Abstract
Candida orthopsilosis represents a closely related cryptic genospecies of Candida parapsilosis complex-misidentified in routine diagnostic assays. This is emerging in settings where central venous catheters, invasive medical interventions, and echinocandin treatments are most likely to be used. A 59-year-old, non-neutropenic male patient, was admitted to an intensive care unit (ICU) due to respiratory distress syndrome, following a partial gastrectomy. As a result of duodenal stump leakage, re-laparotomy was required, abdominal drains were provided and central line catheters were exchanged. Multiple isolates of Candida orthopsilosis drawn from consecutive blood cultures were identified, despite ongoing echinocandin therapy and confirmed in vitro echinocandins susceptibility of the isolated strain. Species identification was verified via ITS region sequencing. Herein, we report the well-documented—per clinical data and relevant laboratory diagnosis—first case of a bloodstream infection caused by Candida orthopsilosis in Poland.
1. Introduction
ICU-associated CBSIs (Candida bloodstream infections) in adults, characterized by a high mortality rate (42–85%) [1,2,3], are commonly caused by the Candida (C.) non-albicans group [4], in which Candida (C.) parapsilosis appears to be increasing and is probably due to overuse of echinocandins [5,6,7,8,9]. This species has been reported as the second or third most prevalent agent of ICU-acquired CBSI in South East Asia, Latin America (Brasil and Chile), refs. [10,11,12] and Southern and Central Europe (Italy, Spain, and Poland) [3,13,14,15]. Although published papers on ICU-associated CBSIs in Poland are scarce, C. parapsilosis has been ranked as the second cause of CBSI [15]. In fact, the C. parapsilosis complex species is a heterogenous group composed of the following closely related cryptic genospecies: C. parapsilosis sensu stricto, Candida (C.) metapsilosis, and Candida (C.) orthopsilosis [16]. C. orthopsilosis first emerged in 2005 as a potential human cryptic pathogen and remains a relatively uncommon aethiological agent of CBSIs with an unknown 30-day mortality rate [17,18,19,20,21]. C. orthopsilosis is found in patients with prolonged use of central venous catheters, previous surgical history, and invasive abdominal interventions with total parenteral nutrition [17,19]. The prevalence and transmission of C. orthopsilosis-caused BSI in ICUs varies geographically and ranges from 1.0 to 28.3% [17,18,20,22].
Epidemiological data are underestimated because broadly implemented biochemical identification systems (VITEK-2 Compact and BD Phoenix) frequently misidentify C. orthopsilosis. The use of diagnostic assays is not highly efficient in cryptic species detection, which results in both delayed diagnosis and delayed antifungal therapy of CBSI. Consequently, this is challenging patient’s survival. Nowadays, mass spectrometry, fungal ITS DNA–barcoding, or automated molecular assays, are required for the effective detection of uncommon C. orthopsilosis genospecies [23,24]. More data with a combined clinical and molecular approach are needed to explain the uncertainty regarding practical implications of C. orthopsilosis identification. On this basis, we herein describe the well-documented case of bloodstream infection caused by C. orthopsilosis in the ICU which originates from our single-centre prospective surveillance of invasive fungal infections in the ICU. To our knowledge, this is the first CBSI case from Poland caused by C. orthopsilosis, as described in this paper.
2. Case Presentation
A non-neutropenic, 59-year-old male patient, was admitted to an ICU at University Hospital in Szczecin, Poland due to shortness of breath and respiratory distress that developed following a partial gastrectomy. The gastrectomy was a result of previous gastric ulcer perforation.
On ICU admission, the patient’s height and weight were 180 cm and 90 kg (body mass index (BMI): 27.8), respectively. His respirations were unstable and mechanical ventilation with oxygen of 40% was administered. Blood pressure of 110/70 mmHg, heart rate of 95 beats/min with regular rhythm, and body temperature of 38.2 °C were observed. Laboratory parameters on ICU admission were the following: elevated white blood cell count (WBC) of 29,000 cells/mm3 (reference range, 4000 to 10,000 cells/mm3), and elevated C-reactive protein (CRP) level of 232 mg/l (reference range, <5 mg/dL), in combination with low procalcitonin level (PRC)—0.6 ng/mL (reference range 0.5–2 ng/mL). Due to pneumonia, diagnosed by chest X-ray, the intravenous empiric therapy with piperacillin/tazobactam (each dose of 4.5 g at a 6 h interval) was applied and maintained for 9 days. Mortality risk ratios assessed with SOFA (Sepsis-related Organ Failure Assessment) and APACHE II (Acute Physiology and Chronic Health Evaluation) scales were 15–20% (8 points) and 29% (18 points), respectively. Fungal colonization had been screened by culturing samples collected from commonly colonized body sites to predict the Candida colonization index (CI). The anal, urine, and bronchoalveolar samples were negative for Candida, whereas swabs from underarm skin, groin skin, and the nasal cavity, were positive for C. parapsilosis sensu stricto. Confirmed multifocal colonization and clinical parameters, such as parenteral nutrition and post-abdominal surgery, resulted in a Candida Score (CS) index ≥ 3. Following CS index and ESCMID (European Society of Clinical Microbiology and Infectious Diseases) guidelines [25], fluconazole prophylaxis was introduced (initial day—400 mg divided into two doses of 200 mg at an interval of 12 h; following days—200 mg each at an interval of 24 h) and continued for 7 days.
