Abstract
In sporadic idiopathic pulmonary fibrosis (sIPF) and pulmonary fibrosis caused by a mutation in telomere (TRG-PF) or surfactant related genes (SRG-PF), there are a number of aberrant cellular processes known that can lead to fibrogenesis. We investigated whether RNA expression of genes involved in these processes differed between sIPF, TRG-PF, and SRG-PF and whether expression levels were associated with survival. RNA expression of 28 genes was measured in lung biopsies of 26 sIPF, 17 TRG-PF, and 6 SRG-PF patients. Significant differences in RNA expression of TGFBR2 (p = 0.02) and SFTPA2 (p = 0.02) were found between sIPF, TRG-PF, and SRG-PF. Patients with low (<median) expression of HSPA5 (p = 0.04), COL1A1 (p = 0.03), and ATF4 (0.005) had significantly longer survival rates than patients with high (≥median) expression of these genes. In addition, we scored for low (0) or high (1) expression of six endoplasmic reticulum (ER) stress genes (HSP90B1, DDIT3, EDEM1, HSPA5, ATF4, and XBP1) and found that patients with high expression in a low number of ER stress genes (total score 0–1) had longer survival rates than patients with high expression in a high number of ER stress genes (total score 2–6) (p = 0.03). In conclusion, there are minor differences between sIPF, TRG-PF, and SRG-PF and high expression in a high number of ER stress genes significantly associated with shorter survival time, suggesting that ER stress may be a target for therapy for PF.
1. Introduction
Idiopathic pulmonary fibrosis (IPF) is characterized by damage to the alveolar epithelium and accumulation of extracellular matrix in the interstitium. IPF has a poor prognosis with a median survival of approximately 3–4 years [,]. The disease is highly heterogeneous, while in the majority of patients etiology is unknown, some of the patients have genetic pulmonary fibrosis. Pathogenic mutations causing pulmonary fibrosis have been identified in two different groups of genes, surfactant related genes (SRG) such as SFTPC and SFTPA2, or telomere related genes (TRG) such as TERT and RTEL1 []. In a study by Snetselaar et al. [], telomere length was measured in white blood cells of patients with sporadic IPF (sIPF) and SRG-PF and TRG-PF. It was shown that some sIPF patients had short telomeres comparable with TRG-PF, whereas others had a telomere length comparable with SRG-PF. This is in congruence with a recent report showing the wide range in telomere length in patients with pulmonary fibrosis who do not carry a likely pathogenic mutation []. This suggests that disease drivers in sIPF may overlap with those in specific genetic groups.
In previous studies, several molecular processes involved in IPF have been identified such as extracellular matrix deposition [,], endoplasmic reticulum (ER) stress [,], senescence [,], and hypoxia []. In addition, different cell types of the bronchoalveolar compartment, and particularly the alveolar type 2 cell (AT2), were found to be involved in the pathogenesis of IPF []. IPF expression studies showed an increase in expression of fibrogenesis related genes such as TGFB1, ACTA2 [,,], and hypoxia genes including HIF1A and EPAS1 [,,], but also involvement of cellular processes such as autophagy [,]. Mutations in surfactant and telomere genes were shown to each lead to different processes in the lung, and it is unknown to what extent these processes overlap. Heterozygous mutations in SRG have a toxic gain of function effect on surfactant processing [,,] and may cause ER stress with upregulation of ER stress associated genes, such as HSPA5 and XBP1 [,,]. On the other hand, heterozygous TRG mutations cause haploinsufficiency leading to excessive shortening of telomeres and were shown to increase DNA damage related processes including upregulation of TP53BP1 and TP53 [] and cause senescence with altered expression of CDKN2A and CDKN1A []. Because disease cause and outcome are so heterogeneous in PF, a better understanding of the involvement of the different processes and their relation to patient survival is warranted and may aid development of therapies targeting specific processes in patients. To determine whether TRG-PF and SRG-PF have distinct expressions of genes involved in disease pathogenesis, we measured RNA expression in diagnostic lung biopsies of sIPF, TRG-PF, and SRG-PF. In addition, we investigated whether different levels of RNA expression are associated with survival.
2. Results
RNA expression of 28 genes involved in IPF pathogenesis was measured in FFPE surgical lung biopsies of three groups of patients. (26 sIPF, 17 TRG-PF, and 6 SRG-PF). Clinical characteristics of the three patient groups are displayed in Table 1. Telomere length in tissue and blood was significantly different between the three groups (p = 0.001 and p < 0.001, respectively). Telomere length was lower in TRG-PF patients than the other two patient groups. In addition, male predominance was present in sIPF (92.3%) and TRG-PF patients (82.4%), but not in SRG-PF patients (50%). The 28 genes are associated with different processes, such as senescence, DNA damage, endoplasmic reticulum (ER) stress, surfactant homeostasis, extracellular matrix (ECM), autophagy, hypoxia, and protein degradation. Statistical analysis showed no difference in RNA expression of 25 genes (expression of TP53, CDKN2A, and CDKN1A was not measured in SRG-PF patients) between TRG-PF and SRG-PF patients. Comparison of RNA expression of the 28 genes between sIPF, TRG-PF, and SRG-PF showed a significant difference in RNA expression of TGFBR2 (p = 0.02) and SFTPA2 (p = 0.02). Post-hoc analysis showed significantly lower expression of TGFBR2 in TRG-PF compared to sIPF patients (p = 0.001) and lower expression of SFTPA2 in SRG-PF compared to sIPF patients (p = 0.002, Figure 1). Two out of three patients with an SFTPC mutation had the lowest levels of SFTPC expression. The median SFTPC expression in the SFTPC mutation carriers was just 0.03. This was not significantly different from the SFPTC level of 0.05 in the patients carrying an SFTPA2 mutation. Similarly, two out of three patients carrying an SFTPA2 mutation had the lowest SFTPA2 expression and the median SFTPA2 expression in the entire SFTPA2 group was just 0.008. However, this was not significantly lower than the SFTPA2 expression in the patients carrying an SFTPC mutation (0.004).

