Abstract
JASMONATE ZIM domain (JAZ) proteins, inhibitors of the jasmonic acid (JA) signaling pathway, are identified in different plants, such as rice and Arabidopsis. These proteins are crucial for growth, development, and abiotic stress responses. However, limited information is available regarding the JAZ family in alfalfa. This study identified 11 JAZ genes (MsJAZs) in the “Zhongmu No.1” reference genome of alfalfa. The physical and chemical properties, chromosome localization, phylogenetic relationships, gene structure, cis-acting elements, and collinearity of the 11 MsJAZ genes were subsequently analyzed. Tissue-specific analysis revealed distinct functions of different MsJAZ genes in growth and development. The expression patterns of MsJAZ genes under salt stress conditions were validated using qRT-PCR. All MsJAZ genes responded to salt stress, with varying levels of upregulation over time, highlighting their role in stress responses. Furthermore, heterogeneous expression of MsJAZ1 in Arabidopsis resulted in significantly lower seed germination and survival rates in OE-2 and OE-4 compared to the WT under 150 mM NaCl treatment. This study establishes a foundation for further exploration of the function of the JAZ family and provides significant insights into the genetic improvement of alfalfa.
1. Introduction
Plants adapt to many stimuli during growth and development, including biotic stress, such as pests and diseases, as well as abiotic stress, such as salt, cold, and drought [1,2,3]. Phytohormones, such as auxins, cytokinins, ethylene, gibberellins, brassinosteroids, and jasmonates, are crucial for regulating physiological and molecular responses to abiotic stresses [4,5]. Among these, jasmonic acid (JA) is a vital signaling molecule. It regulates a wide range of cellular activities [6]. JA and its cyclic precursors and derivatives are collectively known as jasmonates (JAs). They play important roles in regulating various physiological processes in plants, including growth, development, and defense [7,8]. Jasmonate ZIM domain proteins (JAZ) can mediate the switch-like activation and repression of the JA pathway [9].
JAZ proteins are a subfamily of the TIFY superfamily and possess two conserved domains: the TIFY domain, characterized by the amino acid pattern of TIF [F/Y] XG, and the Jas domain, with a characteristic motif of SLX2FX2KRX2RX5PY [10,11]. JAZ is a negative regulator of the JA signaling pathway [12]. The JA content in plants is dynamically regulated [13]. Under normal conditions, JAs are present at low levels, and JAZ proteins inhibit the activity of many transcription factors, such as MYC2, MYC3, and MYC4 [9]. If the JA level is high, JAZ proteins will interact with JA-Ile receptor complexes, promoting the formation of co-receptor complexes consisting of the F-box protein COI1 and degron sequences located in the Jas motifs of JAZ. The hormone-dependent interaction of the COI1-JAZ co-receptor complex leads to JAZ protein degradation through the ubiquitin/26 s proteasome pathway, thereby releasing the JAZ-repressed expression of transcription factors, and many JA response genes are induced to express [14,15,16].
Previous studies have indicated that JAZ proteins are crucial in different growth processes in plants, including root growth, seed germination, and defense [17,18,19,20]. For instance, JAZ proteins can regulate jasmonate-enhanced root growth inhibition by inhibiting RHD6 and RHD6 LIKE1 transcription factors [17]. OsJAZ1 negatively regulates development by repressing the bHLH-MYB complex. However, when the JAZ protein is degraded, it releases the bHLH-MYB complex, and activates downstream gene expression to regulate stamen development [21]. The ABA signaling pathway is vital for seed germination [22]. Studies have shown that the accumulation of JAZ proteins can attenuate ABA signaling, whereas JAZ mutants can enhance ABA responses [23]. Further analysis revealed that JAZ proteins repress the transcription of ABI5 and ABI3, thereby regulating the ABA-mediated seed germination process [24]. Angiosperms can resist various stresses in nature through the COI1-JAZ-DELLA-PIF signaling pattern [25]. JAZ genes not only participate in the growth process of plants, but also respond to some abiotic stresses, including salt and drought stress, which are significant factors impacting plant growth and development [26,27]. In rice, OsJAZ9 interacts with OsNINJA and OsbHLH to form a transcriptional regulatory complex that regulates salt tolerance [28]. OsJAZ1 enhances drought sensitivity through the JA and ABA pathways [29]. PeJAZ2 positively reduces plant sensitivity to drought through ABA-induced stomatal closure by H2O2 production [30].
Alfalfa (Medicago sativa L.) is an important forage crop worldwide, and is known as the “King of Forages” [31]. Alfalfa has high nutritional value, such as rich protein content, and many kinds of vitamins [32]. Mining excellent genes in alfalfa is crucial for improving M. sativa research at the theoretical level, screening, and breeding new varieties. Previously, 12 and 15 JAZ genes were found in Arabidopsis thaliana [33] and rice [10], respectively, and 275 JAZ genes in 29 soybean genomes [34]. Despite its importance, the JAZ gene family in alfalfa remains underexplored. To address this gap, our study aimed to identify and analyze the JAZ gene family in M. sativa. We identified 11 JAZ proteins in the alfalfa genome and conducted a comprehensive analysis of the characteristics of 11 MsJAZ genes, including chromosome localization, phylogenetic analysis, gene structure collinearity analysis, and cis-acting element analysis. The evolutionary relationships between MsJAZs and A. thaliana, Medicago truncatula, and Glycine max were analyzed. To further analyze the potential function of MsJAZs, qRT-PCR was used to detect their expression patterns under salt stress. MsJAZ1 was cloned and heterologously expressed in Arabidopsis, and the phenotype of transgenic Arabidopsis under salt stress was investigated. In summary, we identified and analyzed all the JAZ genes in the alfalfa genome, providing reference for further functional studies.