On the 8th day, fluconazole was replaced with micafungin (each dose equal to 100 mg/day) as a result of deterioration of the patient’s general condition, acute abdominal symptoms, and leakage of intestinal contents through abdominal drains. Due to leakage from the duodenal stump, urgent re-laparotomy was required. After total gastrectomy with esophago-intestinal anastomosis, the wound was stitched, and abdominal drains were provided.
On the 11th day, empiric broad spectral antibacterial therapy with imipenem and vancomycin was provided due to increasing inflammatory parameters (CRP—436 mg/L, PRC—0.7 ng/mL). Multiple blood cultures remained negative and micafungin was continued.
On the 11th and 13th day, two re-relaparotomies accompanied by rinsing of the peritoneal cavity—while leaving the abdominal cavity open—were performed. The patient received total parenteral nutrition as a result of ineffective enteral nutrition previously provided by the Flocar probe, inserted behind Treitz ligament; a leakage of the administered glucose through the open abdominal wall was observed. No microorganisms were cultivated from the blood samples at this stage while laboratory indicators of infection were persistently high (WBC—29,000 cells/mm3, CRP—513 mg/L, PRC—1.03 ng/mL).
On the 13th day, Candida–bloodstream infection (CBSI) was established through multiple isolations of C. orthopsilosis from blood cultures with confirmed susceptibility to all echinocandins [26]. Despite micafungin administration, over the following consecutive days of the patient’s ICU stay, C. orthopsilosis was isolated from peripheral blood and from blood samples collected via the central line catheter. The suspected catheter was removed and submitted to MAKI culture technique to determine the risk of CLABSI (central line associated BSI). Following MAKI testing, C. orthopsilosis was detected and all possible catheters, as well as abdominal drains, were exchanged. According to the microbiological report, micafungin had been continued [25,27,28]. Unfortunately, no further surgical treatment was available, although gastrointestinal leakage was observed. On the 40th day of the ICU stay, the patient died without resolution of CBSI.
2.1. Early Monitoring of the Risk of CBSI by Fungal Cell Wall Biomarkers
Testing of mannan and 1,3 β-D-glucan for early CBSI detection revealed conflicting data. Weekly tested serum mannan antigen was negative with concentrations below the cut-off point for positive samples of <62.5 pg/mL; whereas 1,3 β-D-glucan values were high above the cut-off threshold of 80 pg/mL, ranging from 212.3 to 606.0 pg/mL. Additionally, 1,3 β-D-glucan was detected one day before CBSI diagnosis was established via positive blood culture. Decreasing 1,3 β-D-glucan concentrations were noted after micafungin administration (Table 1).

Table 1.
Clinical and diagnosis data of 59-year-old male patient with CBSI caused by C. orthopsilosis.
2.2. Mass Spectrometry Biotyping of C. orthopsilosis Isolates
Representative protein mass spectra (MSPs) for all particular isolates are presented in Figure 1. Both C. orthopsilosis and C. parapsilosis isolates have been grouped based on their protein MSPs into two clusters (Figure 2). C. orthopsilosis has been misidentified as C. parapilosis or C. famata by biochemical platform VITEK-2 Compact.
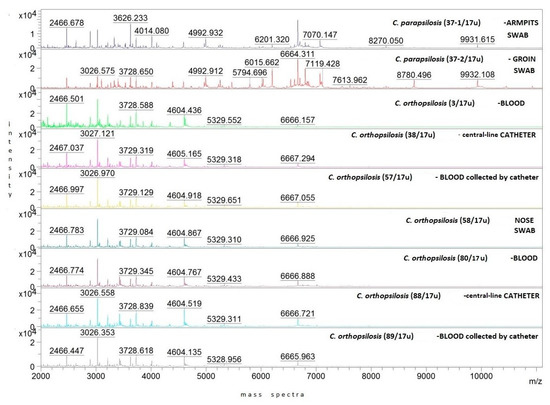
Figure 1.
Representative MSPs for clinical C. parapsilosis and C. orthopsilosis strains isolated from different body locations of 59-year-old male patient with CBSI. Mass spectra intensity displayed in arbitrary units (a.u. = 0, 1, 2, 3, 4 × 104) on the y-axis, whereas mass-to-charge ratio expressed in m/z units on the x-axis; color was assigned to number of strain.
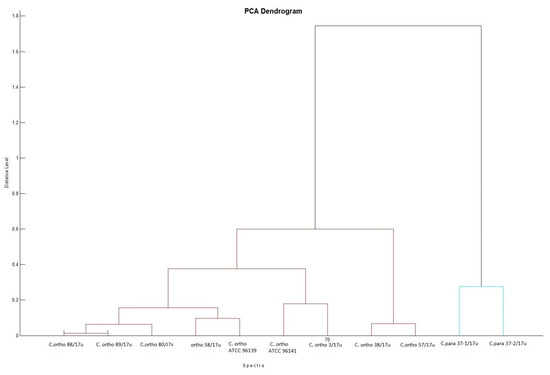
Figure 2.
Biotyper PCA (principal component analysis) dendrogram clustering representative MSPs for isolates of C. orthopsilosis and C. parapsilosis with distances displayed in relative units on the y-axis; MSPs for isolates of C. orthopsilosis are clustered with MSPs for reference C. orthopsilosis ATCC 96139 and ATCC 96141. Protein similarity presented on dendrogram by means of a relative distance level between isolates included into dendrogram, normalized to a maximum value of 1000 (cut-off). All tested isolates are presented by their abbreviated species name (C. ortho and C. para) and the isolation number.
