccine with mutated KRAS peptides. These T cells were able to recognize and attach to cells presenting KRAS peptides, showing promising potential. Some T cells could recognize multiple KRAS mutations, suggesting they could be effective against different tumor types. The study also confirmed that processed KRAS peptides were effectively presented to T cells, supporting the idea that KRAS mutations could be good targets for cancer treatment.
Sonntag et al. [78] reported on a patient with metastasized pancreatic ductal carcinoma who received personalized neoantigen-derived multipeptide vaccines as treatment. These vaccines targeted the patient’s unique neoantigens and were given as therapy. The outcome showed that the four-peptide vaccine triggered a strong and lasting immune response against the neoantigens. Importantly, this immune response was associated with a prolonged period of clinical remission for the patient.
Chen et al. [79] conducted a study to investigate personalized neoantigen-based immunotherapy for advanced pancreatic cancer patients who did not respond to standard treatments. They designed personalized cancer vaccines from neoantigens, with each patient receiving up to 20 neoantigen peptides. These vaccines, called iNeo-Vac-P01, were given to seven patients with low tumor mutation burden. The study found that iNeo-Vac-P01 was safe, with no severe side effects. Patients treated with iNeo-Vac-P01 had a mean overall survival of 24.1 months compared to 8.3 months without the vaccine. Progression-free survival averaged 3.1 months. Notably, one patient who survived for 21 months after vaccination showed a significant increase in antigen-specific TCR clones afterward.
4.3. Glioblastoma Studies
Keskin et al. [80] investigated using personalized neoantigen vaccines to treat glioblastoma, a type of brain tumor. They found that patients who did not receive dexamethasone, a powerful corticosteroid often used for brain swelling, had immune responses both in the body and within the tumor. These responses involved CD4+ and CD8+ T cells targeting neoantigens. However, despite these immune reactions, all ten patients in the study had their tumors come back and eventually passed away due to the disease getting worse. This suggests that while the immune responses were triggered, they faced significant hurdles in effectively fighting the tumors.
Johanns et al. [81] conducted a study primarily to assess the effectiveness of a personalized vaccine treatment plan. This plan involved giving the patient an autologous tumor lysate-dendritic cell vaccine (DCVax-L) first, followed by a neoantigen-based synthetic long peptide vaccine (GBM.PVax). The patient they studied was a 66-year-old woman diagnosed with GBM. After receiving the vaccines, they examined the patient’s blood and tumor-infiltrating lymphocytes (TILs). They found clear responses from both CD8+ and CD4+ T cells specifically targeting the neoantigens induced by the peptide vaccine. By analyzing the tumor’s genetic and transcriptomic characteristics before and after treatment, they discovered evidence of how the tumor evolved and possible ways it evades the immune system. This highlights the complexity of interactions between tumors and the immune system in GBM.
4.4. Lung Cancer Studies
Li et al. [82] presented a detailed case report of an Asian lung cancer patient who received personalized peptide vaccination (PPV) targeting specific neo-epitopes. After starting PPV, the patient showed significant clinical improvement with rapid tumor shrinkage. Impressively, within just three to four months, there was a substantial reduction in tumor size and positive clinical outcomes. Importantly, the personalized neo-epitope vaccination was well tolerated, with no significant treatment-related side effects. These findings highlight the potential effectiveness and safety of personalized neo-epitope vaccination as a treatment option for lung cancer patients. Further research and clinical trials are needed to confirm these results.
Li et al. [83] conducted a phase I clinical trial to evaluate the feasibility, safety, and effectiveness of personalized Neoantigen (NeoAg) peptide vaccination (PPV) in patients with stage III/IV non-small cell lung cancer (NSCLC) and EGFR mutations. Out of 29 enrolled patients, 24 received the vaccine. The median progression-free survival was 6.0 months, and the overall survival was 8.9 months. Notably, within 3-4 months of starting PPV, seven patients showed objective clinical responses, including one complete response. All responders had EGFR-mutated tumors, with four patients also receiving EGFR tyrosine kinase inhibitor (TKI) therapy alongside PPV. Immune monitoring revealed that five responders developed T-cell responses to EGFR NeoAg peptides due to the vaccine. Moreover, four responding patients had increased frequencies of neoantigen-specific CD8+ T cells in their blood during PPV, indicating T-cell responses were triggered. Combining PPV with EGFR inhibitor therapy was well tolerated, suggesting personalized neoantigen-based vaccination could be a promising immunotherapy for NSCLC patients with EGFR mutations.
4.5. Gastrointestinal Cancers Studies
Tran et al. [84] utilized next-generation sequencing and high-throughput immunologic screening to examine tumor-infiltrating lymphocytes (TILs) from 10 patients with metastatic gastrointestinal cancers. Their findings indicated that TILs from 9 out of 10 patients contained CD4+ and/or CD8+ T cells recognizing neo-epitopes derived from mutations in the patient’s own tumor. In total, they identified 17 immunogenic peptide neo-epitopes. This suggests that most patients with these cancers harbor T cells targeting specific tumor mutations, offering promise for tailored vaccines or cell therapies against common epithelial cancers.
4.6. Epithelial Cancer Studies
Cafri et al. [85] used a highly sensitive in vitro stimulation and cell enrichment technique to look for memory T cells in the blood of six patients with metastatic cancer. They found memory T cells capable of targeting both unique and shared mutations present in the blood of people with epithelial cancer. While past studies focused on T cells in tumors, this study shows specific T cells exist in the blood, offering a non-invasive way to find and isolate cells or receptors that react to mutations. This discovery opens up new possibilities for using these T cells to develop personalized immunotherapies against epithelial cancers.
Zeng et al. [86] conducted a case study to explore personalized neoantigen-based immunotherapy for treating collecting duct carcinoma (CDC) of the kidney. They focused on an Asian patient with metastatic CDC, which had progressed despite previous Sorafenib treatment. The researchers identified 13 specific neoantigens in the patient’s tumor based on their genetic profile. Using these neoantigens, they developed a personalized treatment plan involving long peptide vaccines and neoantigen-reactive T cells (NRTs). After six cycles of this treatment, the patient showed significant improvements, with stable disease status in tumor burden and relief from bone pain. Tests on blood cells collected after treatment revealed immune responses to 12 of the 13 neoantigens. Biopsy samples taken from CDC sites after three months showed a reduction in mutant allele frequency related to 92% of the neoantigens, suggesting potential elimination of tumor cells carrying these specific neoantigens.
