Abstract
The mechanistic target of rapamycin (mTOR) kinase is a central regulator of cell growth and metabolism. It is the catalytic subunit of two distinct large protein complexes, mTOR complex 1 (mTORC1) and mTORC2. mTOR activity is subjected to tight regulation in response to external nutrition and growth factor stimulation. As an important mechanism of signaling transduction, the ‘second messenger’ cyclic nucleotides including cAMP and cGMP and their associated cyclic nucleotide-dependent kinases, including protein kinase A (PKA) and protein kinase G (PKG), play essential roles in mediating the intracellular action of a variety of hormones and neurotransmitters. They have also emerged as important regulators of mTOR signaling in various physiological and disease conditions. However, the mechanism by which cAMP and cGMP regulate mTOR activity is not completely understood. In this review, we will summarize the earlier work establishing the ability of cAMP to dampen mTORC1 activation in response to insulin and growth factors and then discuss our recent findings demonstrating the regulation of mTOR signaling by the PKA- and PKG-dependent signaling pathways. This signaling framework represents a new non-canonical regulation of mTOR activity that is independent of AKT and could be a novel mechanism underpinning the action of a variety of G protein-coupled receptors that are linked to the mTOR signaling network. We will further review the implications of these signaling events in the context of cardiometabolic disease, such as obesity, non-alcoholic fatty liver disease, and cardiac remodeling. The metabolic and cardiac phenotypes of mouse models with targeted deletion of Raptor and Rictor, the two essential components for mTORC1 and mTORC2, will be summarized and discussed.
Keywords:
mTOR; cAMP; cGMP; PKA; PKG; Raptor; Rictor; obesity; fatty liver disease; cardiac hypertrophy 1. Introduction
The mechanistic target of rapamycin (mTOR) kinase is a central regulator of cell growth and metabolism. It acts as a signaling node that integrates the external inputs and coordinates the major anabolic and catabolic pathways to maintain metabolic homeostasis [1]. The mTOR kinase is an evolutionarily conserved serine/threonine kinase and a member of the phosphoinositide kinase-related kinase (PIKK) family. It is the catalytic subunit of two distinct large protein complexes, mTOR complex 1 (mTORC1) and mTORC2. These two complexes are distinguished by their accessory proteins, sensitivity to rapamycin, and distinct substrates and functions [1].
The regulatory-associated protein of mTOR (RAPTOR) and the rapamycin-insensitive companion of mTOR (RICTOR) are the defining components for mTORC1 and mTORC2, respectively (Figure 1). Rapamycin forms a complex with FKBP12 (FK506-binding protein of 12 kDa) and prevents the substrate’s entry into the kinase activity site and thus inhibits mTORC1 activity [2,3,4,5,6]. mTORC2 is relatively insensitive to acute rapamycin treatment. However, prolonged rapamycin treatment inhibits mTORC2 signaling due to the sequestering of the cellular mTOR pool into the rapamycin-bound complex [7,8]. It has been well established that mTORC1 plays essential roles to promote protein synthesis, lipid metabolism, mitochondrial biogenesis, nucleotide synthesis, and glycolysis, while suppressing autophagy and lysosomal biogenesis (reviewed in [1]). mTORC2 controls a spectrum of cellular functions including cell survival and cytoskeletal organization (reviewed in [1]).
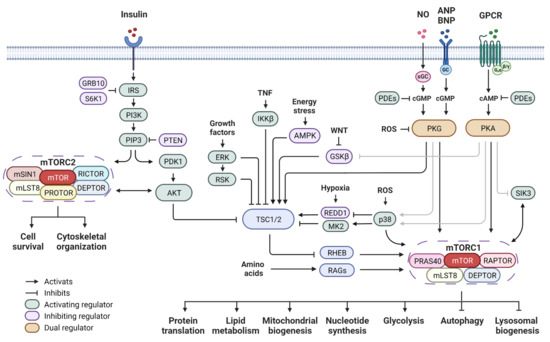
Figure 1.
mTOR signaling network and cyclic nucleotide-dependent protein kinase pathway. Insulin acts through the PI3K pathway and leads to signaling cascades that further activate mTORC1 and mTORC2. Upstream regulators converge at the TSC complex to either activate or inhibit mTORC1 activity. The RAG GTPases are the key component of the amino acid sensing machinery. The second messenger signaling including cAMP and cGMP pathways regulates mTOR activity through both direct and indirect mechanisms. Abbreviations: mTOR: mechanistic target of rapamycin; RAPTOR: regulatory-associated protein of mTOR; RICTOR: rapamycin-insensitive companion of mTOR; DEPTOR: DEP domain-containing mTOR-interacting protein; mLST8: mammalian lethal with SEC13 protein 8; PRAS40: proline-rich AKT substrate of 40 kDa; mSIN1: mammalian stress-activated protein kinase-interaction protein 1; PROTOR: protein observed with RICTOR; TSC1/2: tuberous sclerosis complex 1/2; RHEB: Ras homolog mTORC1 binding; RAGs: Ras-related GTPases; IRS: insulin receptor substrate; PI3K: phosphoinositide 3-kinase; PIP3: phosphatidylinositol 3,4,5-triphosphate; PTEN: phosphatase and tensin homolog; PDK1: phosphoinositide-dependent protein kinase 1; GRB10: growth factor receptor bound protein 10; AKT: protein kinase B; ERK: extracellular signal-regulated kinase; RSK: ribosomal S6 kinase 1; TNF: tumor necrosis factor; IKKβ: inhibitor of nuclear factor kappa-B kinase subunit beta; AMPK: AMP-activated protein kinase; WNT: wingless and lnt-1; GSKβ: glycogen synthase kinase 3β; REDD1: regulated in development and DNA damage responses 1; p38 MAPK: p38 mitogen-activated protein kinases; MK2: MAPK activated protein kinase 2; ROS: reactive oxygen species; NO: nitric oxide; ANP: atrial natriuretic peptide; BNP: B-type natriuretic peptide; GPCR: G protein-coupled receptor; GC: guanylyl cyclase: sGC: soluble guanylyl cyclase; cAMP: cyclic adenosine monophosphate; cGMP: cyclic guanosine monophosphate; PKA: cAMP-dependent protein kinase; PKG: cGMP-dependent protein kinase; SIK3: salt-inducible kinase 3; PDEs: phosphodiesterases.
mTOR activity is tightly regulated to sense the nutrients and growth factors to orchestrate cell growth and metabolic program. The integrated upstream signals that control mTORC1 activity mainly include two sets of small G proteins, RHEB (Ras homolog mTORC1 binding) and RAGs (Ras-related GTPases) (Figure 1). RHEB is located in the lysosomal surface and in its active GTP-bound state it is competent to activate mTORC1 kinase activity [9,10]. The RAGs GTPases are essential components of the amino acid sensing machinery. They promote the subcellular translocation of mTORC1 to lysosomes [11,12], where mTORC1 subsequently interacts with and becomes activated by RHEB. The tuberous sclerosis complex (TSC) is a complex of TSC1-TSC2 (hamartin–tuberin) and it acts as a GTPase activating protein to negatively regulate the lysosomal RHEB [13,14,15,16]. Upstream signaling kinases phosphorylate TSC, thus modifying TSC activity, and in turn stimulate (AKT, ERK, RSK-1, IKKβ) or inhibit (AMPK and GSK-3β) mTORC1 activity (Figure 1). mTORC2 is primarily activated by growth factors through the phosphoinositide 3-kinase (PI3K) pathway (Figure 1). Binding of phosphatidylinositol 3,4,5-triphosphate (PIP3) leads to mTORC2 activation [17,18,19]. The tumor suppressor PTEN (phosphatase and tensin homolog) inhibits mTORC2 activity by promoting the conversion of PIP3 to phosphatidylinositol 4,5-bisphosphate (PIP2) [20,21].
2. Cyclic Nucleotide-Dependent Kinase Regulation of mTOR Signaling
As briefly reviewed in Part 1, the mTOR signaling cascades are tightly regulated in response to external nutrition and growth factors to coordinate cell growth and metabolism. As an important mechanism of signaling transduction, the ‘second messenger’ cyclic nucleotides including cAMP and cGMP and their associated cyclic nucleotide-dependent kinases, including protein kinase A (PKA) and protein kinase G (PKG), have been shown to exert both positive and negative regulation of mTOR signaling in various physiological contexts and disease conditions. In the following part, we will summarize the earlier work establishing the ability of cAMP to dampen mTORC1 activation in response to insulin and growth factors and then discuss our recent findings demonstrating the regulation of mTOR signaling by the PKA- and PKG-dependent signaling pathways that represent a new non-canonical pathway independent of AKT, and we will review the implications of these signaling events in cardiometabolic disease, such as obesity, non-alcoholic fatty liver disease, and cardiac remodeling.
2.1. Regulation of mTOR Signaling by PKA
The seven transmembrane-spanning G protein-coupled receptors (GPCRs) are cell surface proteins that, upon agonist stimulation, can interact with heterotrimeric G proteins that in turn regulate various effector enzymes or ion channels (see [22] for review). GPCRs such as the β-adrenergic receptors (βAR), thyroid-stimulating hormone (TSH) receptor, and glucagon receptor interact with the Gs heterotrimer to stimulate adenylyl cyclase to convert ATP into the ‘second messenger’ cAMP. Other GPCRs can regulate inositol phosphate metabolism or inhibit cAMP production. Downstream events that respond to increases in cAMP include activating the cAMP-dependent protein kinases (PKA) as well as small G proteins such as exchange proteins directly activated by cAMP-1 (EPAC1) and EPAC2. The regulation of mTOR signaling by the cAMP/PKA pathway has been documented in many studies. However, as not appreciated by these previous studies, we have shown that cAMP can either inhibit or activate mTOR activity depending on the cell type, signaling context, and physiological conditions (summarized in Table 1).
Since mTOR was first determined to be involved in growth and responsive to growth factors, the first studies examining the effect of cAMP reported that it acts to antagonize the effect of insulin or growth factors on mTOR activity. These studies were mostly performed in cultured cell lines. Monfar and colleagues reported that reagents that increase cAMP and activate PKA inhibit the cytokine interleukin-2 (IL-2)-dependent activation of S6K1 in IL-2-responsive lymphoid cells [23]. Scott and Lawrence showed that increasing cAMP blocked the effect of insulin in 3T3-L1 adipocyte to activate mTOR, suggesting an antagonizing effect of cAMP on insulin action and mTOR activity [24]. Xie and colleagues further showed that elevation of cAMP in mouse embryonic fibroblasts (MEFs) and human embryonic kidney 293 (HEK293) cells inhibits insulin-stimulated mTORC1 activation via a PKA-dependent mechanism, and prolonged elevation in cAMP can also inhibit mTORC2 [25]. This cAMP-dependent inhibition of mTORC1/2 was proposed to be caused by the dissociation of mTORC1 and 2 and a reduction in mTOR catalytic activity [25]. Rocha and colleagues reported that, in Ret/PTC1-positive thyroid carcinoma cell TPC-1, cAMP markedly inhibits the Raf/ERK cascade, leading to mTOR pathway inhibition [26]. Kim and colleagues showed that, in several cell lines, the cAMP-dependent signaling pathway inhibits AKT activity by blocking the coupling between AKT and its upstream regulators, PDK, in the plasma membrane, leading to the inhibition of mTOR [27].
Studies have also been conducted in primary cell culture or physiologically relevant tissues. Mothe-Satney and colleagues showed that, in primary cultures of hepatocytes, glucagon increases the mTOR phosphorylation at Ser2448 as insulin or amino acids do, but glucagon inhibits the insulin- and amino acid-induced activation of S6K1 and phosphorylation of 4E-BP1 [28]. Another study by Kimball and colleagues showed that glucagon represses both basal and amino acid-induced phosphorylation of 4E-BP1 and S6K1 in perfused rat liver as an experimental model [29]. The repression is associated with the activation of PKA and enhanced phosphorylation of liver kinase B1 (LKB1) and the AMP-activated protein kinase (AMPK). However, this study also showed that glucagon simultaneously enhances phosphorylation of two S6K1 downstream effectors S6 and eukaryotic initiation factor 4B (eIF-4B) [29]. This effect was proposed to act through the activation of the ERK-RSK signaling pathway. In addition, Baum and colleagues showed that glucagon acts in a dominant manner to repress insulin-induced mTORC1 signaling in perfused rat liver [30]. In the presence of a combination of insulin and glucagon, AKT and TSC2 phosphorylation and PKA activity were all increased compared with controls. However, mTORC1 signaling was repressed compared with livers perfused with a medium containing insulin alone, and this effect was proposed to be associated with reduced assembly of the mTORC1-eIF3 complex. In another more recent study, Csukasi and colleagues reported that the parathyroid hormone (PTH) receptor, another GPCR coupled to cAMP and PKA activation, led to mTORC1 inhibition during skeletal development [31]. It is known that PKA can inhibit salt-inducible kinase 3 (SIK3) and the other two SIK family members [32,33]. It was proposed that PTH/PTHrP signaling can inhibit SIK3. In this case, these authors showed that inhibiting SIK3 prevents the proteasomal degradation of DEPTOR, a negative regulator of mTOR. In a recent study, our group identified that SIK3 is an mTORC1 substrate in brown adipocytes that interacts with RAPTOR and can be phosphorylated in a rapamycin-sensitive manner [34]. Thus, this SIK3-mTORC1 connection could be a new layer of the signaling network that might convey PKA action to mTOR signaling and deserves further investigation.