2. Results
2.1. Identification of JAZ Genes in the Alfalfa Genome
Eleven JAZ were identified in the M. sativa genome (Zhongmu No.1) through the hidden Markov model (HMM) and domain analysis, and these members were named MsJAZ1–MsJAZ11 based on their chromosomal locations. The basic information regarding MsJAZ is listed in Table 1. The length of the MsJAZ proteins ranged from 131 aa (MsJAZ2) to 337 aa (MsJAZ11), with a mean length of 240 aa. The MW of the MsJAZ proteins ranged from 15.09 (MsJAZ2) to 38.03 kDa (MsJAZ1). The pI of MsJAZ proteins was between 7.10 (MsJAZ3) and 9.56 (MsJAZ6). These proteins were alkaline (pI > 7.0). All MsJAZs were located in the nucleus by subcellular localization prediction, while MsJAZ1 and MsJAZ3 were also found in the cell membrane.

Table 1.
Characteristics of MsJAZ genes in Medicago sativa.
The 11 JAZ genes were distributed on 5 chromosomes of the alfalfa genome, among which no MsJAZ genes existed on chromosomes 3, 4, and 7 (Figure 1). The greatest number of MsJAZ genes (4 of 11) was distributed on chromosome 2, while three MsJAZ genes were located on chromosome 8. There are two MsJAZ genes on chromosome 5 and only one MsJAZ gene on chromosomes 1 or 6.
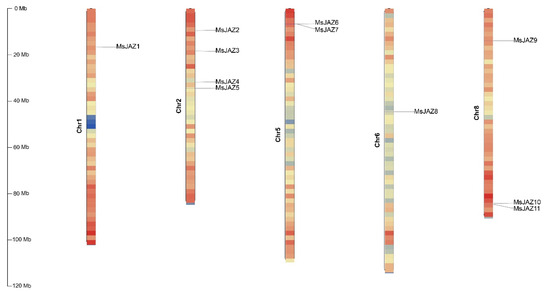
Figure 1.
Chromosomal distribution diagram of alfalfa the JAZ gene. TBtools (v1.108, Chen, C., GZ, China) was used to visualize the position of each MsJAZ on the chromosome. Eleven MsJAZs are mapped onto the five chromosomes of Medicago sativa. The vertical bars represent the chromosomes of alfalfa. The scale on the left indicates the chromosome length (Mb). Color gradients indicate gene density, and red to blue indicate gene density from high to low. Each MsJAZ gene is labeled at its respective position on the chromosomes.
2.2. Phylogenetic Relationships Analysis of MsJAZs
To assess the evolutionary relationship of JAZ between alfalfa and other plants, a maximum likelihood phylogenic tree was generated, containing 11 in alfalfa, 12 in A. thaliana, and 13 in M. truncatula (Figure 2). Based on the known phylogenetic tree grouping of the Arabidopsis JAZ genes, the 36 JAZ proteins were divided into groups I–Ⅴ. Except for group Ⅲ, all other groups contained alfalfa JAZ family members. Group I included the most MsJAZ proteins, with five, accounting for 45% of the total, and group IV had the fewest MsJAZ proteins, with one. According to the grouping of phylogenetic trees, 11 MsJAZ were divided into four branches (Figure 3A), and their gene structure and motif composition were analyzed.
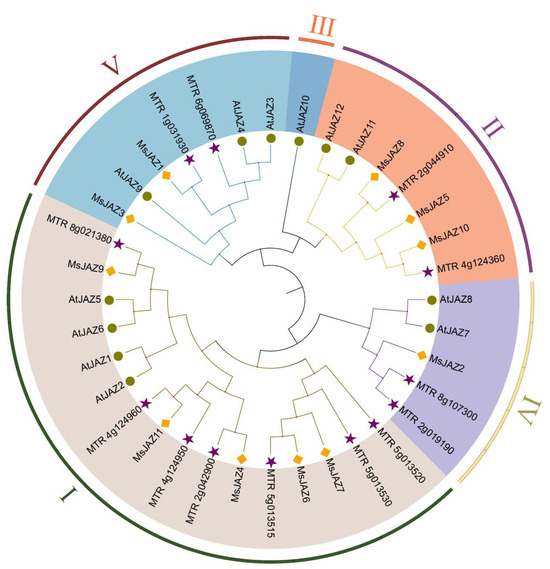
Figure 2.
Phylogenetic tree of the JAZ genes in M.sativa, M. truncatula, and A. thaliana. The phylogenetic tree was constructed using the maximum likelihood method (bootstrap method with 1000 replications was selected) of MEGA 6.0 software. JAZ genes were divided into five groups (I–Ⅴ) according to the known Arabidopsis JAZ family, and were represented using different colors. Using different markers to distinguish each species, yellow diamonds, purple stars, and green circles, respectively, represent M. sativa, M. truncatula, and A. thaliana.
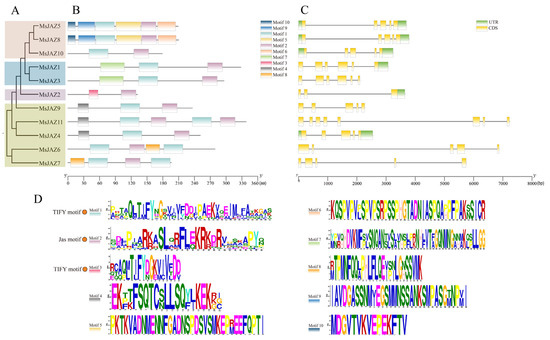
Figure 3.