2.3. Antifungal Susceptibility of C. orthopsilosis Isolates
The minimum concentration of antifungals required to inhibit C. parapsilosis and C. orthopsilosis growth was determined by E-test gradient strips and the automated VITEK-2 system. Skin-colonizing isolates of C. parapsilosis were susceptible to amphotericin B (MIC = 0.03 µg/mL), fluconazole (MIC = 0.38 µg/mL), and echinocandins (MIC range 0.25 to 0.50 µg/mL). The cryptic isolates of C. orthopsilosis were also susceptible to amphotericin B (MIC range 0.03 to 0.25 µg/mL), fluconazole (MIC range 0.19 to 0.50 µg/mL), and all echinocandins—caspofungin (max MIC = 0.50 µg/mL), micafungin (max MIC = 1.00 µg/mL), and anidulafungin (max MIC = 0.50 µg/mL). EUCAST MIC breakpoints and epidemiologic cut-off (ECOFF) values for C. orthopsilosis are still not established in the updated EUCAST guidelines [20], therefore MIC breakpoints for C. parapsilosis were applied to determine antifungal resistance for C. orthopsilosis. Multiple isolates of C. orthopsilosis, from blood and the nasal cavity, were similar in MIC values against echinocandins when compared with reference C. orthopsilosis strain ATCC 96141. C. orthopsilosis strains isolated from catheter tips and from blood collected by catheters had elevated MICs (MIC = 1.0 µg/mL and MIC = 0.75 µg/mL respectively) against micafungin but were still susceptible to this drug, according to EUCAST recommendations (Table 2).

Table 2.
Susceptibility of C. orthopsilosis and C. parapsilosis to antifungal drugs detected by E-test.
2.4. ITS Sequencing of C. orthopsilosis Isolates
The dendrogram based on the shared ITS sequence similarity revealed three clusters for C. orthopsilosis, C. parapsilosis sensu stricto, and C. metapsilosis (ATCC 96143), when included as a mixture of isolates for three genospecies. Our isolates with a high ITS homology were clustered into two well-differentiated groups, assigned to C. orthopsilosis and C. parapsilosis sensu stricto by neighbour-joining analysis (Figure 3). The ITS region sequences for C. orthopsilosis were 96% similar to C. parapsilosis sequences and >99% similar to C. orthopsilosis sequences of blood isolates found in NCBI GenBank. Our blood and catheter isolates from the hospital in Poland were closely related (99.80–100% ITS match) to blood (ATCC 96141) and catheter (ATCC 96139T) strains isolated in San Antonio, TX, USA. A 100% similarity in ITS region between C. orthopsilosis isolated from blood and central line collected blood from our patient can suggest central line associated origin of CBSI. C. orthopsilosis blood isolates derived from different timepoint collections were also highly similar when tested via ITS sequencing. As revealed by ITS sequencing, isolates from skin were assigned to C. parapsilosis sensu stricto and were not detected in other body sites in addition to the patient’s blood samples during his hospital stay. ITS sequences, determined in the study for complex genospecies, were deposited in GenBank with the accession numbers shown in Table 3 (column 3 from left side).
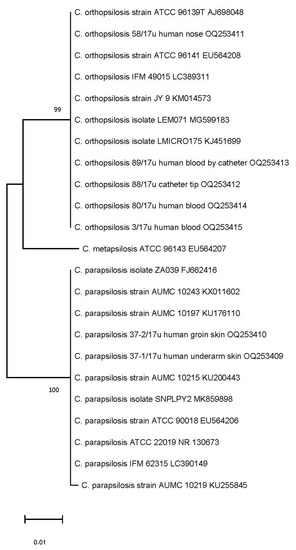
Figure 3.
Phylogenetic neighbour-joining tree reflecting a genetic similarity of ITS sequences. Numbers at each branch indicate similarity percentage, expressed in bootstrap values of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates). The scale bar indicates 0.01 changes/site (number of differences).

Table 3.
Comparative analysis of ITS similarity between isolates derived from patient and GenBank, based on the neighbour-joining method.
2.5. Materials and Methods
2.5.1. Blood Culturing and Species Identification
Multiple blood samples were collected during every episode of fever for culturing in automated microbiological system BacT/ALERT 3D (bioMérieux, Warsaw, Poland). Species identification was performed on the day of indicated yeast growth by means of automated multiplex PCR (Film Array BCID Panel, bioMérieux, Warsaw, Poland). C. orthopsilosis was off—assay panel and finally was not identified within this system. All Candida strains isolated from the patient were identified by VITEK-2 system (bioMérieux, Warsaw, Poland) as C. parapsilosis or C. famata. MALDI-Biotyper (Bruker Daltonics, Bremen, Germany) re-classified the samples from blood, central line catheter, and nasal as C. orthopsilosis with high confidence score value > 2. Each isolate tested six times to submit the high-confidence mass signatures to mass spectrometry strain biotyping. Unique cell-wall components of yeast: mannan (Bio-Rad, Warsaw, Poland) and 1,3 β-D-glucan (Fungitell, Associates of Cape Cod, East Falmouth, MA, USA) were assessed simultaneously directly in serum samples.