4.7. Multiple Tumor Studies
Ott et al. [87] conducted a phase Ib clinical trial to evaluate a personalized neoantigen-based vaccine called NEO-PV-01, combined with the immune checkpoint inhibitor PD-1 blockade (Nivolumab). The trial involved 82 patients with advanced melanoma, non-small cell lung cancer, or bladder cancer. Patients received NEO-PV-01 alongside Nivolumab. After vaccination, all patients showed new neoantigen-specific CD4+ and CD8+ T-cell responses. These T cells were cytotoxic and capable of entering the tumor to kill cells. The study also observed epitope spreading, where T-cell responses expanded to neoantigens not included in the vaccine, potentially boosting the anti-tumor immune response. Importantly, the combination therapy was safe, with no significant treatment-related adverse events reported.
4.8. Breast Tumor Studies
Sha et al. [88] aimed to investigate a neoantigen-based vaccine targeting tumor-specific mutated peptides from gene mutations. They studied a 57-year-old woman with a phyllodes tumor who had undergone left breast mass resection. Through high-throughput gene sequencing and detailed analysis, they tailored the vaccine to target the patient’s unique neoantigens. Remarkably, the vaccine led to a complete pathological response in lung metastasis, demonstrating the safety and effectiveness of this immunization approach.
Li et al. [89] developed neoantigen DNA vaccines by combining elongated epitopes with a mutant ubiquitin variant. This fusion aimed to improve antigen processing and presentation. Epitopes were identified using various computational, laboratory-based, and live organism techniques. The study found that the optimized DNA vaccines were immunogenic, generating strong neoantigen-specific immune responses in mice. These immune reactions were comparable to those induced by synthetic long peptide vaccines targeting the same neoantigens. Additionally, when used alongside immune checkpoint blockade therapy, the optimized DNA vaccines showed potential in triggering anti-tumor immunity in preclinical models.
4.9. T-Cell Recognition
In their study, Smith et al. [90] showed that common modifications used to improve peptide binding can unexpectedly affect T-cell recognition. They focused on the recognition of both the wild-type (WT) and an anchor-modified version of the well-known gp100209 tumor antigen (ITDQVPFSV) presented by the MHC molecule HLA-A2, using three different gp100209-specific TCRs. Interestingly, they found significant variations in functional responses, highlighting the need for caution when using and interpreting outcomes from anchor-modified peptides. These findings are important because they shed light on the unpredictability of tumor neoantigen immunogenicity, especially in specific scenarios.
The recent increase in studies on peptide-based cancer vaccination highlights the growing interest in this field. Human immunization experiments, which are common in many of these studies, are crucial for assessing the safety and effectiveness of peptide vaccines and evaluating the immune response to potential neoantigens. Most of these studies have emerged in the past few years, showing a clear trend of progress in research. However, human immunization experiments involving neoantigens are still relatively rare. Only a small number of neoantigens have been tested in humans so far, making it challenging to develop accurate prediction tools for immunogenicity. Additionally, many types of cancers have not yet been studied using human immunization experiments. Moreover, many studies involve single-case reports, often with patients in advanced stages of the disease. This makes it difficult to track disease progression or regression after vaccination within a limited timeframe. Despite these challenges, the collective efforts in this field offer a promising future for cancer immunotherapy.
5. Databases Containing Human Tumor Antigens
To develop cancer vaccines with optimal safety and efficacy, a thorough investigation of diverse human tumor antigens is imperative. In recent times, remarkable strides have been achieved in this domain, unveiling a plethora of cancer antigens that undergo rigorous validation. This growing repository of validated antigens augments the likelihood of identifying promising candidates for vaccine formulation. Furthermore, the utilization of computational antigen prediction algorithms necessitates access to high-quality experimental data, which is crucial for their effective training. Herein, we delineate some of the prominent human tumor antigen databases currently available, pivotal in facilitating this critical research endeavor.
The Immune Epitope Database (IEDB, https://www.iedb.org/) [94] is a freely available resource funded by NIAID. Although not specific to cancer, the IEDB contains a vast collection of experimentally validated epitopes from various sources, including tumor-associated antigens. As a companion site to the IEDB, The Cancer Epitope Database and Analysis Resource (CEDAR, https://cedar.iedb.org/) [95] provides a freely accessible, comprehensive collection of cancer epitope and receptor data curated from the literature. The Cancer Antigenic Peptide Database (CAPDb, https://caped.icp.ucl.ac.be/) [96] is a comprehensive database of experimentally validated tumor antigens, which includes information on the antigenic peptides presented on major histocompatibility complex (MHC) molecules. The Tumor-Specific Neoantigen Database (TSNAdb v1.0, http://biopharm.zju.edu.cn/tsnadb) [97] is based on pan-cancer immunogenomic analyses of somatic mutation data and human leukocyte antigen (HLA) allele information for 16 tumor types with 7748 tumor samples from The Cancer Genome Atlas (TCGA) and The Cancer Immunome Atlas (TCIA). NEPdb (http://nep.whu.edu.cn/) [98] is a database of T-cell experimentally validated neoantigens and pan-cancer predicted neo-epitopes for cancer immunotherapy. It contains more than 17,000 validated human immunogenic neoantigens and ineffective neo-epitopes within HLAs via curating the published literature with their semi-automatic pipeline. Furthermore, NEPdb also provides pan-cancer level predicted HLA-I neo-epitopes derived from 16,745 shared cancer somatic mutations, using state-of-the-art predictors. The Catalogue of Somatic Mutations in Cancer (COSMIC, https://cancer.sanger.ac.uk/cosmic) [99] is the world’s largest and most comprehensive resource for exploring the impact of somatic mutations in human cancer. It also provides information on tumor antigens resulting from these mutations. A comprehensive online database (TANTIGEN 2.0, http://projects.met-hilab.org/tadb) [100] catalogs 4,296 antigen variants from 403 unique tumor antigens and more than 1500 T-cell epitopes and HLA ligands. It also contains validated TCR sequences specific for cognate T-cell epitopes and tumor antigen gene/mRNA/protein expression information in major human cancers extracted by the Human Pathology Atlas. The cancer antigen database (CAD, http://cad.bio-it.cn/) [101] is a collection of verified cancer antigen peptides crucial to the development of neoantigen-based cancer vaccines. The role of each dataset for algorithm improvement in cancer antigen research, especially neoantigen prediction, is also discussed. A database of T-cell-defined human tumor antigens was developed by Vigneron et al. (www.cancerimmunity.org/peptide) [102]. It compiles all human antigenic peptides described in the literature that fulfill a set of strict criteria needed to ascertain their actual “tumor antigen” nature.