Several other studies have also linked cAMP with mTORC1 activation. For example, Kwon and colleagues reported that the GLP-1 (glucagon-like peptide-1) receptor agonist, Exenatide, dose-dependently enhanced phosphorylation of S6K1 in isolated rat islets through a glucose-dependent and cAMP-mediated mechanism [35]. They also showed that this process was rapamycin-sensitive and results in the alteration of intracellular calcium flux [35]. Brennesvik and colleagues reported that adrenalin potentiated an insulin-stimulated activation of AKT and S6K through a mechanism that depends on cAMP and Epac in skeletal muscle [36]. Moore and colleagues showed that cAMP increasing agents such as GLP-1 and forskolin lead to the S6 activation in the pancreatic beta-cell line MIN6 (mouse insulinoma cell line 6) and islets of Langerhans through a mechanism that depends on PKA [37]. Blancquaert and colleagues reported that TSH/cAMP activates mTORC1 in PC Cl3 rat thyroid cells, leading to phosphorylation of S6K1 and 4E-BP1. This effect was proposed to be caused in part by T246 phosphorylation of PRAS40, which was found as an in vitro substrate of PKA [38]. Palaniappan and Menon reported that human chorionic gonadotropin stimulates theca-interstitial cell proliferation and cell cycle regulatory proteins through a mechanism involved in the cAMP-dependent activation of AKT and the mTORC1 signaling pathway [39]. Kim and colleagues reported that cAMP phosphodiesterase 4D (PDE4D) is a binding partner of RHEB and acts as a cAMP-specific negative regulator of mTORC1 [40]. Under basal conditions, it is reported that PDE4D binds RHEB in a noncatalytic manner that does not require its cAMP-hydrolyzing activity and thereby inhibits the ability of RHEB to activate mTORC1. In this same report, the authors state that elevated cAMP levels disrupt the interaction of PDE4D with RHEB and increase the interaction between RHEB and mTOR. This enhanced RHEB–mTOR interaction induces the activation of mTORC1 and cap-dependent translation. This study proposed a new role for PDE4 as a molecular transducer for cAMP signaling to regulate mTOR activity.
In 2016, our group reported the novel observation that mTORC1 activation is required for the β-adrenergic stimulation of adipose tissue browning [41]. Pharmacological inhibition of mTORC1 or genetic deletion of its defining component RAPTOR in adipocyte impairs the brown and beige adipocyte expansion in response to β-adrenergic agonist treatment or cold temperature exposure [41]. Specifically, our study showed for the first time that PKA can directly phosphorylate in RAPTOR at Ser791 and mTOR at S1276, S1288, and S2112 [41], providing a novel signaling mechanism whereby PKA regulates mTOR signaling, and this is likely a major mechanism responsible for the previous reports of the cAMP activation of mTORC1. A cell-type-specific knock-in of a RAPTOR mutation that changes Ser791 into a phosphorylation-resistant Ala791 in mice has been made in our lab and will be reported on in the future. For these and other studies, the generation of an antibody that recognizes the phosphorylated S791 in RAPTOR will be extremely helpful.
2.2. Regulation of mTOR Signaling by PKG
Newer studies are demonstrating that, like PKA, the cGMP/PKG pathway can also control mTORC1 activity (Table 1). The cardiac natriuretic peptides (NPs) stimulate cGMP/PKG signaling through the action of guanyl cyclase-coupled NP receptor A (NPRA). Our group has shown that, like PKA, PKG phosphorylates Ser791 on RAPTOR [42], and related to this, the NP-stimulated adipose tissue browning [43] is impaired with the pharmacological inhibition of mTORC1 by rapamycin [42,43]. It will be interesting to determine whether the adipose tissue browning effect of NPs is impaired in mice with a phosphorylation-resistant Ser791Ala knock-in mutation. This will provide insight into the in vivo function of the S791 phosphorylation site in the context of the NP-regulated adipose tissue thermogenesis program.
PKG has also been reported to regulate mTOR signaling by phosphorylating other components of the mTOR network. Ranek and colleagues reported that PKG1 activates TSC2 by direct phosphorylation at Ser1365 and Ser1366 in cardiomyocytes, thereby inhibiting mTORC1 and promoting autophagy activity to counter adverse remodeling during cardiac stress [44]. In addition, studies have shown that PKG1α oxidation at cysteine-42 can be induced by oxidative stress [45]. Oeing and colleagues showed that the PKG1α cysteine-42 redox state controls mTORC1 activation in pathological cardiac hypertrophy [46]. The oxidation of PKG1α at C42 reduces its phosphorylation of TSC2, resulting in amplified pressure overload-stimulated mTORC1 activity and adverse cardiac remodeling [46]. As PKG1 is the major effector of both nitric oxide-generated cGMP and natriuretic peptide-stimulated cGMP, these studies provide mechanistic insights into the protective effects of these reagents against heart disease and oxidative stress.
2.3. Indirect Regulation of mTOR Signaling
As the major effector kinases of second messenger signaling, several reports have suggested that PKA and PKG can regulate mTOR activity through indirect mechanisms via several downstream signaling regulators, such as p38 mitogen-activated protein kinases (MAPKs) and glycogen synthase kinase 3 (GSK3) (Table 1).
p38 is a class of MAPKs that is responsive to stress stimuli and plays a central role in a diverse range of stress-induced signaling pathways [47,48]. Li and colleagues showed that p38 MAPK controls TSC2 Ser1210 phosphorylation through its downstream kinase MAPKAPK2 (MK2), which enhances TSC2 interaction with 14-3-3 proteins and in turn inhibits TSC2 function [49]. Cully and colleagues reported that p38-activating stresses, such as H2O2 and anisomycin, were able to activate TORC1 in mammalian and Drosophila tissue culture [50]. This stress-induced TORC1 activation could be blocked by RNAi against mitogen-activated protein kinase kinase 3 and 6 (MKK3/6). p38 was also required for the activation of TORC1 in response to amino acids and growth factors. In another study, Hernández and colleagues reported that reactive oxygen species (ROS) can activate p38 in certain conditions of cell stress, which in turn reduces the level of REDD1 (regulated in development and DNA damage responses 1) [51]. The decline in REDD1 leads to the dissociation of the TSC1/2 via enhanced association of TSC2 with 14-3-3 family members [51]. In addition, González-Terán and colleagues showed that, upon hypertrophy-inducing stimuli, p38γ/δ promote mTORC1 activity and cardiac hypertrophy by phosphorylating DEPTOR and leading its ubiquitination and degradation [52]. Given the well-established crosstalk of p38 with PKA [53,54] and PKG [43,55,56] signaling, it would be interesting to determine whether p38 might also convey the PKA and PKG actions to mTORC1. However, the physiological and pathological context of such regulation are yet to be established.
GSK3β is an established negative regulator of mTORC1 signaling. After the primed phosphorylation of TSC2 by AMPK at S1345, GSK3β phosphorylates TSC2 at several amino acid residues (Thr1329, Ser1333, Ser1337, Ser1341) and results in mTORC1 inhibition [57]. GSK3 activity is inhibited through phosphorylation of serine 21 in GSK3α and serine 9 in GSK3β. These serine residues in GSK3 are within motifs that allow them to be targets of both AKT and PKA [58]. PKA physically associates with, phosphorylates, and inactivates both isoforms of GSK3 [58]. Thus, the activity of GSK3 can be modulated either by growth factors that work through the PI3K-AKT cascade or by hormonal stimulation of G protein-coupled receptors that link to changes in intracellular cAMP levels [58], which will ultimately dictate the mTORC1 activity.
PKA might regulate mTORC2 activity through both direct and indirect mechanisms. As mentioned earlier, a study from our group identified three PKA phosphorylation sites in mTOR, and phosphorylation-resistant mutations of these sites (S1276, S1288, and S2112) resulted in impaired mTORC1 activation as indicated by S6K1 phosphorylation [41]. Thus, it is possible that PKA can regulate mTORC2 activity through the direct phosphorylation of mTOR. In addition, PKA might be able to control mTORC2 activity through inhibiting GSK3. Chen and colleagues reported that GSK-3β phosphorylates the mTORC2 component RICTOR at Serine 1235 in cellular stress conditions [59]. This modification interferes with the binding of AKT to mTORC2 and thus inhibits AKT activity. Koo and colleagues reported that GSK3 is associated with RICTOR and directly phosphorylates the Thr 1695 [60]. This modification results in the recruitment of E3 ubiquitin ligase F-Box and WD repeat domain-containing 7 (FBW7) and leads to RICTOR degradation. In both cases, GSK3 negatively regulates mTORC2 activity. However, in another study, Urbanska and colleagues reported that GSK3 can positively regulate mTORC2 in mature neurons [61], suggesting that there may be important cell-type-specific regulatory pathways.

Table 1.
Regulation of mTOR by cAMP and cGMP signaling pathways.
Table 1.
Regulation of mTOR by cAMP and cGMP signaling pathways.
