Abstract
Legume crops establish symbiosis with nitrogen-fixing rhizobia for biological nitrogen fixation (BNF), a process that provides a prominent natural nitrogen source in agroecosystems; and efficient nodulation and nitrogen fixation processes require a large amount of phosphorus (P). Here, a role of GmPAP4, a nodule-localized purple acid phosphatase, in BNF and seed yield was functionally characterized in whole transgenic soybean (Glycine max) plants under a P-limited condition. GmPAP4 was specifically expressed in the infection zones of soybean nodules and its expression was greatly induced in low P stress. Altered expression of GmPAP4 significantly affected soybean nodulation, BNF, and yield under the P-deficient condition. Nodule number, nodule fresh weight, nodule nitrogenase, APase activities, and nodule total P content were significantly increased in GmPAP4 overexpression (OE) lines. Structural characteristics revealed by toluidine blue staining showed that overexpression of GmPAP4 resulted in a larger infection area than wild-type (WT) control. Moreover, the plant biomass and N and P content of shoot and root in GmPAP4 OE lines were also greatly improved, resulting in increased soybean yield in the P-deficient condition. Taken together, our results demonstrated that GmPAP4, a purple acid phosphatase, increased P utilization efficiency in nodules under a P-deficient condition and, subsequently, enhanced symbiotic BNF and seed yield of soybean.
1. Introduction
Legume plants, such as the common bean (Phaseolus vulgaris), soybean (Glycine max), pea (Pisum sativum), and chickpea (Cicer arietinum L.), are very important legume crops that provide humans and animals with food and feed materials. Legume plants can establish symbiotic associations with nitrogen (N)-fixing rhizobia in soil [1,2]. Symbiotic biological nitrogen fixation (BNF) is processed in nodules and the resulting fixed nitrogen products are in the form of ureides, allantoin, and allantoic acids transported from nodules to shoot for plant growth and development [3,4,5]. In order to meet the food demand for all the population of the world, researchers aim to improve crop production; however, agriculture is highly dependent on the application of excessive industrial nitrogen fertilizer in the field, which not only costs a heavy price, and reduces the utilization efficiency of nitrogen fertilizer, but also results in production reduction, waste of resources, lower environment impacts, and so on [6,7]. Thus, it is of significance to find a natural way of replacement of nitrogen fertilizer and increase nitrogen utilization efficiency. For this purpose, legume plants are of great interest due to their capacity for nitrogen fixation.
Although nitrogen fixation in nodules benefits soybean plants with N nutrient, the nodulation and nitrogen fixation process consumes much energy. Several studies have reported that P was particularly essential for nodulation and energy consumption for N2 fixation in legume plants. P deficiency severely inhibits nodule development, nitrogen fixation, and total N content in legume plants [8,9]. In Medicago truncatula, under P-depleted condition, nodule dry matter (DM) and the resulting total concentration of amino-N compounds in nodules was significantly reduced compared to the normal P growth condition [10]. In soybean, the nodule number, nodule fresh weight and size, and the nitrogenase activity decreased significantly in low P-stress condition [11,12]. Therefore, legume plants have developed several adaptive strategies to maintain P homeostasis for symbiotic nitrogen fixation in nodules under P-starvation condition, including induction and secretion of acid phosphatase (APase; EC 3.1.3.2) activity of roots and nodules [13,14].
Purple acid phosphatases (PAPs; EC 3.1.3.2), a specific group of acid phosphatases, have been found in several plant species, such as Arabidopsis thaliana, common bean (Phaseolus vulgaris), rice (Oryza sativa), Medicago truncatula, and soybean [15,16,17,18]. PAPs have diverse functions in plants and most of them were induced by P starvation. PvPAP3, in common bean (Phaseolus vulgaris), was induced in P starvation and enhanced utilization efficiency of extracellular ATP supplied as the sole P source [18]. AtPAP12 and AtPAP26 in Arabidopsis thaliana were identified as the predominant PAP isozymes secreted by roots under P-deficient condition and played a vital role in improving extracellular P-use efficiency [15]. GmPAP7a/7b were also up-regulated by P starvation and data indicated that GmPAP7a/7b contributed to extracellular ATP utilization in soybean [19]. GmPAP17 had a strong response to low phosphorus stress in root functions in the adaptation of soybean to low P stress, possibly through its involvement in P recycling in plants [20]. All these published research papers suggested that PAP members could be involved in P acquisition and recycling in plants.