Table 1.
Demographics and clinical characteristics of patient groups at time of biopsy.
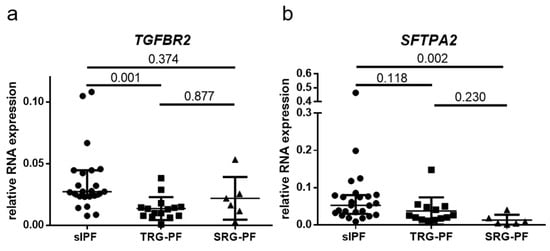
Figure 1.
Relative RNA expression in lung tissue of sporadic IPF (sIPF) patients and pulmonary fibrosis patients with a telomere related gene (TRG-PF) or surfactant related gene (SRG-PF) mutation (a). Relative RNA expression of TGFBR2. A significant difference in RNA expression was found between the three groups (Kruskal–Wallis test p = 0.02). Post hoc analysis showed significantly higher expression in sIPF compared to TRG-PF (p = 0.001). There were no significant differences in expression between sIPF and SRG-PF (0.374) or between TRG-PF and SRG-PF (0.877). (b). Relative RNA expression of SFTPA2. A significant difference in RNA expression was found between the three groups (Kruskal–Wallis test p = 0.02). Post hoc analysis showed significantly higher expression in sIPF compared to SRG-PF patients (p = 0.002). There were no significant differences in expression between sIPF and TRG-PF (0.118) or between TRG-PF and SRG-PF (0.23).
2.1. Clustering of Genes and Patients
Unsupervised two-way hierarchical clustering of RNA expression of the 28 genes in 49 patients demonstrated five major vertical clusters of genes and two major horizontal clusters of patients (Figure 2). The upper cluster of patients contains 10 sIPF, 12 TRG-PF, and 3 SRG-PF patients, whereas the lower cluster contains 16 sIPF, 5 TRG-PF, and 3 SRG-PF patients. The distribution of patient groups was not significantly different between the two clusters (p = 0.12). Analysis of the clinical characteristics of the two clusters showed no differences except for a significantly higher number of deaths in the lower cluster compared to the upper cluster (p = 0.04, Table 2).
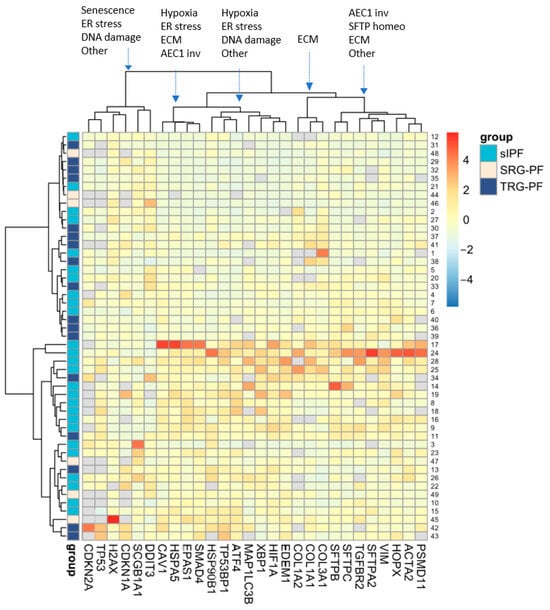
Figure 2.
Unsupervised two-way hierarchical clustering of RNA expression of 28 genes in 49 patients. Rows represent individual patients and columns represent genes. Grey blocks indicate that RNA expression of a certain gene was not measured due to shortage of tissue or undetectable expression. Clustering of process related genes in each of the five main gene clusters is indicated at the top with arrows pointing to the cluster. AEC1 inv: alveolar epithelial type 1 involvement, SFTP homeo: surfactant homeostasis; ECM: extracellular matrix; other: this includes bronchiolar involvement, autophagy, or protein degradation.

Table 2.
Demographics and clinical characteristics of two patient clusters.
The 28 measured genes can be categorized in gene process groups: surfactant homeostasis (SFTPC, SFTPA2, and SFTPB), ER-stress (HSP90B1, EDEM1, DDIT3, ATF4, XBP1, and HSPA5), DNA-damage (TP53BP1, H2AX, and TP53), extracellular matrix (ACTA2, VIM, COL1A1, COL1A2, COL3A1, SMAD4, and TGFBR2), hypoxia (HIF1A and EPAS1), senescence (CDKN2A and CDKN1A), protein degradation (PSMD11) and autophagy (MAP1LC3B), and AEC1 involvement (HOPX and CAV1) and bronchiolar involvement (SCGB1A1). HOPX and CAV1 encode proteins regulating the repair of AEC1 after injury. SCGB1A1 encodes a secretoglobin that exerts an anti-inflammatory function in the small airways. Genes were not randomly distributed over the five major vertical clusters (Figure 2). Both senescence genes were part of one cluster, and all other three surfactant homeostasis genes were part of another cluster. Furthermore, the three collagen encoding genes formed a separate cluster.
2.2. Survival
We investigated whether overall survival was different between patients with high and low RNA expression of a gene. Comparison showed significantly longer survival in patients with low (<median) RNA expression of ER stress genes ATF4 (64 months vs. 22 months; p = 0.005), HSPA5 (64 months vs. 22 months; p = 0.04), and for ECM gene COL1A1 (64 months vs. 22 months; p = 0.03) compared to patients with high (≥median) RNA expression of these genes (Figure 3a–c).
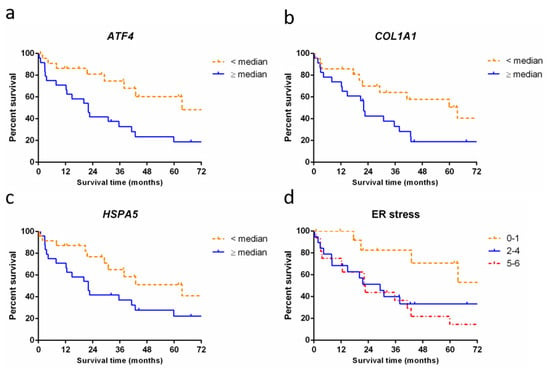
Figure 3.