Phylogenetic relationships, motif compositions, and structures analysis of the MsJAZs family. (A) The phylogenetic tree of MsJAZs of M. sativa was constructed by the maximum likelihood method of MEGA 6.0 software. All MsJAZs were divided into four groups and represented by different colors. (B) The motif composition of MsJAZs was analyzed with the MEME program. A different color is used for each motif. (C) The UTR, CDS, and intron organization of MsJAZs. The green boxes, yellow boxes, and thin black lines represent UTRs, CDSs, and introns, respectively. (D) Sequence logos of motifs 1–10. Motif 1 and motif 3 represent the TIFY motif, and motif 2 represents the Jas motif.
2.3. Gene Structure and Conserved Motif Analysis
The exon and intron structures of the MsJAZs were investigated to understand the structural composition of these genes (Figure 3C). Genetic structure analysis revealed that MsJAZ genes exhibit some similarity in the four groups, but also present many differences between the groups. In total, 64% MsJAZ genes (7 of 11) have five exons. In group Ⅱ, all genes contained five exons, and had UTR sections at both terminals. In group Ⅴ, all genes had six exons. Only one gene (MsJAZ2) had the fewest number of exons in group Ⅳ, with three exons. In group I, MsJAZ11 had eight exons, which was the largest number. All other genes contained five exons, and only MsJAZ4 had UTR sections at both terminals.
In order to further analyze the function of MsJAZs, we analyzed the conserved motifs of 11 proteins. (Figure 3B). A total of ten motifs were detected, and the details of the ten motifs are provided in Table S2. The results showed that all MsJAZ proteins contained the TIFY (motif 1 or motif 3) and Jas motifs (motif 2) (Figure 3D) (Figure S1). Seven additional motifs were present only in specific groups. For example, motifs 5, 6, 9, and 10 were only discovered in group Ⅱ, motif 7 was only identified in group Ⅴ, and motifs 4 and 8 were only present in group Ⅰ. These results indicated that the MsJAZ of the same group had similar gene structures and motif compositions, whereas the MsJAZ in different groups possessed distinct structures and motifs, which might have contributed to the functional diversity among MsJAZs.
2.4. Prediction of Cis-Acting Elements in MsJAZs Promoters
To further investigate the potential functions of MsJAZs, the cis-elements in the promoter regions of the MsJAZs were predicted by PlantCARE. These cis-acting elements are divided into four categories: light, hormone, environmental stress, and development-responsive elements (Figure 4). Light-responsive elements mainly contain Box 4, GT1-motif, and G-box, and all genes contain Box 4 elements. Ten MsJAZ genes contained ABA response elements (ABRE), nine MsJAZ genes contained JA response elements (CGTCA and TGACG motifs), and five MsJAZ genes contained SA response elements (TCA-element). These hormone-responsive elements are crucial for plant biotic and abiotic stress resistance. In addition, several environmental stress-related elements were identified as being responsive to anaerobic conditions (ARE), low temperature (LTR), drought (MBS), and stress (TC-rich repeats). All MsJAZ genes contained ARE elements, with some numbers as high as 13. These results showed that MsJAZ genes have cis-acting elements of different types and numbers in their promoter regions, which may be related to their special functions.
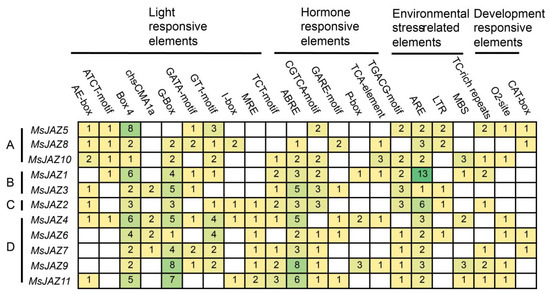
Figure 4.
Prediction of cis-acting elements of MsJAZ genes. (A–D): MsJAZ genes were divided into four groups. Cis-acting elements are divided into light, hormones, environmental stress, and development response elements. The digit represents the number of elements.
2.5. Analysis of the Gene Duplication and Synteny of the MsJAZs
Genome duplication events in this family were investigated to understand the evolution of MsJAZ genes better. A syntenic relationship between the MsJAZ genes was discovered using MCScanX from TBTools. Five segmental duplication events were found in the MsJAZs (Figure 5). This result indicates that segmental duplication plays a crucial role in the evolution of the MsJAZs gene family. To better understand the evolutionary constraints, the Ka, Ks, and Ka/Ks ratios of the MsJAZ gene pairs were calculated. All five gene pairs had Ka/Ks ratios below 1, suggesting that the MsJAZ gene family experienced purification selection (Table 2).
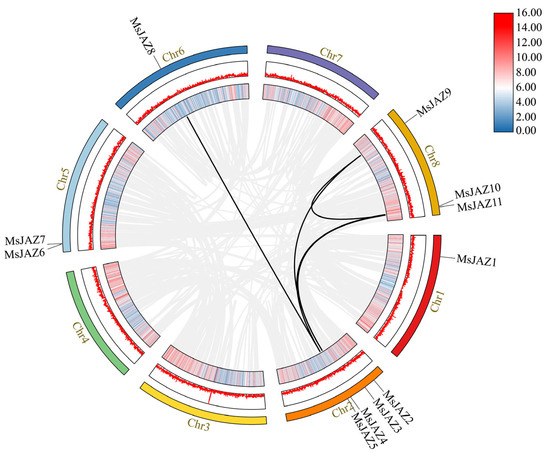
Figure 5.
Schematic diagram of the syntenic relationships of MsJAZ genes in M. sativa. The outermost colored circles represent the eight chromosomes of alfalfa, and the position of the MsJAZ genes on the chromosome is marked on the circle. The gray lines show the syntenic regions in alfalfa genome, and the black lines indicate JAZ gene pairs in segmental duplication events. The innermost and the middle circles represent the density and abundance of genes at that location on the chromosome.

Table 2.
Ks and Ka analysis of duplicated gene pairs.