However, not so many research papers focused on the role of PAPs in nodule development and nitrogen fixation in legume plants under P-deficient condition. Nodulin PvNOD33, a putative phosphatase from the common bean, was highly expressed in the inner cortex of infection cells of mature nodules and was inferred to function in carbon metabolism [21]. A novel PAP gene, GmPAP21, in soybean, was greatly induced by P limitation in soybean nodules, and altered expression of GmPAP21 significantly affected acid phosphatase and nodule growth and development [22]. Another PAP gene, GmPAP12, reported by our team previously, was highly expressed in nodules and promoted nodule development and nitrogen fixation by enhancing P utilization under P-limitation condition [14]. Previously, our research team found that GmPAP4, a new purple acid phosphatase, showed highly induced expression in soybean roots under P-deficient conditions, and overexpression of GmPAP4 in Arabidopsis resulted in significant rises in P acquisition and utilization compared with the wild-type control [17]. Here, we reported that GmPAP4 was also highly expressed in nodules during development in soybean and its expression was induced by low P stress. Systematical experiments were further conducted to explore the function of GmPAP4 in nodule development and nitrogen fixation under P-deficient condition. We found that overexpression of GmPAP4 significantly increased nodule development and nitrogen fixation, and, finally, improved plant growth and seed yield under P-deficient condition.
2. Results
2.1. GmPAP4 Was Preferentially Expressed in Infection Zones of Nodules in Soybean
To investigate the expression pattern, the transcript abundance of GmPAP4 was examined by quantitative real-time PCR (qRT-PCR) in various organs of soybean inoculated with rhizobia in low-nitrogen nutrient condition. We found that GmPAP4 was expressed higher in nodules than in any other organs and expression of GmPAP4 was increased during nodule development, suggesting a role of GmPAP4 in soybean nodulation (Figure 1).
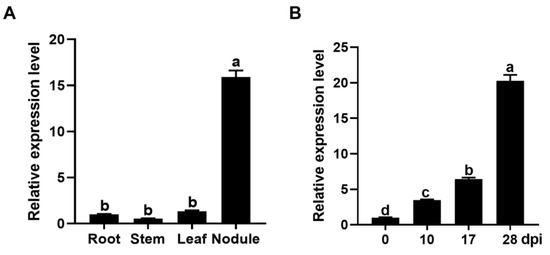
Figure 1.
Spatio-temporal relative expression of GmPAP4 in soybean. (A) Expression profiles of GmPAP4 in various organs of soybean. (B) Time course expression analysis of GmPAP4 in nodules (0, 10, 17, and 28 dpi). The lowercase letters a–d indicate significant differences, p < 0.05.
Next, to precisely determine the tissue-specific localization of GmPAP4, we carried out RNA in situ hybridization analysis in different developmental stages of nodules. We found that the transcript of GmPAP4 was specifically localized in the nitrogen-fixing zones of nodules in all the developmental stages. There was no hybridization signal detected in nodule samples with a sense probe, suggesting the specificity of the antisense probe (Figure 2). Together, these results demonstrate that GmPAP4 genes potentially function in nodule development and nitrogen fixation.
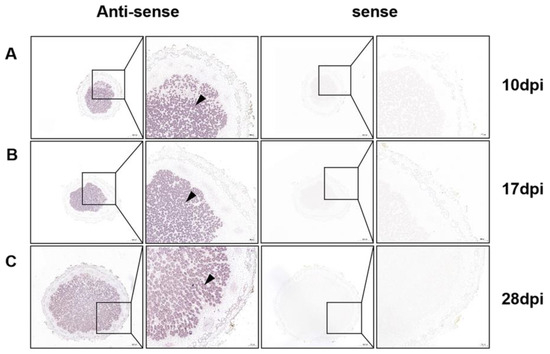
Figure 2.
RNA in situ hybridization analysis of GmPAP4 in different developmental stages of nodules. Soybean nodules were harvested at 10, 17, and 28 dpi (A–C) and digoxigenin-labelled probes specific for GmPAP4 were used for the detection of its transcript. The black arrow head indicates the infected cells in the nodules. Scale bar = 500 µm (left panel of antisense or sense) and 100 µm (right panel of antisense or sense).
2.2. GmPAP4 Was Highly Induced by Low Phosphorus Stress in Soybean Nodules
To determine the response of GmPAP4 to low phosphorus (LP) stress in nodules, we planted the soybean plants inoculated with rhizobia under optimal phosphorus (OP) or LP conditions for 28 days. Our qRT-PCR analysis showed that GmPAP4 could be induced under LP stress compared to OP condition (Figure 3A). To further elucidate the expression of GmPAP4 in nodules under different P conditions, we cloned the promoter of GmPAP4 2000 bp upstream of the translational start codon fused with the GUS reporter gene (pGmPAP4:GUS). And the transgenic composite plants carrying pGmPAP4:GUS were inoculated with rhizobia and the transgenic roots and nodules were harvested at 28 days post inoculation (dpi) for GUS staining under optimal phosphorus (OP) and LP conditions. The results showed that the promoter of GmPAP4 was highly expressed in nodules and GUS activity was increased by 21.6% under LP stress compared with OP condition (Figure 3B,C). All these data indicated that GmPAP4 may be involved in P signaling in nodules of soybean.
