Abstract
Clostridioides difficile (C. difficile) is a global threat and has significant implications for individuals and health care systems. Little is known about host molecular mechanisms and transcriptional changes in peripheral immune cells. This is the first gene expression study in whole blood from patients with C. difficile infection. We took blood and stool samples from patients with toxigenic C. difficile infection (CDI), non-toxigenic C. difficile infection (GDH), inflammatory bowel disease (IBD), diarrhea from other causes (DC), and healthy controls (HC). We performed transcriptome-wide RNA profiling on peripheral blood to identify diarrhea common and CDI unique gene sets. Diarrhea groups upregulated innate immune responses with neutrophils at the epicenter. The common signature associated with diarrhea was non-specific and shared by various other inflammatory conditions. CDI had a unique 45 gene set reflecting the downregulation of humoral and T cell memory functions. Dysregulation of immunometabolic genes was also abundant and linked to immune cell fate during differentiation. Whole transcriptome analysis of white cells in blood from patients with toxigenic C. difficile infection showed that there is an impairment of adaptive immunity and immunometabolism.
1. Introduction
Clostridioides difficile (formerly known as Clostridium difficile and commonly referred to as C. difficile) is an established threat to public health globally and a significant burden to healthcare systems [,,]. Lately, there have been worries about its re-emergence [] (Figure S1). The United Kingdom (UK), which has one of the lowest incidence rates in the world, has now had a 5-year consecutive increase (January 2019–April 2023) corresponding to a 10-year high []. This trend is echoed in reports from Australia and other countries [,]. Even in areas with decreasing health care-associated infections (US and Canada), recurrent and community-acquired infections appear to be on the rise [,,,,]. Lack of centralized surveillance systems in Asia and Africa, reduced testing during the coronavirus-19 pandemic (COVID-19), and underdiagnosis mean that there is an underestimate of the true epidemiological burden globally [,,,].
C. difficile is a resilient, versatile pathogen. Over the years, it has evolved to develop various survival mechanisms, as reflected in its remarkable phylogenetic diversity, with sequenced isolates sharing less than one-fifth of genes []. Transmission sources are abundant given that C. difficile persists in both the environment and hosts; the spores are extremely difficult to eradicate and are responsible for (re-) initiating infection [,,]. It is intrinsically resistant to most antibiotics without compromising fitness, and there is an emergence of strains with decreased susceptibility to vancomycin and metronidazole, which are two of three available anti-microbials for C. difficile [,,]. In a US study, the majority of patients were healthy adults less than 65 years old [], while a Swedish nationwide population-based study showed that mortality increased with infection irrespective of age and co-morbidity []. Asymptomatic carriage has been reported in around 3% of the adult population, but it can be as high as 10% in health-care settings []. Intestine pathogens facilitate C. difficile carriage and shedding [,], but colonization and symptomatic infection are indistinguishable with routine diagnostics, making management difficult [].
Increased recurrence rates of C. difficile infection (~30% for a first episode) remain a major challenge—this increases mortality and subsequent relapses [,]. Fidaxomicin, which has narrow spectrum activity, has been shown to reduce recurrence rate, contrary to metronidazole, which has a wide anti-microbial burden that can have a deleterious effect on the gut microbiome [,]. The effectiveness of fecal microbiota transplant in recurrent but not primary infections underscores the importance of gut microbiome as a protective factor []. More recently, a microbiota-based oral therapeutic of Firmicutes spores (VOWST; VOS, formerly SER-109) has demonstrated promising results in clinical trials [,,]. Another strategy to prevent recurrent infection is the use of monoclonal antibodies against C. difficile toxin B (bezlotoxumab) [,]. Patients who effectively produce neutralizing antibodies are at reduced risk of recurrence [,]. Hence, the development of a vaccine may be feasible. Currently, there is a candidate (PF-06425090) in late-phase clinical trials that does not prevent infection but reduces severity following a three-dosing (at least) regimen to achieve sustained immunity [,].
Overall, the interplay between gut dysbiosis and host immunity is complex and poorly investigated [,]. Thus, mechanistic understanding of C. difficile infection is important as it will help in developing better treatment and prevention strategies. Our study attempted to shed light on immune responses triggered by C. difficile at the gene expression level in peripheral blood. Assuming that gut dysbiosis and C. difficile infection, particularly, have an effect on the transcriptome of circulating cells, it is possible that this may result in a set of commonly expressed genes in patients with diarrhea due to different causes and a unique gene expression signature associated with C. difficile infection.
2. Results
2.1. Patient Characteristics
We collected clinical data and samples from 200 patients with diarrhea and 51 healthy controls (HC) across five sites in the Northwest of England between February 2013 and July 2015 (Figure S2). Patients infected with C. difficile were either cases, CDI (n = 78), or controls, GDH (n = 37), depending on the presence or absence of toxin, respectively. Of the 40 patients with inflammatory bowel disease (IBD), 67% (n = 27) had ulcerative colitis, 25% (n = 10) had Crohn’s disease and 8% (n = 3) had non-specific colitis. Diarrhea controls (DC, n = 45) comprised patients with infective (n = 16, 36%), antibiotic-associated (n = 8, 18%), and non-specific (n = 21, 47%) diarrhea (Figure 1).
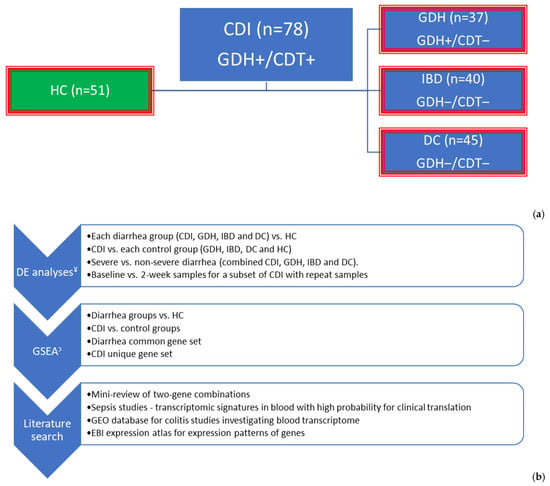
Figure 1.
Graphical summary of methodology: (i) To identify diarrhea common immune responses in peripheral blood, each diarrhea group (blue boxes) was compared with HC (green box), and the overlapped genes in all four differential expression analyses were extracted. (ii) To identify CDI unique genes, CDI was compared with each of the control groups (red outline), and the overlapped genes in all four differential expression analyses were extracted. (iii) Functional annotation of differentially expressed genes and gene sets was performed with GSEA and literature searches. (a) Cases and controls and (b) Analysis plan. ¥ The primary filter for DE analysis was FDR adj. p < 0.05. When gene-sets were required to have a maximum number for GSEA (e.g., String database 3000 proteins max), |log2FC| < 0.5 was added. To reduce further the number of genes for literature searches, we used the top 20 genes with the highest |log2FC|, ↄ: GSEA was performed using R studio (Reactome pathways), IPA (summary and comparison), and String online database. DE: differential expression, CDI: toxigenic C. difficile infection, GDH: non-toxigenic C. difficile infection, IBD: inflammatory bowel disease, DC: diarrhea controls, HC: healthy controls, NICE: National Institute for Health and Care Excellence, GSEA: gene set enrichment analysis, EBI: European Bioinformatics Institute, FDR adj. p: false discovery rate adjusted p-value, |log2FC|: an absolute value of logarithm with base 2 of fold change, IPA: Ingenuity Pathway Analysis by QIAGEN.
Of the CDI patients, 62% (n = 48) had severe disease as per NICE criteria [,,]. CDI patients were older with more co-morbidities, increased frailty, and more likely to have more than one acute illness compared with the IBD and HC groups (Table 1). IBD was the youngest group with the fewest co-morbidities but with the most severe symptoms in relation to the number of bowel movements and duration (Table 1). The diarrhea groups were inpatients, while HC were mainly (n = 28, 55%) attenders at hypertension, clinical biochemistry, and general internal medicine clinics (Figure S3). As a result, HC had a higher incidence of risk factors for cardiovascular disease (CVD), including high blood pressure, hyperlipidemia, obesity, smoking, and alcohol excess (Table 1).

Table 1.
Clinical variables per study group.
The presenting complaints of all IBD patients were diarrhea and/or symptoms consistent with colitis (e.g., abdominal pain, per rectal bleeding), which led to early stool sampling and recruitment (Table 1). Most CDI and GDH patients had an acute illness which was followed by C. difficile infection (Table 1). The presence of entero-colic disease on admission was associated with earlier clinical stool samples and transcriptomic samples (Kruskal–Wallis test p < 0.001). There was no difference for the period between the transcriptomic sample and the worst day of diarrhea or antibiotic initiation among the groups (Table 1).
The distribution of white cell count (WCC) was the same across the groups, but if WCC was combined with temperature and creatinine to reflect severity, CDI patients had significantly more severe disease (Table 1). The overall mortality in the year following recruitment to this study was higher in patients with toxigenic and non-toxigenic C. difficile infection, with the CDI group having higher 28-day all-cause mortality (Table 1).
Half of the CDI (50%, n = 39) and more than half of the GDH (60%, n = 22) patients had a hospital-acquired infection, of which 67% (n = 26) and 73% (n = 16), respectively, had prior use of antibiotics. Around two-thirds of CDI and GDH patients were on a proton pump inhibitor (PPI) at the time of infection, and this figure was similar in DC (Table 1). Of the CDI and GDH stool samples, 68 (87%) and 20 (54%), respectively, were ribotyped: 002, 014, and 078 were the most frequent isolates (Figure S4a). Toxin genes were detected with multiplex PCR in 61% of isolates (n = 11) from a subset (n = 18) of the GDH stool samples (Figure S4b). Although clinical testing for C. difficile was negative (GDH-/CDT-) for the IBD and DC cohorts, stool cultures were positive in 5% (n = 2) and 16% (n = 7), respectively (Figure S5). Asymptomatic carriage was 2% (n = 1) amongst the HC.
2.2. Patients with Different Types of Diarrhea Have Significant Variation at the Gene Expression Level in Peripheral Blood Enriched for Innate Immunity and Neutrophil Activation
Principal component analysis (PCA) of the blood transcriptome showed that 29.7% of the total variance was represented by the first two principal components (Figure 2). Differential expression analysis identified a few thousand genes with a false discovery rate (FDR) adj. p < 0.05, indicating expression in diarrhea groups varied from HC (Figure 3 and Figure 4). One-fifth (n = 2255) of the differentially expressed genes were common in all diarrhea groups (Figure 5a), and the most significant reactome pathways were neutrophil degranulation and innate immune response (Figure 5b).
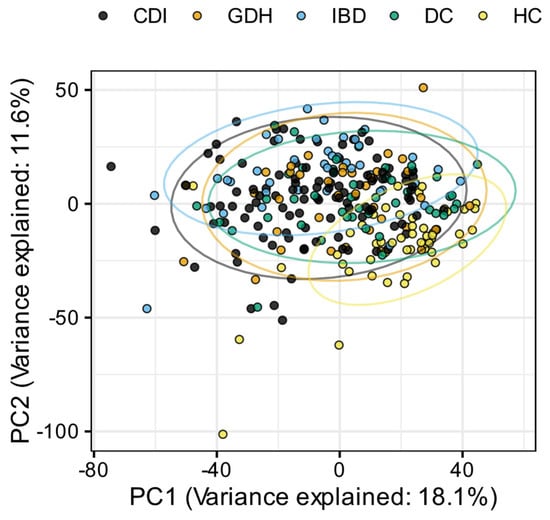
Figure 2.
Principal component analysis (PCA) normalized gene expression data for each sample, colored by participant groups. CDI: toxigenic C. difficile infection, GDH: non-toxigenic C. difficile infection, IBD: inflammatory bowel disease, DC: diarrhea controls, HC: healthy controls, PC1: primary component 1, PC2: primary component 2.
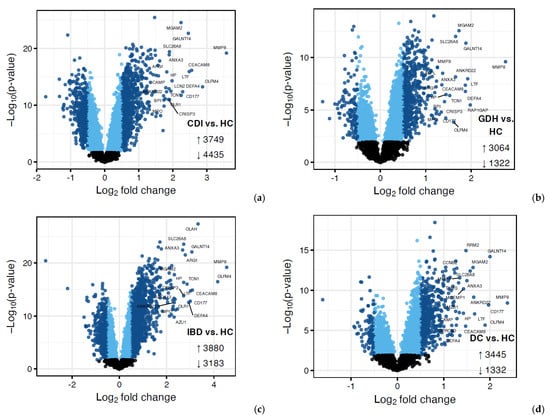
Figure 3.
Volcano plots (a–d) of differential expression (DE) analysis of diarrhea groups vs. HC where genes with FDR adjusted p < 0.05 are displayed in blue, with darker blue points indicating those with |log2FC| > 0.5. CDI: toxigenic C. difficile infection, GDH: non-toxigenic C. difficile infection, IBD: inflammatory bowel disease, DC: diarrhea controls, HC: healthy controls, FDR adj. p: false discovery rate adjusted p-value, |log2FC|: logarithm with base 2 of fold change.
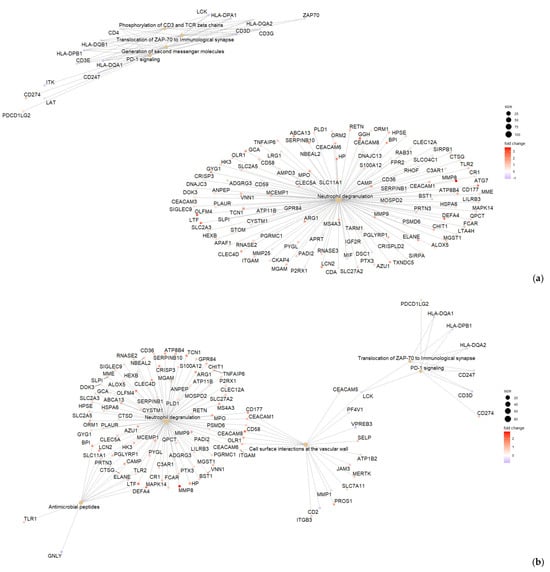

Figure 4.
Enrichment summaries of differentially expressed genes with FDR adjusted p < 0.05 and|log2FC| > 0.5 between CDI vs. HC (a), GDH vs. HC (b), IBD vs. HC (c) and DC vs. HC (d) developed with IPA. CDI: toxigenic C. difficile infection, GDH: non-toxigenic C. difficile infection, IBD: inflammatory bowel disease, DC: diarrhea controls, HC: healthy controls, FDR adj. p: false discovery rate adjusted p-value, |log2FC|: logarithm with base 2 of fold change, IPA: ingenuity pathway analysis.