6. Summary and Future Perspective
The accumulation of knowledge over the past 15 years concerning tumor immunogenicity suggests a promising future for cancer immunotherapy overall. This progress is evident in the development of monoclonal antibodies, immune checkpoint inhibitors, and cancer vaccines. While the current limitation revolves around the scarcity of in vivo validated data concerning tumor immunogenicity, strides are being made as more human immunization experiments with peptide antigens are conducted. Experimentally validated immunogens will play a crucial role in the development of more robust in silico prediction algorithms, thereby facilitating cancer neoantigen discovery—a process that sets off a positive feedback loop. Ultimately, these advancements hold the potential to pave the way for the first FDA-approved therapeutic peptide cancer vaccine in the near future.
Peptide-based neoantigen cancer vaccines represent a promising frontier in the realm of cancer immunotherapy, owing to their personalized nature and capacity to elicit robust and enduring anti-tumor responses. These vaccines are generally acknowledged for their safety and tolerability in clinical settings. Encouraging outcomes have emerged from clinical investigations, revealing extended overall survival and disease control among patients receiving personalized neoantigen vaccines. Their compatibility with other therapeutic modalities, such as immune checkpoint inhibitors or adoptive T-cell therapies, further bolsters the overall anti-tumor response and augments treatment efficacy. Neoantigen-based vaccines have been probed across various cancer types, rendering them versatile with potential applicability in diverse malignancies. Nevertheless, inherent limitations persist. Tumor antigen heterogeneity and challenges in identification and targeting, as well as limited coverage, diminish the vaccines’ overall efficacy. Ongoing challenges in optimizing vaccine formulations, encompassing adjuvant selection and delivery techniques, may impact their potency. Immune evasion and suppression mechanisms further compromise therapeutic responses. Moreover, the personalized nature of these vaccines incurs higher costs and potential restricted access for certain patients, limiting their widespread adoption. Although initial clinical trials have shown promise, their long-term efficacy and broader benefits in larger patient cohorts warrant further evaluation. Despite these constraints, continual research and progress in cancer immunotherapy strive to address these hurdles, upholding peptide-based neoantigen cancer vaccines as a promising avenue for personalized and targeted cancer treatment. As investigations progress, these vaccines hold substantial potential in enhancing cancer treatment outcomes and patient survival.
Author Contributions
Conceptualization, S.S. and I.D.; methodology, I.D.; software, S.S.; validation, S.S.; formal analysis, S.S.; investigation, S.S.; resources, S.S.; data curation, S.S.; writing—original draft preparation, S.S.; writing—review and editing, I.D.; visualization, S.S.; supervision, I.D.; project administration, I.D.; funding acquisition, I.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Bulgarian national plan for recovery and resilience through the Bulgarian National Science Fund, grant number BG-RRP-2.004-0004-C01, and by the Science and Education for Smart Growth Operational Program and co-financed by the European Union through the European Structural and Investment funds (Grant No. BG05M2OP001-1.001-0003).
Acknowledgments
During the preparation of this work, the authors used ChatGPT for English editing. After using this tool, the manuscript underwent a comprehensive editing service. The authors take full responsibility for the content of the publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| AAS | Amino acid-substitution |
| AML | Acute myeloid leukemia |
| APC | Antigen-presenting cell |
| BRAF | B-Raf proto-oncogene |
| CARs | Chimeric antigen receptors |
| CD4+ T cells | CD4-expressing T cells |
| CD8+ T cells | CD8-expressing T cells |
| CDC | Collecting duct carcinoma |
| CGA | Cancer germline antigen |
| CTA | Cancer/testis antigen |
| CLT | Cytotoxic T lymphocyte |
| EGFR | Epidermal growth factor receptor |
| EGFRvIII | Epidermal growth factor receptor variant III |
| gp100 | Glycoprotein 100 |
| HLA | Human leukocyte antigen |
| HPV-E6/E7 | Human papillomavirus oncoproteins E6/E7 |
| HTLV-1 | Human T-cell lymphotropic virus type 1 |
| ITH | Intratumoral heterogeneity |
| KRAS | Kirsten rat sarcoma virus |
| MAGE-A3 | Melanoma antigen-A3 |
| MAPK | Mitogen-activated protein kinases |
| MART-1 | Melanoma antigen recognized by T cells 1 |
| mDC | Mature dendritic cell |
| MHC-I | MHC-Class I |
| MHC-II | MHC-Class II |
| MHC | Major Histocompatibility Complex |
| ML | Machine learning |
| NGS | Next generation sequencing |
| NK cells | Natural killer cells |
| NRT | Neoantigen-reactive T cell |
| NSCLC | Non-small cell lung cancer |
| NY-ESO-1 | New York esophageal squamous cell carcinoma 1 |
| PDX | Patient-derived xenograft |
| PFS | Progression-free survival |
| PSA | Prostate-specific antigen |
| PSL-DA | Partial least squares-Discriminant analysis |
| p-MHC | peptide-MHC complex |
| p53 | Tumor protein 53 |
| RNA-seq | RNA-sequencing |
| SCM | Scoring card method |
| SNV | single nucleotide variant |
| TAA | Tumor-associated antigens |
| TCR | T-cell receptor |
| TIL | Tumor-infiltrating lymphocyte |
| TKI | Tyrosine kinase inhibitor |
| TMB | Tumor mutational burden |
| TSA | Tumor-specific antigens |
| WES | whole-exome sequencing |
