Detection of SARS-CoV-2 Based on Nucleic Acid Amplification Tests (NAATs) and Its Integration into Nanomedicine and Microfluidic Devices as Point-of-Care Testing (POCT)
Abstract
1. Introduction
2. Importance of Diagnosis and the Use of POCT
- -
- Detection of viral RNA: this involves nucleic acid amplification tests (NAATs), such as RT-qPCR.
- -
- Detection of viral antigens: such as immunodiagnostic techniques, including lateral flow assays (LFA).
- -
- Detection of viral antibodies: serological techniques, such as enzyme-linked immunosorbent assays (ELISAs) or chemiluminescent immunoassays (CLIAs).
3. Nucleic Acid Amplification Tests (NAATs)
3.1. RT-qPCR Is the Gold Standard Method
3.2. Digital Droplet Polymerase Chain Reaction (ddPCR)
3.3. Isothermal Amplification
3.4. CRISPR-Based Methods
3.5. Next-Generation Sequencing (NGS)
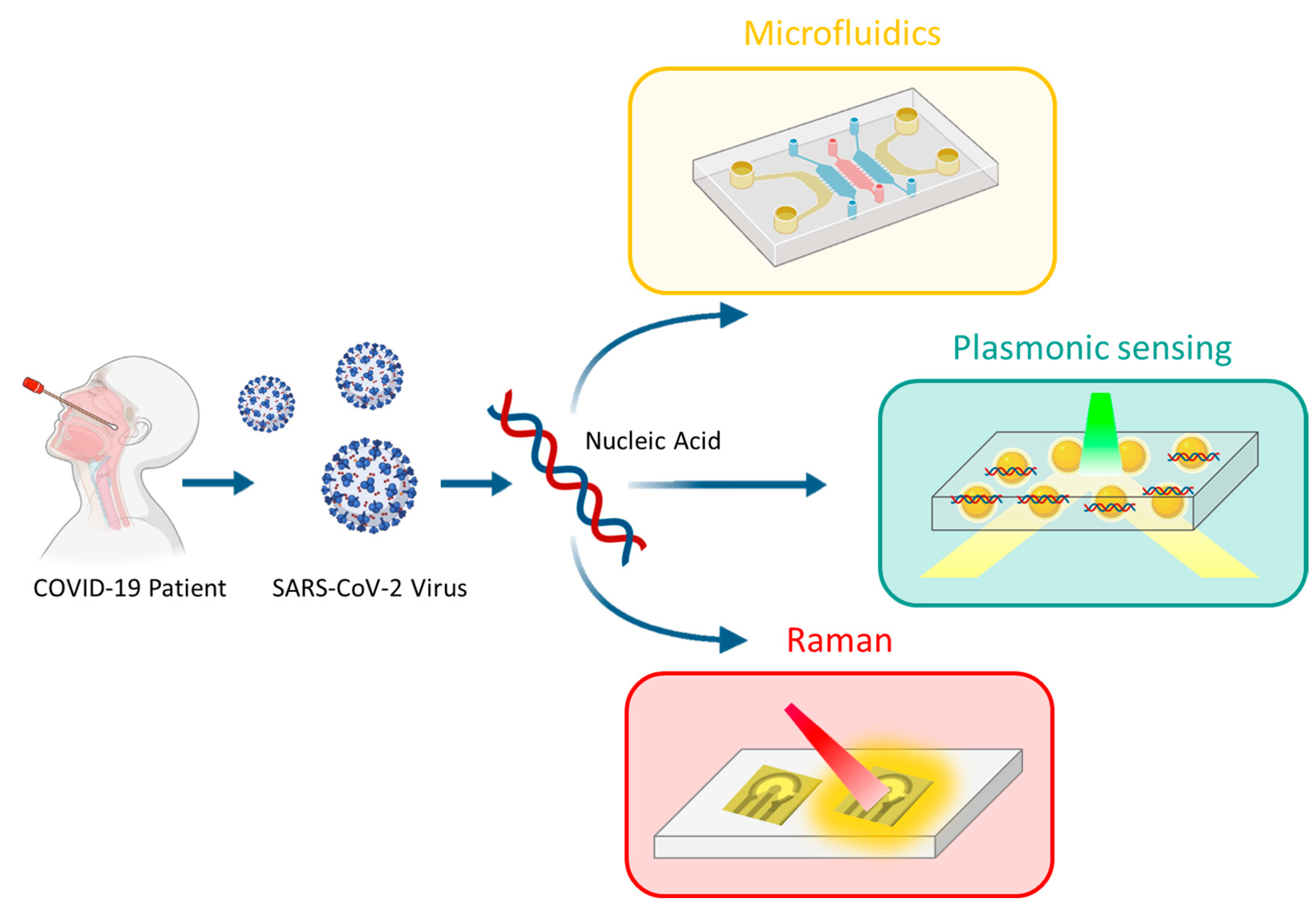
4. Microfluidics Integration and Nanomedicine Advances
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Worldometers Information of COVID-19 Pandemics. Available online: https://www.worldometers.info/coronavirus/ (accessed on 11 February 2023).