| No. | Reagents | Signaling Cascade and Effects | Mechanism | Targets | Readouts | Ref. |
|---|---|---|---|---|---|---|
| Inhibition by cAMP | ||||||
| 1 | Forskolin, IBMX | Inhibits interleukin-2-dependent mTORC1 activation in IL-2-responsive lymphoid cells | Activates PKA and inhibits PI3K and S6K1 activation | PI3K | p-S6K1 | [23] |
| 2 | Forskolin, cAMP analogue | Prevents the effect of insulin on mTORC1 activation in 3T3-L1 adipocyte | Inhibits mTOR phosphorylation and activation | mTOR | p-4E-BP | [24] |
| 3 | Forskolin, IBMX | Inhibits insulin and amino acid-stimulated mTORC1 activation in MEFs and HEK293 cells | Promotes mTOR complex disassembly and inhibits mTOR catalytic activity | mTOR | p-S6K1, p-4E-BP | [25] |
| 4 | Forskolin, IBMX | Inhibits serum-stimulated mTORC2 activation after prolonged cAMP elevation in MEFs and HEK293 cells | Promotes mTOR complex disassembly and inhibits mTOR catalytic activity | mTOR | p-PKB, p-PKC | [25] |
| 5 | Forskolin, cAMP analogue | Inhibits mTORC1 activation in Ret/PTC1-positive thyroid carcinoma cell TPC-1 | Inhibits the Raf/ERK cascade and depends on PKA | Raf/ERK cascade | p-S6K1, p-4E-BP | [26] |
| 6 | cAMP analogue | Inhibits EGF-stimulated mTOR activity in Swiss 3T3, HEK293, COS, and Rat2 cells | Inhibits PI3K activity, PDK1 translocation to plasma membrane, and AKT phosphorylation | PI3K/PDK/AKT cascade | p-S6K1 | [27] |
| 7 | Glucagon, cAMP analogue | Inhibits insulin- and amino acid-induced mTORC1 activation in serum-free primary rat hepatocytes | NA | NA | p-S6K1, p-4E-BP1 | [28] |
| 8 | Glucagon | Represses both basal and amino acid-induced mTORC1 activation in perfused rat liver | Activates PKA and enhances phosphorylation of LKB1 Thr172 and AMPK Ser428 | LKB1/AMPK pathway | p-S6K1, p-4E-BP1 | [29] |
| 9 | Glucagon | Acts in a dominant manner to repress insulin-induced mTORC1 signaling in perfused rat liver | Reduces assembly of the mTORC1-eIF3 complex | mTORC1-eIF3 complex | p-S6K1, p-4E-BP1 | [30] |
| 10 | PTH/PTHrP | Leads to mTORC1 inhibition during skeletal development | Inhibits SIK3 and prevents DEPTOR degradation | DEPTOR | p-S6K, p-S6 | [31] |
| Activation by cAMP | ||||||
| 11 | Forskolin, GLP-1RA exenatide | Activates mTORC1 in isolated rat islets | Mobilizes intracellular Ca2+ influx and upregulates ATP production | KATP channel | p-S6K1 | [35] |
| 12 | Adrenalin, cAMP analogue | Potentiates insulin-stimulated mTORC1 activation in soleus muscles ex vivo | Depends on cAMP and Epac but not PKA | EPAC | p-S6K1 | [36] |
| 13 | GLP-1, Forskolin | Leads to S6 phosphorylation in the pancreatic beta cell line MIN6 and islets | Depends on PKA phosphorylation of S6 at Ser235/Ser236 | S6 | p-S6 | [37] |
| 14 | TSH | Activates mTORC1 in PC Cl3 rat thyroid cells | Acts in part via phosphorylation of PRAS40 Thr246 by PKA and augments 4E-BP1 binding to RAPTOR | PRAS40 | p-S6K1, p-4E-BP1 | [38] |
| 15 | hCG, Forskolin | Activates mTORC1 in theca-interstitial cells | Involved in cAMP-dependent activation of AKT | AKT | p-S6K, p-S6, p-4E-BP1 | [39] |
| 16 | cAMP | Leads mTORC1 activation in HEK293 cells | Disrupts the interaction of PDE4D with RHEB and increases the interaction between RHEB and mTOR | PDE4D and RHEB | p-S6K1, p-4EBP1 | [40] |
| 17 | β-AR agonist | Leads to mTORC1 activation in adipocyte and mouse adipose tissue | Direct PKA phosphorylation of RAPTOR Ser791 and mTOR Ser1276, Ser1288, and Ser2112 | RAPTOR and mTOR | p-S6K1 | [41] |
| 18 | GLP-1RA liraglutide | Promotes mTORC1 activation in CHO cells expressing GLP-1R | Direct PKA phosphorylation of RAPTOR Ser791 | RAPTOR | p-S6K1 | [62] |
| Activation by cGMP | ||||||
| 19 | Natriuretic peptides | Leads to mTORC1 activation in adipocyte and mouse adipose tissue | Direct PKG phosphorylation of RAPTOR Ser791 | RAPTOR | p-S6K, p-S6 | [42] |
| Inhibition by cGMP | ||||||
| 20 | PKG | Inhibits mTORC1 and promotes autophagy activity | Direct PKG1 phosphorylation of TSC2 Ser1365 and Ser1366, PKG1α C42 oxidation reduces TSC2 phosphorylation and amplifies mTORC1 activity | TSC2 | p-S6K1, p-4E-BP1 | [44,46] |
| Indirect Regulation via p38 | ||||||
| 21 | p38 | Activates mTORC1 by inhibiting TSC2 | Controls TSC2 Ser1210 phosphorylation through MAPKAPK2 (MK2) and increases 14-3-3 binding | TSC2 | p-TSC2 | [49] |
| 22 | p38β | Leads to TORC1 activation in stressed mammalian and Drosophila tissue culture | Occurs downstream of or in parallel to TSC2 and upstream of Rag activation | ND | p-S6K1, p-4E-BP1 | [50] |
| 23 | p38 | Leads to mTORC1 activation in reperfused heart and reoxygenated or H2O2-treated cardiomyocytes (NRVMs) | ROS downregulates REDD1 and promotes TSC2/14-3-3 association | TSC2 | p-S6 | [51] |
| 24 | p38γ/δ | Leads to mTORC1 activation in postnatal cardiac hypertrophic growth | Phosphorylates DEPTOR and leads to its degradation and mTOR activation | DEPTOR | p-mTOR, p-S6K1, p-S6 | [52] |
| Indirect Regulation via GSK3β | ||||||
| 25 | GSK3β | Phosphorylates TSC2 and results in mTORC1 inhibition | PKA physically associates with, phosphorylates, and inactivates both isoforms of GSK3 | TSC2 | p-AKT | [57,58] |
| 26 | GSK3β | Required for mTORC2 inhibition during ER stress | Phosphorylates RICTOR at Ser1235, interferes with the binding of AKT to mTORC2 and thus inhibits AKT | RICTOR | p-AKT | [59] |
| 27 | GSK3β | Required for mTORC2 inhibition | Directly phosphorylates RICTOR at Thr1695, recruits FBW7 and leads to RICTOR degradation | RICTOR | p-AKT, p-SGK1, p-PKCα | [60] |
| 28 | GSK3β | Required for mTORC2 activation in response to BDNF or insulin stimulation in cultured neurons and mouse brain | Increases prosurvival signaling | NA | p-S6, p-AKT | [61] |
Abbreviations: IBMX: 3-Isobutyl-1-methylxanthine; MEFs: mouse embryonic fibroblast; PTH/PTHrP: parathyroid hormone/PTH-related peptide; GLP-1: glucagon-like peptide-1; GLP-1RA: GLP-1 receptor agonist; KATP channels: ATP-sensitive K+ channels; TSH: thyroid-stimulating hormone; hCG: human chorionic gonadotropin; β-AR: β-adrenergic receptor; NRVMs: neonatal rat ventricular myocytes; ER: endoplasmic reticulum; GPCRs: G protein-coupled receptors.
3. Role of mTOR Signaling in Cardiometabolic Disease
3.1. Role of mTOR Signaling in Obesity
Obesity develops as increased adiposity in fat mass due to a chronic surplus of calorie intake over energy expenditure. According to the guidelines, a body mass index (BMI) over 25 is considered overweight and over 30 is obese [63]. Body weight and composition, and the storage of energy as triglyceride in adipose tissue, are determined by the interaction between genetic, environmental, and psychosocial factors [64,65]. The energy balance is determined by the two arms—energy intake and energy expenditure—and they are controlled by both central and peripheral mechanisms. The central nervous system regulates energy balance by controlling the feed behavior, autonomic nervous activity, and neuroendocrine system [64,65]. The energy expenditure takes the form of physical activity, basal metabolism, and adaptive thermogenesis [64,65]. This part of the review will summarize the studies describing the role of mTOR signaling in obesity and systemic metabolism, with a focus on the adipose tissue and the neuronal control of food intake.
Insulin acts through the PI3K cascade to activate two downstream pathways to mTORC2 as well as via TSC to mTORC1. The dysregulation of mTOR signaling results in insulin resistance, a hallmark of metabolic syndrome [66]. In the fed state, increased blood glucose levels stimulate the pancreas to secrete insulin. Insulin promotes anabolic processes in target tissues to maintain glucose and lipid homeostasis, including lipogenesis, fatty acid import, and glycogen and protein synthesis (reviewed in [1]). During fasting, the drop in nutrients, growth factors, and insulin facilitates the shift of metabolic balance in favor of lipolysis, ketogenesis, and gluconeogenesis (reviewed in [1]). The negative feedback loop between mTORC1 and mTORC2 is tightly regulated under physiological conditions. In obesity, hyperactivation of mTORC1 induced by excessive nutrition and mitogens can lead to phosphorylation of insulin receptor substrate 1 (IRS-1) by S6K1 [67,68,69] and negative regulation of insulin signaling by GRB10 (growth factor receptor bound protein 10) [70,71]. Following phosphorylation at multiple residues, IRS1 is degraded and the insulin signaling is attenuated, leading to insulin resistance and ectopic lipid accumulation in muscle and the liver [67,68,69]. GRB10 inhibits the insulin signaling pathway by disrupting the association of IRS-1/IRS-2 with the insulin receptor [72]. However, GRB10 can also inhibit mTORC1 through a feedback mechanism. mTOR-mediated phosphorylation at Ser501/503 switches the binding preference of GRB10 from the insulin receptor to RAPTOR, leading to the dissociation of RAPTOR from mTOR and the inhibition of mTORC1 signaling [73].
Studies with adipose tissue-specific Raptor knockout mice have demonstrated that mTORC1 is essential for adipose tissue function and homeostasis (Table 2). Polak and colleagues reported that adipose-specific Raptor knockout mice had substantially less adipose tissue, elevated energy expenditure proposed to be due to mitochondrial uncoupling, were protected against diet-induced obesity, hypercholesterolemia, and insulin resistance [74]. However, this study used the Fabp4/aP2-Cre, which shows a leaky expression in other tissues including the brain [75,76,77], and it is difficult to appreciate whether these phenotypes are caused by the adipose tissue, per se, or by defects in the other tissues. Our group reported that mice with mTORC1 impairment through adipocyte-specific deletion of Raptor by the Adipoq-Cre were refractory to the βAR-dependent induction of uncoupling protein 1 (UCP1) expression and expansion of beige/brite adipocytes in white adipose tissue (WAT) [41]. Mechanistically, PKA directly phosphorylates mTOR and RAPTOR on unique serine residues. Abrogation of the PKA site within RAPTOR disrupted βAR/mTORC1 activation of S6K1 while a phospho-mimetic RAPTOR augmented S6K1 activity [41]. This study suggested that PKA activation of mTORC1 is essential for β-adrenergic stimulation of adipose browning. Lee and colleagues further showed that RAPTOR/mTORC1 loss in adipocytes (Adipoq-Cre) causes progressive lipodystrophy and fatty liver disease [78]. Adipose Raptor knockout (Adipoq-Cre) mice are resistant to high-fat diet (HFD)-induced obesity, but this is due to failed adipose tissue expansion and not increased energy expenditure, which in turn results in severe hepatomegaly associated with hyperphagia and defective dietary lipid absorption [78]. Thus, mTORC1 activity in mature adipocytes is essential for maintaining normal adipose tissue growth and its selective loss in mature adipocytes leads to a progressive lipodystrophy disorder and systemic metabolic disease. In addition, Paolella and colleagues showed that mice with adipocyte Raptor deletion (Adipoq-Cre) failed to completely suppress lipolysis in the fed state and displayed prominent hypertriglyceridemia and hypercholesterolemia [79]. This study suggested that adipose tissue mTORC1 activity is necessary for appropriate suppression of lipolysis and for the maintenance of systemic lipid homeostasis. In another study, Zhang and colleagues reported that adipose-specific depletion of Raptor (Adipoq-Cre) increased prostaglandin (PG) production by cyclooxygenase-2 (COX-2) and promoted differentiation of progenitor cells to beige adipocytes [80]. This study identifies adipocyte-derived PGs as key regulators of white adipocyte browning and this was proposed to occur through an mTORC1-dependent mechanism.
mTORC2 signaling also plays an important role in adipose tissue function and homeostasis (Table 2). Early work by Cybulski and colleagues showed that mice with adipose-specific Rictor deletion by Fabp4/aP2-Cre have increased body size due to an increase in the size of non-adipose organs [81]. These mice have a disproportionately enlarged pancreas and are hyperinsulinemic, but they are glucose-tolerant and display elevated levels of insulin-like growth factor 1 (IGF1) and IGF1 binding protein 3 (IGFBP3) [81]. These findings indicate that mTORC2 in adipose tissue plays an unexpectedly central role in controlling whole-body growth. However, as noted earlier, these results must be interpreted with caution due to the use of the Fabp4/aP2-Cre driver, which can also alter mTORC2 activity in non-adipose tissues due to this leaky expression [75,76,77]. Tang and colleagues showed that mTORC2 functions in white adipose tissue (WAT) to control expression of the lipogenic transcription factor ChREBPβ (carbohydrate response element-binding protein β). Conditionally deleting Rictor in mature adipocytes (Adipoq-Cre) decreases ChREBPβ expression, which reduces DNL in WAT, and impairs hepatic insulin sensitivity [82]. Mechanistically, RICTOR/mTORC2 promotes ChREBPβ expression in part by controlling glucose uptake, but without impairing pan-AKT signaling [82]. Thus, mTORC2 functions in WAT as part of an extra-hepatic nutrient-sensing mechanism to control glucose homeostasis. Hung and colleagues showed that Rictor loss in the Myf5+ precursor lineage (Myf5-Cre) shifts BAT metabolism to a more oxidative and less lipogenic state and protects mice against obesity and metabolic disease [83]. Jung and colleagues showed that conditionally deleting Rictor in murine brown adipocytes (Ucp1-Cre) inhibits de novo lipid synthesis, promotes lipid catabolism and thermogenesis, and protects against diet-induced obesity and hepatic steatosis [84]. Deleting Rictor in brown adipocytes appears to drive lipid catabolism independently of AKT by promoting FoxO1 deacetylation, and this occurs in a pathway distinct from its positive role in anabolic lipid synthesis [84]. These findings suggest a new paradigm of mTORC2 function filling an important gap in our understanding of this less well-understood mTOR complex.
As a crucial arm of the energy balance in obesity, feeding behavior is subjected to short-term and long-term controls for the initiation and termination of meals and the status of energy storage [64,65]. The hypothalamus is a region in the brain that controls appetite and is essential for metabolic homeostasis. The GLP-1 receptor agonists (GLP-1RA) have emerged as the second-generation anti-obesity medications due to their metabolically beneficial effects to stimulate insulin secretion while suppressing food intake [85,86]. Studies by Burmeister and colleagues showed that mTORC1 signaling is also essential in mediating the weight loss effect of GLP-1RA action [87]. Pharmacological GLP-1R activation in the ventromedial hypothalamus (VMH) reduces food intake and body weight, and mTORC1 activity appears necessary for these effects of GLP-1RA [87]. In a recent study, in collaboration with our lab, Le and colleagues showed that GLP-1RA similarly stimulates mTORC1 activity via PKA phosphorylation of RAPTOR at Ser791, and this phosphorylation event is relevant to the weight loss effect of the therapeutic GLP-1RA liraglutide [62]. In addition, knock-in mice expressing Ser791Ala RAPTOR were partially resistant to GLP-1R agonist-induced weight loss. These results support the conclusion that PKA activation downstream of the GLP-1R leads to phosphorylation of RAPTOR at Ser791, which activates mTORC1 signaling in a non-canonical fashion to facilitate the weight-lowering effects of liraglutide. Further studies are needed to investigate whether the RAPTOR S791 phosphorylation is required for the effect of GLP-1RA on insulin secretion.