| WGS | Whole genome sequencing |
| WT1 | Wilms tumor 1 |
| WT | Wild-type |
References
- Tsung, K.; Norton, J.A. In situ vaccine, immunological memory and cancer cure. Hum. Vaccines Immunother. 2016, 12, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.J.; Burgess-Brown, N.A.; Jiang, S. Beyond just peptide antigens: The complex world of peptide-based cancer vaccines. Front. Immunol. 2021, 12, 696791. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.; Chan, T.A. Immunogenic peptide discovery in cancer genomes. Curr. Opin. Genet. Dev. 2015, 30, 7–16. [Google Scholar] [CrossRef]
- Dustin, M.L. The cellular context of T cell signaling. Immunity 2009, 30, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Soam, S.S.; Khan, F.; Bhasker, B.; Mishra, B.N. Prediction of MHC class I binding peptides using probability distribution functions. Bioinformation 2009, 3, 403–408. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bontrop, R.E. Comparative genetics of MHC polymorphisms in different primate species: Duplications and deletions. Hum. Immunol. 2006, 67, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, M.; Del Cid, N.; Rizvi, S.M.; Peters, L.R. MHC class I assembly: Out and about. Trends Immunol. 2008, 29, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Doneva, N.; Doytchinova, I.; Dimitrov, I. Predicting Immunogenicity Risk in Biopharmaceuticals. Symmetry 2021, 13, 388. [Google Scholar] [CrossRef]
- Feltkamp, M.C.; Vierboom, M.P.; Kast, W.M.; Melief, C.J. Efficient MHC class I-peptide binding is required but does not ensure MHC class I-restricted immunogenicity. Mol. Immunol. 1994, 31, 1391–1401. [Google Scholar] [CrossRef]
- Tenzer, S.; Wee, E.; Burgevin, A.; Stewart-Jones, G.; Friis, L.; Lamberth, K.; Chang, C.H.; Harndahl, M.; Weimershaus, M.; Gerstoft, J.; et al. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat. Immunol. 2009, 10, 636–646. [Google Scholar] [CrossRef]
- Klein, L.; Hinterberger, M.; Wirnsberger, G.; Kyewski, B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 2009, 9, 833–844. [Google Scholar] [CrossRef]
- Wang, G.; Wan, H.; Jian, X.; Li, Y.; Ouyang, J.; Tan, X.; Zhao, Y.; Lin, Y.; Xie, L. INeo-Epp: A Novel T-Cell HLA Class-I Immunogenicity or Neoantigenic Epitope Prediction Method Based on Sequence-Related Amino Acid Features. Biomed. Res. Int. 2020, 2020, 5798356. [Google Scholar] [CrossRef]
- Yewdell, J.W.; Bennink, J.R. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 1999, 17, 51–88. [Google Scholar] [CrossRef]
- Brossart, P. The Role of Antigen Spreading in the Efficacy of Immunotherapies. Clin. Cancer Res. 2020, 26, 4442–4447. [Google Scholar] [CrossRef] [PubMed]
- Baxevanis, C.N.; Perez, S.A. Endogenous immunity at the forefront of tumor dormancy. Future Sci. OA 2015, 1, FSO13. [Google Scholar] [CrossRef]
- Kim, S.K.; Cho, S.W. The Evasion Mechanisms of Cancer Immunity and Drug Intervention in the Tumor Microenvironment. Front. Pharmacol. 2022, 13, 868695. [Google Scholar] [CrossRef]
- Bright, R.K.; Bright, J.D.; Byrne, J.A. Overexpressed oncogenic tumor-self antigens. Hum. Vaccin. Immunother. 2014, 10, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Jaigirdar, A.; Rosenberg, S.A.; Parkhurst, M. A High-avidity WT1-reactive T-Cell Receptor Mediates Recognition of Peptide and Processed Antigen but not Naturally Occurring WT1-positive Tumor Cells. J. Immunother. 2016, 39, 105–116. [Google Scholar] [CrossRef]
- Fetsch, P.A.; Marincola, F.M.; Filie, A.; Hijazi, Y.M.; Kleiner, D.E.; Abati, A. Melanoma-associated antigen recognized by T cells (MART-1): The advent of a preferred immunocytochemical antibody for the diagnosis of metastatic malignant melanoma with fine-needle aspiration. Cancer 1999, 87, 37–42. [Google Scholar] [CrossRef]
- Zhao, Y.; Baldin, A.V.; Isayev, O.; Werner, J.; Zamyatnin, A.A., Jr.; Bazhin, A.V. Cancer Vaccines: Antigen Selection Strategy. Vaccines 2021, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Pinzani, P.; Lind, K.; Malentacchi, F.; Nesi, G.; Salvianti, F.; Villari, D.; Kubista, M.; Pazzagli, M.; Orlando, C. Prostate-specific antigen mRNA and protein levels in laser microdissected cells of human prostate measured by real-time reverse transcriptase-quantitative polymerase chain reaction and immuno-quantitative polymerase chain reaction. Hum. Pathol. 2008, 39, 1474–1482. [Google Scholar] [CrossRef]
- Bart, J.; Groen, H.J.; van der Graaf, W.T.; Hollema, H.; Hendrikse, N.H.; Vaalburg, W.; Sleijfer, D.T.; de Vries, E.G. An oncological view on the blood-testis barrier. Lancet. Oncol. 2002, 3, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Gjerstorff, M.F.; Kock, K.; Nielsen, O.; Ditzel, H.J. MAGE-A1, GAGE and NY-ESO-1 cancer/testis antigen expression during human gonadal development. Hum. Reprod. 2007, 22, 953–960. [Google Scholar] [CrossRef]
- Fiszer, D.; Kurpisz, M. Major histocompatibility complex expression on human, male germ cells: A review. Am. J. Reprod. Immunol. 1998, 40, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Al-Khadairi, G.; Roelands, J.; Hendrickx, W.; Dermime, S.; Bedognetti, D.; Decock, J. NY-ESO-1 Based Immunotherapy of Cancer: Current Perspectives. Front. Immunol. 2018, 9, 947. [Google Scholar] [CrossRef] [PubMed]
- Esfandiary, A.; Ghafouri-Fard, S. MAGE-A3: An immunogenic target used in clinical practice. Immunotherapy 2015, 7, 683–704. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Moderna and Merck Expand mRNA Cancer Vaccines Collaboration. Merck. Published 3 May 2018. Available online: https://www.merck.com/news/moderna-and-merck-expand-mrna-cancer-vaccines-collaboration/ (accessed on 8 December 2021).