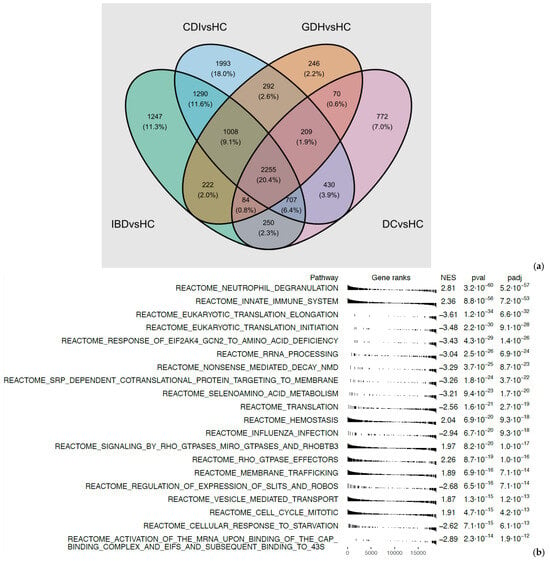
Figure 5.
(a) Venn diagram of diarrhea groups vs. HC (FDR adj. p-value < 0.05) and (b) Reactome pathways from GSEA of pooled t statistics comparing all diarrhea groups vs. HC. CDI: toxigenic C. difficile infection, GDH: non-toxigenic C. difficile infection, IBD: inflammatory bowel disease, DC: diarrhea controls, HC: healthy controls, FDR adj. p: false discovery rate adjusted p-value.
We narrowed down differentially expressed genes by applying an additional criterion of |log2FC| > 0.5. All diarrhea groups co-shared 362 genes, and neutrophil degranulation remained the most significantly upregulated pathway (Figure 4 and Figure S6). Downregulation of adaptive immunity appeared unique to CDI (Figure S6).
To search the literature, we confined differentially expressed genes between each diarrhea group and HC to the top 20 with the highest fold-change. Common genes in all comparisons were 12 (ANKRD22, ANXA3, CD177, CEACAM8, DEFA4, GALNT14, HP, LTF, MGAM2, MMP8, OLFM4, and SLC26A8) and they were all involved in neutrophil functions. A mini-review showed that transcripts from the common 12-gene set were found in the blood of patients with various inflammatory conditions, including sepsis and infections, cardiovascular disease, auto-immune diseases, cancer, mental illness, and pregnancy (Table S1). Although we excluded studies with samples other than whole blood (e.g., PBMC and leucocytes) and non-adjusted p-values, we still encountered some of the common genes in non-eligible work (data not presented). Then, we focused on sepsis studies where members of the common 12-gene set were found in two studies (differentially expressed genes between sepsis endotypes) and three classifiers, two of which were derived from multi-cohort analysis (Table S2). Moreover, the GEO database was searched to retrieve studies investigating the blood transcriptome in colitis (Table S3), and we encountered genes from the 12-gene set in six of the seven eligible publications (Table S4). The most popular genes in all our searches were OLFM4, CEACAM8, HP, and MMP8 and the least frequent (found only in an IBD study) was MGAM2.
To investigate genes that were associated with severity, we modified the CDI criteria for severity and applied them to all the diarrhea groups (Table S5). We found 487 differentially expressed genes with an FDR adj. p < 0.05 between severe vs. non-severe all-cause diarrhea cases, which were enriched for neutrophil degranulation and innate immunity. Of this 487-gene list, 236 were dysregulated significantly in severe CDI, and SLPI was amongst the top 20 upregulated. Of the common 12-gene set, 8 (67%) genes were significantly upregulated in patients with severe diarrhea compared with those with non-severe diarrhea (Table S6).
A subset of CDI patients was sampled 2 weeks following infection (n = 25). Due to the small sample size, we used gene-sets for differential expression analysis. Most genes (n = 10) of the common 12-gene set showed a reduction in expression from baseline (Table S6), and only SLPI in the severity 487-gene set was significantly (FDR adj. p < 0.05) decreased in repeat sampling.
IBD was the only group where transcriptomics in blood had been performed previously. Of the seven studies we found through the GEO database (Table S3), three presented differentially expressed genes between patients with ulcerative colitis and/or Crohn’s disease and healthy controls. We extracted genes with an FDR adj. p < 0.05 and |log2FC| > 0.5 (or |FC| > 1.5) from these studies. There was an overlap with our cohort (1769 differentially expressed genes between IBD and HC with an FDR adj. p < 0.05 and |log2FC| > 0.5), as shown in Figure 6.
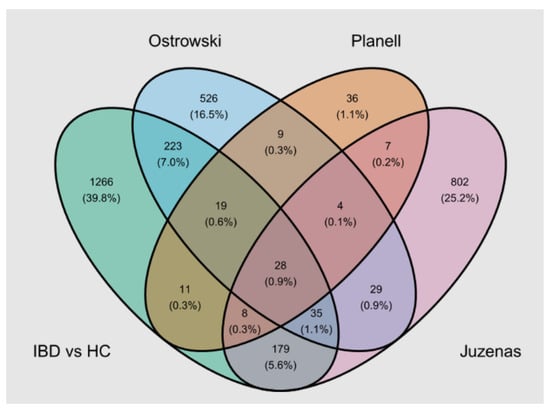
Figure 6.
Venn diagram representing the overlap of differentially expressed genes between IBD and HC (FDR adj. p-value < 0.05 and |log2FC| > 0.5) among our cohort (IBD vs. HC) and published studies [,,]. IBD: inflammatory bowel disease, HC: healthy controls.
2.3. CDI Is Associated with Dysregulation of Adaptive Immunity
We compared CDI with the three diarrhea groups (GDH, IBD, and DC), and we found that gene expression in peripheral blood differed, with most genes being downregulated in CDI (Figure 7a–c). Although neutrophil degranulation was observed between CDI vs. DC, it did not appear when CDI was compared with IBD or GDH (Figure 7d–f). A comparison of CDI vs. all four controls with Ingenuity Pathway Analysis (IPA) revealed that the IL-8 pathway was the only significant canonical pathway (|z-score| > 2) in all comparisons, which was least upregulated in CDI compared with the control groups (Figure 8a).
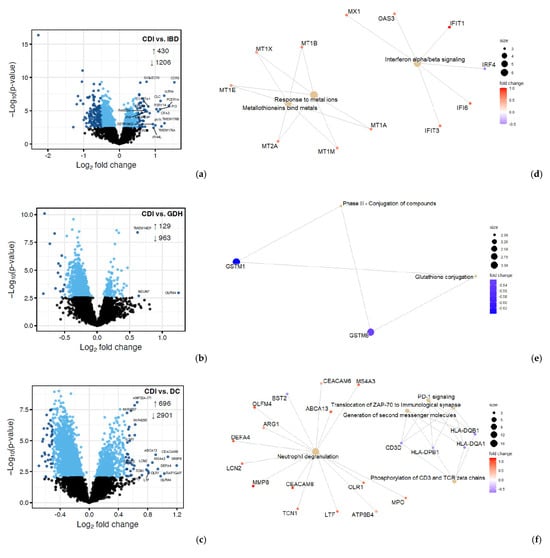
Figure 7.
Differential expression (DE) analysis of CDI vs. diarrhea groups. (a–c) Volcano plots where genes with FDR adjusted p < 0.05 are displayed in blue, with darker blue points indicating those considered differentially expressed for IPA. (d–f) Enrichment summaries of genes with |log2FC| > 0.5. CDI: toxigenic C. difficile infection, GDH: non-toxigenic C. difficile infection, IBD: inflammatory bowel disease, DC: diarrhea controls, FDR adj. p: false discovery rate adjusted p-value, |log2FC|: logarithm with base 2 of fold change, IPA: Ingenuity Pathway Analysis.
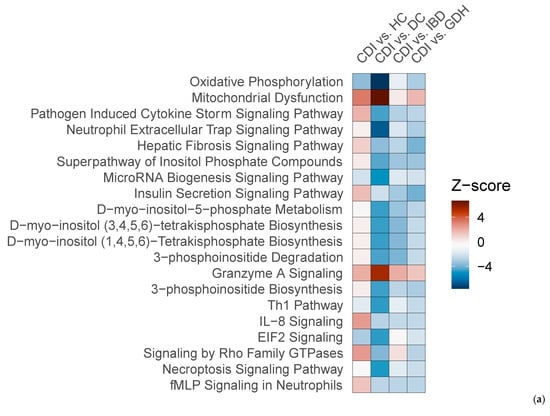
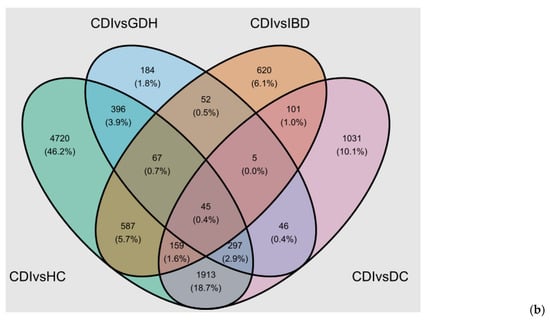
Figure 8.
(a) Top 20 canonical pathways of the IPA comparison of CDI vs. HC, CDI vs. DC, CDI vs. IBD, and CDI vs. GDH (|z-score| > 2) and (b) Venn diagram of CDI vs. all control groups (FDR adj. p-value < 0.05). CDI: toxigenic C. difficile infection, GDH: non-toxigenic C. difficile infection, IBD: inflammatory bowel disease, DC: diarrhea controls, HC: healthy controls, FDR adj. p: false discovery rate adjusted p-value.
We identified 45 genes that were uniquely dysregulated in CDI compared with all four control groups, and all but three were downregulated (Figure 8b). Gene set enrichment analysis (GSEA) did not return any enriched terms with any resource (Reactome and Hallmark database via R studio, Spring, DAVID). We checked the 45 gene list in severe (n = 48) vs. non-severe (n = 30) CDI cases (as defined by NICE) and in the subset of baseline vs. repeat samples (n = 25), and there were no genes with an FDR adj. p < 0.05.
Functions were, therefore, manually curated and summarized for each gene of the 45 unique signature (Table 2). Over one-third of genes (n = 16) were involved in mitochondrial function with downregulation of factors participating in fatty acid oxidation and mitophagy (mitochondrial autophagy). Almost a third of genes (n = 13) were associated with T-cell memory and humoral immunity. The rest had important roles in metabolism and the cytoskeleton or were poorly described in the literature. There were two transcripts, GPAA1 and PIGU, which were directly associated as subunits for the glycosylphosphatidylinositol transamidase (GPI-T) complex. GPI-T is composed of five subunits (GPAA1, PIGU, PIGS, PIGT, and PIGK) [], with PIGS and PIGT also being significantly downregulated in CDI and IBD (vs. HC).

Table 2.
Functions of the CDI unique 45 gene set. Most genes were downregulated in CDI compared with all control groups (GDH, IBD, DC, and HC), and only three genes (ARLNC1, NSUN7, and OLFM4) were upregulated.
3. Discussion
This is the first report of altered gene expression in peripheral blood during C. difficile infection. We identified an innate immune response, where neutrophils played a central role, shared by all diarrhea groups, whereas colitis due to toxigenic C. difficile infection was characterized by the downregulation of genes associated with adaptive immunity.
As expected, our groups of patients were remarkably heterogeneous. The C. difficile infection (toxigenic and non-toxigenic) cohort was typical, mostly older than 65 years and frail with many co-morbidities, had prolonged hospitalization with an acute illness, and demonstrated increased mortality [,]. Subjects with IBD were in their 40s, mildly frail, and had prolonged diarrhea before presentation and sampling. The DC group laid in the middle with advanced age and many co-morbidities but was generally less frail and had shorter hospitalization before recruitment compared with the CDI and GDH groups.
Despite the heterogeneity between groups, we identified a common inflammatory response at the transcription level in blood. Innate immunity and particularly neutrophil-related functions were significantly upregulated in all diarrhea groups compared with our HC, which was consistent with previous reports []. This non-specific response in the blood was shared in most inflammatory conditions, including sepsis. Presumably, there were more than 12 (that our search was confined to) co-shared genes from the big pool of common genes (2255). Circulating neutrophil subpopulations, although they have lower total RNA content per cell compared with other circulating cells, alter their transcriptome to minor environmental changes such as brief exercise, and this enhancement is proportional to the extent of inflammation/disease and migration activity [,,,]. Nevertheless, the literature is scarce regarding the overlap in neutrophil activation signatures amongst the different inflammatory conditions [,].
Among the commonly encountered genes during inflammation, MGAM2, which encodes maltase-glucoamylase 2, was reported in a study in inflammatory bowel disease as a member of a multi-gene set in blood reflecting inflammation in the gut []. It was upregulated in severe cases but did not significantly decrease in the two weeks after the onset of C. difficile infection. MGAM2 is largely uncharacterized and potentially associated with immune responses and glucose metabolism [,]. SLPI, on the other hand, was the only gene that was significantly upregulated in severe cases and downregulated in repeat sampling. SLPI is an anti-microbial peptide with anti-inflammatory properties secreted by epithelial and immune cells [,]. Its high expression in various tumors has been inconsistently linked to prognosis depending on the cancer stage [,,]. Moreover, it is a principal regulator of gut responsiveness to microbial stimuli by ameliorating the destructive effect of inflammation []. Colonic recruitment of monocytes and neutrophils during experimental C. difficile infection in mice has been proposed to be a major producer of SLPI [,].
Our CDI cohort uniquely expressed a 45 gene signature. Although the 45 unique genes were not computationally enriched, we identified important patterns through analysis of the literature. Most genes in CDI play important roles in adaptive immunity and cell metabolism, particularly fatty acid oxidation and mitochondrial homeostasis. Downregulation of immune-related genes such as CD19 (B cell-specific marker), SPIB (B cell memory), STING1 (T cell memory), GPA33 (memory CD4), VHL (memory CD8), and LPAR5 (effector lymphocytes) potentially points towards an impairment in antibody production and differentiation into memory cells.
In concert, metabolic reprogramming and its principal regulators, the mitochondria, directly influence immune cell proliferation, differentiation, homeostasis, and recall responses [,]. Hence, reduced expression of genes involved in the critical steps of catabolism such as fatty acid oxidation and subsequent oxidative phosphorylation (COQ10A, CYB561A3, NDUFAF3, PEX3, SMIM20, ALDH9A1, and COQ6), and upregulation of facilitators of rapid energy production (NSUN7, VHL) portray an active effector phenotype of immune cells rather than quiescent memory. Moreover, disrupted autophagy in memory lymphocytes results in the accumulation of damaged mitochondria, disrupted fatty acid oxidation, and attenuated secondary immune responses [,]. This was observed in our CDI unique signature by downregulation of genes maintaining mitochondrial integrity (CYB561A3, NAA30, PMPCA, and TOP1MT) and promoting mitophagy (EI24, FANCF, and RNF41) or autophagy (GPAA1, PIGU, and SPIB).