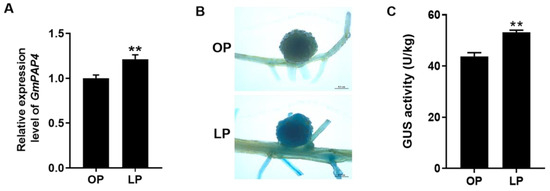
Figure 3.
Response of GmPAP4 to LP stress. (A) The relative expression value of GmPAP4 in nodules under different P conditions. The expression value of GmPAP4 was normalized based on the expression of GmActin11 (Glyma.18g290800) used as a reference gene. (B) Histochemical staining analysis of the promoter of GmPAP4 in transgenic composite soybean roots and nodules at 28 dpi, Scale bar = 0.1 cm. (C) GUS activity of nodules expressing pGmPAP4:GUS. Three independent experiments were performed and images from one representative experiment were shown here (n > 10). Each error bar represents the mean of three biological replicates with ±SE. Asterisks indicate significant differences within a p level in t-tests (**: 0.001 < p ≤ 0.01).
2.3. GmPAP4 Expression Was Positively Associated with Nodulation and Nitrogen Fixation under LP Stress
To further investigate the role of GmPAP4 in nodulation and biological nitrogen fixation under LP stress, stable transgenic soybean plants overexpressing GmPAP4 were generated through Agrobacterium-mediated transformation. Finally, two stable transgenic overexpression lines (OE1 and OE2) were obtained and confirmed by specific PCR and glyphosate resistance tests (Supplemental Figure S1). RT-PCR analysis showed that the transcript accumulation of GmPAP4 was about 10 times higher in OE lines than in nodules of WT control and the expression level of GmPAP4 in nodules of the two stable lines did not show significant differences (Supplemental Figure S2). Next, the structural characteristics of nodules formed on GmPAP4 OE lines were examined. Images of cross-sectioned nodules stained with toluidine blue dye revealed that GmPAP4 overexpression greatly affected nodule morphological development (Supplemental Figure S3A). In comparison with WT nodules, the two GmPAP4 OE lines displayed larger infection zones by 17.0% and 14.4%, respectively (Supplemental Figure S3B). These results further indicated that GmPAP4 was associated with nodule formation and, therefore, was crucial for nitrogen fixation.
Subsequent evaluation was conducted to determine the effect of altered expression of GmPAP4 on nodule development and growth under LP condition at 28 dpi, at which point the nodules were more functional and active. We found that overexpression of GmPAP4 increased nodule development and biological nitrogen fixation ability. The two stable transgenic lines (OE1 and OE2) showed an increase of 33.6% and 27.6% in the total nodule number, an increase of 69.9% and 57.1% in nodule fresh weight, resulting in increased signal nodule weight, and an increase of 47.8% and 32.7% in nitrogenase activity, respectively (Figure 4A–E). On the other hand, the functional and active nodules were always pink due to the presence of the leghemoglobin (GmLbc3) protein, an oxygen carrier, required for nitrogenase activity and biological nitrogen fixation in nodules [23]. We found that the expression of GmLbc3 in nodules of two GmPAP4 OE lines was greatly increased compared with that in WT nodules (Figure 4F). And, finally, the nodule N concentration in the two stable lines showed a 7.2% and 5.0% increase compared with WT control (Figure 4G). All these data suggested that overexpression of GmPAP4 directly increased nodule development as well as the capacity of symbiotic nitrogen fixation in nodules of soybean under LP condition.
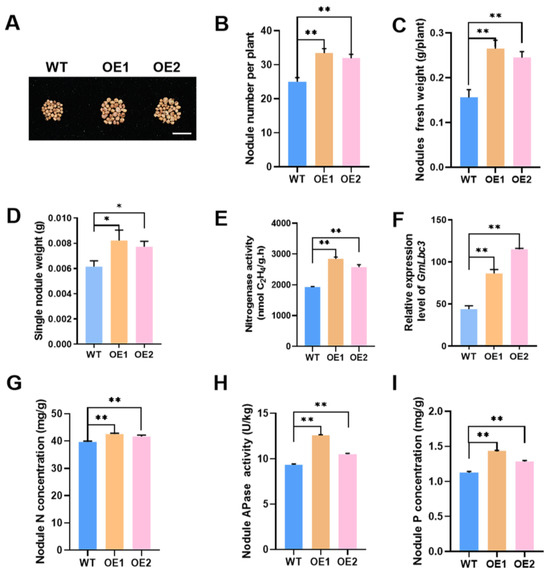
Figure 4.
Phenotypic analysis of nodulation of stable transgenic soybean plants overexpressing (OE) of GmPAP4. (A) Nodule growth performance at 28 dpi. Scale bar in (A) = 1 cm. (B) Nodule number per plant. (C) Nodule fresh weight. (D) Single nodule weight. (E) Nitrogenase activity measured by the acetylene reduction assay. (F) Quantitative real-time PCR analysis of GmLbc3 expression in nodules. (G) Nodule N concentration. (H) APase activities of nodules. (I) Nodule P concentration. Each error bar represents the mean of three biological replicates with ±SE. Asterisks indicate significant differences within a p level in t-tests (*: 0.01 < p ≤ 0.05, **: 0.001 < p ≤ 0.01).