- Mingaleeva, R.N.; Nigmatulina, N.A.; Sharafetdinova, L.M.; Romozanova, A.M.; Gabdoulkhakova, A.G.; Filina, Y.V.; Shavaliyev, R.F.; Rizvanov, A.A.; Miftakhova, R.R. Biology of the SARS-CoV-2 Coronavirus. Biochemistry 2022, 87, 1662–1678. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef] [PubMed]
- Šimičić, P.; Židovec-Lepej, S. A Glimpse on the Evolution of RNA Viruses: Implications and Lessons from SARS-CoV-2. Viruses 2022, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebuhr, J. Discovery of an RNA Virus 3′→5′ Exoribonuclease That Is Critically Involved in Coronavirus RNA Synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef]
- Denison, M.R.; Graham, R.L.; Donaldson, E.F.; Eckerle, L.D.; Baric, R.S. Coronaviruses. RNA Biol. 2011, 8, 270–279. [Google Scholar] [CrossRef]
- Domingo, E. QUASISPECIES. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 1999; pp. 1431–1436. [Google Scholar]
- Ahmad, A.; Fawaz, M.A.M.; Aisha, A. A Comparative Overview of SARS-CoV-2 and Its Variants of Concern. Infez. Med. 2022, 30, 328. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Elbe, S.; Buckland-Merrett, G. Data, Disease and Diplomacy: GISAID’s Innovative Contribution to Global Health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef]
- WHO Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 27 May 2023).
- World Health Organization. WHO Announces Simple, Easy-to-Say Labels for SARS-CoV-2 Variants of Interest and Concern 2021. Available online: https://www.who.int/news/item/31-05-2021-who-announces-simple-easy-to-say-labels-for-sars-cov-2-variants-of-interest-and-concern (accessed on 12 February 2023).
- European Centre for Disease Prevention and Control (ECDC). Available online: https://www.ecdc.europa.eu/en/covid-19/country-overviews (accessed on 12 February 2023).