To further identify the orthologous relationships of MsJAZ genes in different crops, the synteny of M. sativa with A. thaliana, G. max, and M. truncatula was analyzed. The results showed that 11 orthologous gene pairs were identified between Arabidopsis and alfalfa, 16 between M. truncatula and alfalfa, and 31 between Glycine max and alfalfa (Figure 6). Seven JAZ genes (MsJAZ1/2/4/5/9/10/11) were simultaneously identified as orthologous genes between the three species and alfalfa genomes. These genes may play an essential role in the evolution of the JAZ family.
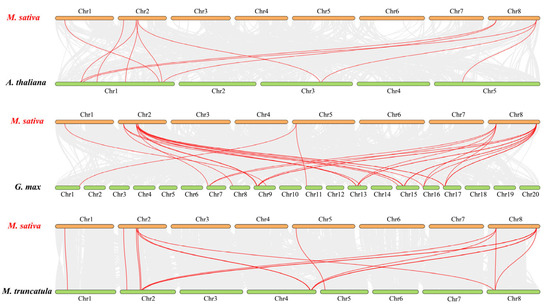
Figure 6.
Synteny analysis of JAZ genes between M. sativa and three representative plant species. The orange boxes represent the chromosomes of M. sativa, and the green boxes represent the chromosomes of A. thaliana, G. max, and M. truncatula. Gray lines in the background indicate collinear blocks between alfalfa and the indicated plant, while the red lines indicate the duplication of JAZ gene pairs.
2.6. Expression Analysis of MsJAZ Genes in Different Tissues
To further understand the expression patterns of MsJAZ genes in different tissues of alfalfa, transcriptome data of different tissues of alfalfa were obtained from public databases, including flowers, roots, leaves, post-elongation stems, elongating stems, and nodules (Figure 7). The original data are presented in Supplementary Table S1. In total, 45% MsJAZ genes (MsJAZ1, MsJAZ6, MsJAZ9, MsJAZ10, and MsJAZ11) were highly expressed in leaves, 27% (MsJAZ3, MsJAZ2 and MsJAZ7) in the elongating stems, and 27% (MsJAZ4, MsJAZ5 and MsJAZ8) in the roots. The expression levels of all MsJAZ genes were low in flowers. These results show that different MsJAZ genes may play different roles in regulating the growth and development of alfalfa.
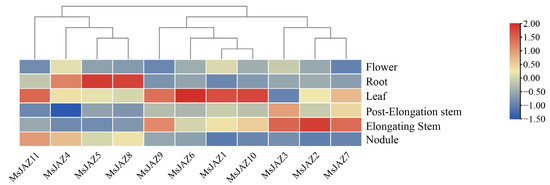
Figure 7.
Expression analysis of MsJAZ genes in different tissues (flowers, roots, leaves, post-elongating stems, elongating stems, and nodules). The legend represents the relative expression levels, blue indicates low expression levels and red indicates high expression levels.
2.7. Expression Analysis of MsJAZ Genes in Salt Stress Treatments
To understand the response of the 11 MsJAZs to salt stress, the expression patterns of MsJAZs under salt stresses were analyzed. Different MsJAZs exhibited different expression patterns under salt stress (Figure 8). Although most MsJAZs were upregulated, the time and degree of upregulation differed slightly. However, the same group of MsJAZ members showed similar trends. For example, group I members (MsJAZ4, MsJAZ6, MsJAZ7, MsJAZ9, and MsJAZ11) were all expressed instantaneously at 1 h or 2 h. Except for MsJAZ9, which had the highest expression level at 24 h, the expression of the remaining members gradually decreased to similar or slightly higher levels than at 0 h.
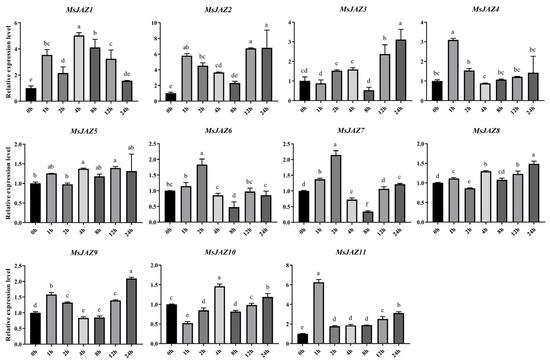
Figure 8.
Expression patterns of 11 MsJAZ genes under 200 mM NaCl treatment. The horizontal coordinates represent processing times, and the vertical coordinates represent relative expression levels. The error bars represent the standard error of the means of three independent replicates. ANOVA with p < 0.05. There are significant differences between different letters (a–f).
2.8. Analysis of Salt Resistance of Arabidopsis with Overexpression of MsJAZ1
Because MsJAZ1 contains more stress-related cis-acting elements and has a significant response to salt stress, MsJAZ1 was heterologously overexpressed in Arabidopsis to explore its function under salt stress. There was no significant difference in the survival rate of Arabidopsis between the WT and transgenic lines on 1/2 MS plates under normal conditions after 20 days of germination (Figure 9A,C). However, under 150 mM NaCl on 1/2 MS plates, the survival rate of WT was 42%, whereas the survival rates of OE-2 and OE-4 were only 11% and 18%, respectively, which were significantly lower than that of WT. The germination rates of the WT and transgenic lines were also explored. Under normal conditions, there was no significant difference in germination rates between the WT and transgenic lines OE-2 and OE-4 except on the second day (Figure 9B). However, under 150 mM NaCl treatment, the seed germination rate of the WT line was significantly faster than that of OE-2 and OE-4 from the third day. On the fourth day, WT was 5.65 and 2.6 times OE-2 and OE-4, respectively. On the seventh day, WT was 1.58 and 1.27 times more than OE-2 and OE-4, respectively. Moreover, when the seedlings grown normally for 7 days were placed on a 150 mM NaCl plate for 3 days, both OE-2 and OE-4 lines were albino (Figure 9D). Although WT was also partially albino, most of the leaves remained green. These results indicate that the overexpression of MsJAZ1 in Arabidopsis makes Arabidopsis more sensitive to salt.