Immunometabolism is a relatively recent concept but also a rapidly evolving field interlinked with oxidation–reduction (redox) reactions [,]. In the colonic mucosa of mice infected with C. difficile, upregulation of glycolytic metabolism has been observed during the peak of inflammation which reverted to mitochondrial metabolism during the recovery phase []. Key players in lymphocyte fate and metabolism are mitochondria [] and mitochondrial autophagy (or mitophagy) is necessary for maintaining long-lived IgG memory B cells []. This could be crucial in the generation of humoral immunity. Indeed, the production of neutralizing antibodies against C. difficile toxins has been shown to be ineffective in various reports [,]. This is also reflected in the increased recurrence rate and low immunogenicity [,]. T cell-mediated immunity is even more elusive, but evidence suggests that CD4 cells may have an important role in C. difficile clearance and reversal of gut microbiome homeostasis [,].
Our unique CDI signature included genes associated with the cytoskeleton and genomic stability, which appear to have important roles in cell homeostasis. Cytoskeleton components play important roles in lymphocyte polarization and communication through the immunological synapse, as well as receptor signaling [,,]. Similarly, DNA damage repair systems are crucial to adaptive immunity for receptor diversity (TCR/BCR) and antibody specificity (V(D)J recombination, class switch recombination, and somatic hypermutation) [].
Interestingly, some of the 45 unique genes could be mediators of C. difficile pathogenesis or targets of toxin A (TcdA) and B (TcdB). TcdA binds to sulfated glycosaminoglycans, which are found on GPI-anchored receptors (GPAA1, PIGU), while low-density lipoprotein receptor (LDLRAP1) assists cell entry []. TcdB binds frizzled receptors inhibiting the Wnt/β-catenin pathway (OLFM4, STING1), which is required for the generation of CD8 memory stem cells [,,]. Both toxins inactivate small Rho GTPases by glycosylation, resulting in cytoskeletal disruption, cell rounding, cell cycle arrest, and death []. Moreover, they induce preliminary Ca2+ influx, disrupt mitochondrial integrity, and generate reactive oxygen species (ROS) with subsequent redox (MOCS1) signaling and DNA damage [,]. C. difficile toxins cause mitochondrial damage in the colonic epithelium []. It is possible that circulating immune cells are infected with C. difficile toxins, but their effect at the molecular level can only be speculated [,].
Our study has several limitations. First, this is a discovery cohort, and we have not performed clinical or technical validation. We regrettably report that top genes have not been confirmed with quantitative or droplet PCR due to inadequate sample volume. As this is the first report of transcriptomic changes in peripheral blood in patients with C. difficile infection, the lack of publicly available datasets prevented in silico validation. Therefore, data should be used cautiously in the future in consideration of the lack of validation; however, it is reassuring from the experimental perspective that common and IBD-related genes were replicated in other work. Second, the findings are based on gene expression products, and we have not explored proteins or proteomics. Functions have been inferred from the protein end-product and even though mRNA and protein levels are correlated in the literature, we have not performed confirmatory experiments in our cohort. Nevertheless, links among CDI-unique genes provide compelling avenues for further investigation. Third, we explored bulk RNA in blood which is justified given that this topic has not been previously approached in the literature. The findings question our understanding of peripheral host immune responses to toxigenic C. difficile and offer directions for further study with more precise and accurate methods, such as single-cell RNA-sequencing in blood and spatial transcriptomics in colonic tissue.
4. Materials and Methods
4.1. Study Population
Patients with diarrhea were recruited from five hospitals (Royal Liverpool Hospital or RLH, University Hospital of Aintree or UHA, St. Helen’s and Knowsley or STK, Whiston Trust Hospital or WTH and Walton Centre for Neurology and Neurosurgery or WNN) in the Northwest of England. Patients who developed diarrhea that led to or during hospital admission and were categorized between grades five and seven on the Bristol stool chart were allocated into four groups: CDI, GDH, IBD, and DC. Stool samples were tested for glutamate dehydrogenase (GDH) and C. difficile toxin (CDT) A and/or B and/or binary toxin with a two-step protocol [] at the local clinical microbiology laboratory. CDI cases were toxin-positive with a double positive test (GDH+/CDT+), and diarrhea controls were toxin-negative (Figure 1). Patients with previously confirmed C. difficile infection were excluded. We also recruited healthy volunteers (HC).
Severe CDI cases were defined as per the NICE and European criteria for C. difficile infection [,]. We used a modification of these criteria to apply to all diarrhea groups. Severe cases were defined if they had one of (1) white cell count (WCC) > 15 × 109 cells/L, (2) serum creatinine > 133 μmol/L, and (3) temperature > 38.5 °C (Table S5).
4.2. Sample Collection and Processing
Stool and blood samples were collected at the time of recruitment (baseline) for all subjects and two weeks following the diagnosis of C. difficile infection (repeat) for a subset of CDI patients.
Stool samples with sufficient yield following routine testing were anonymized and stored at −80 °C. All fecal samples underwent alcohol-shock treatment, followed by a culture for C. difficile on Brazier’s cefoxitin–cycloserine egg yolk agar (EO Labs, Bonnybridge, United Kingdom). Internal core sequences of both toxins A and B genes and a species-specific internal fragment of the triose-phosphate isomerase (TPI) housekeeping gene were targeted by a multiplex PCR assay in control samples, as previously described [].
Whole blood samples were collected in Tempus Blood RNA Tubes (Thermo Fisher Scientific, Loughborough, UK) and stored at −80 °C. RNA was purified with the Tempus Spin RNA Isolation kit (Applied Biosystems, Foster City, USA), which utilizes a glass fiber filter-based technique for RNA isolation as per the manufacturer’s instructions. The quality control (QC) assessment of extracted RNA was performed with the Agilent 2100 Bioanalyzer and Thermo Scientific NanoDrop Spectrophotometer. Samples with RNA integrity number (RIN) ≥ 6.5 (Figure S8) and RNA minimum quantity of 50 ng were processed with the ClariomTM D assay for Humans on the GeneChip console (Affymetrix, Singapore ) at the European (Nottingham) Arabidopsis Stock Centre (NASC) as per manufacturer protocol (GeneChip WT PLUS Reagent Kit User Guide (P/N 703174, Rev A.0, Thermo Fisher Scientific Inc.).
4.3. Microarray Data Processing
All microarray data preprocessing and analyses were performed in R version 4.4.2. The code to perform the R analyses in this paper is available at “https://github.com/CBFLivUni/Gene_expression_dysregulation_in_whole_blood_of_patients_with_C_difficile_infection.git (accessed on 21 November 2024)”.
Clariom D files were imported, and log2-transformed raw data were inspected for outliers using relative log expression plots. One DC sample was excluded from further analysis as all probe intensity values were considerably lower than the remaining microarrays, with many below the limit of detection. Background subtraction, quantile normalization, and summarization were performed using the robust multichip average algorithm at the ‘core’ target level, as implemented by the ‘oligo’ R package.
Prior to statistical analysis, a background intensity threshold was chosen based on histograms of median transcript cluster (TC) intensity across all samples and background control probe sets. TCs with at least 37 samples—the smallest disease group set—possessing greater intensity than the threshold were included in the analyses.
TCs were annotated with gene IDs using the ‘AnnotationDbi’ package. Those that mapped to multiple Entrez gene IDs were excluded, and where multiple TCs mapped to the same gene ID the TC with the greatest mean expression across samples was selected and the remainder filtered out.
4.4. Differential Expression Analyses
Tests for differences in mean RNA abundance between disease groups were performed using the ‘limma’ R package, adjusting for age and sex. To allow for potential non-linear associations between age and gene expression, age was fitted as a natural spline with 3 degrees of freedom. The potential effects of the microarray batch were assessed by principal component analysis (Figure S7). No clear association was apparent, and batch ID was not adjusted for in the differential expression analyses.
The p-values were adjusted for FDR using the Benjamini–Hochberg method, with gene contrasts yielding adjusted p-values < 0.05 considered to be differentially expressed.
4.5. Gene Set Enrichment Analysis
GSEA was performed ranking genes by the mean of t-statistics from differential expression analyses of each diarrhea group vs. HC and CDI vs. each control group using the fgsea package [], with Hallmark and Reactome gene set downloaded from “https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp (accessed on 3 August 2022)”.
4.6. Ingenuity Pathway Analysis
- Common genes: Diarrhea common genes were identified through Venn diagrams at the area of convergence from the differential expression analysis (FDR adj. p < 0.05) of each diarrhea group vs. the HC arm. To narrow down the number of common genes, filtering criteria were applied, including an absolute value of the logarithm with base 2 of fold change (|log2FC|) > 0.5 and the top 20 dysregulated genes with the highest |log2FC|.
- Unique genes: CDI unique genes were identified through Venn diagrams at the area of convergence of differential expression analysis (FDR adj. p < 0.05) of CDI cases vs. each control group (GDH, IBD, DC, and HC).
- Severity genes: Diarrhea severity genes derived from differential expression analysis (FDR adj. p < 0.05) between severe vs. non-severe diarrhea (combined CDI, GDH, IBD, and DC) as defined with the modified criteria.
The IPA by Qiagen, the String v12.0 database (https://string-db.org/), and the Database for Annotation, Visualization, and Integrated Discovery (DAVID) were used for functional annotation of gene sets.
4.7. Literature Search
To answer the question if the common diarrhea gene set is specific to entero-colitis or encountered in other illnesses, the literature was searched for the top common genes from three different aspects: mini-review of the literature, blood transcriptomics in sepsis, and blood transcriptomics in colitis.
For the non-exhaustive mini-review, we conducted PubMed searches with two gene-symbol combinations from the diarrhea common signature. Studies were included if they had at least five subjects per group, whole genome expression in whole blood was investigated with commercial microarrays or RNA-sequencing, and differential gene expression analysis between cases and healthy controls used an adjusted p-value < 0.05. Only data from discovery cohorts were investigated. If a list of differentially expressed genes was not published, we selected important genes from the data presented in the publication. Important genes were hub or crucial genes suggested by the authors and/or genes that ranked highly with annotation tools such as protein-protein interactions (PPI) networks and IPA [,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,].
Sepsis is probably a field in which gene expression signatures will probably be translated into clinical practice in the near future. We searched sepsis studies, as previously summarized by Tsakiroglou et al. [], for the top common diarrhea genes [,,,,]. Finally, we also conducted a Gene Expression Omnibus (GEO) database search for colitis studies that have reported from the top common diarrhea gene set (Table S2) [,,,,,,].
To functionally annotate the CDI unique gene set, we searched PubMed and the European Bioinformatics Institute (EBI) Expression Atlas with the gene symbols and manually curated the most relevant information.
5. Conclusions
C. difficile is an interesting pathogen because of its resilience (extreme spore resistance), adaptation (remarkable phylogenetic variability, pronounced antimicrobial resistance, toxigenic and non-toxigenic isolates), and relationships (unique host and gut microbiome equilibrium, non-pathogenic colonization at an early age and asymptomatic carriers, increased risk for relapse). These unique characteristics, in concert with the lack of adequate therapeutics, impose a global threat and underscore the importance of understanding molecular mechanisms of host immune responses. We propose that future research with a focus on adaptive immunity and immunometabolism will fuel the discovery of novel therapeutic targets for C. difficile. Genes such as MGAM2 and SLPI require further characterization as biomarkers for patient stratification. Last but not least, the co-expression of neutrophil-related genes during various inflammatory conditions is a concept that warrants further investigation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms252312653/s1.
Author Contributions
Conceptualization, M.T., M.P. and F.M.; methodology, M.T., A.E., M.P. and F.M.; software, A.E. and M.T.; validation, M.T.; formal analysis, M.T. and A.E.; investigation, M.T., A.D.-C., P.R., M.L., R.H. and E.Z.; resources, M.P.; data curation, A.E.; writing—original draft preparation, M.T.; writing—review and editing, A.E., M.P. and M.T.; visualization, A.E. and M.T.; supervision, M.P.; project administration, M.T.; funding acquisition, M.P., F.M. and M.T. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by the MRC grant MR/K000551/1 (The Interactions between Clostridium difficile, Intestinal Microbiota, and the Host Response in Hospitalized Patients project).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the National Health Service (NHS) Ethics Committee (08/H1005/32).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Microarray data are available on Gene Expression Omnibus GEO, accession GSE276395, using the access token ‘ihixiuiajbgplar’ without quotes.
Acknowledgments
We wish to thank the NIHR through the Liverpool BRC for funding the work. We also want to thank all the patients who kindly donated their samples.
Conflicts of Interest
M.P. has received partnership funding for the following: MRC Clinical Pharmacology Training Scheme (co-funded by MRC and Roche, UCB, Eli Lilly, and Novartis). He has developed an HLA genotyping panel with MC Diagnostics but does not benefit financially from this. M.P. is part of the IMI Consortium ARDAT. None of this funding is related to the current paper.
References
- Balsells, E.; Shi, T.; Leese, C.; Lyell, I.; Burrows, J.; Wiuff, C.; Campbell, H.; Kyaw, M.H.; Nair, H. Global burden of Clostridium difficile infections: A systematic review and meta-analysis. J. Glob. Health 2019, 9, 010407. [Google Scholar] [CrossRef] [PubMed]
- CDC, Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/clostridioides-difficile-508.pdf (accessed on 3 November 2023).
- WHO. WHO 2023 Data Call is Now Open for Antibacterials in the Preclinical Development Pipeline. Available online: https://www.who.int/news-room/articles-detail/who-2023-data-call-is-now-open-for-antibacterials-in-the-preclinical-development-pipeline (accessed on 2 November 2023).
- Finn, E.; Andersson, F.L.; Madin-Warburton, M. Burden of Clostridioides difficile infection (CDI)—A systematic review of the epidemiology of primary and recurrent CDI. BMC Infect. Dis. 2021, 21, 456. [Google Scholar] [CrossRef] [PubMed]
- UK Health Security Agency. Quarterly Epidemiological Commentary: Mandatory Gram-Negative Bacteraemia, MRSA, MSSA and C. difficile Infections (Data up to January to March 2023). Available online: https://www.gov.uk/government/statistics/mrsa-mssa-gram-negative-bacteraemia-and-cdi-quarterly-report/quarterly-epidemiological-commentary-mandatory-gram-negative-bacteraemia-mrsa-mssa-and-c-difficile-infections-data-up-to-january-to-march-2023 (accessed on 2 November 2023).
- Australian Commission on Safety and Quality in Health Care. Resourse Library—C. difficile. Available online: https://www.safetyandquality.gov.au/publications-and-resources/resource-library?resource_search=c.%20difficile (accessed on 3 November 2023).