Next, to confirm whether the improved nodulation in GmPAP4 OE lines is due to overexpression of GmPAP4, APase activities of nodules in GmPAP4 OE lines were assessed under low P condition and we found that overexpression of GmPAP4 led to 34.6% and 12.6% increases in nodule APase activities relative to WT control (Figure 4H). Consequently, the P concentration of nodules was enhanced significantly, with 27.5% and 14.1% increases in the two GmPAP4 OE lines (Figure 4I). These data indicated that GmPAP4 might facilitate nodule development and nitrogen fixation by enhancing P utilization efficiency in nodules of soybean under P-deficient condition.
2.4. Overexpression of GmPAP4-Enhanced Soybean Plant Growth under P-Deficient Conditions
In the meantime, the role of overexpression of GmPAP4 on soybean plant growth inoculated with rhizobia was evaluated under LP condition and we found that overexpression of GmPAP4 resulted in much better growth relative to the control plants. Overexpression of GmPAP4 led to 21.5% and 17.7% increases in plant height, 38.7% and 16.2% increases in shoot fresh weight, and 20.7% and 13.2% increases in shoot dry weight, respectively. In the case of soybean roots, we found that root fresh weight increased by 24.3% and 15.4% and root dry weight increased by 43.4% and 24.8% in GmPAP4 OE lines (Figure 5A–E). Moreover, the two transgenic lines showed 7.4% and 6.6% increases in N content of the shoots and 14.6% and 12.1% increases in N content of the roots, and increases of 3.3% and 9.9% and 32.1% and 26.1% in P content of the shoots and roots, respectively, relative to the control plants (Figure 5F–I).
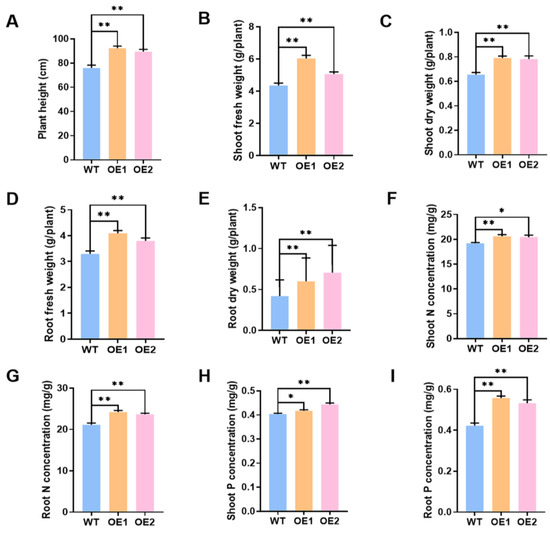
Figure 5.
Phenotypic analysis of stable transgenic soybean plants overexpressing of GmPAP4 at 28 dpi under low P stress. (A) Plant height. (B) Shoot fresh weight. (C) Shoot dry weight. (D) Root fresh weight. (E) Root dry weight. (F) Shoot N concentration. (G) Root N concentration. (H) Shoot P concentration. (I) Root P concentration. Each error bar represents the mean of three biological replicates with ±SE. Asterisks indicate significant differences within a p level in t-tests. (*: 0.01 < p ≤ 0.05, **: 0.001 < p ≤ 0.01).
2.5. Overexpression of GmPAP4 Enhanced Soybean Yield under P-Deficient Conditions
The growth performance of GmPAP4 OE lines subsequently led to higher seed yield compared with the WT control. GmPAP4 OE lines resulted in increases in pod number by 17.0% and 19.0%, increases in seed number by 15.4% and 12.3%, and increases in seed weight by 19.2% and 14.4%, respectively (Figure 6). All these results suggested that GmPAP4 was associated with nodule formation and nitrogen fixation and, subsequently, affected soybean growth, N nutrition, and grain yield under P-deficient stress.
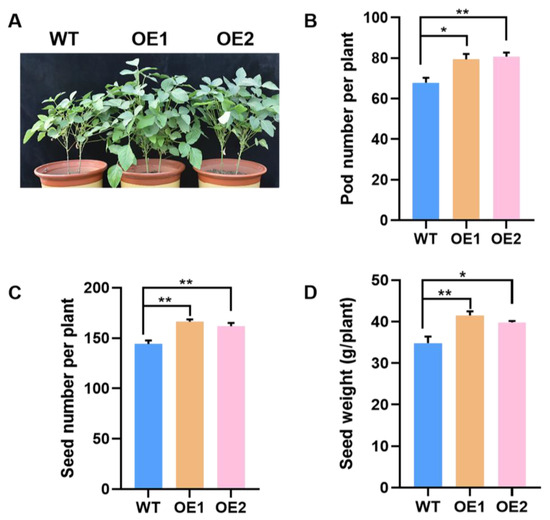
Figure 6.