Survival of patients with pulmonary fibrosis stratified by RNA expression levels. (a) Survival curve showing significantly longer survival in patients with low expression of ATF4 (64 months, n = 23, dashed line) compared to patients with high expression of ATF4 (22 months, n = 24, solid line, p = 0.005); (b) Survival curve showing a significantly longer survival in patients with low expression of COL1A1 (64 months, n = 22, dashed line) compared to patients with high expression of COL1A1 (22 months, n = 23, solid line, p = 0.03). (c) Survival curve showing a significantly longer survival in patients with low RNA expression of HSPA5 (64 months, n = 24, dashed line) compared to patients with high expression of HSPA5 (22 months, n = 24, solid line, p = 0.04). (d) Accumulative score in patients for high (+1 point) or low (0 points) expression in each of six ER stress genes (HSP90B1, DDIT3, EDEM1, HSPA5, ATF4, and XBP1). Survival curve showing significantly longer median survival in patients with high RNA expression of 0–1 ER stress genes (95 months, n = 14, dashed line) compared to patients with high expression of 2–4 ER stress genes (29 months, n = 19, solid line) and high expression of 5–6 ER stress genes (22 months, n = 16, dash-dotted line, p = 0.03).
Next, we grouped three or more genes that play a role in the same processes into process expression groups: surfactant homeostasis (SFTPB, SFTPC, and SFTPA2), DNA damage (TP53BP1, H2AX, and TP53), ECM (ACTA2, VIM, TGFBR2, COL1A1, and SMAD4), and ER stress (HSP90B1, DDIT3, EDEM1, HSPA5, ATF4, and XBP1). For each patient, a process expression score was calculated based on the sum of above (+1) or below (0) median expression for each gene in the process. Comparison of overall survival between high, average, and low scoring patients showed a significant difference for ER stress: patients with low expression of ER stress genes (score 0–1) showed a longer survival time than patients with average or high expression of these genes (score 2–4 or 5–6). Median survival in low expressing ER stress patients was 95 months versus 29 and 22 months in the average and high expressing patients, respectively (p = 0.03, Figure 3d). Survival analysis results of surfactant homeostasis, DNA damage, and extracellular matrix showed no significant differences (see Supplementary Materials). Results for processes represented by only two genes were also included in the Supplementary Materials.
3. Discussion
Several processes have been associated with the development of PF in general and with surfactant or telomere related PF specifically. In this study, we quantified RNA expression of proteins that have previously been shown to be involved in pulmonary fibrosis in lung biopsies of patients with sporadic and genetic IPF and analyzed the effect on survival. There was no significant clustering of patient groups and genes and no significant differences in RNA expression between the groups with TRG-PF and SRG-PF. This is the first study that showed significantly worse survival in PF patients with high expression of two or more ER stress genes compared to PF patients with low expression of five or all six ER stress genes.
Several gene expression studies have been performed on lung tissue from patients with pulmonary fibrosis. First, it was shown that expression in IPF patients differed significantly from control subjects. In a study by Wang et al. [], microarray data from lung tissue of 131 IPF patients with UIP on pathology and 12 controls were used. Comparison of IPF patients and controls resulted in 988 differentially expressed genes. In addition, by ward clustering and principal component analysis of gene expression profiles of 131 IPF patients, six patient clusters were found. The six clusters differed in disease severity and differential expression compared to controls in genes that play a role in processes such as extracellular matrix organization, regulation of cell migration, collagen catabolic process, cilium, cilium assembly, angiogenesis, and lung alveolar morphology. Furthermore, in a whole genome oligonucleotide microarray study by Yang et al. [], RNA expression of 41,000 genes and transcripts was measured in lung tissue of 16 sporadic idiopathic interstitial pneumonia patients, 10 familial idiopathic interstitial pneumonia patients, and 9 normal control subjects. In total, 135 transcripts were found to be upregulated or downregulated more than 1.8-fold in pulmonary fibrosis patients compared to normal control subjects. After hierarchical clustering, four patient clusters were identified; all controls except for two samples clustered together and all familial idiopathic interstitial pneumonia except for three samples clustered together. Comparison between sporadic idiopathic interstitial pneumonia (IIP) and familial IIP patients resulted in 142 transcripts from 62 genes with known functions that were more than 1.8-fold upregulated or downregulated. Interestingly, they found that genes from the same functional categories, such as calcium/potassium binding, cell adhesion, cell proliferation and death, ECM degradation, and cytokines/chemokines, are differentially expressed between sporadic and familial IIP as well as between IIP and normal control subjects, but then to a larger extent in the familial IIP patients. But because no genetic analysis was performed, it remains unclear whether patients carrying a TRG or SRG mutation had been included.
In our study, we included patients with familial disease of known cause and divided them over two groups: patients carrying an SRG and patients carrying a TRG mutation and studied expression of genes involved in mutation driven aberrant processes in PF. To our surprise, we detected no differences in expression of any of the studied genes between the TRG and SRG-PF groups. However, when we compared sIPF, TRG-PF, and SRG-PF, we found significant differences in RNA expression of TGFBR2 and SFTPA2 between the three groups. The cluster analysis did not result in clustering of patients in their respective sIPF, TRG-PF, or SRG-PF patient group. On the other hand, there was clustering of genes belonging to the same processes. As surfactant homeostasis genes clustered together and senescence genes also clustered together, this indicates that the expression levels may correctly inform about the activity of the process.