- Karampatakis, T.; Tsergouli, K.; Kandilioti, E.; Nikopoulou, A.; Katsifa, H.; Kachrimanidou, M. Implication of COVID-19 pandemic on the incidence of Clostridioides difficile infection in a Greek tertiary hospital. J. Med Microbiol. 2023, 72, 001689. [Google Scholar] [CrossRef]
- Du, T.; Choi, K.B.; Silva, A.; Golding, G.R.; Pelude, L.; Hizon, R.; Al-Rawahi, G.N.; Brooks, J.; Chow, B.; Collet, J.C.; et al. Characterization of Healthcare-Associated and Community-Associated Clostridioides difficile Infections among Adults, Canada, 2015-2019. Emerg. Infect. Dis. 2022, 28, 1128–1136. [Google Scholar] [CrossRef]
- Feuerstadt, P.; Theriault, N.; Tillotson, G. The burden of CDI in the United States: A multifactorial challenge. BMC Infect. Dis. 2023, 23, 132. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, Y.; Grinspan, A.M. Epidemiology of community-acquired and recurrent Clostridioides difficile infection. Ther. Adv. Gastroenterol. 2021, 14, 17562848211016248. [Google Scholar] [CrossRef]
- Liu, C.; Monaghan, T.; Yadegar, A.; Louie, T.; Kao, D. Insights into the Evolving Epidemiology of Clostridioides difficile Infection and Treatment: A Global Perspective. Antibiotics 2023, 12, 1141. [Google Scholar] [CrossRef] [PubMed]
- Okafor, C.M.; Clogher, P.; Olson, D.; Niccolai, L.; Hadler, J. Trends in and Risk Factors for Recurrent Clostridioides difficile Infection, New Haven County, Connecticut, USA, 2015–2020. Emerg. Infect. Dis. 2023, 29, 877–887. [Google Scholar] [CrossRef]
- Ramirez, J.A.; Angulo, F.J.; Carrico, R.M.; Furmanek, S.; Oliva, S.P.; Zamparo, J.M.; Gonzalez, E.; Zhang, P.; Parrish, L.A.W.; Marimuthu, S.; et al. Misdiagnosis of Clostridioides difficile Infections by Standard-of-Care Specimen Collection and Testing among Hospitalized Adults, Louisville, Kentucky, USA, 2019–2020(1). Emerg. Infect. Dis. 2023, 29, 919–928. [Google Scholar] [CrossRef]
- Vendrik, K.E.W.; Baktash, A.; Goeman, J.J.; Harmanus, C.; Notermans, D.W.; de Greeff, S.C.; Kuijper, E.J. Comparison of trends in Clostridioides difficile infections in hospitalised patients during the first and second waves of the COVID-19 pandemic: A retrospective sentinel surveillance study. Lancet Reg. Health Eur. 2022, 19, 100424. [Google Scholar] [CrossRef]
- Viprey, V.F.; Davis, G.L.; Benson, A.D.; Ewin, D.; Spittal, W.; Vernon, J.J.; Rupnik, M.; Banz, A.; Allantaz, F.; Cleuziat, P.; et al. A point-prevalence study on community and inpatient Clostridioides difficile infections (CDI): Results from Combatting Bacterial Resistance in Europe CDI (COMBACTE-CDI), July to November 2018. Euro Surveill. 2022, 27, 2100704. [Google Scholar] [CrossRef] [PubMed]
- Schnizlein, M.K.; Young, V.B. Capturing the environment of the Clostridioides difficile infection cycle. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Cizek, A.; Masarikova, M.; Mares, J.; Brajerova, M.; Krutova, M. Detection of Plasmid-Mediated Resistance to Metronidazole in Clostridioides difficile from River Water. Microbiol. Spectr. 2022, 10, e0080622. [Google Scholar] [CrossRef] [PubMed]
- Markovska, R.; Dimitrov, G.; Gergova, R.; Boyanova, L. Clostridioides difficile, a New “Superbug”. Microorganisms 2023, 11, 845. [Google Scholar] [CrossRef]
- Papazyan, R.; Ferdyan, N.; Srinivasan, K.; Gonzalez, C.; Shannon, W.D.; Blount, K.; Fuchs, B.C. Human Fecal Bile Acid Analysis after Investigational Microbiota-Based Live Biotherapeutic Delivery for Recurrent Clostridioides difficile Infection. Microorganisms 2023, 11, 135. [Google Scholar] [CrossRef]
- Buddle, J.E.; Fagan, R.P. Pathogenicity and virulence of Clostridioides difficile. Virulence 2023, 14, 2150452. [Google Scholar] [CrossRef] [PubMed]
- Darkoh, C.; Keita, K.; Odo, C.; Oyaro, M.; Brown, E.L.; Arias, C.A.; Hanson, B.M.; DuPont, H.L. Emergence of Clinical Clostridioides difficile Isolates with Decreased Susceptibility to Vancomycin. Clin. Infect. Dis. 2022, 74, 120–126. [Google Scholar] [CrossRef]
- Greentree, D.H.; Rice, L.B.; Donskey, C.J. Houston, We Have a Problem: Reports of Clostridioides difficile Isolates With Reduced Vancomycin Susceptibility. Clin. Infect. Dis. 2022, 75, 1661–1664. [Google Scholar] [CrossRef]
- Moon, R.C.; Bleak, T.C.; Rosenthal, N.A.; Couturier, B.; Hemmert, R.; Timbrook, T.T.; Brown, H.; Fang, F.C. Epidemiology and Economic Burden of Acute Infectious Gastroenteritis Among Adults Treated in Outpatient Settings in US Health Systems. Am. J. Gastroenterol. 2023, 118, 1069–1079. [Google Scholar] [CrossRef]
- Boven, A.; Vlieghe, E.; Engstrand, L.; Andersson, F.L.; Callens, S.; Simin, J.; Brusselaers, N. Clostridioides difficile infection-associated cause-specific and all-cause mortality: A population-based cohort study. Clin. Microbiol. Infect. 2023, 29, 1424–1430. [Google Scholar] [CrossRef]
- Curry, S.R.; Hecker, M.T.; O’Hagan, J.; Kutty, P.K.; Alhmidi, H.; Ng-Wong, Y.K.; Cadnum, J.L.; Jencson, A.L.; Gonzalez-Orta, M.; Saldana, C.; et al. Natural History of Clostridioides difficile Colonization and Infection Following New Acquisition of Carriage in Healthcare Settings: A Prospective Cohort Study. Clin. Infect. Dis. 2023, 77, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B.; Jenior, M.L.; Keenan, O.; Hart, J.L.; Specker, J.; Abbas, A.; Rangel, P.C.; Di, C.; Green, J.; Bustin, K.A.; et al. Enterococci enhance Clostridioides difficile pathogenesis. Nature 2022, 611, 780–786. [Google Scholar] [CrossRef] [PubMed]
- VanInsberghe, D.; Elsherbini, J.A.; Varian, B.; Poutahidis, T.; Erdman, S.; Polz, M.F. Diarrhoeal events can trigger long-term Clostridium difficile colonization with recurrent blooms. Nat. Microbiol. 2020, 5, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.P.; Chen, X.; Williams, D.; Xu, H.; Cuddemi, C.A.; Daugherty, K.; Barrett, C.; Miller, M.; Foussadier, A.; Lantz, A.; et al. Host Immune Markers Distinguish Clostridioides difficile Infection From Asymptomatic Carriage and Non-C. difficile Diarrhea. Clin. Infect. Dis. 2020, 70, 1083–1093. [Google Scholar] [CrossRef]
- van Rossen, T.M.; Ooijevaar, R.E.; Vandenbroucke-Grauls, C.; Dekkers, O.M.; Kuijper, E.J.; Keller, J.J.; van Prehn, J. Prognostic factors for severe and recurrent Clostridioides difficile infection: A systematic review. Clin. Microbiol. Infect. 2022, 28, 321–331. [Google Scholar] [CrossRef]
- Feuerstadt, P.; Nelson, W.W.; Drozd, E.M.; Dreyfus, J.; Dahdal, D.N.; Wong, A.C.; Mohammadi, I.; Teigland, C.; Amin, A. Mortality, Health Care Use, and Costs of Clostridioides difficile Infections in Older Adults. J. Am. Med. Dir. Assoc. 2022, 23, 1721–1728.e1719. [Google Scholar] [CrossRef]
- Cao, X.; Boyaci, H.; Chen, J.; Bao, Y.; Landick, R.; Campbell, E.A. Basis of narrow-spectrum activity of fidaxomicin on Clostridioides difficile. Nature 2022, 604, 541–545. [Google Scholar] [CrossRef]
- Bishop, E.J.; Tiruvoipati, R. Management of Clostridioides difficile infection in adults and challenges in clinical practice: Review and comparison of current IDSA/SHEA, ESCMID and ASID guidelines. J. Antimicrob. Chemother. 2022, 78, 21–30. [Google Scholar] [CrossRef]
- Singh, T.; Bedi, P.; Bumrah, K.; Gandhi, D.; Arora, T.; Verma, N.; Schleicher, M.; Rai, M.P.; Garg, R.; Verma, B.; et al. Fecal Microbiota Transplantation and Medical Therapy for Clostridium difficile Infection: Meta-analysis of Randomized Controlled Trials. J. Clin. Gastroenterol. 2022, 56, 881–888. [Google Scholar] [CrossRef]
- Feuerstadt, P.; Louie, T.J.; Lashner, B.; Wang, E.E.L.; Diao, L.; Bryant, J.A.; Sims, M.; Kraft, C.S.; Cohen, S.H.; Berenson, C.S.; et al. SER-109, an Oral Microbiome Therapy for Recurrent Clostridioides difficile Infection. N. Engl. J. Med. 2022, 386, 220–229. [Google Scholar] [CrossRef]
- Sims, M.D.; Khanna, S.; Feuerstadt, P.; Louie, T.J.; Kelly, C.R.; Huang, E.S.; Hohmann, E.L.; Wang, E.E.L.; Oneto, C.; Cohen, S.H.; et al. Safety and Tolerability of SER-109 as an Investigational Microbiome Therapeutic in Adults with Recurrent Clostridioides difficile Infection: A Phase 3, Open-Label, Single-Arm Trial. JAMA Netw. Open 2023, 6, e2255758. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, F.; Wright, W.F.; Lee, T.C. Extended Follow-up of Microbiome Therapeutic SER-109 for Recurrent Clostridioides difficile Infection. JAMA 2023, 329, 1032–1033. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, S.; Yamada, A.; Komaki, Y.; Komaki, F.; Micic, D.; Sakuraba, A. Efficacy and Safety of Monoclonal Antibodies Against Clostridioides difficile Toxins for Prevention of Recurrent Clostridioides difficile Infection: A Systematic Review and Meta-Analysis. J. Clin. Gastroenterol. 2021, 55, 43–51. [Google Scholar] [CrossRef]
- Mohamed, M.F.H.; Ward, C.; Beran, A.; Abdallah, M.A.; Asemota, J.; Kelly, C.R. Efficacy, Safety, and Cost-effectiveness of Bezlotoxumab in Preventing Recurrent Clostridioides difficile Infection: Systematic Review and Meta-analysis. J. Clin. Gastroenterol. 2023, 58, 389–401. [Google Scholar] [CrossRef]
- Haddad, N.S.; Nozick, S.; Kim, G.; Ohanian, S.; Kraft, C.S.; Rebolledo, P.A.; Wang, Y.; Wu, H.; Bressler, A.; Le, S.N.T.; et al. Detection of Newly Secreted Antibodies Predicts Nonrecurrence in Primary Clostridioides difficile Infection. J. Clin. Microbiol. 2022, 60, e0220121. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.P.; Poxton, I.R.; Shen, J.; Wilcox, M.H.; Gerding, D.N.; Zhao, X.; Laterza, O.F.; Railkar, R.; Guris, D.; Dorr, M.B. Effect of Endogenous Clostridioides difficile Toxin Antibodies on Recurrence of C. difficile Infection. Clin. Infect. Dis. 2020, 71, 81–86. [Google Scholar] [CrossRef]
- Frost, I.; Sati, H.; Garcia-Vello, P.; Hasso-Agopsowicz, M.; Lienhardt, C.; Gigante, V.; Beyer, P. The role of bacterial vaccines in the fight against antimicrobial resistance: An analysis of the preclinical and clinical development pipeline. Lancet Microbe 2023, 4, e113–e125. [Google Scholar] [CrossRef]
- Remich, S.; Kitchin, N.; Peterson, J.; Li, P.; Pride, M.W.; Brock, L.; Anderson, A.S.; Gruber, W.C.; Jansen, K.U.; Lockhart, S.P.; et al. A Phase 2 Extension Study Evaluating the Immunogenicity, Safety, and Tolerability of 3 or 4 Doses of a Clostridioides difficile Vaccine in Healthy US Adults Aged 65 to 85 Years. J. Infect. Dis. 2023, 229, 367–375. [Google Scholar] [CrossRef]
- Littmann, E.R.; Lee, J.J.; Denny, J.E.; Alam, Z.; Maslanka, J.R.; Zarin, I.; Matsuda, R.; Carter, R.A.; Susac, B.; Saffern, M.S.; et al. Host immunity modulates the efficacy of microbiota transplantation for treatment of Clostridioides difficile infection. Nat. Commun. 2021, 12, 755. [Google Scholar] [CrossRef]
- Naz, F.; Petri, W.A. Host Immunity and Immunization Strategies for Clostridioides difficile Infection. Clin. Microbiol. Rev. 2023, 36, e0015722. [Google Scholar] [CrossRef]
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C.; et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, e1–e48. [Google Scholar] [CrossRef] [PubMed]
- UK Health Security Agency. Clostridioides difficile infection: Updated Guidance on Management and Treatment. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1118277/UKHSA-CDI-guideline-july-2022_DRAFT.pdf (accessed on 30 November 2023).