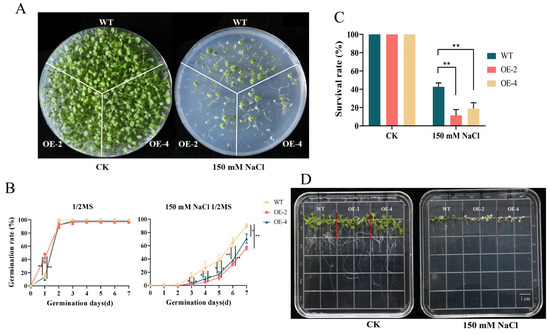
Figure 9.
Analysis of salt resistance overexpressing MsJAZ1 in Arabidopsis. (A–C) The growth of the WT and OE lines of Arabidopsis overexpressing MsJAZ1 on a plate germinated for 20 days treated with CK and 150 mM NaCl. The statistical survival rate is shown in (C). The bottle green, orange, and yellow columns represent WT, OE-2, and OE-4, respectively. The asterisk on the column indicates a significant difference between WT and transgenic lines (* p < 0.05; ** p < 0.01, t-test). (B) Statistical analysis of the germination rate of Arabidopsis WT and OE lines overexpressing MsJAZ1 from 0 to 7 days under CK and 150 mM NaCl treatment. The yellow, orange, and bottle green lines represent WT, OE-2, and OE-4, respectively. The asterisk on the column indicates significant difference between WT and transgenic lines (* p < 0.05; ** p < 0.01, t-test). (D) Salt tolerance analysis of WT and transgenic lines at the seedling stage.
3. Discussion
JAZ is a crucial negative regulator of the JA signaling pathway. It plays a crucial role in plant growth and responses to both biotic and abiotic stresses [21,35]. Genome-wide identification of JAZ family members has been accomplished in many plants [10,33,36,37,38,39,40]. However, a comprehensive analysis and detailed study of the alfalfa JAZ family at the whole-genome level is incomplete. Therefore, we analyzed all JAZ genes in alfalfa to better understand the JAZ family at the molecular level and explore its resistance process.
A total of 11 MsJAZ genes were identified in this study, similar to those in Arabidopsis, suggesting a conserved evolutionary pattern among these species. This similarity may be attributed to using the “Zhongmu NO.1″ haploid reference genome [41]. Based on phylogenetic grouping information from Arabidopsis [42], the JAZ family members of A. thaliana, M. sativa, and M. truncatula were divided into five groups. Notably, group III did not contain any JAZ family members from alfalfa, suggesting that this species lacks a JAZ gene with a function similar to that of AtJAZ10. Exon–intron structures can provide further support for phylogenetic grouping [43]. In this study, most MsJAZs in the same group had similar gene structures, such as all group II members (MsJAZ5, MsJAZ8, and MsJAZ10) containing the same number of exons and similar lengths. However, there are cases in which individual members of the same group have different gene structures. For example, MsJAZ11 in group I contains eight exons, which is different from other members, indicating that it may have functional divergences or new functions in the evolutionary process. Gene replication can also reflect the evolution of gene families and genomes [44]. In this study, five pairs of segmental repetition events were detected, among which the genes involved in the MsJAZ5/MsJAZ8 and MsJAZ5/MsJAZ10 gene pairs were all in the same clade. The genes involved in the MsJAZ4/MsJAZ11 and MsJAZ9/MsJAZ11 gene pairs were also in the same clade. The Ka/Ks ratios of the gene pairs suggested that the alfalfa JAZ gene family experienced purification selection during evolution, which might have resulted in preserved functionality or pseudogenization [40].
In recent years, there have been many studies on the functions of JAZ genes. In soybean, GmJAZ3 interacted with both GmRR18a and GmMYC2a, which can promote seed size, weight, and other organ sizes in stable transgenic soybean plants, increase the protein content of soybean seeds, and reduce the fatty acid content [45]. Through BLASTP, MsJAZ1 and MsJAZ3 were found to have high homology with GmJAZ3, reaching 88% and 73%, respectively. The expression levels of the two MsJAZ genes in leaves and elongating stems were higher than those in other tissues, so we speculated that MsJAZ1 and MsJAZ3 may affect the protein content of alfalfa by regulating the development of leaves and other tissues. Abiotic stresses seriously affect plant growth and productivity [46]. In this study, we found that all MsJAZ genes respond to salt stress, that different MsJAZ genes have various degrees of upregulation at different times, and that the expression patterns of members of the same clade are similar. Previous studies have shown that overexpression of GhJAZ2 can significantly enhance the sensitivity of transgenic cotton to salt stress [47]. Rice salt sensitive 3 (RSS3) interacts with OsJAZ9, OsJAZ11, and non-R/B-like bHLH TFs to form a complex to regulate JA-mediated salt stress response [48]. In this study, MsJAZ1 contained the largest number of stress-related cis-acting elements, and the qRT-PCR results showed that it responded to salt stress. Therefore, MsJAZ1 was cloned and heterologously expressed in A. thaliana to further explore the function of MsJAZ. The results showed that under treatment with 150 mM NaCl, the germination and survival rates of transgenic Arabidopsis were significantly lower than those of the wild type (WT), and their overall growth was also poorer. These results suggest that MsJAZ1 may negatively regulate plant salt tolerance.
Alfalfa is an important forage grass, but various adverse environments limit its growth. In recent years, with the development of various biotechnologies, breeding new varieties of alfalfa has become increasingly common. Therefore, this study provides a reference for further study on the function of the MsJAZ gene and the cultivation of new varieties with stress resistance.