2.5.2. Genetic Analysis
ITS region sequencing was used to verify the outcomes provided by MALDI–Biotyper. Preceding genomic DNA extraction was made using a commercial DNA extraction kit (EURx, Gdańsk, Poland) with several of our own modifications. Preliminarily, the yeasts were incubated for 20 h on Sabouraud dextrose agar at 30 °C. Subsequently, these were then washed, frozen in liquid nitrogen, and crushed via a grinding tool (EURx, Gdansk, Poland). Implementing the same method used for fungal genomic DNA extraction, we directly extracted from Candida-positive blood samples. The DNA amplification was performed with universal primers ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) (Genomed, Warsaw, Poland), and revealed the PCR product corresponding to the ITS1–5.8S–ITS2 rDNA region (shortly ITS). The PCR was carried out with the same conditions as previously described [21,29]; amplicons were then purified and sequenced by Sanger DNA sequencing in both the forward and reverse directions (Genomed, Warsaw, Poland) for molecular identification of Candida species.
2.5.3. Bioinformatic Analysis
ITS nucleotide sequences of particular isolates that were revealed in the study were uploaded to BLAST analysis (http://blast.ncbi.nlm.nih.gov (accessed on 27 February 2023). They were compared with ITS sequences, available in NCBI GenBank, for the reference species of C. orthopsilosis (ATCC 96141 and ATCC 96139T), C. parapsilosis (ATCC 90018 and ATCC 22019), as well as for blood and catheter isolates derived from CBSI-derived samples. Then, those have been submitted to multiple ITS sequence alignments with ClustalW algorithm in Mega11 online software. The aligned sequences (Figure S1 were processed, and the phylogenetic tree as a dendrogram based on the neighbour-joining method was generated, reflecting the percentage of ITS sequence similarity with 1000 bootstrap replicates.
3. Discussion
This report is focused on a clinical and diagnosis approach to discuss the fatal case of C. orthopsilosis BSI. Nowadays, the C. orthopsilosis and cryptic non-albicans species are changing the epidemiology of CBSI worldwide and the overall incidence of CBSI, due to the cryptic species, seems to be increasing [17,22,23]. Although, there are still limited, underrated data on C. orthopsilosis as a causative agent for CBSI in the ICU [8,17]. Clinical studies focused on CBSI in ICU are mostly retrospective and do not discriminate between closely related species [1,3,4]. Simultaneously, laboratory studies mostly skip the clinical implications of the data presented [22]. Improved species identification seems to be crucial to monitor local epidemiology, implement infection control measures, and manage appropriate antifungal therapy [4,30].
In our study, C. orthopsilosis has been misidentified as C. parapsilosis or C. famata by biochemical identification, whereas a mass spectrometry met the daily needs of clinicians as a reliable, cost- and time-effective platform for detection of C. orthopsilosis via blood cultures from the ICU patient [30,31]. C. orthopsilosis and C. parapsilosis shared the epidemiological relationship at ICU; therefore, intraspecies identification with sequencing and matching of ITS region was essential [22,23,32]. The high ITS homology between C. orthopsilosis isolated from blood, central line collected blood, and central line catheter, after its removal at the same timepoint, was indicated. Since C. orthopsilosis has been recovered from intravascular catheters we concluded that the catheter was the most probable site through which C. orthopsilosis reached the bloodstream [22,33]. Previous findings [14] reported that the source of C. parapsilosis CBSI was a vascular catheter in more than 50% of cases.
C. orthopsilosis has not been found as the patient’s commensal fungus prior to the BSI episode; therefore, a per catheter skin colonization has not been strictly associated with the etiology of catheter related CBSI. This was in accordance with other reports [34] in which the blood isolate belonged to a species other than that of the colonizing site in 13% of the cases. The C. orthopsilosis strain, identical to the ones of blood origin, was also found in a nasal swab after CBSI diagnosis, thus a colonization of the patient’s nasal cavity, and a potential strain translocation (by nasogastric tube), cannot be excluded. In-depth analysis of the C. orthopsilosis origin on the patient’s catheters can suggest that this cryptic species, similarly to C. parapsilosis sensu stricto, is associated with exogenous transmission during catheter care in the respective ICU environment [5,14,23,35].