- van Prehn, J.; Reigadas, E.; Vogelzang, E.H.; Bouza, E.; Hristea, A.; Guery, B.; Krutova, M.; Norén, T.; Allerberger, F.; Coia, J.E.; et al. European Society of Clinical Microbiology and Infectious Diseases: 2021 update on the treatment guidance document for Clostridioides difficile infection in adults. Clin. Microbiol. Infect. 2021, 27, S1–S21. [Google Scholar] [CrossRef] [PubMed]
- Juzenas, S.; Hübenthal, M.; Lindqvist, C.M.; Kruse, R.; Steiert, T.A.; Degenhardt, F.; Schulte, D.; Nikolaus, S.; Zeissig, S.; Bergemalm, D.; et al. Detailed Transcriptional Landscape of Peripheral Blood Points to Increased Neutrophil Activation in Treatment-Naïve Inflammatory Bowel Disease. J. Crohns Colitis 2022, 16, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, J.; Dabrowska, M.; Lazowska, I.; Paziewska, A.; Balabas, A.; Kluska, A.; Kulecka, M.; Karczmarski, J.; Ambrozkiewicz, F.; Piatkowska, M.; et al. Redefining the Practical Utility of Blood Transcriptome Biomarkers in Inflammatory Bowel Diseases. J. Crohns Colitis 2019, 13, 626–633. [Google Scholar] [CrossRef]
- Planell, N.; Masamunt, M.C.; Leal, R.F.; Rodríguez, L.; Esteller, M.; Lozano, J.J.; Ramírez, A.; Ayrizono, M.L.S.; Coy, C.S.R.; Alfaro, I.; et al. Usefulness of Transcriptional Blood Biomarkers as a Non-invasive Surrogate Marker of Mucosal Healing and Endoscopic Response in Ulcerative Colitis. J. Crohns Colitis 2017, 11, 1335–1346. [Google Scholar] [CrossRef]
- Zhang, H.; Su, J.; Li, B.; Gao, Y.; Liu, M.; He, L.; Xu, H.; Dong, Y.; Zhang, X.C.; Zhao, Y. Structure of human glycosylphosphatidylinositol transamidase. Nat. Struct. Mol. Biol. 2022, 29, 203–209. [Google Scholar] [CrossRef]
- Brahma, S.; Henikoff, S. The BAF chromatin remodeler synergizes with RNA polymerase II and transcription factors to evict nucleosomes. Nat. Genet. 2024, 56, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Zhang, P.; Zhang, Y.; Sun, L.; Cui, G.; Guo, X.; Wang, H.; Zhang, X.; Shi, Y.; Yu, Z. HIF-1α/Actl6a/H3K9ac axis is critical for pluripotency and lineage differentiation of human induced pluripotent stem cells. FASEB J. 2020, 34, 5740–5753. [Google Scholar] [CrossRef]
- McDonald, B.; Chick, B.Y.; Ahmed, N.S.; Burns, M.; Ma, S.; Casillas, E.; Chen, D.; Mann, T.H.; O’Connor, C.; Hah, N.; et al. Canonical BAF complex activity shapes the enhancer landscape that licenses CD8(+) T cell effector and memory fates. Immunity 2023, 56, 1303–1319.e5. [Google Scholar] [CrossRef]
- Zhong, P.Q.; Zhong, L.; Yao, J.J.; Liu, D.D.; Yuan, Z.; Liu, J.M.; Chen, M.; Yao, S.F.; Zhao, Y.; Liu, L.; et al. ACTL6A interacts with p53 in acute promyelocytic leukemia cell lines to affect differentiation via the Sox2/Notch1 signaling pathway. Cell. Signal. 2019, 53, 390–399. [Google Scholar] [CrossRef]
- Končitíková, R.; Vigouroux, A.; Kopečná, M.; Šebela, M.; Moréra, S.; Kopečný, D. Kinetic and structural analysis of human ALDH9A1. Biosci. Rep. 2019, 39, BSR20190558. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pitchiaya, S.; Cieślik, M.; Niknafs, Y.S.; Tien, J.C.; Hosono, Y.; Iyer, M.K.; Yazdani, S.; Subramaniam, S.; Shukla, S.K.; et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat. Genet. 2018, 50, 814–824. [Google Scholar] [CrossRef]
- Chen, W.; He, W.; Cai, H.; Hu, B.; Zheng, C.; Ke, X.; Xie, L.; Zheng, Z.; Wu, X.; Wang, H. A-to-I RNA editing of BLCAP lost the inhibition to STAT3 activation in cervical cancer. Oncotarget 2017, 8, 39417–39429. [Google Scholar] [CrossRef] [PubMed]
- Gromova, I.; Svensson, S.; Gromov, P.; Moreira, J.M.A. Identification of BLCAP as a novel STAT3 interaction partner in bladder cancer. PLoS ONE 2017, 12, e0188827. [Google Scholar] [CrossRef]
- Han, F.; Hu, M.; Zhang, L.; Fan, X.; Wang, J.; Lou, Z.; Wang, S.; Chen, L.; Ye, Y.; Ding, Y.; et al. A-to-I RNA editing of BLCAP promotes cell proliferation by losing the inhibitory of Rb1 in colorectal cancer. Exp. Cell Res. 2022, 417, 113209. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Robinson, M.E.; Ma, N.; Artadji, D.; Ahmed, M.A.; Xiao, G.; Sadras, T.; Deb, G.; Winchester, J.; Cosgun, K.N.; et al. IFITM3 functions as a PIP3 scaffold to amplify PI3K signalling in B cells. Nature 2020, 588, 491–497. [Google Scholar] [CrossRef]
- Walker, K.; Mistry, A.; Watson, C.M.; Nadat, F.; O’Callaghan, E.; Care, M.; Crinnion, L.A.; Arumugakani, G.; Bonthron, D.T.; Carter, C.; et al. Inherited CD19 Deficiency Does Not Impair Plasma Cell Formation or Response to CXCL12. J. Clin. Immunol. 2023, 43, 1543–1556. [Google Scholar] [CrossRef]
- Barmeyer, C.; Erko, I.; Awad, K.; Fromm, A.; Bojarski, C.; Meissner, S.; Loddenkemper, C.; Kerick, M.; Siegmund, B.; Fromm, M.; et al. Epithelial barrier dysfunction in lymphocytic colitis through cytokine-dependent internalization of claudin-5 and -8. J. Gastroenterol. 2017, 52, 1090–1100. [Google Scholar] [CrossRef]
- Mandel, I.; Paperna, T.; Glass-Marmor, L.; Volkowich, A.; Badarny, S.; Schwartz, I.; Vardi, P.; Koren, I.; Miller, A. Tight junction proteins expression and modulation in immune cells and multiple sclerosis. J. Cell. Mol. Med. 2012, 16, 765–775. [Google Scholar] [CrossRef]
- Wang, M.; Guo, J.; Zhao, Y.Q.; Wang, J.P. IL-21 mediates microRNA-423-5p /claudin-5 signal pathway and intestinal barrier function in inflammatory bowel disease. Aging 2020, 12, 16099–16110. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, L.; Li, Z.; Yang, S.; Tang, L.; Gong, H. Identification of Crucial Genes and Infiltrating Immune Cells Underlying Sepsis-Induced Cardiomyopathy via Weighted Gene Co-Expression Network Analysis. Front. Genet. 2021, 12, 812509. [Google Scholar] [CrossRef] [PubMed]
- Tsui, H.S.; Pham, N.V.B.; Amer, B.R.; Bradley, M.C.; Gosschalk, J.E.; Gallagher-Jones, M.; Ibarra, H.; Clubb, R.T.; Blaby-Haas, C.E.; Clarke, C.F. Human COQ10A and COQ10B are distinct lipid-binding START domain proteins required for coenzyme Q function. J. Lipid Res. 2019, 60, 1293–1310. [Google Scholar] [CrossRef]
- Al-Eitan, L.N.; Tarkhan, A.H.; Alghamdi, M.A.; Al-Qarqaz, F.A.; Al-Kofahi, H.S. Transcriptome analysis of HPV-induced warts and healthy skin in humans. BMC Med Genom. 2020, 13, 35. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, R.; Trudeau, S.J.; Wolinsky, E.; Ast, T.; Liang, J.H.; Jiang, C.; Ma, Y.; Teng, M.; Mootha, V.K.; et al. CYB561A3 is the key lysosomal iron reductase required for Burkitt B-cell growth and survival. Blood 2021, 138, 2216–2230. [Google Scholar] [CrossRef]
- Zhang, J.; Cheng, Y.; Duan, M.; Qi, N.; Liu, J. Unveiling differentially expressed genes upon regulation of transcription factors in sepsis. 3 Biotech 2017, 7, 46. [Google Scholar] [CrossRef]
- Cassioli, C.; Baldari, C.T. Lymphocyte Polarization During Immune Synapse Assembly: Centrosomal Actin Joins the Game. Front. Immunol. 2022, 13, 830835. [Google Scholar] [CrossRef] [PubMed]
- Liedmann, S.; Liu, X.; Guy, C.S.; Crawford, J.C.; Rodriguez, D.A.; Kuzuoğlu-Öztürk, D.; Guo, A.; Verbist, K.C.; Temirov, J.; Chen, M.J.; et al. Localization of a TORC1-eIF4F translation complex during CD8(+) T cell activation drives divergent cell fate. Mol. Cell 2022, 82, 2401–2414.e9. [Google Scholar] [CrossRef]
- Wang, H.; Chen, X.; Jin, Y.; Liu, T.; Song, Y.; Zhu, X.; Zhu, X. The role of DYNLT3 in breast cancer proliferation, migration, and invasion via epithelial-to-mesenchymal transition. Cancer Med. 2023, 12, 15289–15303. [Google Scholar] [CrossRef]
- Devkota, S.; Jeong, H.; Kim, Y.; Ali, M.; Roh, J.I.; Hwang, D.; Lee, H.W. Functional characterization of EI24-induced autophagy in the degradation of RING-domain E3 ligases. Autophagy 2016, 12, 2038–2053. [Google Scholar] [CrossRef]
- Zheng, P.; Chen, Q.; Tian, X.; Qian, N.; Chai, P.; Liu, B.; Hu, J.; Blackstone, C.; Zhu, D.; Teng, J.; et al. DNA damage triggers tubular endoplasmic reticulum extension to promote apoptosis by facilitating ER-mitochondria signaling. Cell Res. 2018, 28, 833–854. [Google Scholar] [CrossRef] [PubMed]
- Sumpter, R.; Sirasanagandla, S.; Fernández, Á.F.; Wei, Y.; Dong, X.; Franco, L.; Zou, Z.; Marchal, C.; Lee, M.Y.; Clapp, D.W.; et al. Fanconi Anemia Proteins Function in Mitophagy and Immunity. Cell 2016, 165, 867–881. [Google Scholar] [CrossRef]
- Tan, W.; Deans, A.J. The ubiquitination machinery of the Fanconi Anemia DNA repair pathway. Prog. Biophys. Mol. Biol. 2021, 163, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, R.; Peralta, C.; Yaseen, A.; Pavletich, N.P. Structure of the FA core ubiquitin ligase closing the ID clamp on DNA. Nat. Struct. Mol. Biol. 2021, 28, 300–309. [Google Scholar] [CrossRef]
- Alderawi, A.; Caramori, G.; Baker, E.H.; Hitchings, A.W.; Rahman, I.; Rossios, C.; Adcock, I.; Cassolari, P.; Papi, A.; Ortega, V.E.; et al. FN3K expression in COPD: A potential comorbidity factor for cardiovascular disease. BMJ Open Respir. Res. 2020, 7, e000714. [Google Scholar] [CrossRef] [PubMed]
- Torres, G.G.; Nygaard, M.; Caliebe, A.; Blanché, H.; Chantalat, S.; Galan, P.; Lieb, W.; Christiansen, L.; Deleuze, J.F.; Christensen, K.; et al. Exome-Wide Association Study Identifies FN3KRP and PGP as New Candidate Longevity Genes. J. Gerontol. A Biol. Sci. Med. Sci. 2021, 76, 786–795. [Google Scholar] [CrossRef]
- Opstelten, R.; de Kivit, S.; Slot, M.C.; van den Biggelaar, M.; Iwaszkiewicz-Grześ, D.; Gliwiński, M.; Scott, A.M.; Blom, B.; Trzonkowski, P.; Borst, J.; et al. GPA33: A Marker to Identify Stable Human Regulatory T Cells. J. Immunol. 2020, 204, 3139–3148. [Google Scholar] [CrossRef]
- Opstelten, R.; Suwandi, J.S.; Slot, M.C.; Morgana, F.; Scott, A.M.; Laban, S.; Nikolic, T.; Turksma, A.W.; Kroeze, A.; Voermans, C.; et al. GPA33 is expressed on multiple human blood cell types and distinguishes CD4(+) central memory T cells with and without effector function. Eur. J. Immunol. 2021, 51, 1377–1389. [Google Scholar] [CrossRef]
- Chen, Z.; Cui, Y.; Yao, Y.; Liu, B.; Yunis, J.; Gao, X.; Wang, N.; Cañete, P.F.; Tuong, Z.K.; Sun, H.; et al. Heparan sulfate regulates IL-21 bioavailability and signal strength that control germinal center B cell selection and differentiation. Sci. Immunol. 2023, 8, eadd1728. [Google Scholar] [CrossRef]
- Schultheis, N.; Becker, R.; Berhanu, G.; Kapral, A.; Roseman, M.; Shah, S.; Connell, A.; Selleck, S. Regulation of autophagy, lipid metabolism, and neurodegenerative pathology by heparan sulfate proteoglycans. Front. Genet. 2022, 13, 1012706. [Google Scholar] [CrossRef]
- Rodríguez-Jiménez, C.; Gómez-Coronado, D.; Frías Vargas, M.; Cerrato, F.; Lahoz, C.; Saban-Ruiz, J.; González-Nieto, D.; Lasunción, M.A.; Mostaza, J.M.; Rodríguez-Nóvoa, S. A new variant (c.1A>G) in LDLRAP1 causing autosomal recessive hypercholesterolemia: Characterization of the defect and response to PCSK9 inhibition. Atherosclerosis 2019, 284, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Oda, S.K.; Shotts, K.; Donovan, E.E.; Strauch, P.; Pujanauski, L.M.; Victorino, F.; Al-Shami, A.; Fujiwara, Y.; Tigyi, G.; et al. Lysophosphatidic acid receptor 5 inhibits B cell antigen receptor signaling and antibody response. J. Immunol. 2014, 193, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Kremer, K.N.; Buser, A.; Thumkeo, D.; Narumiya, S.; Jacobelli, J.; Pelanda, R.; Torres, R.M. LPA suppresses T cell function by altering the cytoskeleton and disrupting immune synapse formation. Proc. Natl. Acad. Sci. USA 2022, 119, e2118816119. [Google Scholar] [CrossRef]
- Turner, J.A.; Fredrickson, M.A.; D’Antonio, M.; Katsnelson, E.; MacBeth, M.; Van Gulick, R.; Chimed, T.S.; McCarter, M.; D’Alessandro, A.; Robinson, W.A.; et al. Lysophosphatidic acid modulates CD8 T cell immunosurveillance and metabolism to impair anti-tumor immunity. Nat. Commun. 2023, 14, 3214. [Google Scholar] [CrossRef]
