Recombinant TP-84 Bacteriophage Glycosylase–Depolymerase Confers Activity against Thermostable Geobacillus stearothermophilus via Capsule Degradation
Abstract
1. Introduction
2. Results and Discussion
2.1. Cloning of the TP84_26 DP and His6-TP84_26 Genes
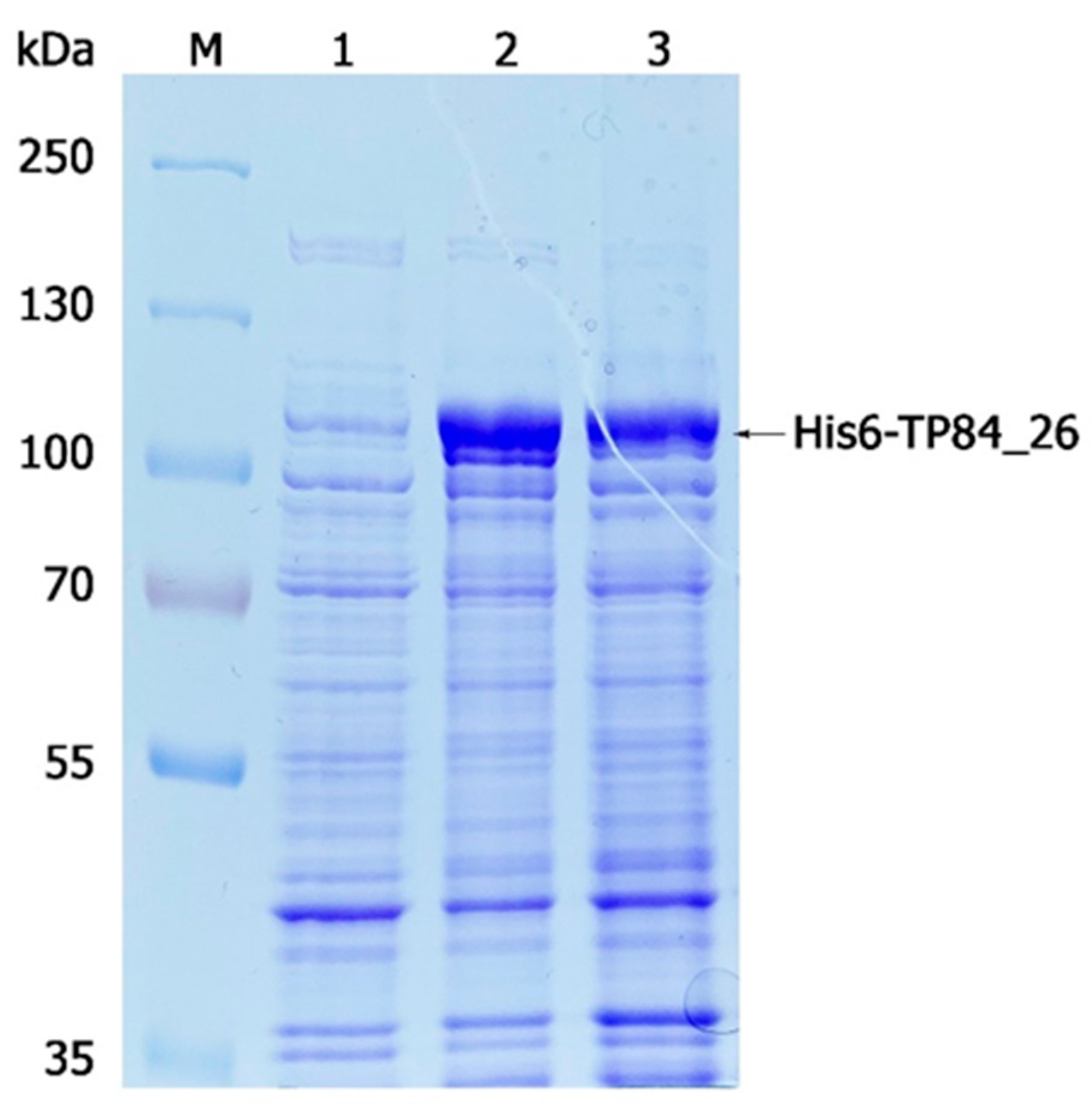
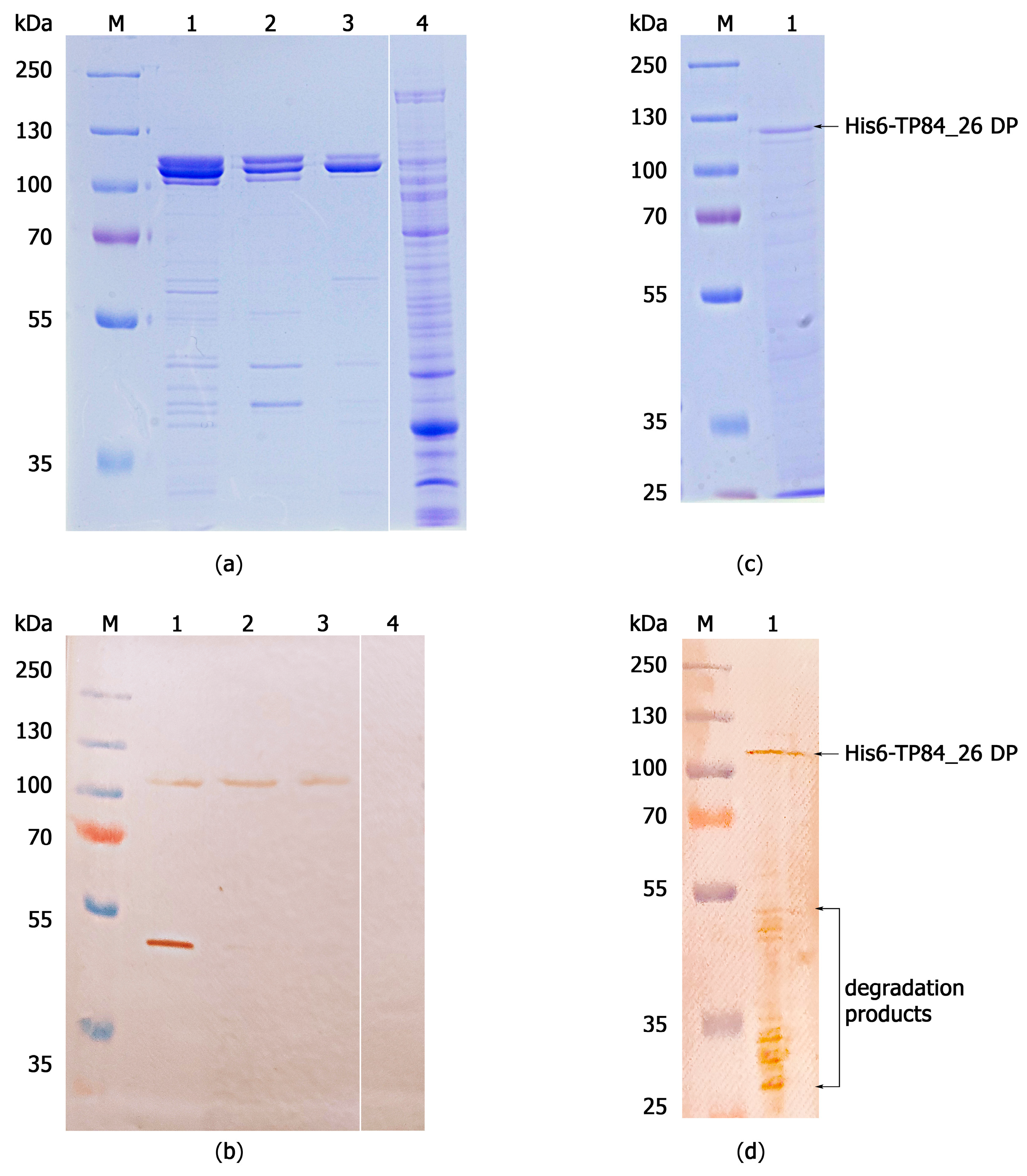
2.2. Gene Expression/Purification of Recombinant His6-TP84_26 DP and TP84_26 DP and Determination of TP84_26 DP Localization within TP-84 Capsid
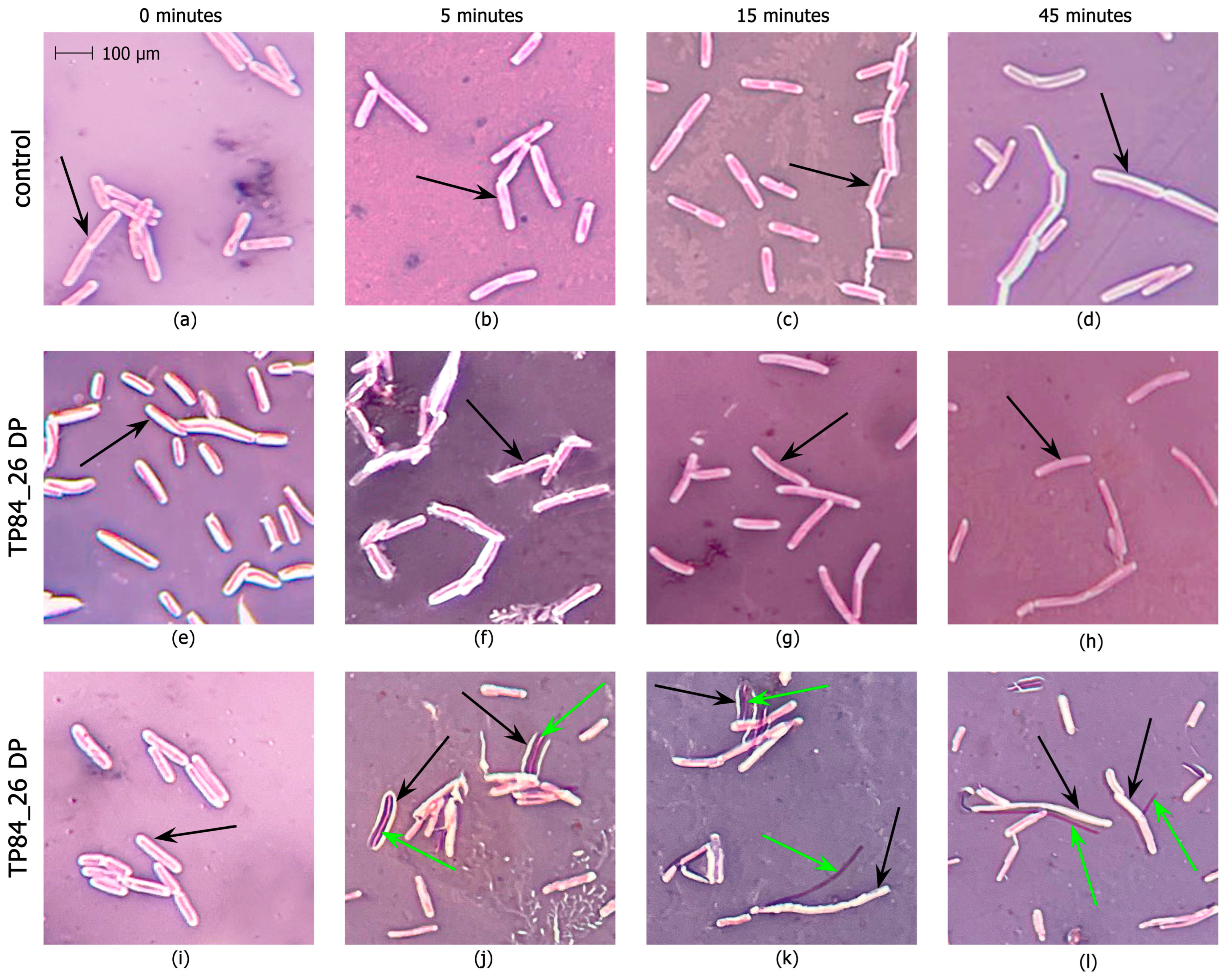
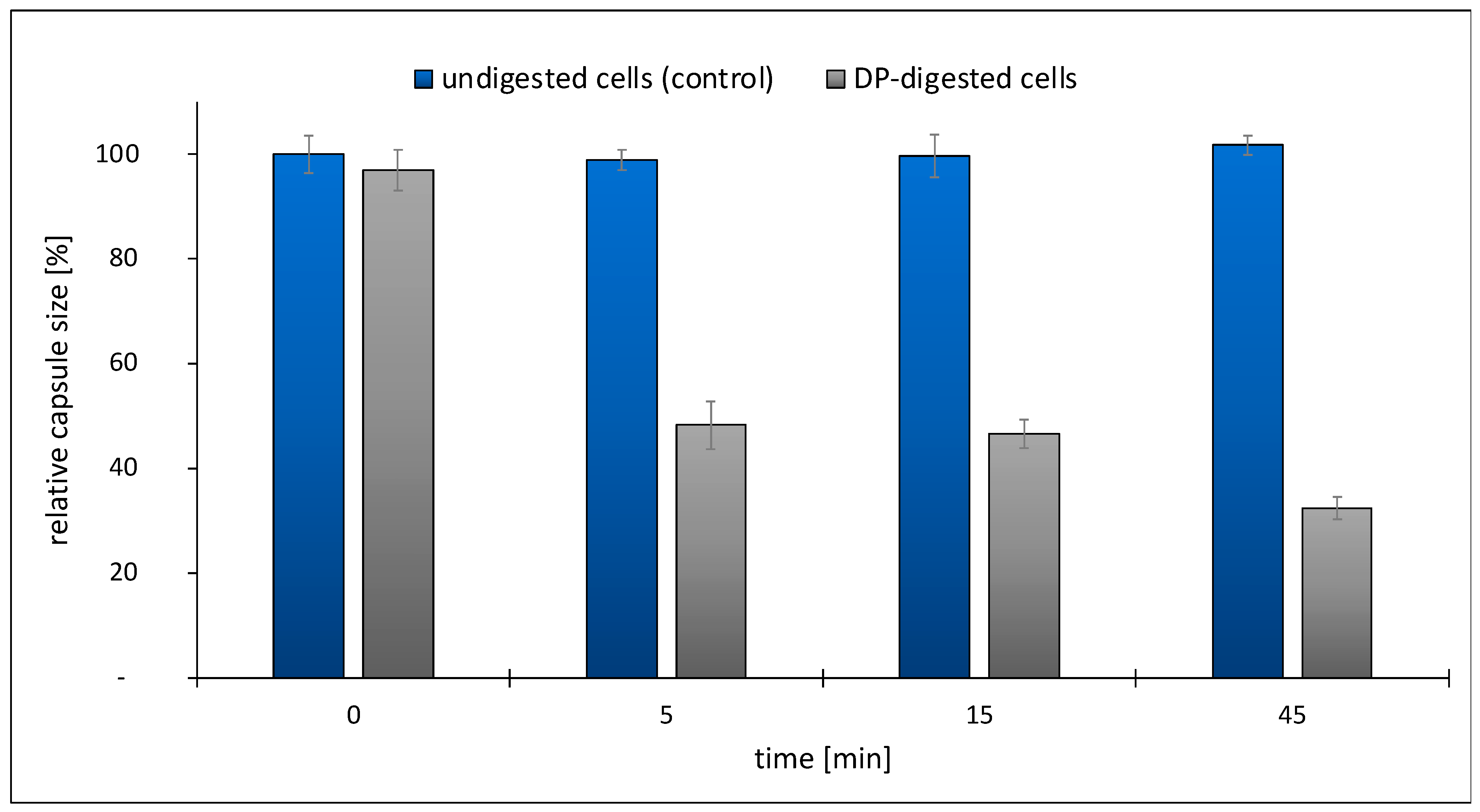
2.3. Development of an Enzymatic Assay for TP84_26 DP and Functional Implications
3. Materials and Methods
3.1. Bacterial Strains, Bacteriophage, Reagents
3.2. Cloning of TP84_26 DP ORF into E. coli
3.3. TP-84 Genetic Expression of His6-TP84_26 and TP84_26 Clones
3.4. Recombinant TP84_26 DP Variant Purification
3.4.1. His6-TP84_26 IMAC Affinity Purification
3.4.2. Universal TP84_26 DP and His6-TP84_26 DP Purification Protocol
3.5. Immunoblotting
3.6. Functional Enzymatic Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skowron, P.M.; Kropinski, A.M.; Zebrowska, J.; Janus, L.; Szemiako, K.; Czajkowska, E.; Maciejewska, N.; Skowron, M.; Łoś, J.; Łoś, M.; et al. Sequence, Genome Organization