4. Methods and Materials
4.1. Plant Materials and Growth Conditions
The alfalfa seeds “Zhongmu No. 1” were obtained from the Institute of Animal Science of the Chinese Academy of Agricultural Sciences. The growth conditions of the seedlings were consistent with the previous method [32]. Four weeks later, the seedlings were treated with 1/2 Hoagland solution containing 200 mM NaCl to simulate salt stress. The treatments were applied for 0, 1, 2, 4, 8, 12, and 24 hours (h), respectively, to mimic abiotic conditions. Every treatment had three biological replicates for each point, and all samples were stored at −80 °C for total RNA extraction.
4.2. Identification of JAZ Family Members in Alfalfa
The alfalfa reference genome sequence (Zhongmu No.1) was downloaded from Figshare (https://figshare.com/, accessed on 15 March 2023) [41]. The hidden Markov model (HMM) profile for the TIFY domain (PF06200) and Jas domain (PF09425) of the JAZ family were extracted from the Pfam database (https://www.ebi.ac.uk/interpro/entry/pfam/#table, accessed on 18 March 2023). Then, HMMER3.0 (http://hmmer.janelia.org/, accessed on 19 March 2023) was used to search alfalfa protein sequences that matched the JAZ HMM profiles, and sequences with an E-value less than 1.0 × 10−5 were selected. These candidates, MsJAZ, were submitted to NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd, accessed on 21 March 2023) to confirm further. The predicted isoelectric (pI) and molecular weights (MWs) of all MsJAZ genes were predicted with the ExPASy website (https://web.expasy.org/compute_pi/, accessed on 29 March 2023). Plant-mPLo (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 30 March 2023) was used to predict subcellular localization.
4.3. Chromosome Location Analysis and Phylogenetic Tree Construction
The genomic position of the identified MsJAZs was confirmed and mapped using the TBtools software (v1.108, Chen, C., GZ, China) [49]. JAZ family protein sequences in A. thaliana were downloaded from the TAIR website (https://www.arabidopsis.org/, accessed on 3 April 2023). The JAZ genes of M. truncatula were screened by BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 2 April 2023) and submitted to NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd, accessed on 2 April 2023). The JAZ protein sequences of M. sativa, A. thaliana, and M. truncatula were aligned using ClustalW with default parameters. A phylogenetic tree was then constructed using MEGA6.0 (Tamura, K., Tokyo, Japan), and the maximum likelihood method (bootstrap method with 1000 replications was selected) [50]. The iTOL website was used to beautify phylogenetic trees (https://itol.embl.de/, accessed on 6 April 2023) [51].
4.4. Gene Structure and Conserved Domains Analysis
The MEME website (https://meme-suite.org/meme/doc/meme.html, accessed on 8 April 2023) was used to analyze conserved motifs in MsJAZ proteins. The parameters in the program are selected: the maximum number of motifs was 10; the minimum was 6; the maximum motif width was 200 [52]. The gene structural information is included in the alfalfa gff file. Visualization using TBtools (v1.108, Chen, C., GZ, China) [35].
4.5. Cis-Acting Elements Analysis of MsJAZ Promoter
The upstream 2 kb sequence of the MsJAZs was extracted. Then, extracted sequences were submitted to the PlantCARE website (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 13 April 2023) for cis-acting element analysis [53].
4.6. Gene Duplication and Synteny Analysis
Genomic data for A. thaliana, M. truncatula, and G. max were downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov/, accessed on 25 April 2023). One-step MCScanX from TBtools (v1.108, Chen, C., GZ, China) with default parameters was used to analyze the pattern of gene duplication of the JAZ genes in Medicago sativa. Ka/Ks values for gene pairs were calculated using a simple Ka/Ks calculator in TBTools (v1.108, Chen, C., GZ, China). The Dual Synteny Plot in TBtools (v1.108, Chen, C., GZ, China) was used to determine the syntenic relationship between orthologous MsJAZ genes of alfalfa and other species.
4.7. Transcriptome Data Analysis
Transcriptomic data for six alfalfa tissues (leaves, nodules, elongating stems, flowers, post-elongation stems, and roots) were retrieved from the NCBI database (SRP055547) [54], and a correlation heatmap was analyzed using TBtools (v1.108, Chen, C., GZ, China) (Table S5).
4.8. Cloning and Expression Vector Construction of MsJAZ1
Total RNA from alfalfa leaves was extracted according to the instructions of the plant total RNA extraction kit (Promega, Madison, WI, USA), and cDNA was obtained by reverse transcription. The upstream primer MsJAZ1-F and downstream primer MsJAZ1-R were designed, and PCR amplification was performed using a cDNA template. The reaction was performed at 95 °C for 3 min. 95 °C, 30 s; 53 °C, 30 s; 72 °C, 1 min; 30 cycles; 72 °C for 5 min. A heterogenic expression vector was constructed, and the upstream primer MsJAZ1-eYFP-F and downstream primer MsJAZ1-eYFP-R were designed for amplification (Table S4). The amplified product was recovered and cloned seamlessly into the expression vector to obtain a recombinant plasmid. The correctly sequenced plasmid was transferred into Agrobacterium tumefaciens GV3101 (Huayueyang Biotechnology, Beijing, China) and transformed into A. thaliana (Cultivar “Col-0”) [55]. Transgenic plants were screened on 1/2 MS medium containing 4 mg/mL phosphinothricin (PPT) (Coolaber, Beijing, China) for PCR detection. Transgenic A. thaliana was planted by a single plant until homozygous plants were obtained.