- Grings, M.; Seminotti, B.; Karunanidhi, A.; Ghaloul-Gonzalez, L.; Mohsen, A.W.; Wipf, P.; Palmfeldt, J.; Vockley, J.; Leipnitz, G. ETHE1 and MOCS1 deficiencies: Disruption of mitochondrial bioenergetics, dynamics, redox homeostasis and endoplasmic reticulum-mitochondria crosstalk in patient fibroblasts. Sci. Rep. 2019, 9, 12651. [Google Scholar] [CrossRef]
- Landgraf, B.J.; McCarthy, E.L.; Booker, S.J. Radical S-Adenosylmethionine Enzymes in Human Health and Disease. Annu. Rev. Biochem. 2016, 85, 485–514. [Google Scholar] [CrossRef] [PubMed]
- Mellis, A.T.; Roeper, J.; Misko, A.L.; Kohl, J.; Schwarz, G. Sulfite Alters the Mitochondrial Network in Molybdenum Cofactor Deficiency. Front. Genet. 2020, 11, 594828. [Google Scholar] [CrossRef]
- Wang, Y.L.; Zhao, W.W.; Bai, S.M.; Feng, L.L.; Bie, S.Y.; Gong, L.; Wang, F.; Wei, M.B.; Feng, W.X.; Pang, X.L.; et al. MRNIP condensates promote DNA double-strand break sensing and end resection. Nat. Commun. 2022, 13, 2638. [Google Scholar] [CrossRef]
- Van Damme, P.; Kalvik, T.V.; Starheim, K.K.; Jonckheere, V.; Myklebust, L.M.; Menschaert, G.; Varhaug, J.E.; Gevaert, K.; Arnesen, T. A Role for Human N-alpha Acetyltransferase 30 (Naa30) in Maintaining Mitochondrial Integrity. Mol. Cell. Proteom. 2016, 15, 3361–3372. [Google Scholar] [CrossRef]
- Varland, S.; Silva, R.D.; Kjosås, I.; Faustino, A.; Bogaert, A.; Billmann, M.; Boukhatmi, H.; Kellen, B.; Costanzo, M.; Drazic, A.; et al. N-terminal acetylation shields proteins from degradation and promotes age-dependent motility and longevity. Nat. Commun. 2023, 14, 6774. [Google Scholar] [CrossRef]
- Vercellino, I.; Sazanov, L.A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell Biol. 2022, 23, 141–161. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Li, S.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1α. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef]
- Gattinoni, L.; Zhong, X.S.; Palmer, D.C.; Ji, Y.; Hinrichs, C.S.; Yu, Z.; Wrzesinski, C.; Boni, A.; Cassard, L.; Garvin, L.M.; et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 2009, 15, 808–813. [Google Scholar] [CrossRef]
- Huber, A.; Jose, S.; Kassam, A.; Weghorn, K.N.; Powers-Fletcher, M.; Sharma, D.; Mukherjee, A.; Mathew, A.; Kulkarni, N.; Chandramouli, S.; et al. Olfactomedin-4 (+) neutrophils exacerbate intestinal epithelial damage and worsen host survival after Clostridioides difficile infection. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wang, X.Y.; Chen, S.H.; Zhang, Y.N.; Xu, C.F. Olfactomedin-4 in digestive diseases: A mini-review. World J. Gastroenterol. 2018, 24, 1881–1887. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.L.M.; van der Klei, I.J. The peroxisome biogenesis factors Pex3 and Pex19: Multitasking proteins with disputed functions. FEBS Lett. 2019, 593, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.A.; Haque, F.; Stanlie, A.; Husain, A.; Mondal, S.; Nakata, M.; Taniguchi, T.; Taniguchi, H.; Honjo, T. Phf5a regulates DNA repair in class switch recombination via p400 and histone H2A variant deposition. EMBO J. 2021, 40, e106393. [Google Scholar] [CrossRef]
- Karmakar, S.; Rauth, S.; Nallasamy, P.; Perumal, N.; Nimmakayala, R.K.; Leon, F.; Gupta, R.; Barkeer, S.; Venkata, R.C.; Raman, V.; et al. RNA Polymerase II-Associated Factor 1 Regulates Stem Cell Features of Pancreatic Cancer Cells, Independently of the PAF1 Complex, via Interactions with PHF5A and DDX3. Gastroenterology 2020, 159, 1898–1915.e6. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, D.; Ruan, G.X.; Wang, R.; Li, Y.; Chen, W.; Huang, H.; Wang, J.; Meng, L.; Zhu, Z.; et al. Spliceosome component PHD finger 5A is essential for early B lymphopoiesis. Development 2024, 151, dev202247. [Google Scholar] [CrossRef]
- Kunová, N.; Havalová, H.; Ondrovičová, G.; Stojkovičová, B.; Bauer, J.A.; Bauerová-Hlinková, V.; Pevala, V.; Kutejová, E. Mitochondrial Processing Peptidases-Structure, Function and the Role in Human Diseases. Int. J. Mol. Sci. 2022, 23, 1297. [Google Scholar] [CrossRef]
- Shi, D.L. Planar cell polarity regulators in asymmetric organogenesis during development and disease. J. Genet. Genom. 2023, 50, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Stiegler, A.L.; Boggon, T.J. The pseudoGTPase group of pseudoenzymes. FEBS J. 2020, 287, 4232–4245. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Song, J.; Ye, D. CRL3s: The BTB-CUL3-RING E3 Ubiquitin Ligases. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei,, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 211–223. [Google Scholar]
- Smits, D.J.; Dekker, J.; Schot, R.; Tabarki, B.; Alhashem, A.; Demmers, J.A.A.; Dekkers, D.H.W.; Romito, A.; van der Spek, P.J.; van Ham, T.J.; et al. CLEC16A interacts with retromer and TRIM27, and its loss impairs endosomal trafficking and neurodevelopment. Hum. Genet. 2023, 142, 379–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, T.; Zhang, J.; Yang, M.; Li, N.; Xu, X.; Cao, X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat. Immunol. 2009, 10, 744–752. [Google Scholar] [CrossRef]
- Yang, M.; Chen, T.; Li, X.; Yu, Z.; Tang, S.; Wang, C.; Gu, Y.; Liu, Y.; Xu, S.; Li, W.; et al. K33-linked polyubiquitination of Zap70 by Nrdp1 controls CD8+ T cell activation. Nat. Immunol. 2015, 16, 1253–1262. [Google Scholar] [CrossRef]
- Tong, Y.; Lear, T.B.; Evankovich, J.; Chen, Y.; Londino, J.D.; Myerburg, M.M.; Zhang, Y.; Popescu, I.D.; McDyer, J.F.; McVerry, B.J.; et al. The RNFT2/IL-3Rα axis regulates IL-3 signaling and innate immunity. JCI Insight 2020, 5, e133652. [Google Scholar] [CrossRef]
- Scheepers, A.; Schmidt, S.; Manolescu, A.; Cheeseman, C.I.; Bell, A.; Zahn, C.; Joost, H.G.; Schürmann, A. Characterization of the human SLC2A11 (GLUT11) gene: Alternative promoter usage, function, expression, and subcellular distribution of three isoforms, and lack of mouse orthologue. Mol. Membr. Biol. 2005, 22, 339–351. [Google Scholar] [CrossRef]
- Dennerlein, S.; Oeljeklaus, S.; Jans, D.; Hellwig, C.; Bareth, B.; Jakobs, S.; Deckers, M.; Warscheid, B.; Rehling, P. MITRAC7 Acts as a COX1-Specific Chaperone and Reveals a Checkpoint during Cytochrome c Oxidase Assembly. Cell Rep. 2015, 12, 1644–1655. [Google Scholar] [CrossRef]
- Horiuchi, S.; Koike, T.; Takebuchi, H.; Hoshino, K.; Sasaki, I.; Fukuda-Ohta, Y.; Kaisho, T.; Kitamura, D. SpiB regulates the expression of B-cell-related genes and increases the longevity of memory B cells. Front. Immunol. 2023, 14, 1250719. [Google Scholar] [CrossRef]
- Miyazaki, R.; Saiga, H.; Kato, T.; Bakoshi, T.; Senba, R.; Shintani, A.; Suzuki, M.; Takao, K.; Sasaki, I.; Iizuka, A.; et al. The mechanism of action of Spi-B in the transcriptional activation of the interferon-α4 gene. Biochem. Biophys. Res. Commun. 2020, 525, 477–482. [Google Scholar] [CrossRef]
- Nagel, S.; Meyer, C.; Kaufmann, M.; Drexler, H.G.; MacLeod, R.A. Aberrant expression of homeobox gene SIX1 in Hodgkin lymphoma. Oncotarget 2015, 6, 40112–40126. [Google Scholar] [CrossRef] [PubMed]
- Quaney, M.J.; Pritzl, C.J.; Luera, D.; Newth, R.J.; Knudson, K.M.; Saxena, V.; Guldenpfennig, C.; Gil, D.; Rae, C.S.; Lauer, P.; et al. STING controls T cell memory fitness during infection through T cell-intrinsic and IDO-dependent mechanisms. Proc. Natl. Acad. Sci. USA 2023, 120, e2205049120. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Lin, Y.; Lin, F.; Yang, M.; Li, J.; Zhang, R.; Huang, Z.; Shen, Q.; Tang, R.; Zheng, C. β-Catenin Is Required for the cGAS/STING Signaling Pathway but Antagonized by the Herpes Simplex Virus 1 US3 Protein. J. Virol. 2020, 94, 13. [Google Scholar] [CrossRef] [PubMed]
- Serna, M.; Carranza, G.; Martín-Benito, J.; Janowski, R.; Canals, A.; Coll, M.; Zabala, J.C.; Valpuesta, J.M. The structure of the complex between α-tubulin, TBCE and TBCB reveals a tubulin dimer dissociation mechanism. J. Cell Sci. 2015, 128, 1824–1834. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.S.; Jun, S.; Kim, M.J.; Lee, S.H.; Suh, H.N.; Lien, E.M.; Jung, H.Y.; Lee, S.; Zhang, J.; Yang, J.I.; et al. TMEM9 promotes intestinal tumorigenesis through vacuolar-ATPase-activated Wnt/β-catenin signalling. Nat. Cell Biol. 2018, 20, 1421–1433. [Google Scholar] [CrossRef]
- Jung, Y.S.; Stratton, S.A.; Lee, S.H.; Kim, M.J.; Jun, S.; Zhang, J.; Zheng, B.; Cervantes, C.L.; Cha, J.H.; Barton, M.C.; et al. TMEM9-v-ATPase Activates Wnt/β-Catenin Signaling Via APC Lysosomal Degradation for Liver Regeneration and Tumorigenesis. Hepatology 2021, 73, 776–794. [Google Scholar] [CrossRef]
- Wei, W.; Jiang, F.; Liu, X.C.; Su, Q. TMEM9 mediates IL-6 and IL-1β secretion and is modulated by the Wnt pathway. Int. Immunopharmacol. 2018, 63, 253–260. [Google Scholar] [CrossRef]
- Al Khatib, I.; Deng, J.; Lei, Y.; Torres-Odio, S.; Rojas, G.R.; Newman, L.E.; Chung, B.K.; Symes, A.; Zhang, H.; Huang, S.N.; et al. Activation of the cGAS-STING innate immune response in cells with deficient mitochondrial topoisomerase TOP1MT. Hum. Mol. Genet. 2023, 32, 2422–2440. [Google Scholar] [CrossRef]
- Al Khatib, I.; Deng, J.; Symes, A.; Kerr, M.; Zhang, H.; Huang, S.N.; Pommier, Y.; Khan, A.; Shutt, T.E. Functional characterization of two variants of mitochondrial topoisomerase TOP1MT that impact regulation of the mitochondrial genome. J. Biol. Chem. 2022, 298, 102420. [Google Scholar] [CrossRef]
- Baechler, S.A.; Factor, V.M.; Dalla Rosa, I.; Ravji, A.; Becker, D.; Khiati, S.; Miller Jenkins, L.M.; Lang, M.; Sourbier, C.; Michaels, S.A.; et al. The mitochondrial type IB topoisomerase drives mitochondrial translation and carcinogenesis. Nat. Commun. 2019, 10, 83. [Google Scholar] [CrossRef]
- Pommier, Y.; Nussenzweig, A.; Takeda, S.; Austin, C. Human topoisomerases and their roles in genome stability and organization. Nat. Rev. Mol. Cell Biol. 2022, 23, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Komatsu, S.; Ohashi, T.; Kiuchi, J.; Kamiya, H.; Shimizu, H.; Arita, T.; Konishi, H.; Shiozaki, A.; Kubota, T.; et al. Overexpression of Tetraspanin31 contributes to malignant potential and poor outcomes in gastric cancer. Cancer Sci. 2022, 113, 1984–1998. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Wurm, J.P.; Sharma, S.; Immer, C.; Pogoryelov, D.; Kötter, P.; Lafontaine, D.L.; Wöhnert, J.; Entian, K.D. Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res. 2016, 44, 4304–4316. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Tang, H.; Yuan, Z.; Zhang, E.; Wang, X. The Dual Role of USP11 in Cancer. J. Oncol. 2022, 2022, 9963905. [Google Scholar] [CrossRef] [PubMed]
- Istomine, R.; Alvarez, F.; Almadani, Y.; Philip, A.; Piccirillo, C.A. The Deubiquitinating Enzyme Ubiquitin-Specific Peptidase 11 Potentiates TGF-β Signaling in CD4(+) T Cells to Facilitate Foxp3(+) Regulatory T and T(H)17 Cell Differentiation. J. Immunol. 2019, 203, 2388–2400. [Google Scholar] [CrossRef]
- Jiao, M.; Bao, X.; Hu, M.; Pan, D.; Liu, X.; Kim, J.; Li, F.; Li, C.Y. VHL loss enables immune checkpoint blockade therapy by boosting type I interferon response. bioRxiv 2023. [Google Scholar] [CrossRef]
- Xu, S.; Huo, J.; Huang, Y.; Aw, M.; Chen, S.; Mak, S.; Yip, L.Y.; Ho, Y.S.; Ng, S.W.; Tan, A.H.; et al. von Hippel-Lindau Protein Maintains Metabolic Balance to Regulate the Survival of Naive B Lymphocytes. iScience 2019, 17, 379–392. [Google Scholar] [CrossRef]
- Zhu, Z.; Johnson, R.L.; Zhang, Z.; Herring, L.E.; Jiang, G.; Damania, B.; James, L.I.; Liu, P. Development of VHL-recruiting STING PROTACs that suppress innate immunity. Cell. Mol. Life Sci. 2023, 80, 149. [Google Scholar] [CrossRef]
- Cho, N.H.; Cheveralls, K.C.; Brunner, A.D.; Kim, K.; Michaelis, A.C.; Raghavan, P.; Kobayashi, H.; Savy, L.; Li, J.Y.; Canaj, H.; et al. OpenCell: Endogenous tagging for the cartography of human cellular organization. Science 2022, 375, eabi6983. [Google Scholar] [CrossRef]