Seed yield analysis of GmPAP4 OE lines in the pot experiment under low P condition. (A) Plant growth performance at pot culture. (B) Pod number per plant. (C) Seed number per plant. (D) Seed weight per plant. Each error bar represents the mean of three biological replicates with ±SE. Asterisks indicate significant differences within a p level in t-tests (*: 0.01 < p ≤ 0.05, **: 0.001 < p ≤ 0.01).
3. Discussion
Symbiotic BNF in root nodules plays a crucial role in sustainable agricultural systems and it is an effective alternative way of chemical N fertilizer to provide N nutrient for legume plants [24]. Many studies demonstrate that P is particularly critical in the rhizobium–legume symbiotic process, not only due to its regulation in nodule development but also due to the energy costs of N fixation [25,26]. Several genes, such as purple acid phosphatases, β-expansins, phosphate transporters, GmSPX members, and some transcription factors (GmPHR1, GmPTF1) in soybean, were reported to be highly upregulated in nodules under low P stress [12,27,28,29]. In the soybean whole genome, a total of 38 GmPAPs were isolated and many of the members in the leaves and roots were upregulated at 16 days of P deficiency [19]. However, the functions of most low P-starved up-regulated GmPAPs in nodules, except of GmPAP12 and GmPAP21, remain largely understood [14,22].
In the present study, we found GmPAP4 was expressed highly and specifically in infection zones of nodules detected by qRT-PCR and RNA in situ hybridization (Figure 1 and Figure 2) and GUS activity driven by the promoter of GmPAP4 in composite transgenic nodules was increased significantly in response to P deficiency, indicating the involvement of GmPAP4 in P uptake and utilization for nodule development and nitrogen fixation (Figure 3). Previously, for tissue localization of GmPAP12 and GmPAP21, only RT-PCR and promoter-GUS staining experiments were conducted and they found GmPAP12 and GmPAP21 were highly expressed in the organs of nodules [14,22]. Here, RNA in situ hybridization was performed to specify the infection zone localization of GmPAP4 inside nodules, making the hypothesis of the important role of GmPAP4 in nodule development more reliable and accurate.
Previously, to study the function of GmPAP12 in nodule development and nitrogen fixation, overexpressed composite transgenic lines were generated and evaluated. In the present study, in order to be more convincing, stable transgenic overexpression lines were conducted for the role of GmPAP4 under low P condition (Supplemental Figure S1). The two OE lines exhibited more increased nodule development and nitrogen fixation efficiency (Figure 4 and Figure 5). In the adaptation of the symbiosis process to P starvation, legumes can enhance P utilization within the nodules to tolerate P deficiency [30]. In this study, we found that GmPAP4-OE lines increase APase activity and total P and N acquisition, suggesting that GmPAP4 overexpression enhanced biological nitrogen fixation by more P acquisition (Figure 5). These findings were consistent with the results observed by the overexpression of GmPAP12 under P deficiency. While overexpression of GmPAP21 reduced nodule dry weight, P and N contents in nodules of stable transgenic plants showed contrary results with GmPAP4. All these results indicated that different GmPAP members may cooperate in nodulation by balancing the P acquisition and nitrogen fixation efficiency in soybean nodules.
In the meantime, we found that the two OE lines improved not only nodule development and nitrogen fixation but also plant development and seed yield under P-stress condition (Figure 6). This might be due to the contributions of enhanced N and P acquisition and plant development through symbiotic nitrogen fixation. Taken together, the functional study results presented here strongly suggest that GmPAP4 was involved in plant growth and development regulation that was realized through increased nitrogen and phosphate efficiency, and it played a role in the cross-talk between nitrogen and phosphate pathways in soybean nodules.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The soybean (Glycine max (L.) Merr.) seeds used here were originally obtained from the State Key Laboratory for North China Crop Improvement and Regulation, Hebei Agricultural University. Soybean cultivar Jidou12 and two GmPAP4 overexpression lines in the Jidou12 background were used for phenotypic and functional analysis in all experiments in this study.
Soybean seeds were planted in the vermiculite for 5 days in a controlled chamber (16 h light: 8 h dark cycle at 28 °C). The 5-day-old seedlings were inoculated with rhizobia Bradyrhizobium diazoefficiens USDA110 suspension media (OD600 = 0.08) and, thereafter, watered with nitrogen-free Hoagland nutrient solution comprising the following components: 2.5 mM K2SO4, 2 mM MgSO4·7H2O, 1 mM KH2PO4, 0.15 mM FeCl2, 1.5 mM CaSO4·2H2O, 4.6 × 10−2 mM H3BO3, 9.1 × 10−3 mM MnCl2·4H2O, 7.5 × 10−4 mM ZnSO4, 5 × 10−4 mM CuSO4, 1.1 × 10−4 mM MoO3, 9.4 × 10−5 mM CoCl2·6H2O, and 500 μM (P-sufficient conditions: OP) or 5 μM (P-deficient conditions: LP) of KH2PO4 (pH = 5.8) once a week. Soybean plants and nodules were separately harvested at 28 dpi (days after inoculation) to measure fresh weight, dry weight, height of shoot, total P and N contents, nodule number, and nitrogenase and Apase activity.