All C. orthopsilosis grouped by ITS neighbour-joining in the same clade with strains isolated in San Antonio, TX, USA (ATCC 96141 and ATCC 96139) could suggest that our isolates represent alternative type 1 subspecies of C. orthopsilosis [21,36]. Conversely, as reported by genomic analysis [37], strain ATCC 96141 (MCO456), although closely related to our isolates, when tested by ITS, was a hybrid between two subspecies represented by type 1 and type 2. The relationship between our C. orthopsilosis and these clinical isolates from other geographical locations could be explained by advanced metagenomic analysis [37,38,39]. Serological investigation of fungal antigens has been implemented in our study as well and the fungal cell wall components 1,3 β-D-glucan and mannan have been determined. Mannan antigenemia is commonly determined, however 1,3 β-D-glucan assay is now also available in Poland. Simultaneous labelling and monitoring of different biomarkers is considered to increase the sensitivity of testing. In this study, 1,3 β-D-glucan has been detected as a predictor of CBSI while mannan assay showed to be of lower usefulness during the course of C. orthopsilosis BSI. Increased 1,3 β-D-glucan content and significantly less mannan were observed for C. parapsilosis and C. orthopsilosis [40]. This could explain why mannan antigenemia was not detected, despite positive blood cultures. A previous study [41] has shown that the sensitivity of mannan detection in C. parapsilosis BSI is low, thus our report corresponds with this finding. C. orthopsilosis is considered to be naturally less susceptible to echinocandins, when compared to other non-albicans species [8,42]. A decreased susceptibility is especially presented for micafungin as a result of an intrinsic polymorphism in the Fks1 gene [42]. Thus, the emerging C. orthopsilosis, shown in this study, might have expanded by selection resulting from micafungin usage in extended empirical therapy of CBSI. Increased echinocandin usage and its correlation with increased prevalence of BSI caused by C. parapsilosis has already been reported [6]. Therefore, we hypothesize that pressure of micafungin–mediated therapy could lead to a selection toward C. orthopsilosis and a generation of isolates with elevated values of MICs as well [8,43]. As observed in this study, C. orthopsilosis with the higher MICs of micafungin (0.75–1.00 µg/µL) have been isolated from the central line catheter and from the blood collected by the catheter. In our opinion, similarly to other authors [8,43], micafungin–adapted isolates with elevated MIC values, especially on the surface of intravascular catheters, need to be recorded and it also points to the need for increased care of those catheters in ICUs. Kartsonis et al., [44] proved that C. parapsilosis- infected patients respond well to standard caspofungin therapy when MIC values for C. parapsilosis are 0.50 µg/µL. Another trial [45] revealed that a standard micafungin dosing regimen (100 mg daily) was more effective in therapy of C. parapsilosis—mediated candidemia (75.9%) when compared with caspofungin (64.3%). These data support an assumption of potential activity of echinocandins against C. parapsilosis. Nevertheless, the standard doses of micafungin were sufficient to overcome the higher MICs of C. parapsilosis [45], but it was not sufficient against C. orthopsilosis. This cryptic species appeared in the patient’s samples and was persistently isolated from blood. Data regarding the activity of echinocandins against C. orthopsilosis are still limited and contradictory. So far, the results of antifungal susceptibility testing to echinocandins, if available, have not been commonly implemented into clinical practice, due to the lack of a clear relationship between the in vitro determined MIC value and the clinical outcome [44,46]. The authors of this study postulate that as long as C. orthopsilosis and its antifungal susceptibility data are not included in the epidemiology of CBSI, it will not be possible to obtain comprehensive data to optimize the proper management of CBSI. This may also apply to other crypto species.
Moreover, some ICU patients may not benefit from micafungin therapy due to multiple concomitant risk factors which could impede the success of CBSI treatment [47,48]. High APACHE II scores predicted increased mortality, whereas other risk factors such as the necessity to maintain both central venous catheters and post-surgery abdominal drains, corresponded with factors that have been of great importance in the persistence of infection [49]. Central line catheters infected with C. orthopsilosis could be a potential niche for biofilm formation occurring in higher MIC of C. orthopsilosis to echinocandin. Echinocandins have been proposed as effective against biofilm as they inhibit polysaccharide production and consequently reduce extracellular matrix [50], whereas other studies have shown antifungal therapy as rarely successful without catheter and drain removals [25,47]. In our study, all catheters had been changed but there was still a need to maintain the access for parenteral nutrition, in addition to catecholamine administration. Consequently, micafungin therapy appeared to be unsuccessful.
In summary, C. orthopsilosis was isolated and multiplied in a primarily pneumonic ICU patient, with repeated abdominal surgery, and deteriorating condition. The isolates were obtained via peripheral blood as well as catheter obtained samples. Proper pathogen identification required in-depth analysis as current diagnostic assays remain incapable in distinguishing between most recently reported Candida genospecies. In contrast to mannan antigenemia, 1,3 β-D-glucan levels corresponded with microbiological diagnostic outcomes. Precise guidelines to assess C. orthopsilosis drug-susceptibility remain unavailable and the patient was treated with micafungin according to the most valid version of EUCAST recommendations dedicated to C. parapsilosis. Interestingly, despite genetic identity, MIC values for echinocandins differed in strains isolated from the bloodstream and catheter. This issue requires further molecular studies. Despite potential micafungin activity against C. orthopsilosis and observed decrease in 1,3 β-D-glucan concentration, clinical outcome of the treatment appeared poor. Discrepancies between in vitro data on C. orthopsilosis drug susceptibility and the clinical outcomes have already been reported and possibly results from other risk factors overlapping in an intensive care patient. The exact origin of the isolated C. orthopsilosis strain remains unknown.
4. Conclusions
The investigation of CBSI in the range of its etiology, therapy, and its origin, requires a multidisciplinary approach beyond routine assays and strategies. Precise diagnostics of C. orthopsilosis and other emerging Candida genospecies remains challenging. C. orthopsilosis should be considered in CBSI epidemiology. There is an urgent need for clear recommendations regarding C. orthopsilosis drug susceptibility breakpoints and further treatment. Genetic similarity of all C. orthopsilosis isolates, identified in our patient and highly related to type 1 of the genospecies, suggests catheter origin of the bloodstream infection. Long-term vascular catheter management must consider the risk of fungal transmission and adaptation. This should focus on the emerging genospecies detection, potential of strain selective pressure by antifungals–mediated therapy, and biofilm-producing activity of the genospecies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24076541/s1.