- Virok, D.P.; Simon, D.; Bozsó, Z.; Rajkó, R.; Datki, Z.; Bálint, É.; Szegedi, V.; Janáky, T.; Penke, B.; Fülöp, L. Protein array based interactome analysis of amyloid-β indicates an inhibition of protein translation. J. Proteome. Res. 2011, 10, 1538–1547. [Google Scholar] [CrossRef]
- Garratt, L.W. Current Understanding of the Neutrophil Transcriptome in Health and Disease. Cells 2021, 10, 2406. [Google Scholar] [CrossRef] [PubMed]
- Liew, P.X.; Kubes, P. The Neutrophil’s Role During Health and Disease. Physiol. Rev. 2019, 99, 1223–1248. [Google Scholar] [CrossRef] [PubMed]
- Wigerblad, G.; Cao, Q.; Brooks, S.; Naz, F.; Gadkari, M.; Jiang, K.; Gupta, S.; O’Neil, L.; Dell’Orso, S.; Kaplan, M.J.; et al. Single-Cell Analysis Reveals the Range of Transcriptional States of Circulating Human Neutrophils. J. Immunol. 2022, 209, 772–782. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Shi, Q.; Wu, P.; Zhang, X.; Kambara, H.; Su, J.; Yu, H.; Park, S.Y.; Guo, R.; Ren, Q.; et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat. Immunol. 2020, 21, 1119–1133. [Google Scholar] [CrossRef]
- Najjar, R.; Rogel, N.; Pineda, J.M.B.; Wang, X.; Tran, M.; Bays, A.; Mustelin, T. Large overlap in neutrophil transcriptome between lupus and COVID-19 with limited lupus-specific gene expression. Lupus Sci. Med. 2024, 11, e001059. [Google Scholar] [CrossRef]
- Schimke, L.F.; Marques, A.H.C.; Baiocchi, G.C.; de Souza Prado, C.A.; Fonseca, D.L.M.; Freire, P.P.; Rodrigues Plaça, D.; Salerno Filgueiras, I.; Coelho Salgado, R.; Jansen-Marques, G.; et al. Severe COVID-19 Shares a Common Neutrophil Activation Signature with Other Acute Inflammatory States. Cells 2022, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Argmann, C.; Hou, R.; Ungaro, R.C.; Irizar, H.; Al-Taie, Z.; Huang, R.; Kosoy, R.; Venkat, S.; Song, W.M.; Di’Narzo, A.F.; et al. Biopsy and blood-based molecular biomarker of inflammation in IBD. Gut 2023, 72, 1271–1287. [Google Scholar] [CrossRef]
- Li, M.; Maruthur, N.M.; Loomis, S.J.; Pietzner, M.; North, K.E.; Mei, H.; Morrison, A.C.; Friedrich, N.; Pankow, J.S.; Nauck, M.; et al. Genome-wide association study of 1,5-anhydroglucitol identifies novel genetic loci linked to glucose metabolism. Sci. Rep. 2017, 7, 2812. [Google Scholar] [CrossRef]
- Xu, S.; Feng, Y.; Zhao, S. Proteins with Evolutionarily Hypervariable Domains are Associated with Immune Response and Better Survival of Basal-like Breast Cancer Patients. Comput. Struct. Biotechnol. J. 2019, 17, 430–440. [Google Scholar] [CrossRef]
- Muthas, D.; Reznichenko, A.; Balendran, C.A.; Böttcher, G.; Clausen, I.G.; Kärrman Mårdh, C.; Ottosson, T.; Uddin, M.; MacDonald, T.T.; Danese, S.; et al. Neutrophils in ulcerative colitis: A review of selected biomarkers and their potential therapeutic implications. Scand. J. Gastroenterol. 2017, 52, 125–135. [Google Scholar] [CrossRef]
- Richter, K.; Amati, A.L.; Padberg, W.; Grau, V. Negative regulation of ATP-induced inflammasome activation and cytokine secretion by acute-phase proteins: A mini review. Front. Pharmacol. 2022, 13, 981276. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, T.; Kojima, Y.; Fujimoto, N. Significance of secretory leukocyte peptidase inhibitor in pleural fluid for the diagnosis of benign asbestos pleural effusion. Sci. Rep. 2021, 11, 12965. [Google Scholar] [CrossRef] [PubMed]
- Nugteren, S.; den Uil, S.H.; Delis-van Diemen, P.M.; Simons-Oosterhuis, Y.; Lindenbergh-Kortleve, D.J.; van Haaften, D.H.; Stockmann, H.; Sanders, J.; Meijer, G.A.; Fijneman, R.J.A.; et al. High expression of secretory leukocyte protease inhibitor (SLPI) in stage III micro-satellite stable colorectal cancer is associated with reduced disease recurrence. Sci. Rep. 2022, 12, 12174. [Google Scholar] [CrossRef] [PubMed]
- Zhai, X.; Chen, X.; Wan, Z.; Ge, M.; Ding, Y.; Gu, J.; Hua, J.; Guo, D.; Tan, M.; Xu, D. Identification of the novel therapeutic targets and biomarkers associated of prostate cancer with cancer-associated fibroblasts (CAFs). Front. Oncol. 2023, 13, 1136835. [Google Scholar] [CrossRef] [PubMed]
- Menckeberg, C.L.; Hol, J.; Simons-Oosterhuis, Y.; Raatgeep, H.R.; de Ruiter, L.F.; Lindenbergh-Kortleve, D.J.; Korteland-van Male, A.M.; El Aidy, S.; van Lierop, P.P.; Kleerebezem, M.; et al. Human buccal epithelium acquires microbial hyporesponsiveness at birth, a role for secretory leukocyte protease inhibitor. Gut 2015, 64, 884–893. [Google Scholar] [CrossRef]
- McDermott, A.J.; Falkowski, N.R.; McDonald, R.A.; Frank, C.R.; Pandit, C.R.; Young, V.B.; Huffnagle, G.B. Role of interferon-γ and inflammatory monocytes in driving colonic inflammation during acute Clostridium difficile infection in mice. Immunology 2017, 150, 468–477. [Google Scholar] [CrossRef]
- McDermott, A.J.; Frank, C.R.; Falkowski, N.R.; McDonald, R.A.; Young, V.B.; Huffnagle, G.B. Role of GM-CSF in the inflammatory cytokine network that regulates neutrophil influx into the colonic mucosa during Clostridium difficile infection in mice. Gut Microbes 2014, 5, 476–484. [Google Scholar] [CrossRef]
- Clarke, A.J.; Simon, A.K. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat. Rev. Immunol. 2019, 19, 170–183. [Google Scholar] [CrossRef]
- Deretic, V. Autophagy in inflammation, infection, and immunometabolism. Immunity 2021, 54, 437–453. [Google Scholar] [CrossRef]
- Mills, E.L.; Kelly, B.; O’Neill, L.A.J. Mitochondria are the powerhouses of immunity. Nat. Immunol. 2017, 18, 488–498. [Google Scholar] [CrossRef]
- Muri, J.; Kopf, M. Redox regulation of immunometabolism. Nat. Rev. Immunol. 2021, 21, 363–381. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Tubau-Juni, N.; Bassaganya-Riera, J.; Leber, A.J.; Alva, S.S.; Baker, R.; Hontecillas, R. Modulation of colonic immunometabolic responses during Clostridioides difficile infection ameliorates disease severity and inflammation. Sci. Rep. 2023, 13, 14708. [Google Scholar] [CrossRef] [PubMed]
- Angajala, A.; Lim, S.; Phillips, J.B.; Kim, J.H.; Yates, C.; You, Z.; Tan, M. Diverse Roles of Mitochondria in Immune Responses: Novel Insights Into Immuno-Metabolism. Front. Immunol. 2018, 9, 1605. [Google Scholar] [CrossRef]
- Kodali, S.; Li, M.; Budai, M.M.; Chen, M.; Wang, J. Protection of Quiescence and Longevity of IgG Memory B Cells by Mitochondrial Autophagy. J. Immunol. 2022, 208, 1085–1098. [Google Scholar] [CrossRef]
- Hernández Del Pino, R.E.; Barbero, A.M.; Español, L.; Morro, L.S.; Pasquinelli, V. The adaptive immune response to Clostridioides difficile: A tricky balance between immunoprotection and immunopathogenesis. J. Leukoc. Biol. 2021, 109, 195–210. [Google Scholar] [CrossRef]
- Shah, H.B.; Smith, K.; Scott, E.J., II; Larabee, J.L.; James, J.A.; Ballard, J.D.; Lang, M.L. Human C. difficile toxin–specific memory B cell repertoires encode poorly neutralizing antibodies. JCI Insight 2020, 5, e138137. [Google Scholar] [CrossRef]
- Bauer, M.P.; Nibbering, P.H.; Poxton, I.R.; Kuijper, E.J.; van Dissel, J.T. Humoral immune response as predictor of recurrence in Clostridium difficile infection. Clin. Microbiol. Infect. 2014, 20, 1323–1328. [Google Scholar] [CrossRef]
- Sehgal, K.; Khanna, S. Immune response against Clostridioides difficile and translation to therapy. Ther. Adv. Gastroenterol. 2021, 14, 17562848211014817. [Google Scholar] [CrossRef]
- Cook, L.; Rees, W.D.; Wong, M.Q.; Peters, H.; Levings, M.K.; Steiner, T.S. Fecal Microbiota Transplantation for Recurrent Clostridioides difficile Infection Enhances Adaptive Immunity to C difficile Toxin B. Gastroenterology 2021, 160, 2155–2158.e4. [Google Scholar] [CrossRef]
- Hamada, Y.; Tsurumi, Y.; Nozaki, S.; Katoh, Y.; Nakayama, K. Interaction of WDR60 intermediate chain with TCTEX1D2 light chain of the dynein-2 complex is crucial for ciliary protein trafficking. Mol. Biol. Cell 2018, 29, 1628–1639. [Google Scholar] [CrossRef] [PubMed]
- Mattila, P.K.; Feest, C.; Depoil, D.; Treanor, B.; Montaner, B.; Otipoby, K.L.; Carter, R.; Justement, L.B.; Bruckbauer, A.; Batista, F.D. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity 2013, 38, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Manolakou, T.; Verginis, P.; Boumpas, D.T. DNA Damage Response in the Adaptive Arm of the Immune System: Implications for Autoimmunity. Int. J. Mol. Sci. 2021, 22, 5842. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Tian, S.; Zhang, J.; Liu, Z.; Robinson-McCarthy, L.; Miyashita, S.I.; Breault, D.T.; Gerhard, R.; Oottamasathien, S.; Whelan, S.P.J.; et al. Sulfated glycosaminoglycans and low-density lipoprotein receptor contribute to Clostridium difficile toxin A entry into cells. Nat. Microbiol. 2019, 4, 1760–1769. [Google Scholar] [CrossRef]
- Martins, C.S.; Costa, D.V.S.; Lima, B.B.; Leitäo, R.F.C.; Freire, G.E.; Silva, G.F.M.; Pacífico, D.M.; Abreu, J.G.; Brito, G.A.C. Clostridioides difficile Toxin A-Induced Wnt/β-Catenin Pathway Inhibition Is Mediated by Rac1 Glucosylation. Front. Microbiol. 2020, 11, 1998. [Google Scholar] [CrossRef]
- Tao, L.; Zhang, J.; Meraner, P.; Tovaglieri, A.; Wu, X.; Gerhard, R.; Zhang, X.; Stallcup, W.B.; Miao, J.; He, X.; et al. Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 2016, 538, 350–355. [Google Scholar] [CrossRef]
- Aktories, K.; Schwan, C.; Jank, T. Clostridium difficile Toxin Biology. Annu. Rev. Microbiol. 2017, 71, 281–307. [Google Scholar] [CrossRef]
- Bassotti, G.; Fruganti, A.; Stracci, F.; Marconi, P.; Fettucciari, K. Cytotoxic synergism of Clostridioides difficile toxin B with proinflammatory cytokines in subjects with inflammatory bowel diseases. World J. Gastroenterol. 2023, 29, 582–596. [Google Scholar] [CrossRef]
- Frädrich, C.; Beer, L.A.; Gerhard, R. Reactive Oxygen Species as Additional Determinants for Cytotoxicity of Clostridium difficile Toxins A and B. Toxins 2016, 8, 25. [Google Scholar] [CrossRef]
- Soto Ocaña, J.; Bayard, N.U.; Hart, J.L.; Thomas, A.K.; Furth, E.E.; Lacy, D.B.; Aronoff, D.M.; Zackular, J.P. Nonsteroidal anti-inflammatory drugs sensitize epithelial cells to Clostridioides difficile toxin-mediated mitochondrial damage. Sci. Adv. 2023, 9, eadh5552. [Google Scholar] [CrossRef]
- Fettucciari, K.; Marconi, P.; Marchegiani, A.; Fruganti, A.; Spaterna, A.; Bassotti, G. Invisible steps for a global endemy: Molecular strategies adopted by Clostridioides difficile. Ther. Adv. Gastroenterol. 2021, 14, 17562848211032797. [Google Scholar] [CrossRef] [PubMed]
- Modi, N.; Gulati, N.; Solomon, K.; Monaghan, T.; Robins, A.; Sewell, H.F.; Mahida, Y.R. Differential binding and internalization of Clostridium difficile toxin A by human peripheral blood monocytes, neutrophils and lymphocytes. Scand. J. Immunol. 2011, 74, 264–271. [Google Scholar] [CrossRef] [PubMed]
- DH/HCAI/Infectious Disease. Update Guidance on the Diagnosis and Report of Clostridium Difficile. Available online: https://assets.publishing.service.gov.uk/media/5a7cc0c1e5274a2f304efdb0/dh_133016.pdf (accessed on 16 November 2023).
- Miyajima, F.; Roberts, P.; Swale, A.; Price, V.; Jones, M.; Horan, M.; Beeching, N.; Brazier, J.; Parry, C.; Pendleton, N.; et al. Characterisation and Carriage Ratio of Clostridium difficile Strains Isolated from a Community-Dwelling Elderly Population in the United Kingdom. PLoS ONE 2011, 6, e22804. [Google Scholar] [CrossRef] [PubMed]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2021, 060012. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC). Clostridioides difficile Infections. Annual Epidemiological Report for 2018−2020. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/AER-Clostridium-difficile-2018-2020.pdf (accessed on 14 May 2024).