- Ai, Q.; Li, F.; Zou, S.; Zhang, Z.; Jin, Y.; Jiang, L.; Chen, H.; Deng, X.; Peng, C.; Mou, N.; et al. Targeting KRASG12V mutations with HLA class II-restricted TCR for the immunotherapy in solid tumors. Front. Immunol. 2023, 14, 1161538. [Google Scholar] [CrossRef]
- Welti, M.; Dimitriou, F.; Gutzmer, R.; Dummer, R. Triple Combination of Immune Checkpoint Inhibitors and BRAF/MEK Inhibitors in BRAFV600 Melanoma: Current Status and Future Perspectives. Cancers 2022, 14, 5489. [Google Scholar] [CrossRef]
- Veatch, J.R.; Lee, S.M.; Fitzgibbon, M.; Chow, I.T.; Jesernig, B.; Schmitt, T.; Kong, Y.Y.; Kargl, J.; Houghton, A.M.; Thompson, J.A.; et al. Tumor-infiltrating BRAFV600E-specific CD4+ T cells correlated with complete clinical response in melanoma. J. Clin. Investig. 2018, 128, 1563–1568. [Google Scholar] [CrossRef]
- Liu, Q.; Zhu, H.; Liu, Y.; Musetti, S.; Huang, L. BRAF peptide vaccine facilitates therapy of murine BRAF-mutant melanoma. Cancer Immunol. Immunother. 2018, 67, 299–310. [Google Scholar] [CrossRef]
- Platten, M. EGFRvIII vaccine in glioblastoma-InACT-IVe or not ReACTive enough? Neuro Oncol. 2017, 19, 1425–1426. [Google Scholar] [CrossRef] [PubMed]
- Goff, S.L.; Morgan, R.A.; Yang, J.C.; Sherry, R.M.; Robbins, P.F.; Restifo, N.P.; Feldman, S.A.; Lu, Y.C.; Lu, L.; Zheng, Z.; et al. Pilot Trial of Adoptive Transfer of Chimeric Antigen Receptor-transduced T Cells Targeting EGFRvIII in Patients With Glioblastoma. J. Immunother. 2019, 42, 126–135. [Google Scholar] [CrossRef]
- Yim, E.K.; Park, J.S. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res. Treat. 2005, 37, 319–324. [Google Scholar] [CrossRef]
- Niemi, J.V.L.; Sokolov, A.V.; Schiöth, H.B. Neoantigen Vaccines; Clinical Trials, Classes, Indications, Adjuvants and Combinatorial Treatments. Cancers 2022, 14, 5163. [Google Scholar] [CrossRef]
- Tay, B.Q.; Wright, Q.; Ladwa, R.; Perry, C.; Leggatt, G.; Simpson, F.; Wells, J.W.; Panizza, B.J.; Frazer, I.H.; Cruz, J.L.G. Evolution of Cancer Vaccines-Challenges, Achievements, and Future Directions. Vaccines 2021, 9, 535. [Google Scholar] [CrossRef] [PubMed]
- Kalaora, S.; Barnea, E.; Merhavi-Shoham, E.; Qutob, N.; Teer, J.K.; Shimony, N.; Schachter, J.; Rosenberg, S.A.; Besser, M.J.; Admon, A.; et al. Use of HLA peptidomics and whole exome sequencing to identify human immunogenic neo-antigens. Oncotarget 2016, 7, 5110–5117. [Google Scholar] [CrossRef]
- Feldman, A.L.; Vasmatzis, G.; Asmann, Y.W.; Davila, J.; Middha, S.; Eckloff, B.W.; Johnson, S.H.; Porcher, J.C.; Ansell, S.M.; Caride, A. Novel TRAF1-ALK fusion identified by deep RNA sequencing of anaplastic large cell lymphoma. Genes. Chromosomes Cancer 2013, 52, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Flower, D.R. In silico prediction of cancer immunogens: Current state of the art. BMC Immunol. 2018, 19, 11. [Google Scholar] [CrossRef]
- Boland, C.R.; Ricciardiello, L. How many mutations does it take to make a tumor? Proc. Natl. Acad. Sci. USA 1999, 96, 14675–14677. [Google Scholar] [CrossRef]
- Charneau, J.; Suzuki, T.; Shimomura, M.; Fujinami, N.; Mishima, Y.; Hiranuka, K.; Watanabe, N.; Yamada, T.; Nakamura, N.; Nakatsura, T. Development of antigen-prediction algorithm for personalized neoantigen vaccine using human leukocyte antigen transgenic mouse. Cancer Sci. 2022, 113, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.Q.T.; Tran, T.P.D.; Nguyen, H.T.; Nguyen, T.N.; Pham, T.M.Q.; Nguyen, H.T.P.; Tran, D.H.; Nguyen, V.; Tran, T.S.; Pham, T.N.; et al. Improvement in neoantigen prediction via integration of RNA sequencing data for variant calling. Front. Immunol. 2023, 14, 1251603. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Huber, F.; Arnaud, M.; Kraemer, A.I.; Altimiras, E.R.; Michaux, J.; Taillandier-Coindard, M.; Chiffelle, J.; Murgues, B.; Gehret, T.; et al. Machine learning methods and harmonized datasets improve immunogenic neoantigen prediction. Immunity 2023, 56, 2650–2663.e6. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, R.; Gao, S.; Li, W.; Liu, Y.; Su, G.; Song, M.; Jiang, M.; Jiang, C.; Zhang, X. Artificial intelligence applied in neoantigen identification facilitates personalized cancer immunotherapy. Front. Oncol. 2023, 12, 1054231. [Google Scholar] [CrossRef]
- Badillo, S.; Banfai, B.; Birzele, F.; Davydov, I.I.; Hutchinson, L.; Kam-Thong, T.; Siebourg-Polster, J.; Steiert, B.; Zhang, J.D. An Introduction to Machine Learning. Clin. Pharmacol. Ther. 2020, 107, 871–885. [Google Scholar] [CrossRef]
- Orenbuch, R.; Filip, I.; Rabadan, R. HLA Typing from RNA Sequencing and Applications to Cancer. Methods Mol. Biol. 2020, 2120, 71–92. [Google Scholar] [CrossRef]