Author Contributions
Conceptualization, M.M.-P. and M.A.; methodology, M.M.-P.; validation, M.M.-P.; formal analysis, M.M.-P.; investigation, M.M.-P.; data curation, M.M.-P.; writing—original draft preparation, M.M.-P., M.A., K.J., I.W.-K., A.K. and B.W.; sequencing results analysis and visualization, M.M.-P.; A.K.; supervision, M.M.-P., M.A. and K.J.; funding acquisition, M.M.-P. and B.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Pomeranian Medical University in Szczecin (approval number: KB-0012/279/06/16; KB-0012/150/03/18).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
All relevant data and materials have been included in the manuscript. Anonymous medical data used and/or analyzed during this study are available upon reasonable request and with the consent of the hospital.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Martin-Loeches, I.; Antonelli, M.; Cuenca-Estrella, M.; Dimopoulos, G.; Einav, S.; De Waele, J.J.; Garnacho-Montero, J.; Kanj, S.S.; Machado, F.R.; Montravers, P.; et al. ESICM/ESCMID task force on practical management of invasive candidiasis in critically ill patients. Intensive Care Med. 2019, 45, 789–805. [Google Scholar] [CrossRef]
- Doi, A.M.; Pignatari, A.C.C.; Edmond, M.B.; Marra, A.R.; Camargo, L.F.; Siqueira, R.A.; da Mota, V.P.; Colombo, A.L. Epidemiology and microbiologic characterization of nosocomial candidemia from a Brazilian National Surveillance Program. PLoS ONE 2016, 11, e0146909. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Vena, A.; Trucchi, C.; Ansaldi, F.; Antonelli, M.; Adamkova, V.; Alicino, R.; Almyroudi, M.-P.; Atchade, E.; et al. Incidence and outcome of invasive candidiasis in intensive care units (icus) in Europe: Results of the EUCANDICU project. Crit. Care 2019, 23, 219. [Google Scholar] [CrossRef]
- Logan, C.; Martin-Loeches, I.; Bicanic, T. Invasive candidiasis in critical care: Challenges and future directions. Intensive Care Med. 2020, 46, 2001–2014. [Google Scholar] [CrossRef]
- Trofa, D.; Gácser, A.; Nosanchuk, J.D. Candida parapsilosis, an emerging fungal pathogen. Clin. Microbiol. Rev. 2008, 21, 606–625. [Google Scholar] [CrossRef]
- Forrest, G.N.; Weekes, E.; Johnson, J.K. Increasing incidence of Candida parapsilosis candidemia with caspofungin usage. J. Infect. 2008, 56, 126–129. [Google Scholar] [CrossRef]
- Arendrup, M.C. Epidemiology of invasive candidiasis. Curr. Opin. Crit. Care 2010, 16, 445–452. [Google Scholar] [CrossRef]
- Papp, C.; Kocsis, K.; Tóth, R.; Bodai, L.; Willis, J.R.; Ksiezopolska, E.; Lozoya-Pérez, N.E.; Vágvölgyi, C.; Mora Montes, H.; Gabaldón, T.; et al. Echinocandin-induced microevolution of Candida parapsilosis influences virulence and abiotic stress tolerance. mSphere 2018, 3, e00547-18. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty years of the SENTRY Antifungal Surveillance Program: Results for Candida species from 1997–2016. Open Forum Infect. Dis. 2019, 6, S79–S94. [Google Scholar] [CrossRef]
- Santolaya, M.E.; Thompson, L.; Benadof, D.; Tapia, C.; Legarraga, P.; Cortés, C.; Rabello, M.; Valenzuela, R.; Rojas, P.; Rabagliati, R.; et al. A prospective, multi-center study of Candida bloodstream infections in Chile. PLoS ONE 2019, 14, e0212924. [Google Scholar] [CrossRef]
- Trabasso, P.; Matsuzawa, T.; Fagnani, R.; Muraosa, Y.; Tominaga, K.; Resende, M.R.; Kamei, K.; Mikami, Y.; Schreiber, A.Z.; Moretti, M.L. Isolation and drug susceptibility of Candida parapsilosis sensu lato and other species of C. parapsilosis complex from patients with bloodstream infections and proposal of a novel LAMP identification method for the species. Mycopathologia 2015, 179, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Nucci, M.; Queiroz-Telles, F.; Alvarado-Matute, T.; Tiraboschi, I.N.; Cortes, J.; Zurita, J.; Guzman-Blanco, M.; Santolaya, M.E.; Thompson, L.; Sifuentes-Osornio, J.; et al. Epidemiology of candidemia in Latin America: A laboratory-based survey. PLoS ONE 2013, 8, e59373. [Google Scholar] [CrossRef] [PubMed]