- Ahmad, S.; Singh, P.; Sharma, A.; Arora, S.; Shriwash, N.; Rahmani, A.H.; Almatroodi, S.A.; Manda, K.; Dohare, R.; Syed, M.A. Transcriptome Meta-Analysis Deciphers a Dysregulation in Immune Response-Associated Gene Signatures during Sepsis. Genes 2019, 10, 1005. [Google Scholar] [CrossRef]
- Almansa, R.; Heredia-Rodríguez, M.; Gomez-Sanchez, E.; Andaluz-Ojeda, D.; Iglesias, V.; Rico, L.; Ortega, A.; Gomez-Pesquera, E.; Liu, P.; Aragón, M.; et al. Transcriptomic correlates of organ failure extent in sepsis. J. Infect. 2015, 70, 445–456. [Google Scholar] [CrossRef]
- Gong, F.; Li, R.; Zheng, X.; Chen, W.; Zheng, Y.; Yang, Z.; Chen, Y.; Qu, H.; Mao, E.; Chen, E. OLFM4 Regulates Lung Epithelial Cell Function in Sepsis-Associated ARDS/ALI via LDHA-Mediated NF-κB Signaling. J. Inflamm. Res. 2021, 14, 7035–7051. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Shao, J.; Chen, J.; Zhang, T.; Meng, X.; Zong, R.; Jin, G.; Wu, F. Bioinformatics Analysis for Identifying Pertinent Pathways and Genes in Sepsis. Comput. Math. Methods Med. 2021, 2021, 2085173. [Google Scholar] [CrossRef]
- Lu, J.; Li, Q.; Wu, Z.; Zhong, Z.; Ji, P.; Li, H.; He, C.; Feng, J.; Zhang, J. Two gene set variation indexes as potential diagnostic tool for sepsis. Am. J. Transl. Res. 2020, 12, 2749–2759. [Google Scholar]
- Zhang, W.Y.; Chen, Z.H.; An, X.X.; Li, H.; Zhang, H.L.; Wu, S.J.; Guo, Y.Q.; Zhang, K.; Zeng, C.L.; Fang, X.M. Analysis and validation of diagnostic biomarkers and immune cell infiltration characteristics in pediatric sepsis by integrating bioinformatics and machine learning. World J. Pediatr. 2023, 19, 1094–1103. [Google Scholar] [CrossRef]
- Martínez-Paz, P.; Aragón-Camino, M.; Gómez-Sánchez, E.; Lorenzo-López, M.; Gómez-Pesquera, E.; Fadrique-Fuentes, A.; Liu, P.; Tamayo-Velasco, Á.; Ortega-Loubon, C.; Martín-Fernández, M.; et al. Distinguishing septic shock from non-septic shock in postsurgical patients using gene expression. J. Infect. 2021, 83, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Zhu, Y.; Xie, X.; Wu, J.; Qian, M. Six potential biomarkers in septic shock: A deep bioinformatics and prospective observational study. Front. Immunol. 2023, 14, 1184700. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Jiang, M.; Bai, X.; Ding, L.; Dong, P.; Jiang, M. Identification and Verification of Potential Core Genes in Pediatric Septic Shock. Comb. Chem. High. Throughput Screen. 2022, 25, 2228–2239. [Google Scholar]
- Martínez-Paz, P.; Aragón-Camino, M.; Gómez-Sánchez, E.; Lorenzo-López, M.; Gómez-Pesquera, E.; López-Herrero, R.; Sánchez-Quirós, B.; de la Varga, O.; Tamayo-Velasco, Á.; Ortega-Loubon, C.; et al. Gene Expression Patterns Distinguish Mortality Risk in Patients with Postsurgical Shock. J. Clin. Med. 2020, 9, 1276. [Google Scholar] [CrossRef]
- Li, Z.; Wang, C.; Zhang, X.; Xu, X.; Wang, M.; Dong, L. Crosstalk between septic shock and venous thromboembolism: A bioinformatics and immunoassay analysis. Front. Cell Infect. Microbiol. 2023, 13, 1235269. [Google Scholar] [CrossRef]
- Lévy, Y.; Wiedemann, A.; Hejblum, B.P.; Durand, M.; Lefebvre, C.; Surénaud, M.; Lacabaratz, C.; Perreau, M.; Foucat, E.; Déchenaud, M.; et al. CD177, a specific marker of neutrophil activation, is associated with coronavirus disease 2019 severity and death. iScience 2021, 24, 102711. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, T.; Zhang, Z.; Dai, X.; Zeng, B.; Li, Z.; Li, Z. Bioinformatics and system biology approach to identify the influences among COVID-19, ARDS and sepsis. Front. Immunol. 2023, 14, 1152186. [Google Scholar]
- Liu, S.; Huang, Z.; Deng, X.; Zou, X.; Li, H.; Mu, S.; Cao, B. Identification of key candidate biomarkers for severe influenza infection by integrated bioinformatical analysis and initial clinical validation. J. Cell. Mol. Med. 2021, 25, 1725–1738. [Google Scholar] [CrossRef]
- Shao, M.; Wu, F.; Zhang, J.; Dong, J.; Zhang, H.; Liu, X.; Liang, S.; Wu, J.; Zhang, L.; Zhang, C.; et al. Screening of potential biomarkers for distinguishing between latent and active tuberculosis in children using bioinformatics analysis. Medicine 2021, 100, e23207. [Google Scholar] [CrossRef]
- Li, X.; Xie, Y.; Zhang, D.; Wang, Y.; Wu, J.; Sun, X.; Wu, Q. Identification of Significant Genes in HIV/TB via Bioinformatics Analysis. Ann. Clin. Lab. Sci. 2020, 50, 600–610. [Google Scholar]
- Chen, P.; Liu, Y.; Lin, X.; Yu, B.; Chen, B.; Lin, F. The Underlying Molecular Basis and Mechanisms of Venous Thrombosis in Patients with Osteomyelitis: A Data-Driven Analysis. Genet. Res. 2022, 2022, 5672384. [Google Scholar] [CrossRef] [PubMed]
- Almansa, R.; Socias, L.; Sanchez-Garcia, M.; Martín-Loeches, I.; del Olmo, M.; Andaluz-Ojeda, D.; Bobillo, F.; Rico, L.; Herrero, A.; Roig, V.; et al. Critical COPD respiratory illness is linked to increased transcriptomic activity of neutrophil proteases genes. BMC Res. Notes 2012, 5, 401. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.H.; Othman, Z.; Noor, H.M.; Arshad, S.S.; Yusof, A.K.; Jamal, R.; Rahman, A.R. Peripheral blood gene expression profile of atherosclerotic coronary artery disease in patients of different ethnicity in Malaysia. J. Cardiol. 2012, 60, 192–203. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, X.; Wang, P.; Yan, S.; Wang, G. Study on potential differentially expressed genes in stroke by bioinformatics analysis. Neurol. Sci. 2022, 43, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cadenas, I.; Del Rio-Espinola, A.; Domingues-Montanari, S.; Mendioroz, M.; Fernandez-Morales, J.; Penalba, A.; Rubiera, M.; Hernandez-Guillamon, M.; Rosell, A.; Delgado, P.; et al. Genes involved in hemorrhagic transformations that follow recombinant t-PA treatment in stroke patients. Pharmacogenomics 2013, 14, 495–504. [Google Scholar] [CrossRef]
- Yan, Z.; Wu, Q.; Cai, W.; Xiang, H.; Wen, L.; Zhang, A.; Peng, Y.; Zhang, X.; Wang, H. Identifying critical genes associated with aneurysmal subarachnoid hemorrhage by weighted gene co-expression network analysis. Aging 2021, 13, 22345–22360. [Google Scholar] [CrossRef]
- Xiao, L.; Xiao, W.; Lin, S. Potential biomarkers for active renal involvement in systemic lupus erythematosus patients. Front. Med. 2022, 9, 995103. [Google Scholar] [CrossRef]
- Wang, L.; Yang, Z.; Yu, H.; Lin, W.; Wu, R.; Yang, H.; Yang, K. Predicting diagnostic gene expression profiles associated with immune infiltration in patients with lupus nephritis. Front. Immunol. 2022, 13, 839197. [Google Scholar] [CrossRef]
- Ren, Y.; Labinsky, H.; Palmowski, A.; Bäcker, H.; Müller, M.; Kienzle, A. Altered molecular pathways and prognostic markers in active systemic juvenile idiopathic arthritis: Integrated bioinformatic analysis. Bosn. J. Basic Med. Sci. 2022, 22, 247–260. [Google Scholar] [CrossRef]
- Ward, J.M.; Ambatipudi, M.; O'Hanlon, T.P.; Smith, M.A.; de Los Reyes, M.; Schiffenbauer, A.; Rahman, S.; Zerrouki, K.; Miller, F.W.; Sanjuan, M.A.; et al. Shared and Distinctive Transcriptomic and Proteomic Pathways in Adult and Juvenile Dermatomyositis. Arthritis Rheumatol. 2023, 75, 2014–2026. [Google Scholar] [CrossRef]
- Kvist-Hansen, A.; Kaiser, H.; Wang, X.; Krakauer, M.; Gørtz, P.M.; McCauley, B.D.; Zachariae, C.; Becker, C.; Hansen, P.R.; Skov, L. Neutrophil Pathways of Inflammation Characterize the Blood Transcriptomic Signature of Patients with Psoriasis and Cardiovascular Disease. Int. J. Mol. Sci. 2021, 22, 10818. [Google Scholar] [CrossRef] [PubMed]
- Cordiglieri, C.; Baggi, F.; Bernasconi, P.; Kapetis, D.; Faggiani, E.; Consonni, A.; Andreetta, F.; Frangiamore, R.; Confalonieri, P.; Antozzi, C.; et al. Identification of a gene expression signature in peripheral blood of multiple sclerosis patients treated with disease-modifying therapies. Clin. Immunol. 2016, 173, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; You, K.; Zhong, J.; Ruan, Z.; Wang, B. Identification of potential core genes in Kawasaki disease using bioinformatics analysis. J. Int. Med. Res. 2019, 47, 4051–4058. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.V.; Luna, L.G.; Cotter, J.; Talbert, J.; Leach, S.M.; Kidd, R.; Turner, J.; Kummer, N.; Kervitsky, D.; Brown, K.K.; et al. The peripheral blood transcriptome identifies the presence and extent of disease in idiopathic pulmonary fibrosis. PLoS ONE 2012, 7, e37708. [Google Scholar] [CrossRef]
- Molyneaux, P.L.; Willis-Owen, S.A.G.; Cox, M.J.; James, P.; Cowman, S.; Loebinger, M.; Blanchard, A.; Edwards, L.M.; Stock, C.; Daccord, C.; et al. Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2017, 195, 1640–1650. [Google Scholar] [CrossRef]
- Urbarova, I.; Skogholt, A.H.; Sun, Y.Q.; Mai, X.M.; Grønberg, B.H.; Sandanger, T.M.; Sætrom, P.; Nøst, T.H. Increased expression of individual genes in whole blood is associated with late-stage lung cancer at and close to diagnosis. Sci. Rep. 2023, 13, 20760. [Google Scholar] [CrossRef]
- Hasselbalch, H.C.; Skov, V.; Stauffer Larsen, T.; Thomassen, M.; Hasselbalch Riley, C.; Jensen, M.K.; Bjerrum, O.W.; Kruse, T.A. Transcriptional profiling of whole blood identifies a unique 5-gene signature for myelofibrosis and imminent myelofibrosis transformation. PLoS ONE 2014, 9, e85567. [Google Scholar] [CrossRef]
- Ma, H.; Liu, J.; Li, Z.; Xiong, H.; Zhang, Y.; Song, Y.; Lai, J. Expression profile analysis reveals hub genes that are associated with immune system dysregulation in primary myelofibrosis. Hematology 2021, 26, 478–490. [Google Scholar] [CrossRef]
- Skov, V.; Burton, M.; Thomassen, M.; Stauffer Larsen, T.; Riley, C.H.; Brinch Madelung, A.; Kjær, L.; Bondo, H.; Stamp, I.; Ehinger, M.; et al. A 7-Gene Signature Depicts the Biochemical Profile of Early Prefibrotic Myelofibrosis. PLoS ONE 2016, 11, e0161570. [Google Scholar] [CrossRef]
- Gomez-Lopez, N.; Romero, R.; Hassan, S.S.; Bhatti, G.; Berry, S.M.; Kusanovic, J.P.; Pacora, P.; Tarca, A.L. The Cellular Transcriptome in the Maternal Circulation During Normal Pregnancy: A Longitudinal Study. Front. Immunol. 2019, 10, 2863. [Google Scholar] [CrossRef]
- Heung, M.M.; Jin, S.; Tsui, N.B.; Ding, C.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.; Lo, Y.M. Placenta-derived fetal specific mRNA is more readily detectable in maternal plasma than in whole blood. PLoS ONE 2009, 4, e5858. [Google Scholar] [CrossRef] [PubMed]
- Chaiworapongsa, T.; Romero, R.; Whitten, A.; Tarca, A.L.; Bhatti, G.; Draghici, S.; Chaemsaithong, P.; Miranda, J.; Hassan, S.S. Differences and similarities in the transcriptional profile of peripheral whole blood in early and late-onset preeclampsia: Insights into the molecular basis of the phenotype of preeclampsia. J. Perinat. Med. 2013, 41, 485–504. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, A.; Brattbakk, H.R.; Trentani, A.; Holdhus, R.; Stansberg, C.; Bartz-Johannessen, C.A.; Hughes, T.; Steen, N.E.; Melle, I.; Djurovic, S.; et al. Patients with schizophrenia and bipolar disorder display a similar global gene expression signature in whole blood that reflects elevated proportion of immature neutrophil cells with association to lipid changes. Transl. Psychiatry 2023, 13, 147. [Google Scholar] [CrossRef] [PubMed]
- Tsakiroglou, M.; Evans, A.; Pirmohamed, M. Leveraging transcriptomics for precision diagnosis: Lessons learned from cancer and sepsis. Front. Genet. 2023, 14, 1100352. [Google Scholar] [CrossRef]
- Davenport, E.E.; Burnham, K.L.; Radhakrishnan, J.; Humburg, P.; Hutton, P.; Mills, T.C.; Rautanen, A.; Gordon, A.C.; Garrard, C.; Hill, A.V.; et al. Genomic landscape of the individual host response and outcomes in sepsis: A prospective cohort study. Lancet Respir. Med. 2016, 4, 259–271. [Google Scholar] [CrossRef]
- Burnham, K.L.; Davenport, E.E.; Radhakrishnan, J.; Humburg, P.; Gordon, A.C.; Hutton, P.; Svoren-Jabalera, E.; Garrard, C.; Hill, A.V.S.; Hinds, C.J.; et al. Shared and Distinct Aspects of the Sepsis Transcriptomic Response to Fecal Peritonitis and Pneumonia. Am. J. Respir. Crit. Care Med. 2016, 196, 328–339. [Google Scholar] [CrossRef]
- Sweeney, T.E.; Shidham, A.; Wong, H.R.; Khatri, P. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci. Transl. Med. 2015, 7, 287ra71. [Google Scholar] [CrossRef]
- Sweeney, T.E.; Azad, T.D.; Donato, M.; Haynes, W.A.; Perumal, T.M.; Henao, R.; Bermejo-Martin, J.F.; Almansa, R.; Tamayo, E.; Howrylak, J.A.; et al. Unsupervised Analysis of Transcriptomics in Bacterial Sepsis Across Multiple Datasets Reveals Three Robust Clusters. Crit. Care Med. 2018, 46, 915–925. [Google Scholar] [CrossRef]
- Scicluna, B.P.; van Vught, L.A.; Zwinderman, A.H.; Wiewel, M.A.; Davenport, E.E.; Burnham, K.L.; Nürnberg, P.; Schultz, M.J.; Horn, J.; Cremer, O.L.; et al. Classification of patients with sepsis according to blood genomic endotype: A prospective cohort study. Lancet Respir. Med. 2017, 5, 816–826. [Google Scholar] [CrossRef]
- Palmer, N.P.; Silvester, J.A.; Lee, J.J.; Beam, A.L.; Fried, I.; Valtchinov, V.I.; Rahimov, F.; Kong, S.W.; Ghodoussipour, S.; Hood, H.C.; et al. Concordance between gene expression in peripheral whole blood and colonic tissue in children with inflammatory bowel disease. PLoS ONE 2019, 14, e0222952. [Google Scholar] [CrossRef]
- Kabakchiev, B.; Turner, D.; Hyams, J.; Mack, D.; Leleiko, N.; Crandall, W.; Markowitz, J.; Otley, A.R.; Xu, W.; Hu, P.; et al. Gene expression changes associated with resistance to intravenous corticosteroid therapy in children with severe ulcerative colitis. PLoS ONE 2010, 5, e13085. [Google Scholar] [CrossRef] [PubMed]
- Mo, A.; Marigorta, U.M.; Arafat, D.; Chan, L.H.K.; Ponder, L.; Jang, S.R.; Prince, J.; Kugathasan, S.; Prahalad, S.; Gibson, G. Disease-specific regulation of gene expression in a comparative analysis of juvenile idiopathic arthritis and inflammatory bowel disease. Genome Med. 2018, 10, 48. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).