annotation and Proteomics of the Thermophilic, 47.7-Kb Geobacillus stearothermophilus Bacteriophage TP-84 and Its Classification in the New Tp84virus Genus. PLoS ONE 2018, 13, e0195449. [Google Scholar] [CrossRef]
- Łubkowska, B.; Jeżewska-Frąckowiak, J.; Sobolewski, I.; Skowron, P.M. Bacteriophages of Thermophilic ‘Bacillus Group’ Bacteria—A Review. Microorganisms 2021, 9, 1522. [Google Scholar] [CrossRef] [PubMed]
- Łubkowska, B.; Czajkowska, E.; Stodolna, A.; Sroczyński, M.; Zylicz Stachula, A.; Sobolewski, I.; Skowron, M.P. A Novel Thermostable TP-84 Capsule Depolymerase: A Method for Rapid Polyethyleneimine Processing of a Bacteriophage-Expressed Proteins. Microb. Cell Fact. 2023, 22, 80. [Google Scholar] [CrossRef] [PubMed]
- Logan, N.A.; De Vos, P.; Dinsdale, A. Geobacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 1–26. [Google Scholar]
- Shapton, D.; Hindes, W. The Standardization of a Spore Count Technique. Chem. Ind. 1963, 41, 230–234. [Google Scholar]
- Ren, X.-Y.; Cai, H.-S.; Lin, X.-J.; Qiu, H.-D.; Chen, J.-C. Optimum Medium Composition for Geobacillus Stearothermophilus CHB1 Fermentation. Fujian J. Agric. Sci. 2007, 22, 54–57. [Google Scholar]