For spatial expression analysis, nodules at different developmental stages were separately harvested at 10, 17, and 28 dpi. All tissues were frozen in liquid nitrogen and stored at −80 °C for further RNA extraction and qRT-PCR analysis.
Pot experiments in phytotron were conducted for the effect of overexpression of GmPAP4 on the seed yield of soybean plants. Selected soybean seeds were planted in ceramic pots (diameter: 45 cm; height: 30 cm; bottom diameter: 25 cm) containing vermiculite with a total of 10 pots prepared for each P treatment under optimal growth conditions. Fifty milliliters of rhizobia USDA110 solution (OD600 = 0.08) was applied to each pot with two uniform 5-day-old seedlings. In the meantime, the plants were watered with OP or LP nutrient solution as above once a week and the pod number, seed number and weight were determined at the soybean maturation stage (n = 20).
4.2. Plant mRNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Total mRNA was extracted using the RNAprep pure plant kit (Tiangen, Beijing, China) and the resulting cDNA was synthesized using a PrimeScriptTM RT reagent kit with a gDNA eraser (Takara, Otsu, Shiga Prefecture, Japan). Our qRT-PCR analysis was run on a CFX96™ real-time system (Bio-Rad, Berkeley, CA, USA) using the SYBR Premix EX TaqTM (Takara, Otsu, Shiga Prefecture, Japan). The specific primers used are listed in Table 1. The cycle threshold (CT) values of each sample were standardized to the reference housekeeping gene, GmActin11, and the relative fold change (FC) of gene expression was calculated based on the 2−ΔΔCT method [31].

Table 1.
All primers used in this study.
4.3. Histochemical GUS Staining Analysis
To elucidate the expression pattern of GmPAP4 in soybean nodules, we cloned 2000 bp promoter sequences of GmPAP4 upstream of the translation start codon ATG, fused with β-glucuronidase (GUS) reporter gene (pGmPAP4:GUS). The resulting construct was introduced into the soybean hairy roots by Agrobacterium rhizogenes K599 and the transgenic hairy roots were then inoculated with rhizobia Bradyrhizobium diazoefficiens USDA110 for nodule development. After 4 weeks of rhizobia inoculation, transgenic hairy roots and nodules were stained as described previously and captured with a light microscope (Olympus U-TV0.5XC-3, Tokyo, Japan) [14]. For GUS activity, total nodule proteins were extracted and incubated in a mixture containing 10 mM 4-methylumbelliferyl β-D-glucuronide (MUG; Sigma Chemical Co., St.Louis, MO, USA) for 1 h at 37 °C. The fluorescence product of 4-methylumbelliferone (4-MU) was monitored using a VersaFluor™fluorometer (Bio-Rad, Hercules, CA, USA) with excitation at 365 nm and emission at 455 nm.
4.4. Generation of Stable Transgenic Soybean Plants
A full-length CDS (coding sequence) of GmPAP4 was cloned into a modified overexpression vector pCAMBIA3300 and the resulting construct was introduced into the Agrobacterium tumefaciens strain, EHA105, for stable transgenic soybean transformation using the variety Jidou12 as the background. The positive transgenic plants were identified by bar resistance and, then, the bar gene was amplified using bar-specific primers (Table 1).
4.5. RNA In Situ Hybridization Analysis
RNA in situ hybridization analysis was performed exactly as previously described [32]. The digoxigenin-labelled antisense and sense RNA probes for GmPAP4 were 5′-DIG-AAAGAAGCGUGAGAAUCAGAACAAGAAGGAGU-3′ and 5′-DIG-ACUCCUUCUUGUUCUGAUUCUCACGCUUCUUU-3′, respectively. The hybridization signals were detected by alkaline phosphatase-catalysed color reaction with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Roche; catalogue no. 11175041910, Basel, Switzerland).
4.6. Toluidine Blue Staining
Toluidine blue staining for the observation of infection cells in nodules was performed as described in our previous study [33]. The percentage of the area of infection cells to total cells in a nodule section and the surface area of 100 infection cells were calculated using Image-Pro Plus 6.0 software.
4.7. Measurement of N and P Contents
The measurements of total N and P contents were conducted according to our previous study [14]. Fresh plant samples were dried in an oven at 80 °C first and 0.3 g dried samples were ground and digested with concentrated H2SO4 in a microwave oven. The P content was measured by the color reaction of P-molybdate blue at the absorbance of 700 nm and the N content was determined using the semimicro-Kjeldahl determination method in a nitrogen analyzer.