3.2. Role of mTOR Signaling in Fatty Liver Disease
Nonalcoholic fatty liver disease (NAFLD) is a spectrum of liver disease ranging from nonalcoholic fatty liver (NAFL) to nonalcoholic hepatic steatosis (NASH), the latter of which could advance to cirrhosis and hepatocellular carcinoma (HCC) (reviewed in [88]). NAFL is defined by the presence of fat accumulation in >5% of hepatocytes as assessed by histology or non-invasive imaging [89]. NASH is a more severe and progressive form of the disease and is histologically characterized by hepatocyte injury, inflammation, and fibrosis as observed in liver biopsy [90]. The disease progression from NAFL to NASH is involved in multiplex factors, including metabolic dysfunction, genetic variants, age, ethnicity, and diet [91]. In the US, the incidence of NAFLD as of 2017 was estimated at ~25% of the population and the prevalence of NASH was about 3–4% [92], and this number is growing. Of note, NASH is related to a number of different factors and is closely associated with an increased risk of cardiovascular disease, a leading cause of mortality in patients with the disease [93,94]. Despite its prevalence, no medication is currently approved to treat this condition [88].
After feeding, glucose taken up by the liver is converted into acetyl-CoA through a stepwise process of glycolysis, tricarboxylic acid (TCA) cycle and citrate lyase [95]. Hepatic de novo lipogenesis (DNL) is involved in the synthesis of fatty acids (FAs) from acetyl-CoA and their further processing into triacylglycerols (TAG). DNL is controlled by the rate-limiting enzyme acetyl-CoA carboxylase (ACC), which converts acetyl-CoA to malonyl-CoA, and fatty acid synthesis (FASN), which processes acetyl-CoA and malonyl-CoA into palmitate [95]. Palmitate will be further esterified to produce TAG. Of note, malonyl-CoA inhibits carnitine acyltransferase [96], the rate-limiting step in fatty acid oxidation, and it thus diverts metabolites to lipid synthesis. The expression of ACC and FASN are transcriptionally controlled by various transcriptional regulators including sterol response element-binding protein (SREBP), carbohydrate response element-binding protein (ChREBP), and nuclear receptors such as PPARγ, FXR, and LXR [95]. During fasting, hepatic glucose production is increased through the induction of both glycogenolysis and gluconeogenesis. Fasting also stimulates lipolysis in adipose tissue and thus the release of nonesterified FAs (NEFAs). NEFAs are further converted into ketone bodies in the liver via β-oxidation and ketogenesis [97].
mTOR signaling plays an essential role in hepatic lipid metabolism (reviewed in [97]). As the primary hormone that drives hepatic lipogenesis, insulin acts through PI3K-AKT signaling and depends on mTORC1. mTORC1 regulates the expression of a panel of lipogenic genes, including the master regulator SREBPs. The SREBPs belong to the basic helix–loop–helix leucine zipper transcription factor family, and consist of three members, SREBP1a, SREBP1c, and SREBP2. Despite the functional overlap among SREBPs, SREBP1c is primarily responsible for the lipogenic gene expression program in the liver [95,97]. Upon insulin stimulation, SREBPs are translocated from the endoplasmic reticulum (ER) to Golgi, followed by proteomic N-terminus cleavage and shuttle to the nuclei to drive the expression of genes that control cholesterol and FA synthesis [95,97].
mTORC1 controls the activation of SREBPs through both S6K1-dependent and S6K1-independent mechanisms (reviewed in [97]). The S6K1-dependent mechanism of SREBPs’ regulation by mTORC1 is not well understood; however, Lipin1 and CRTC2 (CREB regulated transcription coactivator 2) are two major regulators known to be involved in the S6K1-independent regulation of SREBPs [97]. Lipin1 can shuttle to nuclei in its dephosphorylated form and promote the binding of SREBPs with the nuclear matrix, thus preventing SREBPs from binding and activating the SRE-containing lipogenic genes [98]. Once phosphorylated by mTOR, Lipin1 is restricted in the cytoplasm and is unable to inhibit the SREBPs’ action in the nuclei [98]. CRTC2 can bind to COPII subunit Sec31A and thus disrupt the transport of SREBPs from the ER to Golgi [99]. Upon feeding, CRTC2 is phosphorylated and inactivated by mTOR, thereby attenuating the inhibition effect of CRTC2 on SREBPs’ maturation [99].
Autophagy plays a critical role in shuttling lipid droplets in the hepatocyte to the lysosome for hydrolysis, a process termed lipophagy [100]. mTORC1 controls lipophagy by inhibitory phosphorylation of the machinery of autophagy, including ULK1/2 (UNC-51-like autophagy activating kinase 1/2), ATG13 (autophagy-related 13), ATG14L (autophagy related 14), and the transcriptional regulators of lysosome biogenesis, including TFEB (transcription factor EB) and TFE3 (transcription factor binding to IGHM enhancer 3). Under fed conditions, TFEB is phosphorylated by mTOR and restricted in the cytoplasm by binding to 14-3-3 [101,102,103,104]. During fasting, TFEB is dephosphorylated and translocated to nuclei to promote the expression of autophagic and lysosomal genes [105,106,107]. TFEB also promotes lipid oxidation by upregulating the expression of PPARγ and PGC1α [108]. In addition, TFE3, another member of the MITF/TFE family that is phosphorylated by mTORC1, also regulates autophagy and lysosomal biogenesis gene expression during starvation [109,110].
Given its critical role in lipid metabolism, the role of mTORC1 in NAFLD is well appreciated but turns out to be complicated (Table 2). Deletion of Raptor in the liver suppresses DNL through the inhibition of the SREBP-1c-regulated lipogenic gene program and protects mice from hepatic steatosis [98]. However, mTORC1 activation through the hepatocyte deletion of TSC1 also protects the mice from liver steatosis [111,112]. And yet additional studies showed that mice lacking Raptor in the liver have increased steatosis [113] and liver injuries [114]. Thus, the role mTORC1 in liver steatosis is still not well understood, which might be due to the complexity of mTORC1 signaling in the context of disease progression [115]. In a recent study, Gosis and colleagues reported that selective inhibition of mTORC1, through the deletion of the RagC/D guanosine triphosphatase-activating protein folliculin (FLCN), protects against NAFLD and NASH [115]. This disease protection is mediated by the activation of TFE3, without affecting the signaling arms mediated by S6K1 and 4E-BP1 [115]. TFE3 inhibits lipogenesis by suppressing proteolytic processing and activation of SREBP-1c and by interacting with SREBP-1c on chromatin [115]. In another study, Uehara and colleagues sought to define the role of hepatic mTORC1 activity in mouse models of NASH and investigate the mTORC1-dependent mechanisms responsible for protection against liver damage in NASH [116]. Utilizing two rodent NASH-promoting diets, they demonstrated that hepatic mTORC1 activity was reduced in mice with NASH, whereas under conditions of insulin resistance and benign fatty liver, mTORC1 activity was elevated [116]. Mice with acute liver-specific loss of TSC1 have an improved profile of NASH and are protected against hepatic inflammation and fibrosis by constitutive mTORC1 activity. In another study, Kim and colleagues reported a mechanism where mTORC1-independent (‘free’) RAPTOR negatively regulates hepatic AKT activity and lipogenesis through the β-TrCP-mediated degradation of the AKT phosphatase, PHLPP2 [117]. Forced PHLPP2 expression ameliorates hepatic steatosis in diet-induced obese mice. These data suggest that the balance of free and mTORC1-associated RAPTOR governs hepatic lipid accumulation and uncovers the potentially therapeutic role of PHLPP2 activators in NAFLD. Taken together, these studies indicate that the role of mTORC1 in the pathophysiology of NAFLD and NASH needs to be interpreted with caution.
Studies also demonstrated that mTORC2 plays important roles in liver lipid homeostasis (Table 2). Hagiwara and colleagues reported that fed mice with liver-specific Rictor deletion displayed a loss in AKT Ser473 phosphorylation and reduced glucokinase and SREBP1c activity in the liver, leading to constitutive gluconeogenesis and impaired glycolysis and lipogenesis [118]. These liver-specific defects resulted in systemic hyperglycemia, hyperinsulinemia, and hypolipidemia. Expression of constitutively active AKT2 in mTORC2-deficient hepatocytes restored both glucose flux and lipogenesis, whereas glucokinase overexpression rescued glucose flux but not lipogenesis. Thus, mTORC2 regulates hepatic glucose and lipid metabolism via insulin-induced AKT signaling to control whole-body metabolic homeostasis. Yuan and colleagues reported that mice lacking Rictor in the liver (LrictorKO) failed to inhibit hepatic glucose output in response to insulin [119]. LrictorKO mice also resisted the development of hepatic steatosis on a high-fat diet and manifested lower levels of serum cholesterol, expression of SREBP-1c and SREBP-2, and genes of fatty acid and cholesterol biosynthesis. LrictorKO mice had defects in insulin-stimulated AKT Ser-473 and Thr-308 phosphorylation, leading to decreased phosphorylation of AKT substrates. LrictorKO mice also manifested defects in insulin-activated mTORC1 activity, evidenced by decreased S6 kinase and Lipin1 phosphorylation.
Dysregulation of mTOR signaling in the liver also results in cancer progression. Guri and colleagues reported that mice lacking Tsc1 and Pten specifically in the liver (termed L-dKO mice) exhibited concomitant activation of mTORC1 and mTORC2 signaling and displayed hepatosteatosis progressing to hepatocellular carcinoma (HCC) [120]. Longitudinal proteomic, lipidomics, and metabolomic analyses revealed that hepatic mTORC2 promotes de novo fatty acid and lipid synthesis, leading to steatosis and tumor development. In particular, mTORC2 stimulated sphingolipid (glucosylceramide) and glycerophospholipid (cardiolipin) synthesis. Inhibition of fatty acid or sphingolipid synthesis prevented tumor development, indicating a causal effect in tumorigenesis. Increased levels of cardiolipin were associated with tubular mitochondria and enhanced oxidative phosphorylation. Furthermore, increased lipogenesis correlated with elevated mTORC2 activity and HCC in human patients. Thus, mTORC2 promotes cancer via the formation of lipids essential for growth and energy production.
3.3. Role of mTOR Signaling in Cardiac Remodeling
The mTOR pathway regulates both physiological and pathological processes in the heart (reviewed in [121,122]). It is needed for embryonic cardiac development and for postnatal cardiac remodeling in conditions of pressure overload. Under unstressed conditions, mice with constitutive cardiac deletion of mTOR die in utero or during the perinatal period [123]. Mice with cardiac-specific deletion of mTOR in the adult stage showed a reduced life span, along with fatal dilated cardiomyopathy, molecular features of mitochondrial dysfunction, and increased apoptosis and autophagy [123].
mTORC1 is a critical regulator of cardiac hypertrophy and remodeling (Table 2). Cardiac hypertrophy is induced by mechanical stress, such as pressure or volume overload, and by neurohormonal factors, such as angiotensin II (Ang-II) and adrenergic stimulation [121]. mTOR signaling is activated by the stimuli that trigger cardiac hypertrophy and promote both the adaptive and maladaptive cardiac growth [121]. In cardiomyocytes, rapamycin inhibits the Ang-II-induced upregulation of S6K1 and suppresses the increases in S6K1 activation and protein synthesis in response to isoproterenol or phenylephrine treatment [124]. Thus, the G protein-coupled receptor (GPCR) signaling has a major impact on mTORC1 activation during hypertrophy development [121]. Mice with inducible cardiac deletion of RAPTOR in adults develop marked cardiac dysfunction in response to pressure overload induced by transverse aortic constriction (TAC) without the development of compensatory hypertrophy [125,126]. Constitutive mTORC1 activation promotes the transition from adaptive to maladaptive hypertrophy [121]. In mouse models of pressure or volume overload, pharmacological mTORC1 inhibition is protective and improves cardiac remodeling [127,128,129]. RAPTOR haploinsufficiency attenuates heart failure induced by pressure overload or Gαq overexpression [130]. This protective effect is mediated by 4E-BP1-independent mechanisms, such as the effect of mTORC1 on mitochondria and metabolism [130]. The heart undergoes structural and functional changes during aging [121]. Rapamycin administration has been reported to increase lifespan, improve cardiac function, and results in a reduced gene expression signature involving inflammation, hypertrophy, and contractile function [131,132].