- Lee, Y.H.; Brown, M.R.W.; Cheung, H.Y. Defined Minimal Media for the Growth of Prototrophic and Auxotrophic Strains of Bacillus stearothermophilus. J. Appl. Bacteriol. 1982, 53, 179–187. [Google Scholar] [CrossRef]
- Zeigler, D.R. The Geobacillus Paradox: Why Is a Thermophilic Bacterial Genus so Prevalent on a Mesophilic Planet? Microbiology 2014, 160, 1–11. [Google Scholar] [CrossRef]
- Setlow, P. Spores of Bacillus subtilis: Their Resistance to and Killing by Radiation, Heat and Chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef]
- Novik, G.; Savich, V.; Meerovskaya, O.; Novik, G.; Savich, V.; Meerovskaya, O. Geobacillus Bacteria: Potential Commercial Applications in Industry, Bioremediation, and Bioenergy Production. In Growing and Handling of Bacterial Cultures; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef]
- Maniloff, J.; Ackermann, H.W. Taxonomy of Bacterial Viruses: Establishment of Tailed Virus Genera and the Order Caudovirales. Arch. Virol. 1998, 143, 2051–2063. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of Morphology-Based Taxa and Change to Binomial Species Names: 2022 Taxonomy Update of the ICTV Bacterial Viruses Subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and Function of Phage Encoded Depolymerases. Front. Microbiol. 