- Mazzanti, S.; Brescini, L.; Morroni, G.; Orsetti, E.; Pocognoli, A.; Donati, A.; Cerutti, E.; Munch, C.; Montalti, R.; Barchiesi, F. Candidemia in intensive care units over nine years at a large Italian university hospital: Comparison with other wards. PLoS ONE 2021, 16, e0252165. [Google Scholar] [CrossRef]
- Almirante, B.; Rodríguez, D.; Cuenca-Estrella, M.; Almela, M.; Sanchez, F.; Ayats, J.; Alonso-Tarres, C.; Rodriguez-Tudela, J.L.; Pahissa, A. Epidemiology, risk factors, and prognosis of Candida parapsilosis bloodstream infections: Case-control population-based surveillance study of patients in Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 2006, 44, 1681–1685. [Google Scholar] [CrossRef] [PubMed]
- Nawrot, U.; Pajaczkowska, M.; Fleischer, M.; Przondo-Mordarska, H.; Samet, A.; Piasecka-Pazik, D.; Komarnicka, J.; Sulik-Tyszka, B.; Swoboda-Kopeć, E.; Cieślik, J.; et al. Candidaemia in Polish hospitals—A multicentre survey. Mycoses 2013, 56, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Tavanti, A.; Davidson, A.D.; Gow, N.A.R.; Maiden, M.C.J.; Odds, F.C. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis Groups II and III. J. Clin. Microbiol. 2005, 43, 284–292. [Google Scholar] [CrossRef]
- Cantón, E.; Pemán, J.; Quindós, G.; Eraso, E.; Miranda-Zapico, I.; Álvarez, M.; Merino, P.; Campos-Herrero, I.; Marco, F.; de la Pedrosa, E.G.G.; et al. Prospective multicenter study of the epidemiology, molecular identification, and antifungal susceptibility of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis isolated from patients with candidemia. Antimicrob. Agents Chemother. 2011, 55, 5590–5596. [Google Scholar] [CrossRef]
- Puig-Asensio, M.; Pemán, J.; Zaragoza, R.; Garnacho-Montero, J.; Martín-Mazuelos, E.; Cuenca-Estrella, M.; Almirante, B. Impact of therapeutic strategies on the prognosis of candidemia in the ICU. Crit. Care Med. 2014, 42, 1423–1432. [Google Scholar] [CrossRef]
- Tóth, R.; Nosek, J.; Mora-Montes, H.M.; Gabaldon, T.; Bliss, J.M.; Nosanchuk, J.D.; Turner, S.A.; Butler, G.; Vágvölgyi, C.; Gácser, A. Candida parapsilosis: From genes to the bedside. Clin. Microbiol. Rev. 2019, 32, e00111-18. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Messer, S.A.; Pfaller, M.A.; Diekema, D.J. Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis. J. Clin. Microbiol. 2008, 46, 2659–2664. [Google Scholar] [CrossRef]
- Tay, S.T.; Na, S.L.; Chong, J. Molecular differentiation and antifungal susceptibilities of Candida parapsilosis isolated from patients with bloodstream infections. J. Med. Microbiol. 2009, 58, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Barbedo, L.S.; Vaz, C.; Pais, C.; Figueiredo-Carvalho, M.H.G.; Muniz, M.M.; Zancope-Oliveira, R.M.; Sampaio, P. Different scenarios for Candida parapsilosis fungaemia reveal high numbers of mixed C. parapsilosis and Candida orthopsilosis infections. J. Med. Microbiol. 2015, 64, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Khodavaisy, S.; Daneshnia, F.; Najafzadeh, M.-J.; Mahmoudi, S.; Charsizadeh, A.; Salehi, M.-R.; Zarrinfar, H.; Raeisabadi, A.; Dolatabadi, S.; et al. Molecular identification, genotypic diversity, antifungal susceptibility, and clinical outcomes of infections caused by clinically underrated yeasts, Candida orthopsilosis, and Candida metapsilosis: An Iranian Multicenter Study (2014–2019). Front. Cell. Infect. Microbiol. 2019, 9, 264. [Google Scholar] [CrossRef]
- Irinyi, L.; Serena, C.; Garcia-Hermoso, D.; Arabatzis, M.; Desnos-Ollivier, M.; Vu, D.; Cardinali, G.; Arthur, I.; Normand, A.-C.; Giraldo, A.; et al. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database—The quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol. 2015, 53, 313–337. [Google Scholar] [CrossRef]
- Cornely, O.A.; Bassetti, M.; Calandra, T.; Garbino, J.; Kullberg, B.J.; Lortholary, O.; Meersseman, W.; Akova, M.; Arendrup, M.C.; Arikan-Akdagli, S.; et al. ESCMID guideline for the diagnosis and management of Candida diseases 2012: Non-neutropenic adult patients. Clin. Microbiol. Infect. 2012, 18, 19–37. [Google Scholar] [CrossRef]
- EUCAST. Clinical Breakpoints for Fungi, Version 10. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Clinical_breakpoints/AFST_BP_v10.0_200204_updatd_links_200924.pdf (accessed on 16 January 2023).