The activity of mTORC1 can be modified by other pathways during cardiac stress conditions. Studies showed that p38 MAPK contributes to the maladaptive hypertrophy during stress conditions by enhancing mTORC1 activity. As described earlier, reactive oxygen species (ROS) can activate p38, which in turn reduces the level of REDD1 [51]. The decline in REDD1 leads to the dissociation of the TSC1/2 via the enhanced association of TSC2 with 14-3-3 family members [51]. In addition, p38γ/δ MAPKs can phosphorylate DEPTOR and promote its ubiquitination and degradation [52]. p38γ/δ MAPK knockout mice show reduced mTOC1 activity and reduced postnatal heart growth; however, they are protected from angiotensin II-induced hypertrophy [52]. Recent studies revealed that PKG signaling can negatively regulate mTORC1 signaling in pathological cardiac remodeling. As described earlier, PKG activation reduces cardiac hypertrophy in mice who underwent pressure overload [44]. Mechanistically, PKG activates TSC2 by direct phosphorylation at Ser1365 and 1366 and results in mTORC1 inhibition and enhanced autophagy [44]. Gain- or loss-of-function mutations at either of the two adjacent serine residues in TSC2 can bidirectionally control mTORC1 activity stimulated by growth factors or hemodynamic stress and consequently modulate cell growth and autophagy [44]. Homozygous knock-in mice that express a phosphorylation-silencing mutation in TSC2 develop worse heart disease and have higher mortality after sustained pressure overload of the heart, owing to mTORC1 hyperactivity that cannot be rescued by PKG1 stimulation. In addition, PKG activity is reduced in response to hypertrophic stimuli due to the oxidation at cystine 42 [45]. The PKG1α cysteine-42 redox state controls mTORC1 activation in pathological cardiac hypertrophy [46]. Mice with a redox-dead cysteine 42 to serine mutation show improved cardiac hypertrophy and dysfunction in response to pressure overload due to the TSC2 activation and mTORC1 inhibition [46]. Furthermore, soluble guanylyl cyclase-1 (sGC-1) activation and phosphodiesterase (PDE) 9 inhibition but not PDE5 inhibition can offset the growth-hormone-stimulated mTORC1 activity regardless of PKG-1α redox state and phosphorylation of TSC2 S1365 [46].
Myocardial ischemia or energy stress leads to mTORC1 inhibition and protective adaption such as autophagy, which in turn limits myocardial infarction [121]. RHEB is inhibited in response to ischemia or glucose deprivation, leading to mTORC1 inhibition [133]. Forced expression of RHEB induces cell death and ER stress, inhibits autophagy, and increases infarct size. In addition, GSK3β inhibition also leads to mTORC1 activation and thus restricts autophagy, resulting in increased myocardial ischemic injury [134]. Restoration of autophagy rescued the deleterious effect of RHEB upregulation or GSK3β inhibition. Of note, mTORC1 shows differential roles during ischemia and reperfusion [121]. mTORC1 inhibition by rapamycin before ischemia reduces the ischemia/reperfusion (I/R) injury while it does not confer any protection in the reperfusion phase [134,135]. In fact, endogenous mTORC1 is activated during the reperfusion phase and limits I/R injury [134,136]. mTORC1 activation is detrimental during chronic ischemia. In rat models undergoing myocardial infarction (MI), mTORC1 inhibition by everolimus reduced adverse remodeling and infarct size and promoted autophagy [129]. The mTORC1 substrate S6K1 is activated in the heart during MI in mice and the pharmacological inhibition of S6K1 attenuated myocardial remodeling [137].
Unlike the adverse effect of mTORC1, studies have shown that mTORC2 activation promotes compensatory cardiac growth and limits maladaptive hypertrophy (Table 2). Constitutive deletion of Rictor in cardiomyocytes induces cardiac dysfunction and dilation in response to pressure overload [138]. Rictor deletion in adults also leads to cardiac dysfunction in mice undergoing TAC [138]. In addition, activation of mTORC2 protects against age-induced cardiac abnormalities. Systemic heterozygous deletion of Rictor decreases lifespan in male mice, suggesting a protective role of mTORC2 against aging [139]. Furthermore, mTORC2 activation also has a protective role in chronic ischemia. In mice undergoing MI, loss of RICTOR increases myocardial damage and remodeling [140]. Of note, PRAS40 inhibition improves cardiac function and post-infarction remodeling through the mTORC2-induced AKT activation [140]. Thus, induction of a shift from mTORC1 to mTORC2 activation is protective for heart ischemic disease [121].

Table 2.
Metabolic and cardiac phenotypes of Raptor and Rictor knockout mice.
Table 2.
Metabolic and cardiac phenotypes of Raptor and Rictor knockout mice.
| No. | Genes | Tissues | Models | Phenotypes | Ref. |
|---|---|---|---|---|---|
| Adipose tissue | |||||
| 1 | Raptor | Adipocyte Fabp4/aP2-Cre | HFD | Substantially less adipose tissue, protected against diet-induced obesity, hypercholesterolemia, and insulin resistance | [74] |
| 2 | Raptor | Adipocyte Adipoq-Cre | HFD | Progressive lipodystrophy with hepatic steatosis and insulin intolerance, resistant to diet-induced obesity, severe hepatomegaly | [75] |
| 3 | Raptor | Adipocyte Adipoq-Cre | HFD | Fails to completely suppress lipolysis in the fed state and displays prominent hypertriglyceridemia and hypercholesterolemia | [79] |
| 4 | Raptor | Adipocyte Adipoq-Cre | Cold, βAR agonist | Refractory to the β-adrenergic stimulation of UCP1 expression and expansion of beige/brite adipocytes in WAT | [41] |
| 5 | Raptor | Adipocyte Adipoq-Cre | Cold, HFD | Increased prostaglandin (PG) production by COX-2 and promotes differentiation of progenitor cells to beige adipocytes | [80] |
| 6 | Raptor | Global S791A knock-in | GLP-1R agonist | Partially resistant to GLP-1R agonist liraglutide-induced weight loss, lesser reduction in fat mass, lower energy expenditure | [62] |
| 7 | Rictor | Adipocyte Fabp4/aP2-Cre | HFD | Increased body (non-adipose organ) size, hyperinsulinemia but glucose-tolerant, elevated levels of IGF1 | [81] |
| 8 | Rictor | Adipocyte Adipoq-Cre | HFD | Normal body growth, insulin intolerance, impaired adipose insulin signaling results in reduced glucose uptake and de novo lipogenesis | [82] |
| 9 | Rictor | Brown adipocyte Myf5-Cre | HFD | Decreased fat mass and adipocyte size, decreased lipogenesis but elevated mitochondrial activity in brown fat, protected against obesity and metabolic disease | [83] |
| 10 | Rictor | Brown adipocyte Ucp1-Cre | Cold, βAR agonist, HFD | Increased cold tolerance, inhibits de novo lipid synthesis, promotes lipid catabolism and thermogenesis, and protects against diet-induced obesity | [84] |
| Liver | |||||
| 11 | Raptor | Hepatocyte Albumin-Cre | Western diet | Gained less weight, reduced fasted liver size, suppresses hepatic de novo lipogenesis, protects mice from hepatic steatosis and hypercholesterolemia | [98] |
| 12 | Raptor | Hepatocyte AAV-TBG-Cre | Fasting and refeeding | Smaller liver with increased steatosis, higher hepatic TAG and decreased TAG secretion under fasting conditions | [113] |
| 13 | Raptor | Hepatocyte Albumin-Cre | HFD, hepatic carcinogen | Increased liver injuries, aberrant regeneration, enhanced fibrosis, inflammation, and hepatocarcinogenesis | [114] |
| 14 | Rictor | Hepatocyte Albumin-Cre | HFD | Smaller liver with lower glycogen and TG content, hepatic insulin resistance, dysregulated hepatic gluconeogenesis, glycolysis and de novo lipogenesis | [118] |
| 15 | Rictor | Hepatocyte Albumin-Cre | HFD | Failed to inhibit hepatic glucose output, glucose intolerance and insulin resistance, resistant to hepatic steatosis on a high-fat diet | [119] |
| Heart | |||||
| 16 | Raptor | Cardiomyocyte MHC-Cre | TAC | Acute cardiac dysfunction in response to overload, metabolic switch from fatty acid to glucose oxidation, abnormal mitochondria, increased apoptosis, and autophagy | [126] |
| 17 | Raptor | Cardiomyocyte MHC-Cre heterozygote | TAC | Attenuates heart failure induced by pressure overload or Gαq overexpression | [130] |
| 18 | Rictor | Cardiomyocyte MHC-Cre | TAC | Accelerates cardiac dysfunction after aortic constriction, reduces PKC protein levels, does not affect hypertrophy, fibrosis, or metabolic gene expression | [138] |
| 19 | Rictor | Cardiomyocyte AAV9 shRNA | MI | Increases myocardial damage and remodeling | [140] |
Abbreviations: TBG: thyroxine binding globulin; TAC: transverse aortic constriction; MHC: α-myosin heavy chain; MI: myocardial infarction.
4. Conclusions
In this review, we summarize the recent findings demonstrating the regulation of the mTOR signaling network by cAMP and cGMP, as well as by their associated cyclic nucleotide-dependent protein kinases PKA and PKG. Although these signaling crosstalks could be mediated by various mechanisms as reported in previous studies, further effort is needed to elucidate the role of these signaling events in physiological and disease conditions.
Author Contributions
Writing—original draft preparation, F.S.; writing—review and editing, F.S. and S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. F.S. is supported by the American Heart Association Career Development Award 23CDA1048341.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created.
Acknowledgments
The figure was created with BioRender.com.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| mTOR | mechanistic target of rapamycin; |
| RAPTOR | regulatory-associated protein of mTOR; |
| RICTOR | rapamycin-insensitive companion of mTOR; |
| DEPTOR | DEP domain-containing mTOR-interacting protein; |
| mLST8 | mammalian lethal with SEC13 protein 8; |
| PRAS40 | proline-rich AKT substrate of 40 kDa; |
| mSIN1 | mammalian stress-activated protein kinase-interaction protein 1; |
| PROTOR | protein observed with RICTOR; |
| TSC1/2 | tuberous sclerosis complex 1/2; |
| RHEB | Ras homolog mTORC1 binding; |
| RAGs | Ras-related GTPases; |
| IRS | insulin receptor substrate; |
| PI3K | phosphoinositide 3-kinase; |
| PIP3 | phosphatidylinositol 3,4,5-triphosphate; |
| PTEN | phosphatase and tensin homolog; |
| PDK1 | phosphoinositide-dependent protein kinase 1; |
| GRB10 | growth factor receptor bound protein 10; |
| AKT; | protein kinase B; |
| ERK | extracellular signal-regulated kinase; |
| RSK | ribosomal S6 kinase 1; |
| TNF | tumor necrosis factor; |
| IKKβ | inhibitor of nuclear factor kappa-B kinase subunit beta; |
| AMPK | AMP-activated protein kinase; |
| WNT | wingless and lnt-1; |
| GSKβ | glycogen synthase kinase 3β; |
| REDD1 | regulated in development and DNA damage responses 1; |
| p38 MAPK | p38 mitogen-activated protein kinases; |
| MK2 | MAPK activated protein kinase 2; |
| ROS | reactive oxygen species; |
| NO | nitric oxide; |
| ANP | atrial natriuretic peptide; |
| BNP | B-type natriuretic peptide; |
| GPCR | G protein-coupled receptor; |
| GC | guanylyl cyclase; |
| sGC | soluble guanylyl cyclase; |
| cAMP | cyclic adenosine monophosphate; |
| cGMP | cyclic guanosine monophosphate; |
| PKA | cAMP-dependent protein kinase; |
| PKG | cGMP-dependent protein kinase; |