Lawson et al. studied protein expression in AECs in lung tissue of three patients with pulmonary fibrosis and an SFTPC mutation, ten patients with familial interstitial pneumonia without an SFTPC mutation, and ten patients with sIPF. They observed expression of the ER stress proteins BiP, EDEM, and XBP1 in lung tissues of all patients []. In addition, Carleo et al. investigated protein patterns in bronchoalveolar lavage (BAL) fluid of 10 familial and 17 sporadic IPF patients. In total, 22 proteins were found to be differentially expressed between familial and sporadic IPF. The upregulated proteins in sIPF played a role in oxidative stress response and the upregulated proteins in familial IPF (including SP-A2 encoded by SFTPA2) played a role in immune response, coagulation system, wounding response, and ion homeostasis []. The increase of SP-A2 in BAL fluid of familial IPF patients in Carleo et al. is in contrast with the lower RNA expression of SFTPA2 in lung tissue of the SRG-PF patients in our study. Although their small familial cohort was not analyzed for carriage of genetic mutations, it is unlikely that their familial cohort included any patients with an SRG mutation. SRG mutations occur in 3–8% of patients with familial disease, while TRG mutations are present in approximately 35% of familial patients []. However, we did not observe a difference in SFTPA2 RNA expression between our sIPF and familial TRG-PF patients. The level of RNA expression may therefore not reflect protein expression, while increased SP-A2 in BAL fluid may aid identification of familial patients, and the lower RNA expression of SFTPA2 in lung tissue may aid identification of patients with surfactant-related pathology.
Furthermore, the SP-A2 protein level in blood was shown to be a predictive marker for progressive disease [,]. Although it remained unclear what the source is of SP-A2 in blood, and how increased levels in blood mechanistically related to progression of disease in the lung. When analyzing the survival of patients, TRG-PF showed the shortest survival (Table 1), but the survival of patients with SRG-PF could not be calculated due to the low number. However, in a recent study, survival of SRG-PF patients was shown to be comparable with that of sIPF []. Our study suggests that in patients with SRG-PF, levels may be influenced by the presence of surfactant related mutations, and thus future investigations into biomarkers for PF may benefit from stratification of patients by genetic cause of disease.
When we studied the impact of RNA expression of each gene on survival in patients, we found that patients with low expression of HSPA5, COL1A1, or ATF4 showed a significantly longer survival rate than patients with high expression of one of these genes. In a study by Tsitoura et al. [], they observed increased COL1A1 RNA expression in BAL cells of patients with 53 IPF and 62 non-IPF ILD compared to 19 controls. In addition, they also found that high levels of COL1A1 expression were associated with worse survival. However, this was only found in the non-IPF ILD patients and not in the IPF cohort. For ER stress gene ATF4, as far as we know, no association with survival was investigated before in other studies, although it was found that ATF4 expression was increased in IPF lung tissue and colocalized with apoptosis markers CHOP and cleaved caspase 3, and encoded by DDIT3 and CASP3, respectively, in alveolar epithelial cells overlying fibroblast foci []. For ER stress marker GRP78/BiP encoded by HSPA5, conflicting results have been found; in one study elevated GRP94 and CHOP, encoded by HSP90B1 and DDIT3, respectively, and reduced expression of GRP78 were observed in alveolar epithelial cells type 2 (AEC2s) from IPF lung tissue compared to AEC2s from normal donors [], whereas in another study increased GRP78/BiP expression was observed in IPF lung tissue compared to control lung tissue. In the latter study, strong GRP78/BiP staining was found in alveolar epithelial cells in regions of fibroblasts foci []. Fibroblasts foci are aggregates of proliferating fibroblasts and myofibroblasts thought to represent areas of active fibrosis. Therefore, the proximity of strong BiP staining (encoded by HSPA5) to fibroblast foci suggests a role in fibrogenesis. Together with our finding that high expression of ER stress genes, including ATF4 and HSPA5, results in low survival, the evidence suggests that ER stress in IPF may be an important factor for progression of fibrogenesis. It is known that the cause of ER stress may be different in each patient []. However, the absence of differences in expression between our groups of patients and the presence of a correlation with survival suggest that ER stress is an important factor in the progression of pulmonary fibrosis independent of underlying genetic cause. Two recent studies show the feasibility of targeting ER stress in pulmonary fibrosis. Kropski et al. [] showed that ER stress may be targeted by pharmacological chaperones such as sodium phenylbutyrate, which resulted in reduced ER stress in preclinical disease models or by drugs that selectively inhibit oligomerized IRE1α, and which have been investigated in animal models. Chen et al. [] showed that inhibition of IRE1α by an endoribonuclease inhibitor alleviates CS-induced pulmonary inflammation and fibrogenesis in a mouse model.
A limitation of our study is that only bulk RNA expression was measured. Differences in RNA expression between cell types, such as in single cell RNA sequencing, were not investigated. In addition, no spatial transcriptomics and immunohistochemistry was used to localize which cell populations were involved in the regulation of RNA expression in PF. Furthermore, the results were not verified at the protein level and therefore effects of posttranscriptional regulation of gene expression during fibrogenesis was not investigated. The number of patients, especially those with SRG-PF, was quite low. However, the low number of SRG-PF cases was limited by the rarity of these patients compared to those with sIPF. Inclusions of more cases might have shown additional significant differences in the clinical characteristics between the two clusters. In addition, the number of genes measured may be too small to create distinct clusters. The low number of differences in RNA expression between sIPF, TRG-PF, and SRG-PF may also explain why based on most recent finding these patients appear to benefit from the same treatment [,,]. In addition, although all samples were diagnostic lung biopsies, a considerably low DLCO % predicted in especially TRG-PF and SRG-PF patients was present, which suggests already advanced lung disease. It remains possible that more differences, particularly mutation associated differences, would be present if early disease samples had been analyzed. While screening family members at risk may aid early disease detection, genetic analysis is likely to reduce the need for biopsies. In a recent survey, 72% of pulmonologists reported that they might modify their diagnostic work-up according to the results of genetic testing of whom 78% would postpone or exclude surgical lung biopsy [].
In conclusion, analysis of expression of fibrosis related genes in sIPF and TRG-PF and SRG-PF patients showed almost similar gene expression between the groups. The finding that high expression of ER stress genes leads to worse survival time supports development of therapies targeting ER stress that may be beneficial to all PF patients, including those with genetic pulmonary fibrosis. Further studies are needed to investigate this association in more detail.