- Callaway, E. Heavily Mutated Omicron Variant Puts Scientists on Alert. Nature 2021, 600, 21. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, L.; Mo, M.; Liu, T.; Wu, C.; Gong, C.; Lu, K.; Gong, L.; Zhu, W.; Xu, Z. SARS-CoV-2 Omicron RBD Shows Weaker Binding Affinity than the Currently Dominant Delta Variant to Human ACE2. Signal Transduct. Target. Ther. 2022, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 Usage by SARS-CoV-2 Omicron Impacts Infectivity and Fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Woo, H.G. Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies. Front. Immunol. 2022, 12, 6031. [Google Scholar] [CrossRef] [PubMed]
- Tallei, T.E.; Alhumaid, S.; AlMusa, Z.; Fatimawali Kusumawaty, D.; Alynbiawi, A.; Alshukairi, A.N.; Rabaan, A.A. Update on the Omicron Sub-variants BA.4 and BA.5. Rev. Med. Virol. 2023, 33, e2391. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Notarte, K.I.; Fernandez, R.A.; Lippi, G.; Gromiha, M.M.; Henry, B.M. In Silico Evaluation of the Impact of Omicron Variant of Concern Sublineage BA.4 and BA.5 on the Sensitivity of RT-qPCR Assays for SARS-CoV-2 Detection Using Whole Genome Sequencing. J. Med. Virol. 2023, 95, e28241. [Google Scholar] [CrossRef]
- Salehi-Vaziri, M.; Fazlalipour, M.; Seyed Khorrami, S.M.; Azadmanesh, K.; Pouriayevali, M.H.; Jalali, T.; Shoja, Z.; Maleki, A. The Ins and Outs of SARS-CoV-2 Variants of Concern (VOCs). Arch. Virol. 2022, 167, 327–344. [Google Scholar] [CrossRef]
- World Health Organization. Methods for the Detection and Characterisation of SARS-CoV-2 Variants-Second Update; Regional Office for Europe: Copenhagen, Denmark, 2022. [Google Scholar]
- Koczula, K.M.; Gallotta, A. Lateral Flow Assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [CrossRef]
- Dinnes, J.; Sharma, P.; Berhane, S.; van Wyk, S.S.; Nyaaba, N.; Domen, J.; Taylor, M.; Cunningham, J.; Davenport, C.; Dittrich, S.; et al. Rapid, Point-of-Care Antigen Tests for Diagnosis of SARS-CoV-2 Infection. Cochrane Database Syst. Rev. 2022, 7, CD013705. [Google Scholar] [CrossRef]
- Luppa, P.B.; Müller, C.; Schlichtiger, A.; Schlebusch, H. Point-of-Care Testing (POCT): Current Techniques and Future Perspectives. TrAC Trends Anal. Chem. 2011, 30, 887–898. [Google Scholar] [CrossRef]
- Otoo, J.A.; Schlappi, T.S. REASSURED Multiplex Diagnostics: A Critical Review and Forecast. Biosensors 2022, 12, 124. [Google Scholar] [CrossRef]
- Farré, M.; Kantiani, L.; Barceló, D. Chapter 7—Microfluidic Devices: Biosensors. In Chemical Analysis of Food: Techniques and Applications; Picó, Y., Ed.; Academic Press: Boston, MA, USA, 2012; pp. 177–217. ISBN 978-0-12-384862-8. [Google Scholar]
- Joo, J. Diagnostic and Therapeutic Nanomedicine. Adv. Exp. Med. Biol. 2021, 1310, 401–447. [Google Scholar] [CrossRef]
- Tröger, V.; Niemann, K.; Gärtig, C.; Kuhlmeier, D. Isothermal Amplification and Quantification of Nucleic Acids and Its Use in Microsystems. J. Nanomed. Nanotechnol. 2015, 6, 1. [Google Scholar] [CrossRef]
- Kokkoris, V.; Vukicevich, E.; Richards, A.; Thomsen, C.; Hart, M.M. Challenges Using Droplet Digital PCR for Environmental Samples. Appl. Microbiol. 2021, 1, 74–88. [Google Scholar] [CrossRef]
- Smith, C.J.; Osborn, A.M. Advantages and Limitations of Quantitative PCR (Q-PCR)-Based Approaches in Microbial Ecology. FEMS Microbiol. Ecol. 2009, 67, 6–20. [Google Scholar] [CrossRef]