4.8. Statistical Analysis
Means and SE values were calculated by using GraphPad Prism version 8.0.2 (263) (GraphPad Software Inc., San Diego, CA, USA). The two-tailed Student’s t-test was used to calculate the significance between the samples.
5. Conclusions
Overall, in our study, we characterized a purple acid phosphatase, GmPAP4, that was specifically expressed in infected cells of soybean nodules and it could be induced by low P stress. Functional analysis showed that overexpression of GmPAP4 greatly increased nodule number and nitrogen fixation activity, thus enhancing the acquisition of nitrogen and phosphate under low P condition and, as a consequence, enhancing the seed yield of soybean. This study provided a new gene with a high nitrogen-fixing ability for soybean molecular breeding.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25073649/s1.
Author Contributions
C.Z., H.D. and X.S. designed the research and managed the projects. X.S., Z.Y. and H.Z. were responsible for the gene function investigation and data analysis. X.X. and Z.F. performed the ceramic pot culture experiments. C.Z. and H.D. wrote the manuscript. X.L., Y.K. and W.L. provided suggestions during all the processes of the experiments. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Natural Science Foundation of Hebei Province (C2022204144), the S&T Program of Hebei (23567601H), and the Project of Hebei Province Science and Technology Support Program (17927670H).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare they have no conflicts of interest.
References
- Masson-Boivin, C.; Sachs, J.L. Symbiotic nitrogen fixation by rhizobia—The roots of a success story. Curr. Opin. Plant Biol. 2018, 44, 7–15. [Google Scholar] [CrossRef]
- Werner, G.D.A.; Cornwell, W.K.; Sprent, J.I.; Kattge, J.; Kiers, E.T. A single evolutionary innovation drives the deep evolution of symbiotic N-fixation in angiosperms. Nat. Commun. 2014, 5, 4087. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Indrasumunar, A.; Hayashi, S.; Lin, M.H.; Lin, Y.H.; Reid, D.E.; Gresshoff, P.M. Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 2010, 52, 61–76. [Google Scholar] [CrossRef]
- Oldroyd, G.E.; Downie, J.A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.; Tegeder, M. Soybean ureide transporters play a critical role in nodule development, function and nitrogen export. Plant J. 2012, 72, 355–367. [Google Scholar] [CrossRef]
- Marx, H.; Minogue, C.E.; Jayaraman, D.; Richards, A.L.; Kwiecien, N.W.; Siahpirani, A.F.; Rajasekar, S.; Maeda, J.; Garcia, K.; Del Valle-Echevarria, A.R.; et al. A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat. Biotechnol. 2016, 34, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Yates, R.J.; Howieson, J.G.; Reeve, W.G.; O’Hara, G.W. A re-appraisal of the biology and terminology describing rhizobial strain success in nodule occupancy of legumes in agriculture. Plant Soil 2011, 348, 255–267. [Google Scholar] [CrossRef]
- Makoudi, B.; Kabbadj, A.; Mouradi, M.; Amenc, L.; Domergue, O.; Blair, M.; Drevon, J.J.; Ghoulam, C. Phosphorus deficiency increases nodule phytase activity of faba bean–rhizobia symbiosis. Acta Physiol. Plant. 2018, 40, 63. [Google Scholar] [CrossRef]
- Valentine, A.J.; Kleinert, A.; Benedito, V.A. Adaptive strategies for nitrogen metabolism in phosphate deficient legume nodules. Plant Sci. 2017, 256, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Cabeza, R.A.; Liese, R.; Lingner, A.; von Stieglitz, I.; Neumann, J.; Salinas-Riester, G.; Pommerenke, C.; Dittert, K.; Schulze, J. RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N-2 fixation before emerging P deficiency reaches the nodules. J. Exp. Bot. 2014, 65, 6035–6048. [Google Scholar] [CrossRef]
- Chen, L.; Qin, L.; Zhou, L.; Li, X.; Chen, Z.; Sun, L.; Wang, W.; Lin, Z.; Zhao, J.; Yamaji, N.; et al. A nodule-localized phosphate transporter GmPT7 plays an important role in enhancing symbiotic N2 fixation and yield in soybean. New Phytol. 2019, 221, 2013–2025. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, J.; Tan, Z.; Zeng, R.; Liao, H. GmEXPB2, a Cell Wall beta-Expansin, Affects Soybean Nodulation through Modifying Root Architecture and Promoting Nodule Formation and Development. Plant Physiol. 2015, 169, 2640–2653. [Google Scholar] [PubMed]