4. Materials and Methods
4.1. Patients and Tissue Selection
Diagnostic surgical lung biopsies and blood obtained between 1997 and 2016 of 26 sIPF, 17 TRG-PF (RTEL n = 3, TERT n = 14), and 6 SRG-PF (SFTPC n = 3, SFTPA2 n = 3) patients were included in this study. Diagnoses were based on the ATS/ERS/JRS/ALAT guidelines []. All subjects signed written consent for the study approved by the Medical Research Ethics Committees United (MEC-U) of the St. Antonius hospital (R05-08A).
4.2. DNA/RNA Isolation from Lung Tissue and Blood
After removal of paraffin with paraffin dissolver (Macherey-Nagel, Düren, Germany), RNA and DNA was isolated from formalin-fixed paraffin embedded tissue using AllPrep DNA/RNA FFPE kit (Qiagen Benelux BV, Venlo, The Netherlands). DNA and RNA concentration and purity were measured using a NanoDrop spectrophotometer. For isolation of genomic DNA from peripheral white blood cells, a magnetic beads-based method (chemagic DNA blood 10k kit; Perkin Elmer Inc., Waltham, MA, USA) was used.
4.3. Telomere Length Measurements in Lung Tissue and Blood
T/S ratio was measured in isolated DNA from tissue and blood by monochrome multiplex quantitative polymerase chain reaction (MMqPCR) as described before in [,,]. The T/S ratio is a measure for telomere length. Telomere length adjusted for age was calculated by the difference between the observed T/S ratio and the age-adjusted normal value (T/S expected).
4.4. Real-Time PCR
cDNA (6ng per reaction), prepared from RNA using i-script (Bio-Rad GmbH laboratories B.V), was amplified using iQ SYBR Green Supermix (Bio-Rad GmbH laboratories B.V, Lunteren, The Netherlands) and gene specific primers (see Supplementary Materials) in a CFX96 Bio-Rad qPCR machine with the following run conditions: 3 min at 95 °C followed by 45 cycles of 10 s at 95 °C, 20 s at 61 °C, and 25 s at 72 °C. RNA expression was calculated by delta Ct method using the mean of three reference genes: ACTB, RPL13A, and EEF1A1. RNA expression was investigated for genes involved in the following processes: bronchiolar involvement (SCGB1A1), alveolar epithelial cell type 1 (AEC1) involvement (HOPX and CAV1), surfactant homeostasis (SFTPC, SFTPA2, SFTPB), hypoxia (HIF1A, EPAS1), protein degradation (PSMD11), autophagy (MAP1LC3B), senescence (CDKN2A, CDKN1A), ER-stress (HSP90B1, EDEM1, DDIT3, ATF4, XBP1, HSPA5), DNA-damage (TP53BP1, H2AX, TP53) and extracellular matrix (ACTA2, VIM, COL1A1, COL1A2, COL3A1, SMAD4, TGFBR2). List of used primers can be found in Table S1.
4.5. Statistical Analysis
For analysis of differences in demographics and clinical characteristics between sIPF, TRG-PF, and SRG-PF patients, the chi-square test was used for discrete variables and the Kruskal–Wallis test was used for continuous variables, as appropriate with a small sample size. The Mann–Whitney U test was used to compare RNA expression between TRG-PF and SRG-PF followed by false discovery rate (FDR) to correct for multiple comparisons. The Kruskal–Wallis test was used to analyze differences in RNA expression between sIPF, TRG-PF, and SRG-PF followed by FDR to correct for multiple comparisons. Unsupervised two-way hierarchical clustering of RNA expression using Ward’s clustering method with Euclidean distance metric was used to create a heatmap. Chi-square and Mann–Whitney U test were used for discrete and continuous variables, respectively, to compare demographics and clinical characteristics between the two horizontal clusters in the heatmap. For survival analyses, median RNA expression was calculated for each gene and patients were divided into two groups containing patients with <median expression of that gene and patients with ≥median expression of that gene. Additionally, when three or more genes play a role in the same process, these were also analyzed together as one process-group, surfactant homeostasis (SFTPB, SFTPC, SFTPA2), DNA damage (TP53BP1, TP53, H2AX), ECM (ACTA2, VIM, TGFBR2, COL1A1, SMAD4), and ER stress (HSP90B1, DDIT3, EDEM1, HSPA5, ATF4, XBP1). For ECM, only one, COL1A1, of the three collagen genes was included in the survival analysis to prevent overrepresentation of collagen genes. For each process-group of genes, we determined the score for each patient. Per gene, RNA expression ≥median is +1 point and <median is 0 points. The sum of these scores resulted in an accumulative score per process per patient. For each process, we performed survival analysis dividing the patients into groups. Survival was compared using the Kaplan–Meier method with log-rank tests. Survival time was determined from time of lung biopsy until death. Patients were censored when lost to follow-up or when they underwent lung transplantation or at the last contact date. Statistical analyses were performed in SPSS v.26 and R v.4.2.2 (including the following packages: tidyverse version 1.3.2, ggsci v. 2.9, readxl v. 1.4.1, pheatmap v. 1.0.12, ggpubr v. 0.5.0).
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms242316748/s1.
Author Contributions
Conceptualization, D.K., K.M.K., J.J.v.d.V. and C.H.M.v.M.; Data curation, D.K. and K.M.K.; Formal analysis, D.K., K.M.K. and H.M.S.; Funding acquisition, D.K., J.J.v.d.V., J.C.G. and C.H.M.v.M.; Investigation, D.K., K.M.K. and J.J.v.d.V.; Methodology, D.K., K.M.K., J.J.v.d.V. and C.H.M.v.M.; Supervision, C.H.M.v.M.; Validation, K.M.K.; Visualization, D.K. and H.M.S.; Writing—original draft, D.K.; Writing—review and editing, D.K., K.M.K., J.J.v.d.V., H.M.S., J.C.G. and C.H.M.v.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by ZonMW–TopZorg St. Antonius Science Corner grant (Topzorg grant number 842002001), ZonMW TZO vision grant (grant number 10070012010004), and the Prof. Dr. Jaap Swierenga foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Medical Research Ethics Committees United (MEC-U) of the St. Antonius Hospital (R05-08A).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
Data can be requested from the corresponding author.