- Bassetti, M.; Azoulay, E.; Kullberg, B.J.; Ruhnke, M.; Shoham, S.; Vazquez, J.; Giacobbe, D.R.; Calandra, T. EORTC/MSGERC Definitions of invasive fungal diseases: Summary of activities of the intensive care unit Working Group. Clin. Infect. Dis. 2021, 72 (Suppl. S2), S121–S127. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 62, e1–e50. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal rRNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; Volume 38, pp. 315–322. [Google Scholar] [CrossRef]
- Criseo, G.; Scordino, F.; Romeo, O. Current methods for identifying clinically important cryptic Candida species. J. Microbiol. Methods 2015, 111, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Wickes, B.L.; Wiederhold, N.P. Molecular diagnostics in medical mycology. Nat. Commun. 2018, 9, 5135. [Google Scholar] [CrossRef]
- Alves, J.; Alonso-Tarrés, C.; Rello, J. How to identify invasive candidemia in ICU—A narrative review. Antibiotics 2022, 11, 1804. [Google Scholar] [CrossRef]
- Yamin, D.H.; Husin, A.; Harun, A. risk factors of Candida parapsilosis catheter-related bloodstream infection. Front. Public Health 2021, 9, 631865. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Sulim, S.; Holm, A.; Nielsen, L.; Nielsen, S.D.; Knudsen, J.D.; Drenck, N.E.; Christensen, J.J.; Johansen, H.K. Diagnostic issues, clinical characteristics, and outcomes for patients with fungemia. J. Clin. Microbiol. 2011, 49, 3300–3308. [Google Scholar] [CrossRef] [PubMed]
- Delfino, D.; Scordino, F.; Pernice, I.; Lo Posso, C.; David, A.; Barberi, I.; Criseo, G.; Casacio, A.; Romeo, O. Potential association of specific Candida parapsilosis genotypes, bloodstream infections and colonization of health workers’ hands. Clin. Microbiol. Infect. 2014, 20, O946–O951. [Google Scholar] [CrossRef]
- Sai, S.; Holland, L.M.; McGee, C.F.; Lynch, D.B.; Butler, G. Evolution of mating within the Candida parapsilosis species group. Eukaryot. Cell 2011, 10, 578–587. [Google Scholar] [CrossRef]
- Pryszcz, L.P.; Németh, T.; Gácser, A.; Gabaldón, T. Unexpected genomic variability in clinical and environmental strains of the pathogenic yeast Candida parapsilosis. Genome Biol. Evol. 2013, 5, 2382–2392. [Google Scholar] [CrossRef] [PubMed]
- Schröder, M.S.; Martinez de San Vicente, K.; Prandini, T.H.; Hammel, S.; Higgins, D.G.; Bagagli, E.; Wolfe, K.H.; Butler, G. Multiple origins of the pathogenic yeast Candida orthopsilosis by separate hybridizations between two parental species. PLoS Genet. 2016, 12, e1006404. [Google Scholar] [CrossRef]
- Mixão, V.; Gabaldón, T. Hybridization and emergence of virulence in opportunistic human yeast pathogens. Yeast 2018, 35, 5–20. [Google Scholar] [CrossRef]
- Estrada-Mata, E.; Navarro-Arias, M.J.; Pérez-García, L.A.; Mellado-Mojica, E.; López, M.G.; Csonka, K.; Gacser, A.; Mora-Montes, H.M. Members of the Candida parapsilosis complex and Candida albicans are differentially recognized by human peripheral blood mononuclear cells. Front. Microbiol. 2016, 6, 1527. [Google Scholar] [CrossRef]
- Held, J.; Kohlberger, I.; Rappold, E.; Busse Grawitz, A.; Häcker, G. Comparison of (1->3)-β-D-glucan, mannan/anti-mannan antibodies, and Cand-Tec Candida antigen as serum biomarkers for candidemia. J. Clin. Microbiol. 2013, 51, 1158–1164. [Google Scholar] [CrossRef]
- Garcia-Effron, G.; Katiyar, S.K.; Park, S.; Edlind, T.D.; Perlin, D.S. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 2008, 52, 2305–2312. [Google Scholar] [CrossRef]
- Daneshnia, F.; de Almeida Júnior, J.N.; Arastehfar, A.; Lombardi, L.; Shor, E.; Moreno, L.; Verena Mendes, A.; Goreth Barberino, M.; Thomaz Yamamoto, D.; Butler, G.; et al. Determinants of fluconazole resistance and echinocandin tolerance in C. parapsilosis isolates causing a large clonal candidemia outbreak among COVID-19 patients in a Brazilian ICU. Emerg. Microbes. Infect. 2022, 11, 2264–2274. [Google Scholar] [CrossRef] [PubMed]
- Kartsonis, N.; Killar, J.; Mixon, L.; Hoe, C.-M.; Sable, C.; Bartizal, K.; Motyl, M. Caspofungin susceptibility testing of isolates from patients with esophageal candidiasis or invasive candidiasis: Relationship of MIC to treatment outcome. Antimicrob. Agents Chemother. 2005, 49, 3616–3623. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pappas, P.G.; Rotstein, C.M.F.; Betts, R.F.; Nucci, M.; Talwar, D.; De Waele, J.J.; Vazquez, J.A.; Dupont, B.F.; Horn, D.L.; Ostrosky-Zeichner, L.; et al. Micafungin versus caspofungin for treatment of candidemia and other forms of invasive candidiasis. Clin. Infect. Dis. 2007, 45, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Yang, H.; Shan, Z.; Jiang, L.; Zhang, Q. Clinical efficacy and safety of antifungal drugs for the treatment of Candida parapsilosis infections: A systematic review and network meta-analysis. J. Med. Microbiol. 2021, 70, 001434. [Google Scholar] [CrossRef]
- Horn, D.L.; Ostrosky-Zeichner, L.; Morris, M.I.; Ullmann, A.J.; Wu, C.; Buell, D.N.; Kovanda, L.L.; Cornely, O.A. Factors related to survival and treatment success in invasive candidiasis or candidemia: A pooled analysis of two large, prospective, micafungin trials. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Shin, J.H.; Park, K.H.; Shin, M.G.; Suh, S.P.; Ryang, D.W. A fatal case of Candida orthopsilosis fungemia. Korean J. Clin. Microbiol. 2010, 13, 140. [Google Scholar] [CrossRef]
- Paiva, J.A.; Pereira, J.M.; Tabah, A.; Mikstacki, A.; de Carvalho, F.B.; Koulenti, D.; Ruckly, S.; Çakar, N.; Misset, B.; Dimopoulos, G.; et al. Characteristics and risk factors for 28-day mortality of hospital acquired fungemias in ICUs: Data from the EUROBACT study. Crit. Care 2016, 20, 53. [Google Scholar] [CrossRef]
- D’Enfert, C. Biofilms and their role in the resistance of pathogenic Candida to antifungal agents. CDT 2006, 7, 465–670. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).