| SIK3 | salt-inducible kinase 3; |
| FKBP12 | FK506-binding protein of 12 kDa; |
| PIP2 | phosphatidylinositol 4,5-bisphosphate |
| PKA | protein kinase A; |
| PKG | protein kinase G; |
| GPCRs | G protein-coupled receptors; |
| EPAC | exchange protein directly activated by cAMP (EPAC); |
| LKB1 | liver kinase B1; |
| PDE | phosphodiesterase; |
| NPs | natriuretic peptides; |
| TBG | thyroxine binding globulin; |
| TAC | transverse aortic constriction; |
| MHC | α-myosin heavy chain; |
| MI | myocardial infarction; |
| IBMX | 3-Isobutyl-1-methylxanthine; |
| MEFs | Mouse embryonic fibroblast; |
| PTH/PTHrP | parathyroid hor-mone/PTH-related peptide; |
| GLP-1 | Glucagon-like peptide-1; |
| GLP-1RA | GLP-1 receptor agonist; |
| KATP channels | ATP-sensitive K+ channels; |
| TSH | thyroid-stimulating hormone; |
| hCG | human chorionic gonadotropin; |
| β-AR | β-adrenergic receptor; |
| NRVMs | neonatal rat ventricular myocytes; |
| ER | endoplasmic reticulum; |
| GPCRs | G protein-coupled receptors. |
| DNL | de novo lipogenesis; |
| NAFLD | nonalcoholic fatty liver disease; |
| NAFL | nonalcoholic fatty liver; |
| NASH | nonalcoholic hepatic steatosis; |
| HCC | hepatocellular carcinoma; |
| UCP1 | uncoupling protein 1; |
| WAT | white adipose tissue. |
References
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin–receptor complex. Nature 1994, 369, 756–758. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994, 78, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Sabers, C.J.; Martin, M.M.; Brunn, G.J.; Williams, J.M.; Dumont, F.J.; Wiederrecht, G.J.; Abraham, R.T. Isolation of a Protein Target of the FKBP12-Rapamycin Complex in Mammalian Cells. J. Biol. Chem. 1995, 270, 815–822. [Google Scholar] [CrossRef]
- Bierer, B.E.; Mattila, P.S.; Standaert, R.F.; Herzenberg, L.A.; Burakoff, S.J.; Crabtree, G.R.; Schreiber, S.L. Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin. Proc. Natl. Acad. Sci. USA 1990, 87, 9231–9235. [Google Scholar] [CrossRef]
- Chung, J.; Kuo, C.J.; Crabtree, G.R.; Blenis, J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 1992, 69, 1227–1236. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Ali, S.M.; Sengupta, S.; Sheen, J.H.; Hsu, P.P.; Bagley, A.F.; Markhard, A.L.; Sabatini, D.M. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 2006, 22, 159–168. [Google Scholar] [CrossRef]
- Lamming, D.W.; Ye, L.; Katajisto, P.; Goncalves, M.D.; Saitoh, M.; Stevens, D.; Davis, J.G.; Salmon, A.B.; Richardson, A.; Ahima, R.S.; et al. Rapamycin-Induced Insulin Resistance Is Mediated by mTORC2 Loss and Uncoupled from Longevity. Science 2012, 335, 1638–1643. [Google Scholar] [CrossRef]
- Inoki, K.; Li, Y.; Xu, T.; Guan, K.-L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003, 17, 1829–1834. [Google Scholar] [CrossRef]
- Long, X.; Lin, Y.; Ortiz-Vega, S.; Yonezawa, K.; Avruch, J. Rheb Binds and Regulates the mTOR Kinase. Curr. Biol. CB 2005, 15, 702–713. [Google Scholar] [CrossRef]
- Kim, E.; Goraksha-Hicks, P.; Li, L.; Neufeld, T.P.; Guan, K.-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008, 10, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- van Slegtenhorst, M.; Nellist, M.; Nagelkerken, B.; Cheadle, J.; Snell, R.; van den Ouweland, A.; Reuser, A.; Sampson, J.; Halley, D.; van der Sluijs, P. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum. Mol. Genet. 1998, 7, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Inoki, K.; Zhu, T.; Guan, K.-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003, 115, 577–590. [Google Scholar] [CrossRef]
- Huang, J.; Manning, B.D. The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem. J. 2008, 412, 179–190. [Google Scholar] [CrossRef]
- Tee, A.R.; Manning, B.D.; Roux, P.P.; Cantley, L.C.; Blenis, J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. CB 2003, 13, 1259–1268. [Google Scholar] [CrossRef]
- Gan, X.; Wang, J.; Su, B.; Wu, D. Evidence for direct activation of mTORC2 kinase activity by phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 2011, 286, 10998–11002. [Google Scholar] [CrossRef]
- Yuan, H.-X.; Guan, K.-L. The SIN1-PH Domain Connects mTORC2 to PI3K. Cancer Discov. 2015, 5, 1127–1129. [Google Scholar] [CrossRef]
- Liu, P.; Gan, W.; Chin, Y.R.; Ogura, K.; Guo, J.; Zhang, J.; Wang, B.; Blenis, J.; Cantley, L.C.; Toker, A.; et al. PtdIns(3,4,5)P3-Dependent Activation of the mTORC2 Kinase Complex. Cancer Discov. 2015, 5, 1194–1209. [Google Scholar] [CrossRef]
- Maehama, T.; Dixon, J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998, 273, 13375–13378. [Google Scholar] [CrossRef]
- Stambolic, V.; Suzuki, A.; de la Pompa, J.L.; Brothers, G.M.; Mirtsos, C.; Sasaki, T.; Ruland, J.; Penninger, J.M.; Siderovski, D.P.; Mak, T.W. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 1998, 95, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Calebiro, D.; Koszegi, Z.; Lanoiselée, Y.; Miljus, T.; O’Brien, S. G protein-coupled receptor-G protein interactions: A single-molecule perspective. Physiol. Rev. 2021, 101, 857–906. [Google Scholar] [CrossRef] [PubMed]
- Monfar, M.; Lemon, K.P.; Grammer, T.C.; Cheatham, L.; Chung, J.; Vlahos, C.J.; Blenis, J. Activation of pp70/85 S6 kinases in interleukin-2-responsive lymphoid cells is mediated by phosphatidylinositol 3-kinase and inhibited by cyclic AMP. Mol. Cell. Biol. 1995, 15, 326–337. [Google Scholar] [CrossRef]
- Scott, P.H.; Lawrence, J.C., Jr. Attenuation of mammalian target of rapamycin activity by increased cAMP in 3T3-L1 adipocytes. J. Biol. Chem. 1998, 273, 34496–34501. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Ponuwei, G.A.; Moore, C.E.; Willars, G.B.; Tee, A.R.; Herbert, T.P. cAMP inhibits mammalian target of rapamycin complex-1 and -2 (mTORC1 and 2) by promoting complex dissociation and inhibiting mTOR kinase activity. Cell. Signal. 2011, 23, 1927–1935. [Google Scholar] [CrossRef]
- Rocha, A.; Paternot, S.; Coulonval, K.; Dumont, J.E.; Soares, P.; Roger, P.P. Cyclic AMP Inhibits the Proliferation of Thyroid Carcinoma Cell Lines through Regulation of CDK4 Phosphorylation. Mol. Biol. Cell 2008, 19, 4814–4825. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jee, K.; Kim, D.; Koh, H.; Chung, J. Cyclic AMP inhibits Akt activity by blocking the membrane localization of PDK1. J. Biol. Chem. 2001, 276, 12864–12870. [Google Scholar] [CrossRef]
- Mothe-Satney, I.; Gautier, N.; Hinault, C.; Lawrence, J.C.; Van Obberghen, E. In rat hepatocytes glucagon increases mammalian target of rapamycin phosphorylation on serine 2448 but antagonizes the phosphorylation of its downstream targets induced by insulin and amino acids. J. Biol. Chem. 2004, 279, 42628–42637. [Google Scholar] [CrossRef]
- Kimball, S.R.; Siegfried, B.A.; Jefferson, L.S. Glucagon represses signaling through the mammalian target of rapamycin in rat liver by activating AMP-activated protein kinase. J. Biol. Chem. 2004, 279, 54103–54109. [Google Scholar] [CrossRef]
- Baum, J.; Kimball, S.R.; Jefferson, L.S. Glucagon acts in a dominant manner to repress insulin-induced mammalian target of rapamycin complex 1 signaling in perfused rat liver. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E410–E415. [Google Scholar] [CrossRef]
- Csukasi, F.; Duran, I.; Barad, M.; Barta, T.; Gudernova, I.; Trantirek, L.; Martin, J.H.; Kuo, C.Y.; Woods, J.; Lee, H.; et al. The PTH/PTHrP-SIK3 pathway affects skeletogenesis through altered mTOR signaling. Sci. Transl. Med. 2018, 10, eaat9356. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Bultot, L.; Goransson, O. The Salt-Inducible Kinases: Emerging Metabolic Regulators. Trends Endocrinol. Metab. TEM 2018, 29, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Wein, M.N.; Foretz, M.; Fisher, D.E.; Xavier, R.J.; Kronenberg, H.M. Salt-Inducible Kinases: Physiology, Regulation by cAMP, and Therapeutic Potential. Trends Endocrinol. Metab. TEM 2018, 29, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; de Fatima Silva, F.; Liu, D.; Patel, H.U.; Xu, J.; Zhang, W.; Türk, C.; Krüger, M.; Collins, S. Salt-inducible kinase inhibition promotes the adipocyte thermogenic program and adipose tissue browning. Mol. Metab. 2023, 74, 101753. [Google Scholar] [CrossRef]
- Kwon, G.; Marshall, C.A.; Pappan, K.L.; Remedi, M.S.; McDaniel, M.L. Signaling Elements Involved in the Metabolic Regulation of mTOR by Nutrients, Incretins, and Growth Factors in Islets. Diabetes 2004, 53 (Suppl. S3), S225–S232. [Google Scholar] [CrossRef]
- Brennesvik, E.O.; Ktori, C.; Ruzzin, J.; Jebens, E.; Shepherd, P.R.; Jensen, J. Adrenaline potentiates insulin-stimulated PKB activation via cAMP and Epac: Implications for cross talk between insulin and adrenaline. Cell. Signal. 2005, 17, 1551–1559. [Google Scholar] [CrossRef]
- Moore, C.E.; Xie, J.; Gomez, E.; Herbert, T.P. Identification of cAMP-Dependent Kinase as a Third in Vivo Ribosomal Protein S6 Kinase in Pancreatic β-Cells. J. Mol. Biol. 2009, 389, 480–494. [Google Scholar] [CrossRef]
- Blancquaert, S.; Wang, L.; Paternot, S.; Coulonval, K.; Dumont, J.E.; Harris, T.E.; Roger, P.P. cAMP-dependent activation of mammalian target of rapamycin (mTOR) in thyroid cells. Implication in mitogenesis and activation of CDK4. Mol. Endocrinol. 2010, 24, 1453–1468. [Google Scholar] [CrossRef]
- Palaniappan, M.; Menon, K.M. Human chorionic gonadotropin stimulates theca-interstitial cell proliferation and cell cycle regulatory proteins by a cAMP-dependent activation of AKT/mTORC1 signaling pathway. Mol. Endocrinol. 2010, 24, 1782–1793. [Google Scholar] [CrossRef]
- Kim, H.W.; Ha, S.H.; Lee, M.N.; Huston, E.; Kim, H.; Jang, S.K.; Suh, P.-G.; Houslay, M.D.; Ryu, S.H. Cyclic AMP Controls mTOR through Regulation of the Dynamic Interaction between Rheb and Phosphodiesterase 4D. Mol. Cell. Biol. 2010, 30, 5406–5420. [Google Scholar] [CrossRef]
- Liu, D.; Bordicchia, M.; Zhang, C.; Fang, H.; Wei, W.; Li, J.L.; Guilherme, A.; Guntur, K.; Czech, M.P.; Collins, S. Activation of mTORC1 is essential for beta-adrenergic stimulation of adipose browning. J. Clin. Investig. 2016, 126, 1704–1716. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Ceddia, R.P.; Collins, S. Cardiac natriuretic peptides promote adipose ‘browning’ through mTOR complex-1. Mol. Metab. 2018, 9, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Bordicchia, M.; Liu, D.; Amri, E.Z.; Ailhaud, G.; Dessi-Fulgheri, P.; Zhang, C.; Takahashi, N.; Sarzani, R.; Collins, S. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J. Clin. Investig. 2012, 122, 1022–1036. [Google Scholar] [CrossRef]
- Ranek, M.J.; Kokkonen-Simon, K.M.; Chen, A.; Dunkerly-Eyring, B.L.; Vera, M.P.; Oeing, C.U.; Patel, C.H.; Nakamura, T.; Zhu, G.; Bedja, D.; et al. PKG1-modified TSC2 regulates mTORC1 activity to counter adverse cardiac stress. Nature 2019, 566, 264–269. [Google Scholar] [CrossRef]
- Burgoyne, J.R.; Madhani, M.; Cuello, F.; Charles, R.L.; Brennan, J.P.; Schröder, E.; Browning, D.D.; Eaton, P. Cysteine redox sensor in PKGIa enables oxidant-induced activation. Science 2007, 317, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Oeing, C.U.; Nakamura, T.; Pan, S.; Mishra, S.; Dunkerly-Eyring, B.L.; Kokkonen-Simon, K.M.; Lin, B.L.; Chen, A.; Zhu, G.; Bedja, D.; et al. PKG1α Cysteine-42 Redox State Controls mTORC1 Activation in Pathological Cardiac Hypertrophy. Circ. Res. 2020, 127, 522–533. [Google Scholar] [CrossRef]