Acknowledgments
We would like to thank the patients who participated in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Strongman, H.; Kausar, I.; Maher, T.M. Incidence, Prevalence, and Survival of Patients with Idiopathic Pulmonary Fibrosis in the UK. Adv. Ther. 2018, 35, 724. [Google Scholar] [CrossRef]
- Kaunisto, J.; Salomaa, E.-R.; Hodgson, U.; Kaarteenaho, R.; Kankaanranta, H.; Koli, K.; Vahlberg, T.; Myllärniemi, M. Demographics and survival of patients with idiopathic pulmonary fibrosis in the FinnishIPF registry. ERJ Open Res. 2019, 5, 00170–2018. [Google Scholar] [CrossRef] [PubMed]
- van Moorsel, C.H.M.; van der Vis, J.J.; Grutters, J.C. Genetic disorders of the surfactant system: Focus on adult disease. Eur. Respir. Rev. 2021, 30, 200085. [Google Scholar] [CrossRef] [PubMed]
- Snetselaar, R.; van Moorsel, C.H.M.; Kazemier, K.M.; van der Vis, J.J.; Zanen, P.; van Oosterhout, M.F.M.; Grutters, J.C. Telomere length in interstitial lung diseases. Chest 2015, 148, 1011–1018. [Google Scholar] [CrossRef]
- Zhang, D.; Newton, C.A.; Wang, B.; Povysil, G.; Noth, I.; Martinez, F.J.; Raghu, G.; Goldstein, D.; Garcia, C.K. Utility of whole genome sequencing in assessing risk and clinically-relevant outcomes for pulmonary fibrosis. Eur. Respir. J. 2022, 1, 2200577. [Google Scholar] [CrossRef] [PubMed]
- Chien, J.W.; Richards, T.J.; Gibson, K.F.; Zhang, Y.; Lindell, K.O.; Shao, L.; Lyman, S.K.; Adamkewicz, J.I.; Smith, V.; Kaminski, N.; et al. Serum lysyl oxidase-like 2 levels and idiopathic pulmonary fibrosis disease progression. Eur. Respir. J. 2014, 43, 1430–1438. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, C.; Boldt, J.; King, T.E.; Crouch, E.; Vartio, T.; McDonald, J.A. An immunohistochemical study of architectural remodeling and connective tissue synthesis in pulmonary fibrosis. Am. Rev. Respir. Dis. 1989, 140, 1693–1703. [Google Scholar] [CrossRef]
- Lawson, W.E.; Crossno, P.F.; Polosukhin, V.V.; Roldan, J.; Cheng, D.-S.; Lane, K.B.; Blackwell, T.R.; Xu, C.; Markin, C.; Ware, L.B.; et al. Endoplasmic reticulum stress in alveolar epithelial cells is prominent in IPF: Association with altered surfactant protein processing and herpesvirus infection. Am. J. Physiol. Lung Cell Mol. Physiol. 2008, 294, L1119–L1126. [Google Scholar] [CrossRef]
- Korfei, M.; Ruppert, C.; Mahavadi, P.; Henneke, I.; Markart, P.; Koch, M.; Lang, G.; Fink, L.; Bohle, R.-M.; Seeger, W.; et al. Epithelial endoplasmic reticulum stress and apoptosis in sporadic idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2008, 178, 838–846. [Google Scholar] [CrossRef]
- Minagawa, S.; Araya, J.; Numata, T.; Nojiri, S.; Hara, H.; Yumino, Y.; Kawaishi, M.; Odaka, M.; Morikawa, T.; Nishimura, S.L.; et al. Accelerated epithelial cell senescence in IPF and the inhibitory role of SIRT6 in TGF-β-induced senescence of human bronchial epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2011, 300, L391–L401. [Google Scholar] [CrossRef]
- Disayabutr, S.; Kim, E.K.; Cha, S.-I.; Green, G.; Naikawadi, R.P.; Jones, K.D.; Golden, J.A.; Schroeder, A.; Matthay, M.A.; Kukreja, J.; et al. miR-34 miRNAs Regulate Cellular Senescence in Type II Alveolar Epithelial Cells of Patients with Idiopathic Pulmonary Fibrosis. PLoS ONE 2016, 11, e0158367. [Google Scholar] [CrossRef]
- Aquino-Gálvez, A.; González-Ávila, G.; Jiménez-Sánchez, L.L.; Maldonado-Martínez, H.A.; Cisneros, J.; Toscano-Marquez, F.; Castillejos-López, M.; Torres-Espíndola, L.M.; Velázquez-Cruz, R.; Rodríguez, V.H.O.; et al. Dysregulated expression of hypoxia-inducible factors augments myofibroblasts differentiation in idiopathic pulmonary fibrosis. Respir. Res. 2019, 20, 130. [Google Scholar] [CrossRef]
- Xu, Y.; Mizuno, T.; Sridharan, A.; Du, Y.; Guo, M.; Tang, J.; Wikenheiser-Brokamp, K.A.; Perl, A.-K.T.; Funari, V.A.; Gokey, J.J.; et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. J. Clin. Investig. Insight. 2017, 1, e90558. [Google Scholar] [CrossRef]
- Guiot, J.; Henket, M.; Corhay, J.L.; Moermans, C.; Louis, R. Sputum biomarkers in IPF: Evidence for raised gene expression and protein level of IGFBP-2, IL-8 and MMP-7. PLoS ONE 2017, 12, e0171344. [Google Scholar] [CrossRef]