- Liu, P.; Yao, M.; Gong, Y.; Song, Y.; Chen, Y.; Ye, Y.; Liu, X.; Li, F.; Dong, H.; Meng, R.; et al. Benchmarking the Human Leukocyte Antigen Typing Performance of Three Assays and Seven Next-Generation Sequencing-Based Algorithms. Front. Immunol. 2021, 12, 652258. [Google Scholar] [CrossRef]
- Reimann, H.; Nguyen, A.; Sanborn, J.Z.; Vaske, C.J.; Benz, S.C.; Niazi, K.; Rabizadeh, S.; Spilman, P.; Mackensen, A.; Ruebner, M.; et al. Identification and validation of expressed HLA-binding breast cancer neoepitopes for potential use in individualized cancer therapy. J. Immunother. Cancer 2021, 9, e002605. [Google Scholar] [CrossRef] [PubMed]
- Kote, S.; Pirog, A.; Bedran, G.; Alfaro, J.; Dapic, I. Mass Spectrometry-Based Identification of MHC-Associated Peptides. Cancers 2020, 12, 535. [Google Scholar] [CrossRef]
- Thakur, S.; Jain, M.; Zhang, C.; Major, C.; Bielamowicz, K.J.; Lacayo, N.J.; Vaske, O.; Lewis, V.; Murguia-Favela, L.; Narendran, A. Identification and in vitro validation of neoantigens for immune activation against high-risk pediatric leukemia cells. Hum. Vaccin. Immunother. 2021, 17, 5558–5562. [Google Scholar] [CrossRef]
- Chiang, C.L.; Kandalaft, L.E. In vivo cancer vaccination: Which dendritic cells to target and how? Cancer Treat Rev. 2018, 71, 88–101. [Google Scholar] [CrossRef]
- Rojas, L.A.; Sethna, Z.; Soares, K.C.; Olcese, C.; Pang, N.; Patterson, E.; Lihm, J.; Ceglia, N.; Guasp, P.; Chu, A.; et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature 2023, 618, 144–150. [Google Scholar] [CrossRef]
- Zhou, W.; Yu, J.; Li, Y.; Wang, K. Neoantigen-specific TCR-T cell-based immunotherapy for acute myeloid leukemia. Exp. Hematol. Oncol. 2022, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Poorebrahim, M.; Mohammadkhani, N.; Mahmoudi, R.; Gholizadeh, M.; Fakhr, E.; Cid-Arregui, A. TCR-like CARs and TCR-CARs targeting neoepitopes: An emerging potential. Cancer Gene Ther. 2021, 28, 581–589. [Google Scholar] [CrossRef]
- Vensko, S.P.; Olsen, K.; Bortone, D.; Smith, C.C.; Chai, S.; Beckabir, W.; Fini, M.; Jadi, O.; Rubinsteyn, A.; Vincent, B.G. LENS: Landscape of Effective Neoantigens Software. Bioinformatics 2023, 39, btad322. [Google Scholar] [CrossRef]
- Kodysh, J.; Rubinsteyn, A. OpenVax: An open-source computational pipeline for cancer neoantigen prediction. In Bioinformatics for Cancer Immunotherapy. Methods in Molecular Biology; Boegel, S., Ed.; Humana: New York, NY, USA, 2020; Volume 2120. [Google Scholar] [CrossRef]
- Hundal, J.; Kiwala, S.; McMichael, J.; Miller, C.A.; Xia, H.; Wollam, A.T.; Liu, C.J.; Zhao, S.; Feng, Y.Y.; Graubert, A.P.; et al. pVACtools: A Computational Toolkit to Identify and Visualize Cancer Neoantigens. Cancer Immunol. Res. 2020, 8, 409–420. [Google Scholar] [CrossRef]
- Rieder, D.; Fotakis, G.; Ausserhofer, M.; René, G.; Paster, W.; Trajanoski, Z.; Finotello, F. nextNEOpi: A comprehensive pipeline for computational neoantigen prediction. Bioinformatics 2022, 38, 1131–1132. [Google Scholar] [CrossRef]
- Lissabet, J.F.B.; Belén, L.H.; Farias, J.G. TTAgP 1.0: A computational tool for the specific prediction of tumor T cell antigens. Comput. Biol. Chem. 2019, 83, 107103. [Google Scholar] [CrossRef]
- Tappeiner, E.; Finotello, F.; Charoentong, P.; Mayer, C.; Rieder, D.; Trajanoski, Z. TIminer: NGS data mining pipeline for cancer immunology and immunotherapy. Bioinformatics 2017, 33, 3140–3141. [Google Scholar] [CrossRef] [PubMed]
- Charoenkwan, P.; Nantasenamat, C.; Hasan, M.M.; Shoombuatong, W. iTTCA-Hybrid: Improved and robust identification of tumor T cell antigens by utilizing hybrid feature representation. Anal. Biochem. 2020, 599, 113747. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Zou, Q.; Guo, H.; Shi, L. iTTCA-RF: A random forest predictor for tumor T cell antigens. J. Transl. Med. 2021, 19, 449. [Google Scholar] [CrossRef]
- Herrera-Bravo, J.; Herrera Belén, L.; Farias, J.G.; Beltrán, J.F. TAP 1.0: A robust immunoinformatic tool for the prediction of tumor T-cell antigens based on AAindex properties. Comput. Biol. Chem. 2021, 91, 107452. [Google Scholar] [CrossRef] [PubMed]