- Zarubin, T.; Han, J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005, 15, 11–18. [Google Scholar] [CrossRef]
- Canovas, B.; Nebreda, A.R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 346–366. [Google Scholar] [CrossRef]
- Li, Y.; Inoki, K.; Vacratsis, P.; Guan, K.L. The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14-3-3. J. Biol. Chem. 2003, 278, 13663–13671. [Google Scholar] [CrossRef]
- Cully, M.; Genevet, A.; Warne, P.H.; Treins, C.; Liu, T.; Bastien, J.; Baum, B.; Tapon, N.; Leevers, S.J.; Downward, J. A Role for p38 Stress-Activated Protein Kinase in Regulation of Cell Growth via TORC1. Mol. Cell. Biol. 2009, 30, 481–495. [Google Scholar] [CrossRef]
- Hernández, G.; Lal, H.; Fidalgo, M.; Guerrero, A.; Zalvide, J.; Force, T.; Pombo, C.M. A novel cardioprotective p38-MAPK/mTOR pathway. Exp. Cell Res. 2011, 317, 2938–2949. [Google Scholar] [CrossRef] [PubMed]
- González-Terán, B.; López, J.A.; Rodríguez, E.; Leiva, L.; Martínez-Martínez, S.; Bernal, J.A.; Jiménez-Borreguero, L.J.; Redondo, J.M.; Vazquez, J.; Sabio, G. p38γ and δ promote heart hypertrophy by targeting the mTOR-inhibitory protein DEPTOR for degradation. Nat. Commun. 2016, 7, 10477. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Daniel, K.W.; Robidoux, J.; Puigserver, P.; Medvedev, A.V.; Bai, X.; Floering, L.M.; Spiegelman, B.M.; Collins, S. p38 Mitogen-Activated Protein Kinase Is the Central Regulator of Cyclic AMP-Dependent Transcription of the Brown Fat Uncoupling Protein 1 Gene. Mol. Cell. Biol. 2004, 24, 3057–3067. [Google Scholar] [CrossRef] [PubMed]
- Hattori, K.; Naguro, I.; Okabe, K.; Funatsu, T.; Furutani, S.; Takeda, K.; Ichijo, H. ASK1 signalling regulates brown and beige adipocyte function. Nat. Commun. 2016, 7, 11158. [Google Scholar] [CrossRef] [PubMed]
- Komalavilas, P.; Shah, P.K.; Jo, H.; Lincoln, T.M. Activation of mitogen-activated protein kinase pathways by cyclic GMP and cyclic GMP-dependent protein kinase in contractile vascular smooth muscle cells. J. Biol. Chem. 1999, 274, 34301–34309. [Google Scholar] [CrossRef]
- Doronzo, G.; Viretto, M.; Russo, I.; Mattiello, L.; Di Martino, L.; Cavalot, F.; Anfossi, G.; Trovati, M. Nitric oxide activates PI3-K and MAPK signalling pathways in human and rat vascular smooth muscle cells: Influence of insulin resistance and oxidative stress. Atherosclerosis 2011, 216, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Inoki, K.; Ouyang, H.; Zhu, T.; Lindvall, C.; Wang, Y.; Zhang, X.; Yang, Q.; Bennett, C.N.; Harada, Y.; Stankunas, K.; et al. TSC2 Integrates Wnt and Energy Signals via a Coordinated Phosphorylation by AMPK and GSK3 to Regulate Cell Growth. Cell 2006, 126, 955–968. [Google Scholar] [CrossRef]
- Fang, X.; Yu, S.X.; Lu, Y.; Bast, R.C., Jr.; Woodgett, J.R.; Mills, G.B. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc. Natl. Acad. Sci. USA 2000, 97, 11960–11965. [Google Scholar] [CrossRef]
- Chen, C.H.; Shaikenov, T.; Peterson, T.R.; Aimbetov, R.; Bissenbaev, A.K.; Lee, S.W.; Wu, J.; Lin, H.K.; Sarbassov, D.D. ER Stress Inhibits mTORC2 and Akt Signaling through GSK-3β–Mediated Phosphorylation of Rictor. Sci. Signal. 2011, 4, ra10. [Google Scholar] [CrossRef]
- Koo, J.; Wu, X.; Mao, Z.; Khuri, F.R.; Sun, S.-Y. Rictor Undergoes Glycogen Synthase Kinase 3 (GSK3)-dependent, FBXW7-mediated Ubiquitination and Proteasomal Degradation. J. Biol. Chem. 2015, 290, 14120–14129. [Google Scholar] [CrossRef]
- Urbanska, M.; Gozdz, A.; Macias, M.; Cymerman, I.A.; Liszewska, E.; Kondratiuk, I.; Devijver, H.; Lechat, B.; Van Leuven, F.; Jaworski, J. GSK3β Controls mTOR and Prosurvival Signaling in Neurons. Mol. Neurobiol. 2017, 55, 6050–6062. [Google Scholar] [CrossRef]
- Le, T.D.V.; Liu, D.; Ellis, B.J.; Collins, S.; Ayala, J.E. Glucagon-Like Peptide-1 Receptor Activation Stimulates PKA-Mediated Phosphorylation of Raptor and this Contributes to the Weight Loss Effects of Liraglutide. bioRxiv 2022. [Google Scholar] [CrossRef]
- Garvey, W.T.; Mechanick, J.I.; Brett, E.M.; Garber, A.J.; Hurley, D.L.; Jastreboff, A.M.; Nadolsky, K.; Pessah-Pollack, R.; Plodkowski, R. American Association of Clinical Endocrinologists and American College of Endocrinology Comprehensive Clinical Practice Guidelines for Medical Care of Patients with Obesity. Endocr. Pract. 2016, 22 (Suppl. S3), 1–203. [Google Scholar] [CrossRef] [PubMed]
- Spiegelman, B.M.; Flier, J.S. Obesity and the regulation of energy balance. Cell 2001, 104, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Flier, J.S. Obesity wars: Molecular progress confronts an expanding epidemic. Cell 2004, 116, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.L. A comprehensive definition for metabolic syndrome. Dis. Model. Mech. 2009, 2, 231–237. [Google Scholar] [CrossRef]
- Harrington, L.S.; Findlay, G.M.; Gray, A.; Tolkacheva, T.; Wigfield, S.; Rebholz, H.; Barnett, J.; Leslie, N.R.; Cheng, S.; Shepherd, P.R.; et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 2004, 166, 213–223. [Google Scholar] [CrossRef]
- Shah, O.J.; Wang, Z.-Y.; Hunter, T. Inappropriate Activation of the TSC/Rheb/mTOR/S6K Cassette Induces IRS1/2 Depletion, Insulin Resistance, and Cell Survival Deficiencies. Curr. Biol. CB 2004, 14, 1650–1656. [Google Scholar] [CrossRef]
- Um, S.H.; Frigerio, F.; Watanabe, M.; Picard, F.; Joaquin, M.; Sticker, M.; Fumagalli, S.; Allegrini, P.R.; Kozma, S.C.; Auwerx, J.; et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 2004, 431, 200–205. [Google Scholar] [CrossRef]
- Hsu, P.P.; Kang, S.A.; Rameseder, J.; Zhang, Y.; Ottina, K.; Lim, D.; Peterson, T.R.; Choi, Y.; Gray, N.S.; Yaffe, M.B.; et al. The mTOR-Regulated Phosphoproteome Reveals a Mechanism of mTORC1-Mediated Inhibition of Growth Factor Signaling. Science 2011, 332, 1317–1322. [Google Scholar] [CrossRef]
- Yu, Y.; Yoon, S.-O.; Poulogiannis, G.; Yang, Q.; Max, X.; Villén, J.; Kubica, N.; Hoffman, G.R.; Cantley, L.C.; Gygi, S.P.; et al. Phosphoproteomic Analysis Identifies Grb10 as an mTORC1 Substrate That Negatively Regulates Insulin Signaling. Science 2011, 332, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Wick, K.R.; Werner, E.D.; Langlais, P.; Ramos, F.J.; Dong, L.Q.; Shoelson, S.E.; Liu, F. Grb10 inhibits insulin-stimulated insulin receptor substrate (IRS)-phosphatidylinositol 3-kinase/Akt signaling pathway by disrupting the association of IRS-1/IRS-2 with the insulin receptor. J. Biol. Chem. 2003, 278, 8460–8467. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Bai, J.; He, S.; Villarreal, R.; Hu, D.; Zhang, C.; Yang, X.; Liang, H.; Slaga, T.J.; Yu, Y.; et al. Grb10 promotes lipolysis and thermogenesis by phosphorylation-dependent feedback inhibition of mTORC1. Cell Metab. 2014, 19, 967–980. [Google Scholar] [CrossRef] [PubMed]
- Polak, P.; Cybulski, N.; Feige, J.N.; Auwerx, J.; Rüegg, M.A.; Hall, M.N. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008, 8, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Russell, S.J.; Ussar, S.; Boucher, J.; Vernochet, C.; Mori, M.A.; Smyth, G.; Rourk, M.; Cederquist, C.; Rosen, E.D.; et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 2013, 62, 864–874. [Google Scholar] [CrossRef]
- Mullican, S.E.; Tomaru, T.; Gaddis, C.A.; Peed, L.C.; Sundaram, A.; Lazar, M.A. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol. Endocrinol. 2013, 27, 127–134. [Google Scholar] [CrossRef]
- Jeffery, E.; Berry, R.; Church, C.D.; Yu, S.; Shook, B.A.; Horsley, V.; Rosen, E.D.; Rodeheffer, M.S. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte 2014, 3, 206–211. [Google Scholar] [CrossRef]
- Lee, P.L.; Tang, Y.; Li, H.; Guertin, D.A. Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Mol. Metab. 2016, 5, 422–432. [Google Scholar] [CrossRef]
- Paolella, L.M.; Mukherjee, S.; Tran, C.M.; Bellaver, B.; Hugo, M.; Luongo, T.S.; Shewale, S.V.; Lu, W.; Chellappa, K.; Baur, J.A. mTORC1 restrains adipocyte lipolysis to prevent systemic hyperlipidemia. Mol. Metab. 2020, 32, 136–147. [Google Scholar] [CrossRef]
- Zhang, X.; Luo, Y.; Wang, C.; Ding, X.; Yang, X.; Wu, D.; Silva, F.; Yang, Z.; Zhou, Q.; Wang, L.; et al. Adipose mTORC1 Suppresses Prostaglandin Signaling and Beige Adipogenesis via the CRTC2-COX-2 Pathway. Cell Rep. 2018, 24, 3180–3193. [Google Scholar] [CrossRef]
- Cybulski, N.; Polak, P.; Auwerx, J.; Rüegg, M.A.; Hall, M.N. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc. Natl. Acad. Sci. USA 2009, 106, 9902–9907. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wallace, M.; Sanchez-Gurmaches, J.; Hsiao, W.Y.; Li, H.; Lee, P.L.; Vernia, S.; Metallo, C.M.; Guertin, D.A. Adipose tissue mTORC2 regulates ChREBP-driven de novo lipogenesis and hepatic glucose metabolism. Nat. Commun. 2016, 7, 11365. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.M.; Calejman, C.M.; Sanchez-Gurmaches, J.; Li, H.; Clish, C.B.; Hettmer, S.; Wagers, A.J.; Guertin, D.A. Rictor/mTORC2 loss in the Myf5 lineage reprograms brown fat metabolism and protects mice against obesity and metabolic disease. Cell Rep. 2014, 8, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.M.; Hung, C.M.; Hildebrand, S.R.; Sanchez-Gurmaches, J.; Martinez-Pastor, B.; Gengatharan, J.M.; Wallace, M.; Mukhopadhyay, D.; Martinez Calejman, C.; Luciano, A.K.; et al. Non-canonical mTORC2 Signaling Regulates Brown Adipocyte Lipid Catabolism through SIRT6-FoxO1. Mol. Cell 2019, 75, 807–822.e8. [Google Scholar] [CrossRef] [PubMed]
- Holst, J.J. The physiology of glucagon-like peptide 1. Physiol. Rev. 2007, 87, 1409–1439. [Google Scholar] [CrossRef]
- Müller, T.D.; Blüher, M.; Tschöp, M.H.; DiMarchi, R.D. Anti-obesity drug discovery: Advances and challenges. Nat. Rev. Drug Discov. 2022, 21, 201–223. [Google Scholar] [CrossRef]
- Burmeister, M.A.; Brown, J.D.; Ayala, J.E.; Stoffers, D.A.; Sandoval, D.A.; Seeley, R.J.; Ayala, J.E. The glucagon-like peptide-1 receptor in the ventromedial hypothalamus reduces short-term food intake in male mice by regulating nutrient sensor activity. Am. J. Physiol. Endocrinol. Metab. 2017, 313, E651–E662. [Google Scholar] [CrossRef]
- Bence, K.K.; Birnbaum, M.J. Metabolic drivers of non-alcoholic fatty liver disease. Mol. Metab. 2021, 50, 101143. [Google Scholar] [CrossRef]
- Reeder, S.B.; Sirlin, C.B. Quantification of Liver Fat with Magnetic Resonance Imaging. Magn. Reson. Imaging Clin. N. Am. 2010, 18, 337–357. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.E.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Eslam, M.; George, J. Genetic contributions to NAFLD: Leveraging shared genetics to uncover systems biology. Nat. Rev. Gastroenterol. Hepatol. 2019, 17, 40–52. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Anstee, Q.M.; Marietti, M.; Hardy, T.; Henry, L.; Eslam, M.; George, J.; Bugianesi, E. Global burden of NAFLD and NASH: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2017, 15, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Scorletti, E.; Mosca, A.; Alisi, A.; Byrne, C.D.; Targher, G. Complications, morbidity and mortality of nonalcoholic fatty liver disease. Metab. Clin. Exp. 2020, 111, 154170. [Google Scholar] [CrossRef] [PubMed]