- Sulieman, S.; Tran, L.S. Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci. 2015, 239, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Z.; Kong, Y.; Li, X.; Li, W.; Du, H.; Zhang, C. GmPAP12 Is Required for Nodule Development and Nitrogen Fixation under Phosphorus Starvation in Soybean. Front. Plant Sci. 2020, 11, 450. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Qian, W.Q.; Hurley, B.A.; She, Y.M.; Wang, D.W.; Plaxton, W.C. Biochemical and molecular characterization of AtPAP12 and AtPAP26: The predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ. 2010, 33, 1789–1803. [Google Scholar] [CrossRef] [PubMed]
- Hur, Y.J.; Jin, B.R.; Nam, J.; Chung, Y.S.; Lee, J.H.; Choi, H.K.; Yun, D.J.; Yi, G.; Kim, Y.H.; Kim, D.H. Molecular characterization of OsPAP2: Transgenic expression of a purple acid phosphatase up-regulated in phosphate-deprived rice suspension cells. Biotechnol. Lett. 2010, 32, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Li, X.; Ma, J.; Li, W.; Yan, G.; Zhang, C. GmPAP4, a novel purple acid phosphatase gene isolated from soybean (Glycine max), enhanced extracellular phytate utilization in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Tian, J.; Lam, H.M.; Lim, B.L.; Yan, X.; Liao, H. Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol. 2010, 152, 854–865. [Google Scholar] [CrossRef]
- Zhu, S.; Chen, M.; Liang, C.; Xue, Y.; Lin, S.; Tian, J. Characterization of Purple Acid Phosphatase Family and Functional Analysis of GmPAP7a/7b Involved in Extracellular ATP Utilization in Soybean. Front. Plant Sci. 2020, 11, 661. [Google Scholar] [CrossRef]
- Xu, H.Q.; Zhang, H.Y.; Fan, Y.K.; Wang, R.Y.; Cui, R.F.; Liu, X.Q.; Chu, S.S.; Jiao, Y.Q.; Zhang, X.G.; Zhang, D. The purple acid phosphatase GmPAP17 predominantly enhances phosphorus use efficiency in soybean. Plant Sci. 2022, 320, 111283. [Google Scholar] [CrossRef]
- Roussis, A.; Flemetakis, E.; Dimou, M.; Kavroulakis, N.; Venieraki, A.; Aivalakis, G.; Katinakis, P. Nodulin PvNOD33, a putative phosphatase whose expression is induced during Phaseolus vulgaris nodule development. Plant Physiol. Biochem. 2003, 41, 719–725. [Google Scholar] [CrossRef]
- Li, C.; Li, C.; Zhang, H.; Liao, H.; Wang, X. The purple acid phosphatase GmPAP21 enhances internal phosphorus utilization and possibly plays a role in symbiosis with rhizobia in soybean. Physiol. Plant 2017, 159, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Ott, T.; van Dongen, J.T.; Gunther, C.; Krusell, L.; Desbrosses, G.; Vigeolas, H.; Bock, V.; Czechowski, T.; Geigenberger, P.; Udvardi, M.K. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 2005, 15, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Sulieman, S.; Ha, C.V.; Schulze, J.; Tran, L.S. Growth and nodulation of symbiotic Medicago truncatula at different levels of phosphorus availability. J. Exp. Bot. 2013, 64, 2701–2712. [Google Scholar] [CrossRef] [PubMed]
- Thuynsma, R.; Valentine, A.; Kleinert, A. Phosphorus deficiency affects the allocation of below-ground resources to combined cluster roots and nodules in Lupinus albus. J. Plant Physiol. 2014, 171, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Li, C.C.; Gui, S.H.; Yang, T.; Walk, T.; Wang, X.R.; Liao, H. Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann. Bot. 2012, 109, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Zhao, J.; Tian, J.; Chen, L.; Sun, Z.; Guo, Y.; Lu, X.; Gu, M.; Xu, G.; Liao, H. The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 2012, 159, 1634–1643. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, X.; Xu, Y.; Yao, M.; Xie, F.; Gai, J.; Li, Y.; Yang, S. Soybean SPX1 is an important component of the response to phosphate deficiency for phosphorus homeostasis. Plant Sci. 2016, 248, 82–91. [Google Scholar] [CrossRef]
- Araújo, A.P.; Plassard, C.; Drevon, J.J. Phosphatase and phytase activities in nodules of common bean genotypes at different levels of phosphorus supply. Plant Soil 2008, 312, 129–138. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, Z.W.; Du, H.; Xing, X.Z.; Li, W.L.; Kong, Y.B.; Li, X.H.; Zhang, C.Y. A small heat shock protein, GmHSP17.9, from nodule confers symbiotic nitrogen fixation and seed yield in soybean. Plant Biotechnol. J. 2021, 20, 103–115. [Google Scholar] [CrossRef]
- Li, X.; Zheng, J.; Yang, Y.; Liao, H. INCREASING NODULE SIZE1 Expression Is Required for Normal Rhizobial Symbiosis and Nodule Development. Plant Physiol. 2018, 178, 1233–1248. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).