- Charoenkwan, P.; Pipattanaboon, C.; Nantasenamat, C.; Hasan, M.M.; Moni, M.A.; Lio’, P.; Shoombuatong, W. PSRTTCA: A new approach for improving the prediction and characterization of tumor T cell antigens using propensity score representation learning. Comput. Biol. Med. 2023, 152, 106368. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Schaduangrat, N.; Shoombuatong, W. StackTTCA: A stacking ensemble learning-based framework for accurate and high-throughput identification of tumor T cell antigens. BMC Bioinform. 2023, 24, 301. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Chen, I.; Chen, M.Y.; Goedegebuure, S.P.; Gillanders, W.E. Challenges targeting cancer neoantigens in 2021: A systematic literature review. Expert Rev. Vaccines 2021, 20, 827–837. [Google Scholar] [CrossRef]
- Khong, H.T.; Yang, J.C.; Topalian, S.L.; Sherry, R.M.; Mavroukakis, S.A.; White, D.E.; Rosenberg, S.A. Immunization of HLA-A*0201 and/or HLA-DPbeta1*04 patients with metastatic melanoma using epitopes from the NY-ESO-1 antigen. J. Immunother. 2004, 27, 472–477. [Google Scholar] [CrossRef]
- Corbière, V.; Chapiro, J.; Stroobant, V.; Ma, W.; Lurquin, C.; Lethé, B.; van Baren, N.; Van den Eynde, B.J.; Boon, T.; Coulie, P.G. Antigen spreading contributes to MAGE vaccination-induced regression of melanoma metastases. Cancer Res. 2011, 71, 1253–1262. [Google Scholar] [CrossRef]
- Carreno, B.M.; Magrini, V.; Becker-Hapak, M.; Kaabinejadian, S.; Hundal, J.; Petti, A.A.; Ly, A.; Lie, W.R.; Hildebrand, W.H.; Mardis, E.R.; et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015, 348, 803–808. [Google Scholar] [CrossRef]
- Cohen, C.J.; Gartner, J.J.; Horovitz-Fried, M.; Shamalov, K.; Trebska-McGowan, K.; Bliskovsky, V.V.; Parkhurst, M.R.; Ankri, C.; Prickett, T.D.; Crystal, J.S.; et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Investig. 2015, 125, 3981–3991. [Google Scholar] [CrossRef]
- Linette, G.P.; Becker-Hapak, M.; Skidmore, Z.L.; Baroja, M.L.; Xu, C.; Hundal, J.; Spencer, D.H.; Fu, W.; Cummins, C.; Robnett, M.; et al. Immunological ignorance is an enabling feature of the oligo-clonal T cell response to melanoma neoantigens. Proc. Natl. Acad. Sci. USA 2019, 116, 23662–23670. [Google Scholar] [CrossRef]
- Hu, Z.; Leet, D.E.; Allesøe, R.L.; Oliveira, G.; Li, S.; Luoma, A.M.; Liu, J.; Forman, J.; Huang, T.; Iorgulescu, J.B.; et al. Personal neoantigen vaccines induce persistent memory T cell responses and epitope spreading in patients with melanoma. Nat. Med. 2021, 27, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Löwer, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrörs, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Chapman, P.B.; Feilchenfeldt, J.; Brennan, M.F.; Capanu, M.; Gansukh, B.; Jacobs, G.; Levin, A.; Neville, D.; Kelsen, D.P.; et al. Targeting mutated K-ras in pancreatic adenocarcinoma using an adjuvant vaccine. Am. J. Clin. Oncol. 2011, 34, 321–325. [Google Scholar] [CrossRef]
- Dillard, P.; Casey, N.; Pollmann, S.; Vernhoff, P.; Gaudernack, G.; Kvalheim, G.; Wälchli, S.; Inderberg, E.M. Targeting KRAS mutations with HLA class II-restricted TCRs for the treatment of solid tumors. Oncoimmunology 2021, 10, 1936757. [Google Scholar] [CrossRef]
- Sonntag, K.; Hashimoto, H.; Eyrich, M.; Menzel, M.; Schubach, M.; Döcker, D.; Battke, F.; Courage, C.; Lambertz, H.; Handgretinger, R.; et al. Immune monitoring and TCR sequencing of CD4 T cells in a long term responsive patient with metastasized pancreatic ductal carcinoma treated with individualized, neoepitope-derived multipeptide vaccines: A case report. J. Transl. Med. 2018, 16, 23. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, S.; Han, N.; Jiang, J.; Xu, Y.; Ma, D.; Lu, L.; Guo, X.; Qiu, M.; Huang, Q.; et al. A Neoantigen-Based Peptide Vaccine for Patients With Advanced Pancreatic Cancer Refractory to Standard Treatment. Front. Immunol. 2021, 12, 691605. [Google Scholar] [CrossRef]
- Keskin, D.B.; Anandappa, A.J.; Sun, J.; Tirosh, I.; Mathewson, N.D.; Li, S.; Oliveira, G.; Giobbie-Hurder, A.; Felt, K.; Gjini, E.; et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 2019, 565, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Johanns, T.M.; Miller, C.A.; Liu, C.J.; Perrin, R.J.; Bender, D.; Kobayashi, D.K.; Campian, J.L.; Chicoine, M.R.; Dacey, R.G.; Huang, J.; et al. Detection of neoantigen-specific T cells following a personalized vaccine in a patient with glioblastoma. Oncoimmunology 2019, 8, e1561106. [Google Scholar] [CrossRef]
- Li, F.; Chen, C.; Ju, T.; Gao, J.; Yan, J.; Wang, P.; Xu, Q.; Hwu, P.; Du, X.; Lizée, G. Rapid tumor regression in an Asian lung cancer patient following personalized neo-epitope peptide vaccination. Oncoimmunology 2016, 5, e1238539. [Google Scholar] [CrossRef]
- Li, F.; Deng, L.; Jackson, K.R.; Talukder, A.H.; Katailiha, A.S.; Bradley, S.D.; Zou, Q.; Chen, C.; Huo, C.; Chiu, Y.; et al. Neoantigen vaccination induces clinical and immunologic responses in non-small cell lung cancer patients harboring EGFR mutations. J. Immunother. Cancer 2021, 9, e002531, Erratum in J. Immunother. Cancer 2021, 9, 1. [Google Scholar] [CrossRef]