4.9. Detection of Salt Tolerance in Transgenic Arabidopsis
Seeds of homozygous lines of transgenic and wild-type plants were disinfected and seeded on 1/2 MS and 150 mM NaCl 1/2 MS, respectively. The germination rate was counted every day from 0 to 7. The phenotype was observed, and the survival rate was calculated on the 20th day. The seedlings growing normally for 7 days were cultured in 1/2 MS and 150 mM NaCl 1/2 MS for 3 days, respectively. Each medium contained four Arabidopsis seedlings. Each experiment was performed in triplicates.
4.10. Real-Time Quantitative PCR (RT-qPCR) Analysis
The reagents, instruments, and methods used in RT-qPCR were consistent with the previous methods [56]. RT-qPCR-specific primers for 11 MsJAZ genes were designed using the Primer5.0 software (Premierbiosoft, San Francisco, USA). Primer sequences are shown in Table S3. The data from three biological replicates and three technical replicates were analyzed statistically. RT-qPCR was used to analyze the relative gene expression levels using the 2−∆∆Ct method.
4.11. Statistical Analysis
SPSS 25 software (IBM Inc., Chicago, IL, USA) was used for statistical analysis. The Duncan multiple range test was used to analyze differences between groups (ANOVA with p < 0.05). GraphPad Prism 8.0.2 software (GraphPad Software, Boston, FL, USA) was used to plot the data. Adobe Illustrator CC 2020 (ADOBE, San Jose, CA, USA) was used for graphic editing.
5. Conclusions
In this study, the JAZ gene family members of M. sativa were analyzed using bioinformatics tools, and the function of MsJAZ1 under salt stress was explored. A total of 11 MsJAZ genes were identified, which were distributed on five chromosomes. The phylogenetic tree divided MsJAZs into five groups. The members of each branch contained similar gene structures and motifs, indicating that members of the same branch may contain similar functions. Segmental replication was the main driving force for evolution in the MsJAZs gene family. During evolution, the MsJAZ gene family has experienced purification selection. The expression pattern of the MsJAZ gene in six different tissues of alfalfa showed that 45% of MsJAZ genes were highly expressed in leaves, and some genes were highly expressed in roots and elongating stems. RT-qPCR results showed that all MsJAZ genes responded to salt stress. Under salt stress, the germination rate and survival rate of overexpressing MsJAZ1 Arabidopsis were significantly decreased, affecting seedling growth, indicating that MsJAZ1 negatively regulates plant salt tolerance. In summary, the results of this study lay a foundation for further exploration of the function of MsJAZs.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms251910589/s1.
Author Contributions
Conceptualization, J.C. and X.J.; data curation, X.W., M.L. and F.H.; funding acquisition, J.K.; validation, Y.L., L.Z. and Y.Z.; visualization, J.C.; writing—original draft, J.C.; writing—review and editing, J.C., T.Z. and J.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32371770) and the National Key Research and Development Program of China (2022YFF1003203).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176. [Google Scholar] [CrossRef]
- Salvi, P.; Manna, M.; Kaur, H.; Thakur, T.; Gandass, N.; Bhatt, D.; Muthamilarasan, M. Phytohormone signaling and crosstalk in regulating drought stress response in plants. Plant Cell Rep. 2021, 40, 1305–1329. [Google Scholar] [CrossRef]
- Afrin, S.; Huang, J.J.; Luo, Z.Y. JA-mediated transcriptional regulation of secondary metabolism in medicinal plants. Sci. Bull. 2015, 60, 1062–1072. [Google Scholar] [CrossRef]
- Pauwels, L.; Inzé, D.; Goossens, A. Jasmonate-inducible gene: What does it mean? Trends Plant Sci. 2009, 14, 87–91. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate Passes Muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef]
- Zhou, X.E.; Zhang, Y.; Yao, J.; Zheng, J.; Zhou, Y.; He, Q.; Moreno, J.; Lam, V.Q.; Cao, X.; Sugimoto, K.; et al. Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling. Plant Commun. 2023, 4, 100639. [Google Scholar] [CrossRef]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef]
- Wager, A.; Browse, J. Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front. Plant Sci. 2012, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Major, I.T.; Yoshida, Y.; Campos, M.; Kapali, G.; Xin, X.F.; Sugimoto, K.; Ferreira, D.O.; He, S.Y.; Howe, G.A. Regulation of growth–defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Li, Y.; Tang, G.; Hui, S.; Yang, Z.; Zhao, J.; Liu, H.; Cao, J.; Yuan, M. Dynamic phytohormone profiling of rice upon rice black-streaked dwarf virus invasion. J. Plant Physiol. 2018, 228, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Thireault, C.; Shyu, C.; Yoshida, Y.; Aubin, B.S.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef]
- Kaji, T.; Matsumoto, K.; Okumura, T.; Nakayama, M.; Hoshino, S.; Takaoka, Y.; Wang, J.; Ueda, M. Two distinct modes of action of molecular glues in the plant hormone co-receptor COI1-JAZ system. iScience 2023, 27, 108625. [Google Scholar] [CrossRef]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef]
- Han, X.; Kui, M.; He, K.; Yang, M.; Du, J.; Jiang, Y.; Hu, Y. Jasmonate-regulated root growth inhibition and root hair elongation. J. Exp. Bot. 2023, 74, 1176–1185. [Google Scholar] [CrossRef]
- Varshney, V.; Majee, M. JA shakes hands with ABA to delay seed germination. Trends Plant Sci. 2021, 8, 764–766. [Google Scholar] [CrossRef]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Johnson, L.Y.D.; Major, L.T.; Chen, Y.; Yang, C.; Vanegas-Cano, L.J.; Howe, G.A. Diversification of JAZ-MYC signaling function in immune metabolism. New Phytol. 2023, 239, 2277–2291. [Google Scholar] [CrossRef]