- Roach, K.M.; Wulff, H.; Feghali-Bostwick, C.; Amrani, Y.; Bradding, P. Increased constitutive αSMA and Smad2/3 expression in idiopathic pulmonary fibrosis myofibroblasts is KCa3.1-dependent. Respir. Res. 2014, 15, 155. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, D.; Liang, J.; Meltzer, E.B.; Gray, A.; Miura, R.; Wogensen, L.; Yamaguchi, Y.; Noble, P.W. Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J. Exp. Med. 2011, 208, 1459. [Google Scholar] [CrossRef]
- Shochet, G.E.; Bardenstein-Wald, B.; McElroy, M.; Kukuy, A.; Surber, M.; Edelstein, E.; Pertzov, B.; Kramer, M.R.; Shitrit, D. Hypoxia Inducible Factor 1A Supports a Pro-Fibrotic Phenotype Loop in Idiopathic Pulmonary Fibrosis. Int. J. Mol. Sci. 2021, 22, 3331. [Google Scholar] [CrossRef]
- Delbrel, E.; Soumare, A.; Naguez, A.; Label, R.; Bernard, O.; Bruhat, A.; Fafournoux, P.; Tremblais, G.; Marchant, D.; Gille, T.; et al. HIF-1α triggers ER stress and CHOP-mediated apoptosis in alveolar epithelial cells, a key event in pulmonary fibrosis. Sci. Rep. 2018, 8, 17939. [Google Scholar] [CrossRef]
- Patel, A.S.; Lin, L.; Geyer, A.; Haspel, J.A.; An, C.H.; Cao, J.; Rosas, I.O.; Morse, D. Autophagy in Idiopathic Pulmonary Fibrosis. PLoS ONE 2012, 7, e41394. [Google Scholar] [CrossRef]
- Im, J.; Hergert, P.; Nho, R.S. Translational Research in Acute Lung Injury and Pulmonary Fibrosis: Reduced FoxO3a expression causes low autophagy in idiopathic pulmonary fibrosis fibroblasts on collagen matrices. Am. J. Physiol.-Lung Cell Mol. Physiol. 2015, 309, L552. [Google Scholar] [CrossRef]
- Legendre, M.; Butt, A.; Borie, R.; Debray, M.-P.; Bouvry, D.; Filhol-Blin, E.; Desroziers, T.; Nau, V.; Copin, B.; Moal, F.D.-L.; et al. Functional assessment and phenotypic heterogeneity of SFTPA1 and SFTPA2 mutations in interstitial lung diseases and lung cancer. Eur. Respir. J. 2020, 56, 2002806. [Google Scholar] [CrossRef]
- Nogee, L.M.; Dunbar, A.E.; Wert, S.E.; Askin, F.; Hamvas, A.; Whitsett, J.A. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N. Engl. J. Med. 2001, 344, 573–579. [Google Scholar] [CrossRef]
- Klay, D.; Hoffman, T.W.; Harmsze, A.M.; Grutters, J.C.; van Moorsel, C.H.M. Systematic review of drug effects in humans and models with surfactant-processing disease. Eur. Respir. Rev. 2018, 27, 170135. [Google Scholar] [CrossRef]
- Maguire, J.A.; Mulugeta, S.; Beers, M.F. Endoplasmic reticulum stress induced by surfactant protein C BRICHOS mutants promotes proinflammatory signaling by epithelial cells. Am. J. Respir. Cell Mol. Biol. 2011, 44, 404–414. [Google Scholar] [CrossRef]
- Maitra, M.; Wang, Y.; Gerard, R.D.; Mendelson, C.R.; Garcia, C.K. Surfactant protein A2 mutations associated with pulmonary fibrosis lead to protein instability and endoplasmic reticulum stress. J. Biol. Chem. 2010, 285, 22103–22113. [Google Scholar] [CrossRef]
- Nguyen, H.; Uhal, B.D. The unfolded protein response controls ER stress-induced apoptosis of lung epithelial cells through angiotensin generation. Am. J. Physiol.-Lung Cell Mol. Physiol. 2016, 311, L846–L854. [Google Scholar] [CrossRef]
- Alder, J.K.; Barkauskas, C.E.; Limjunyawong, N.; Stanley, S.E.; Kembou, F.; Tuder, R.M.; Hogan, B.L.M.; Mitzner, W.; Armanios, M. Telomere dysfunction causes alveolar stem cell failure. Proc. Natl. Acad. Sci. USA 2015, 112, 5099–5104. [Google Scholar] [CrossRef]
- Liu, T.; De Los Santos, F.G.; Zhao, Y.; Wu, Z.; Rinke, A.E.; Kim, K.K.; Phan, S.H. Telomerase reverse transcriptase ameliorates lung fibrosis by protecting alveolar epithelial cells against senescence. J. Biol. Chem. 2019, 294, 8861–8871. [Google Scholar] [CrossRef]
- Wang, Y.; Yella, J.; Chen, J.; McCormack, F.X.; Madala, S.K.; Jegga, A.G. Unsupervised gene expression analyses identify IPF-severity correlated signatures, associated genes and biomarkers. BMC Pulm. Med. 2017, 17, 133. [Google Scholar] [CrossRef]
- Yang, I.V.; Burch, L.H.; Steele, M.P.; Savov, J.D.; Hollingsworth, J.W.; McElvania-Tekippe, E.; Berman, K.G.; Speer, M.C.; Sporn, T.A.; Brown, K.K.; et al. Gene Expression Profiling of Familial and Sporadic Interstitial Pneumonia. Am. J. Respir. Crit. Care Med. 2007, 175, 45. [Google Scholar] [CrossRef]
- Carleo, A.; Bargagli, E.; Landi, C.; Bennett, D.; Bianchi, L.; Gagliardi, A.; Carnemolla, C.; Perari, M.G.; Cillis, G.; Armini, A.; et al. Comparative proteomic analysis of bronchoalveolar lavage of familial and sporadic cases of idiopathic pulmonary fibrosis. J. Breath Res. 2016, 10, 26007. [Google Scholar] [CrossRef]