- Tran, E.; Ahmadzadeh, M.; Lu, Y.C.; Gros, A.; Turcotte, S.; Robbins, P.F.; Gartner, J.J.; Zheng, Z.; Li, Y.F.; Ray, S.; et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015, 350, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Cafri, G.; Yossef, R.; Pasetto, A.; Deniger, D.C.; Lu, Y.C.; Parkhurst, M.; Gartner, J.J.; Jia, L.; Ray, S.; Ngo, L.T.; et al. Memory T cells targeting oncogenic mutations detected in peripheral blood of epithelial cancer patients. Nat. Commun. 2019, 10, 449. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zhang, W.; Li, Z.; Zheng, Y.; Wang, Y.; Chen, G.; Qiu, L.; Ke, K.; Su, X.; Cai, Z.; et al. Personalized neoantigen-based immunotherapy for advanced collecting duct carcinoma: Case report. J. Immunother. Cancer 2020, 8, e000217. [Google Scholar] [CrossRef] [PubMed]
- Ott, P.A.; Hu-Lieskovan, S.; Chmielowski, B.; Govindan, R.; Naing, A.; Bhardwaj, N.; Margolin, K.; Awad, M.M.; Hellmann, M.D.; Lin, J.J.; et al. A Phase Ib Trial of Personalized Neoantigen Therapy Plus Anti-PD-1 in Patients with Advanced Melanoma, Non-small Cell Lung Cancer, or Bladder Cancer. Cell 2020, 183, 347–362.e24. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.; Liu, Q.; Xie, L.; Shao, J.; Yu, L.; Cen, L.; Li, L.; Liu, F.; Qian, H.; Wei, J.; et al. Case Report: Pathological Complete Response in a Lung Metastasis of Phyllodes Tumor Patient Following Treatment Containing Peptide Neoantigen Nano-Vaccine. Front. Oncol. 2022, 12, 800484. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, X.; Wang, X.; Kim, S.W.; Herndon, J.M.; Becker-Hapak, M.K.; Carreno, B.M.; Myers, N.B.; Sturmoski, M.A.; McLellan, M.D.; et al. Optimized polyepitope neoantigen DNA vaccines elicit neoantigen-specific immune responses in preclinical models and in clinical translation. Genome Med. 2021, 13, 56. [Google Scholar] [CrossRef]
- Smith, A.R.; Alonso, J.A.; Ayres, C.M.; Singh, N.K.; Hellman, L.M.; Baker, B.M. Structurally silent peptide anchor modifications allosterically modulate T cell recognition in a receptor-dependent manner. Proc. Natl. Acad. Sci. USA 2021, 118, e2018125118. [Google Scholar] [CrossRef]
- Fang, X.; Guo, Z.; Liang, J.; Wen, J.; Liu, Y.; Guan, X.; Li, H. Neoantigens and their potential applications in tumor immunotherapy. Oncol. Lett. 2022, 23, 88. [Google Scholar] [CrossRef]
- Ye, L.; Creaney, J.; Redwood, A.; Robinson, B. The Current Lung Cancer Neoantigen Landscape and Implications for Therapy. J. Thorac. Oncol. 2021, 16, 922–932. [Google Scholar] [CrossRef]
- Łuksza, M.; Sethna, Z.M.; Rojas, L.A.; Lihm, J.; Bravi, B.; Elhanati, Y.; Soares, K.; Amisaki, M.; Dobrin, A.; Hoyos, D.; et al. Neoantigen quality predicts immunoediting in survivors of pancreatic cancer. Nature 2022, 606, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A.; et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef]
- Koşaloğlu-Yalçın, Z.; Blazeska, N.; Vita, R.; Carter, H.; Nielsen, M.; Schoenberger, S.; Sette, A.; Peters, B. The Cancer Epitope Database and Analysis Resource (CEDAR). Nucleic Acids Res. 2023, 51, D845–D852. [Google Scholar] [CrossRef]
- Available online: https://www.cancerresearch.org/peptide-database (accessed on 14 November 2023).
- Wu, J.; Zhao, W.; Zhou, B.; Su, Z.; Gu, X.; Zhou, Z.; Chen, S. TSNAdb: A Database for Tumor-specific Neoantigens from Immunogenomics Data Analysis. Genom. Proteom. Bioinform. 2018, 16, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Bai, P.; Fan, W.; Li, Q.; Li, Y.; Wang, D.; Yin, L.; Zhou, Y. NEPdb: A Database of T-Cell Experimentally-Validated Neoantigens and Pan-Cancer Predicted Neoepitopes for Cancer Immunotherapy. Front. Immunol. 2021, 12, 644637. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chitkushev, L.; Olsen, L.R.; Keskin, D.B.; Brusic, V. TANTIGEN 2.0: A knowledge base of tumor T cell antigens and epitopes. BMC Bioinform. 2021, 22 (Suppl. S8), 40. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, L.; Kong, X.; Cao, Y.; Zhang, M.; Sun, Z.; Liu, Y.; Wang, J.; Shen, B.; Bo, X.; et al. CAD v1.0: Cancer Antigens Database Platform for Cancer Antigen Algorithm Development and Information Exploration. Front. Bioeng. Biotechnol. 2022, 10, 819583. [Google Scholar] [CrossRef]
- Vigneron, N.; Stroobant, V.; Van den Eynde, B.J.; van der Bruggen, P. Database of T cell-defined human tumor antigens: The 2013 update. Cancer Immun. 2013, 13, 15. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).