- Qi, T.; Huang, H.; Song, S.; Xie, D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Qanmber, G.; Li, F.; Wang, Z. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 2021, 35, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J.T.; Figueroa, C.R. Jasmonates and plant salt stress: Molecular players, physiological effects, and improving tolerance by using genome-associated tools. Int. J. Mol. Sci. 2021, 22, 3082. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Hu, Y.; Wang, H.; Guo, Q.; Chen, Y.; Howe, G.A.; Yu, D. Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2020, 32, 3846–3865. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.L.; Yao, J.; Mei, C.S.; Tong, X.H.; Zeng, L.J.; Li, Q.; Xiao, L.T.; Sun, T.; Li, J.; Deng, X.W.; et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar]
- Wu, H.; Ye, H.; Yao, R.; Zhang, T.; Xiong, L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar] [CrossRef]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef]
- Rao, S.; Tian, Y.; Zhang, C.; Qin, Y.; Liu, M.; Niu, S.; Li, Y.; Chen, Y. The JASMONATE ZIM-domain–OPEN STOMATA1 cascade integrates jasmonic acid and abscisic acid signaling to regulate drought tolerance by mediating stomatal closure in poplar. J. Exp. Bot. 2023, 74, 443–457. [Google Scholar] [CrossRef]
- Chen, L.; He, F.; Long, R.; Zhang, F.; Li, M.; Wang, Z.; Kang, J.; Yang, Q. A global alfalfa diversity panel reveals genomic selection signatures in Chinese varieties and genomic associations with root development. J. Integr. Plant Bio. 2021, 63, 1937–1951. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, F.; Zhao, G.; Li, M.; Long, R.; Kang, L.; Yang, Q.; Chen, L. Genome-wide identification and phylogenetic and expression analyses of the PLATZ gene family in Medicago sativa L. Int. J. Mol. Sci. 2023, 24, 2388. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The tify family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zheng, H.; Wu, H.; Wang, C.; Liang, Z. Recent genome-wide replication promoted expansion and functional differentiation of the JAZs in soybeans. Int. J. Bio. Macromol. 2023, 238, 124064. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transciption. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Luthe, D. Identification and evolution analysis of the JAZ gene family in maize. BMC Genom. 2021, 22, 256. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Liu, Z.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Genome-wide identification, characterization and expression analysis of the JAZ gene family in resistance to gray leaf spots in tomato. Int. J. Mol. Sci. 2021, 22, 9974. [Google Scholar] [CrossRef]
- Shen, J.; Zou, Z.; Xing, H.; Duan, Y.; Zhu, Y.; Ma, Y.; Wang, Y.; Fang, W. Genome-wide analysis reveals stress and hormone responsive patterns of JAZ family genes in Camellia Sinensis. Int. J. Mol. Sci. 2020, 21, 2433. [Google Scholar] [CrossRef]
- Song, H.; Duan, Z.; Wang, Z.; Li, Y.; Wang, Y.; Li, C.; Mao, W.; Que, Q.; Chen, X.; Li, P. Genome-wide identification, expression pattern and subcellular localization analysis of the JAZ gene family in Toona ciliata. Ind. Crop Prod. 2022, 178, 114582. [Google Scholar] [CrossRef]
- Zheng, L.; Wan, Q.; Wang, H.; Guo, C.; Niu, X.; Zhang, X.; Zhang, R.; Chen, Y.; Luo, K. Genome-wide identification and expression of TIFY family in cassava (Manihot esculenta Crantz). Front. Plant Sci. 2022, 13, 1017840. [Google Scholar] [CrossRef]
- Shen, C.; Du, H.; Chen, Z.; Lu, H.; Zhu, F.; Chen, H.; Meng, X.; Liu, Q.; Liu, P.; Zheng, L.; et al. The chromosome-level genome sequence of the autotetraploid alfalfa and resequencing of core germplasms provide genomic resources for alfalfa research. Mol. Plant 2020, 13, 1250–1261. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Meng, Y.; Huang, D.; Qi, Y.; Chen, M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Bleecker, A.B. Expansion of the receptor-like kinase/pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Bansal, M.S.; Eulenstein, O. The multiple gene duplication problem revisited. Bioinformatics 2008, 24, i132–i138. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, Y.; Tao, J.J.; Lu, L.; Jiang, Z.H.; Wei, J.J.; Wu, C.M.; Yin, C.C.; Li, W.; Bi, Y.D.; et al. GmJAZ3 interacts with GmRR18a and GmMYC2a to regulate seed traits in soybean. J. Integr. Plant Bio. 2023, 65, 1983–2000. [Google Scholar] [CrossRef]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Bio. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Sun, H.; Chen, L.; Li, J.; Hu, M.; Ulah, A.; He, X.; Yang, X.; Zhang, X. The JASMONATE ZIM-Domain Gene Family Mediates JA Signaling and Stress Response in Cotton. Plant Cell Physiol. 2017, 12, 2139–2154. [Google Scholar] [CrossRef]
- Toda, Y.; Tanaka, M.; Ogawa, D.; Kurata, K.; Kurotani, K.I.; Habu, Y.; Ando, T.; Sugimoto, K.; Mitsuda, N.; Katoh, E.; et al. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 2013, 5, 1709–1725. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Wiliams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.; Rouzé, P.; Rombatus, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.A.; Fu, F.; Bucciarelli, B.; Yang, S.S.; Samac, D.A.; Lamb, J.F.S.; Monteros, M.J.; Graham, M.A.; Gronwald, J.W.; Krom, N.; et al. The Medicago sativ gene index 1.2: A web-accessible gene expression atlas for investigating expression differences between Medicago sativa subspecies. BMC Genomics 2015, 16, 502. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Cui, J.; Wang, X.; Li, M.; Zhang, L.; Kang, J. Genome-wide identification, phylogenetic and expression analysis of expansin gene family in Medicago sativa L. Int. J. Mol. Sci. 2024, 25, 4700. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).