2020, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-Encoded Depolymerases: Their Diversity and Biotechnological Applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Bai, C.; Leung, S.S.Y. Translating Bacteriophage-Derived Depolymerases into Antibacterial Therapeutics: Challenges and Prospects. Acta Pharm. Sin. B 2024, 14, 155–169. [Google Scholar] [CrossRef]
- Maszewska, A. Phage Associated Polysaccharide Depolymerases—Characteristics and Application. Postepy Hig. Med. Dosw. 2015, 69, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.Y.; Lin, J.T.; Ricker, N.; Anany, H. The Age of Phage: Friend or Foe in the New Dawn of Therapeutic and Biocontrol Applications? Pharmaceuticals 2021, 14, 199. [Google Scholar] [CrossRef]
- Epstein, I.; Campbell, L.L. Production and Purification of the Thermophilic Bacteriophage TP-84. Appl. Microbiol. 1975, 29, 219–223. [Google Scholar] [CrossRef]
- Bornhorst, J.A.; Falke, J.J. Purification of Proteins Using Polyhistidine Affinity Tags. Methods Enzymol. 2010, 2000, 245–254. [Google Scholar]
- Debeljak, N.; Feldman, L.; Davis, K.L.; Komel, R.; Sytkowski, A.J. Variability in the Immunodetection of His-Tagged Recombinant Proteins. Anal. Biochem. 2006, 359, 216–223. [Google Scholar] [CrossRef]