- Deprince, A.; Haas, J.T.; Staels, B. Dysregulated lipid metabolism links NAFLD to cardiovascular disease. Mol. Metab. 2020, 42, 101092. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Viscarra, J.; Kim, S.J.; Sul, H.S. Transcriptional regulation of hepatic lipogenesis. Nat. Rev. Mol. Cell Biol. 2015, 16, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Dyck, J.R.; Cheng, J.F.; Stanley, W.C.; Barr, R.; Chandler, M.P.; Brown, S.; Wallace, D.; Arrhenius, T.; Harmon, C.; Yang, G.; et al. Malonyl coenzyme a decarboxylase inhibition protects the ischemic heart by inhibiting fatty acid oxidation and stimulating glucose oxidation. Circ. Res. 2004, 94, e78–e84. [Google Scholar] [CrossRef]
- Han, J.; Wang, Y. mTORC1 signaling in hepatic lipid metabolism. Protein Cell 2018, 9, 145–151. [Google Scholar] [CrossRef]
- Peterson, T.R.; Sengupta, S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef]
- Han, J.; Li, E.; Chen, L.; Zhang, Y.-Y.; Wei, F.; Liu, J.; Deng, H.; Wang, Y. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature 2015, 524, 243–246. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef]
- Peña-Llopis, S.; Vega-Rubin-de-Celis, S.; Schwartz, J.C.; Wolff, N.C.; Tran, T.A.T.; Zou, L.; Xie, X.J.; Corey, D.R.; Brugarolas, J. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 2011, 30, 3242–3258. [Google Scholar] [CrossRef] [PubMed]
- Martina, J.A.; Chen, Y.; Gucek, M.; Puertollano, R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012, 8, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Roczniak-Ferguson, A.; Petit, C.S.; Froehlich, F.; Qian, S.; Ky, J.; Angarola, B.; Walther, T.C.; Ferguson, S.M. The Transcription Factor TFEB Links mTORC1 Signaling to Transcriptional Control of Lysosome Homeostasis. Sci. Signal. 2012, 5, ra42. [Google Scholar] [CrossRef]
- Settembre, C.; Zoncu, R.; Medina, D.L.; Vetrini, F.; Erdin, S.; Erdin, S.; Huynh, T.; Ferron, M.; Karsenty, G.; Vellard, M.C.; et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012, 31, 1095–1108. [Google Scholar] [CrossRef]
- Sardiello, M.; Palmieri, M.; di Ronza, A.; Medina, D.L.; Valenza, M.; Gennarino, V.A.; Di Malta, C.; Donaudy, F.; Embrione, V.; Polishchuk, R.S.; et al. A Gene Network Regulating Lysosomal Biogenesis and Function. Science 2009, 325, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Arencibia, M.G.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.L.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef]
- Settembre, C.; Fraldi, A.; Medina, D.L.; Ballabio, A. Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013, 14, 283–296. [Google Scholar] [CrossRef]
- Settembre, C.; De Cegli, R.; Mansueto, G.; Saha, P.K.; Vetrini, F.; Visvikis, O.; Huynh, T.; Carissimo, A.; Palmer, D.; Klisch, T.J.; et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013, 15, 647–658. [Google Scholar] [CrossRef]
- Martina, J.A.; Diab, H.I.; Lishu, L.; Jeong-A, L.; Patange, S.; Raben, N.; Puertollano, R. The Nutrient-Responsive Transcription Factor TFE3 Promotes Autophagy, Lysosomal Biogenesis, and Clearance of Cellular Debris. Sci. Signal. 2014, 7, ra9. [Google Scholar] [CrossRef]
- Raben, N.; Puertollano, R. TFEB and TFE3: Linking Lysosomes to Cellular Adaptation to Stress. Annu. Rev. Cell Dev. Biol. 2016, 32, 255–278. [Google Scholar] [CrossRef]
- Kenerson, H.L.; Yeh, M.M.; Yeung, R.S. Tuberous Sclerosis Complex-1 Deficiency Attenuates Diet-Induced Hepatic Lipid Accumulation. PLoS ONE 2011, 6, e18075. [Google Scholar] [CrossRef]
- Yecies, J.L.; Zhang, H.H.; Menon, S.; Liu, S.; Yecies, D.; Lipovsky, A.; Görgün, C.Z.; Kwiatkowski, D.J.; Hotamisligil, G.S.; Lee, C.-H.; et al. Akt Stimulates Hepatic SREBP1c and Lipogenesis through Parallel mTORC1-Dependent and Independent Pathways. Cell Metab. 2011, 14, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Quinn, W.J.; Wan, M.; Shewale, S.V.; Gelfer, R.; Rader, D.J.; Birnbaum, M.J.; Titchenell, P.M. mTORC1 stimulates phosphatidylcholine synthesis to promote triglyceride secretion. J. Clin. Investig. 2017, 127, 4207–4215. [Google Scholar] [CrossRef] [PubMed]
- Umemura, A.; Park, E.J.; Taniguchi, K.; Lee, J.H.; Shalapour, S.; Valasek, M.A.; Aghajan, M.; Nakagawa, H.; Seki, E.; Hall, M.N.; et al. Liver damage, inflammation, and enhanced tumorigenesis after persistent mTORC1 inhibition. Cell Metab. 2014, 20, 133–144. [Google Scholar] [CrossRef]
- Gosis, B.S.; Wada, S.; Thorsheim, C.; Li, K.; Jung, S.; Rhoades, J.H.; Yang, Y.; Brandimarto, J.; Li, L.; Uehara, K.; et al. Inhibition of nonalcoholic fatty liver disease in mice by selective inhibition of mTORC1. Science 2022, 376, eabf8271. [Google Scholar] [CrossRef] [PubMed]
- Uehara, K.; Sostre-Colón, J.; Gavin, M.; Santoleri, D.; Leonard, K.A.; Jacobs, R.L.; Titchenell, P.M. Activation of Liver mTORC1 Protects against NASH via Dual Regulation of VLDL-TAG Secretion and De Novo Lipogenesis. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 1625–1647. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Qiang, L.; Hayden, M.S.; Sparling, D.P.; Purcell, N.H.; Pajvani, U.B. mTORC1-independent Raptor prevents hepatic steatosis by stabilizing PHLPP2. Nat. Commun. 2016, 7, 10255. [Google Scholar] [CrossRef]
- Hagiwara, A.; Cornu, M.; Cybulski, N.; Polak, P.; Betz, C.; Trapani, F.; Terracciano, L.; Heim, M.H.; Rüegg, M.A.; Hall, M.N. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012, 15, 725–738. [Google Scholar] [CrossRef]
- Yuan, M.; Pino, E.C.; Wu, L.; Kacergis, M.C.; Soukas, A.A. Identification of Akt-independent Regulation of Hepatic Lipogenesis by Mammalian Target of Rapamycin (mTOR) Complex 2. J. Biol. Chem. 2012, 287, 29579–29588. [Google Scholar] [CrossRef]
- Guri, Y.; Colombi, M.; Dazert, E.; Hindupur, S.K.; Roszik, J.; Moes, S.; Jenoe, P.; Heim, M.H.; Riezman, I.; Riezman, H.; et al. mTORC2 Promotes Tumorigenesis via Lipid Synthesis. Cancer Cell 2017, 32, 807–823.e12. [Google Scholar] [CrossRef]
- Sciarretta, S.; Forte, M.; Frati, G.; Sadoshima, J. The complex network of mTOR signalling in the heart. Cardiovasc. Res. 2022, 118, 424–439. [Google Scholar] [CrossRef] [PubMed]
- Sciarretta, S.; Forte, M.; Frati, G.; Sadoshima, J. New Insights into the Role of mTOR Signaling in the Cardiovascular System. Circ. Res. 2018, 122, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Pires, K.M.P.; Whitehead, K.J.; Olsen, C.D.; Wayment, B.; Zhang, Y.C.; Bugger, H.; Ilkun, O.; Litwin, S.E.; Thomas, G.; et al. Mechanistic target of rapamycin (Mtor) is essential for murine embryonic heart development and growth. PLoS ONE 2013, 8, e54221. [Google Scholar] [CrossRef] [PubMed]
- Sadoshima, J.; Izumo, S. Rapamycin Selectively Inhibits Angiotensin II–Induced Increase in Protein Synthesis in Cardiac Myocytes In Vitro: Potential Role of 70-kD S6 Kinase in Angiotensin II– Induced Cardiac Hypertrophy. Circ. Res. 1995, 77, 1040–1052. [Google Scholar] [CrossRef]
- Zhang, D.; Contu, R.; Latronico, M.V.G.; Zhang, J.L.; Rizzi, R.; Catalucci, D.; Miyamoto, S.; Huang, K.; Ceci, M.; Gu, Y.; et al. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J. Clin. Investig. 2010, 120, 2805–2816. [Google Scholar] [CrossRef]
- Shende, P.S.; Plaisance, I.; Morandi, C.; Pellieux, C.; Berthonneche, C.; Zorzato, F.; Krishnan, J.; Lerch, R.; Hall, M.N.; Rüegg, M.A.; et al. Cardiac Raptor Ablation Impairs Adaptive Hypertrophy, Alters Metabolic Gene Expression, and Causes Heart Failure in Mice. Circulation 2011, 123, 1073–1082. [Google Scholar] [CrossRef]
- Shioi, T.; McMullen, J.R.; Tarnavski, O.; Converso, K.L.; Sherwood, M.C.; Manning, W.J.; Izumo, S. Rapamycin Attenuates Load-Induced Cardiac Hypertrophy in Mice. Circulation 2003, 107, 1664–1670. [Google Scholar] [CrossRef]
- McMullen, J.R.; Sherwood, M.C.; Tarnavski, O.; Zhang, L.; Dorfman, A.L.; Shioi, T.; Izumo, S. Inhibition of mTOR Signaling with Rapamycin Regresses Established Cardiac Hypertrophy Induced by Pressure Overload. Circulation 2004, 109, 3050–3055. [Google Scholar] [CrossRef]
- Buss, S.J.; Muenz, S.; Riffel, J.; Malekar, P.; Hagenmueller, M.; Weiss, C.S.; Bea, F.; Bekeredjian, R.; Schinke-Braun, M.; Izumo, S.; et al. Beneficial effects of Mammalian target of rapamycin inhibition on left ventricular remodeling after myocardial infarction. J. Am. Coll. Cardiol. 2009, 54, 2435–2446. [Google Scholar] [CrossRef]
- Dai, D.-F.; Liu, Y.; Basisty, N.; Karunadharma, P.P.; Dastidar, S.G.; Chiao, Y.A.; Chen, T.; Beyer, R.P.; Chin, M.T.; MacCoss, M.J.; et al. Differential effects of various genetic mouse models of the mechanistic target of rapamycin complex I inhibition on heart failure. GeroScience 2019, 41, 847–860. [Google Scholar] [CrossRef]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef]
- Flynn, J.M.; O’Leary, M.N.; Zambataro, C.A.; Academia, E.C.; Presley, M.P.; Garrett, B.J.; Zykovich, A.; Mooney, S.D.; Strong, R.; Rosen, C.J.; et al. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell 2013, 12, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Sciarretta, S.; Zhai, P.; Shao, D.; Maejima, Y.; Robbins, J.M.; Volpe, M.; Condorelli, G.; Sadoshima, J. Rheb is a Critical Regulator of Autophagy during Myocardial Ischemia: Pathophysiological Implications in Obesity and Metabolic Syndrome. Circulation 2012, 125, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Zhai, P.; Sciarretta, S.; Galeotti, J.; Volpe, M.; Sadoshima, J. Differential Roles of GSK-3β During Myocardial Ischemia and Ischemia/Reperfusion. Circ. Res. 2011, 109, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Salloum, F.N.; Durrant, D.; Ockaili, R.A.; Kukreja, R.C. Rapamycin protects against myocardial ischemia-reperfusion injury through JAK2-STAT3 signaling pathway. J. Mol. Cell. Cardiol. 2012, 53, 858–869. [Google Scholar] [CrossRef] [PubMed]
- Matsui, Y.; Takagi, H.; Qu, X.; Abdellatif, M.; Sakoda, H.; Asano, T.; Levine, B.; Sadoshima, J. Distinct Roles of Autophagy in the Heart During Ischemia and Reperfusion: Roles of AMP-Activated Protein Kinase and Beclin 1 in Mediating Autophagy. Circ. Res. 2007, 100, 914–922. [Google Scholar] [CrossRef]
- Di, R.-M.; Wu, X.; Chang, Z.; Zhao, X.; Feng, Q.; Lu, S.; Luan, Q.; Hemmings, B.A.; Li, X.; Yang, Z. S6K inhibition renders cardiac protection against myocardial infarction through PDK1 phosphorylation of Akt. Biochem. J. 2011, 441, 199–207. [Google Scholar] [CrossRef]
- Shende, P.S.; Xu, L.; Morandi, C.; Pentassuglia, L.; Heim, P.; Lebboukh, S.; Berthonneche, C.; Pedrazzini, T.; Kaufmann, B.A.; Hall, M.N.; et al. Cardiac mTOR complex 2 preserves ventricular function in pressure-overload hypertrophy. Cardiovasc. Res. 2015, 109, 103–114. [Google Scholar] [CrossRef]
- Lamming, D.W.; Mihaylova, M.M.; Katajisto, P.; Baar, E.L.; Yilmaz, Ö.H.; Hutchins, A.W.; Gultekin, Y.; Gaither, R.P.; Sabatini, D.M. Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell 2014, 13, 911–917. [Google Scholar] [CrossRef]
- Völkers, M.; Konstandin, M.H.; Doroudgar, S.; Toko, H.; Quijada, P.; Din, S.; Joyo, A.Y.; Ornelas, L.; Samse, K.; Thuerauf, D.J.; et al. Mechanistic Target of Rapamycin Complex 2 Protects the Heart from Ischemic Damage. Circulation 2013, 128, 2132–2144. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).