- Takahashi, H.; Fujishima, T.; Koba, H.; Murakami, S.; Kurokawa, K.; Shibuya, Y.; Shiratori, M.; Kuroki, Y.; Abe, S. Serum surfactant proteins A and D as prognostic factors in idiopathic pulmonary fibrosis and their relationship to disease extent. Am. J. Respir. Crit. Care Med. 2000, 162, 1109–1114. [Google Scholar] [CrossRef]
- Greene, K.E.; King, T.E.; Kuroki, Y.; Bucher-Bartelson, B.; Hunninghake, G.W.; Newman, L.S.; Nagae, H.; Mason, R.J. Serum surfactant proteins-A and -D as biomarkers in idiopathic pulmonary fibrosis. Eur. Respir. J. 2002, 19, 439–446. [Google Scholar] [CrossRef]
- Klay, D.; Grutters, J.C.; van der Vis, J.J.; Platenburg, M.G.; Kelder, J.C.; Tromp, E.; van Moorsel, C.H. Progressive Disease With Low Survival in Adult Patients With Pulmonary Fibrosis Carrying Surfactant-Related Gene Mutations: An Observational Study. Chest 2023, 163, 870–880. [Google Scholar] [CrossRef]
- Tsitoura, E.; Trachalaki, A.; Vasarmidi, E.; Mastrodemou, S.; Margaritopoulos, G.A.; Kokosi, M.; Fanidis, D.; Galaris, A.; Aidinis, V.; Renzoni, E.; et al. Collagen 1a1 Expression by Airway Macrophages Increases In Fibrotic ILDs and Is Associated With FVC Decline and Increased Mortality. Front. Immunol. 2021, 12, 645548. [Google Scholar] [CrossRef]
- Borok, Z.; Horie, M.; Flodby, P.; Wang, H.; Liu, Y.; Ganesh, S.; Firth, A.L.; Minoo, P.; Li, C.; Beers, M.F.; et al. Grp78 loss in epithelial progenitors reveals an age-linked role for endoplasmic reticulum stress in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2020, 201, 198–211. [Google Scholar] [CrossRef]
- Baek, H.A.; Kim, D.S.; Park, H.S.; Jang, K.Y.; Kang, M.J.; Lee, D.G.; Moon, W.S.; Chae, H.J.; Chung, M.J. Involvement of endoplasmic reticulum stress in myofibroblastic differentiation of lung fibroblasts. Am. J. Respir. Cell Mol. Biol. 2012, 46, 731–739. [Google Scholar] [CrossRef]
- Tanjore, H.; Blackwell, T.S.; Lawson, W.E. Emerging evidence for endoplasmic reticulum stress in the pathogenesis of idiopathic pulmonary fibrosis. Am. J. Physiol.-Lung Cell Mol. Physiol. 2012, 302, 721–729. [Google Scholar] [CrossRef]
- Kropski, J.A.; Blackwell, T.S. Endoplasmic reticulum stress in the pathogenesis of fibrotic disease. J. Clin. Invest. 2018, 128, 64–73. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Liu, J.; He, Y.; Wei, Y.; Chen, J. Inhibition of ER stress by targeting the IRE1α-TXNDC5 pathway alleviates crystalline silica-induced pulmonary fibrosis. Int. Immunopharmacol. 2021, 95, 107519. [Google Scholar] [CrossRef]
- Justet, A.; Klay, D.; Porcher, R.; Cottin, V.; Ahmad, K.; Molina, M.M.; Nunes, H.; Reynaud-Gaubert, M.; Naccache, J.M.; Manali, E.; et al. Safety and efficacy of pirfenidone and nintedanib in patients with idiopathic pulmonary fibrosis and carrying a telomere-related gene mutation. Eur. Respir. J. 2021, 57, 2003198. [Google Scholar] [CrossRef]
- Borie, R.; Kannengiesser, C.; Antoniou, K.; Bonella, F.; Crestani, B.; Fabre, A.; Froidure, A.; Galvin, L.; Griese, M.; Grutters, J.C.; et al. European Respiratory Society Statement on Familial Pulmonary Fibrosis. Eur. Respir. J. 2022, 61, 2201383. [Google Scholar] [CrossRef]
- Terwiel, M.; Borie, R.; Crestani, B.; Galvin, L.; Bonella, F.; Fabre, A.; Froidure, A.; Griese, M.; Grutters, J.C.; Johannson, K.; et al. Genetic testing in interstitial lung disease: An international survey. Respirology 2022, 27, 747–757. [Google Scholar] [CrossRef]
- Raghu, G.; Collard, H.R.; Egan, J.J.; Martinez, F.J.; Behr, J.; Brown, K.K.; Colby, T.V.; Cordier, J.-F.; Flaherty, K.R.; Lasky, J.A.; et al. An Official ATS/ERS/JRS/ALAT Statement: Idiopathic pulmonary fibrosis: Evidence-based guidelines for diagnosis and management. Am. J. Respir. Crit. Care Med. 2011, 183, 788–824. [Google Scholar] [CrossRef]
- Cawthon, R.M. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009, 37, e21. [Google Scholar] [CrossRef]
- Snetselaar, R.; Van Batenburg, A.A.; Van Oosterhout, M.F.M.; Kazemier, K.M.; Roothaan, S.M.; Peeters, T.; Van Der Vis, J.J.; Goldschmeding, R.; Grutters, J.C.; Van Moorsel, C.H.M. Short telomere length in IPF lung associates with fibrotic lesions and predicts survival. PLoS ONE 2017, 12, e0189467. [Google Scholar] [CrossRef]
- van Batenburg, A.A.; Kazemier, K.M.; van Oosterhout, M.F.M.; van der Vis, J.J.; van Es, H.W.; Grutters, J.C.; Goldschmeding, R.; van Moorsel, C.H.M. From organ to cell: Multi-level telomere length assessment in patients with idiopathic pulmonary fibrosis. PLoS ONE 2020, 15, e0226785. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).