- Squeglia, F.; Maciejewska, B.; Łątka, A.; Ruggiero, A.; Briers, Y.; Drulis-Kawa, Z.; Berisio, R. Structural and Functional Studies of a Klebsiella Phage Capsule Depolymerase Tailspike: Mechanistic Insights into Capsular Degradation. Structure 2020, 28, 613–624.e4. [Google Scholar] [CrossRef]
- Ghosh, R.; Gilda, J.E.; Gomes, A.V. The Necessity of and Strategies for Improving Confidence in the Accuracy of Western Blots. Expert Rev. Proteom. 2014, 11, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Fast, R.; Eberhard, T.H.; Ruusala, T.; Kurland, C.G. Does Streptomycin Cause an Error Catastrophe? Biochimie 1987, 69, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Suresh, A.; Pan, C.; Ng, W.J. Sodium Azide Inhibition of Microbial Activities and Impact on Sludge Floc Destabilization. Chemosphere 2020, 244, 125452. [Google Scholar] [CrossRef] [PubMed]
- Doss, R.K.; Palmer, M.; Mead, D.A.; Hedlund, B.P. Functional Biology and Biotechnology of Thermophilic Viruses. Essays Biochem. 2023, 67, 671–684. [Google Scholar]
- Zhao, A.; Sun, J.; Liu, Y. Understanding Bacterial Biofilms: From Definition to Treatment Strategies. Front. Cell. Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Łubkowska, B.; Sobolewski, I.; Adamowicz, K.; Zylicz-Stachula, A.; Skowron, P.M. Recombinant TP-84 Bacteriophage Glycosylase–Depolymerase Confers Activity against Thermostable Geobacillus stearothermophilus via Capsule Degradation. Int. J. Mol. Sci. 2024, 25, 722. https://doi.org/10.3390/ijms25020722
Łubkowska B, Sobolewski I, Adamowicz K, Zylicz-Stachula A, Skowron PM. Recombinant TP-84 Bacteriophage Glycosylase–Depolymerase Confers Activity against Thermostable Geobacillus stearothermophilus via Capsule Degradation. International Journal of Molecular Sciences. 2024; 25(2):722. https://doi.org/10.3390/ijms25020722
Chicago/Turabian StyleŁubkowska, Beata, Ireneusz Sobolewski, Katarzyna Adamowicz, Agnieszka Zylicz-Stachula, and Piotr M. Skowron. 2024. "Recombinant TP-84 Bacteriophage Glycosylase–Depolymerase Confers Activity against Thermostable Geobacillus stearothermophilus via Capsule Degradation" International Journal of Molecular Sciences 25, no. 2: 722. https://doi.org/10.3390/ijms25020722
APA StyleŁubkowska, B., Sobolewski, I., Adamowicz, K., Zylicz-Stachula, A., & Skowron, P. M. (2024). Recombinant TP-84 Bacteriophage Glycosylase–Depolymerase Confers Activity against Thermostable Geobacillus stearothermophilus via Capsule Degradation. International Journal of Molecular Sciences, 25(2), 722. https://doi.org/10.3390/ijms25020722