- Jalandra, R.; Yadav, A.K.; Verma, D.; Dalal, N.; Sharma, M.; Singh, R.; Kumar, A.; Solanki, P.R. Strategies and Perspectives to Develop SARS-CoV-2 Detection Methods and Diagnostics. Biomed. Pharmacother. 2020, 129, 110446. [Google Scholar] [CrossRef]
- Serrano-Cumplido, A.; Ruiz Garcia, A.; Segura-Fragoso, A.; Olmo-Quintana, V.; Micó Pérez, R.M.; Barquilla-García, A.; Morán-Bayón, A. Aplicación Del Valor Umbral Del Número de Ciclos (Ct) de PCR En La COVID-19. Med. De Familia. Semer. 2021, 47, 337–341. [Google Scholar] [CrossRef]
- Tom, M.R.; Mina, M.J. To Interpret the SARS-CoV-2 Test, Consider the Cycle Threshold Value. Clin. Infect. Dis. 2020, 71, 2252–2254. [Google Scholar] [CrossRef]
- Dahdouh, E.; Lázaro-Perona, F.; Romero-Gómez, M.P.; Mingorance, J.; García-Rodriguez, J. Ct Values from SARS-CoV-2 Diagnostic PCR Assays Should Not Be Used as Direct Estimates of Viral Load. J. Infect. 2021, 82, 414–451. [Google Scholar] [CrossRef]
- Russo, A.; Minichini, C.; Starace, M.; Astorri, R.; Calò, F.; Coppola, N. Current Status of Laboratory Diagnosis for COVID-19: A Narrative Review. Infect Drug Resist. 2020, 13, 2657–2665. [Google Scholar] [CrossRef]
- Rong, X.; Yang, L.; Chu, H.; Fan, M. Effect of Delay in Diagnosis on Transmission of COVID-19. Math. Biosci. Eng. 2020, 17, 2725–2740. [Google Scholar] [CrossRef]
- Protocol: Real-Time RT-PCR Assays for the Detection of SARS-CoV-2 Institut Pasteur, Paris. Available online: https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf (accessed on 16 February 2023).
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drugs Administration (FDA). SARS-CoV-2 In Vitro Diagnostics EUAs—Molecular Diagnostic Tests for SARS-CoV-2. Available online: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-molecular-diagnostic-tests-sars-cov-2 (accessed on 14 February 2023).
- Bruijns, B.; Folkertsma, L.; Tiggelaar, R. FDA Authorized Molecular Point-of-Care SARS-CoV-2 Tests: A Critical Review on Principles, Systems and Clinical Performances. Biosens. Bioelectron. X 2022, 11, 100158. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. DPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zheng, J.; Li, Z.; Liu, Y.; Jing, F.; Wan, X.; Yamaguchi, Y.; Zhuang, S. A Rapid Nucleic Acid Concentration Measurement System with Large Field of View for a Droplet Digital PCR Microfluidic Chip. Lab A Chip 2021, 21, 3742–3747. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Özay, B.; McCalla, S.E. A Review of Reaction Enhancement Strategies for Isothermal Nucleic Acid Amplification Reactions. Sens. Actuators Rep. 2021, 3, 100033. [Google Scholar] [CrossRef]
- Amaral, C.; Antunes, W.; Moe, E.; Duarte, A.G.; Lima, L.M.P.; Santos, C.; Gomes, I.L.; Afonso, G.S.; Vieira, R.; Teles, H.S.S.; et al. A Molecular Test Based on RT-LAMP for Rapid, Sensitive and Inexpensive Colorimetric Detection of SARS-CoV-2 in Clinical Samples. Sci. Rep. 2021, 11, 16430. [Google Scholar] [CrossRef]
- Kitagawa, Y.; Orihara, Y.; Kawamura, R.; Imai, K.; Sakai, J.; Tarumoto, N.; Matsuoka, M.; Takeuchi, S.; Maesaki, S.; Maeda, T. Evaluation of Rapid Diagnosis of Novel Coronavirus Disease (COVID-19) Using Loop-Mediated Isothermal Amplification. J. Clin. Virol. 2020, 129, 104446. [Google Scholar] [CrossRef]
- Jang, W.S.; Lim, D.H.; Yoon, J.; Kim, A.; Lim, M.; Nam, J.; Yanagihara, R.; Ryu, S.-W.; Jung, B.K.; Ryoo, N.-H.; et al. Development of a Multiplex Loop-Mediated Isothermal Amplification (LAMP) Assay for on-Site Diagnosis of SARS CoV-2. PLoS ONE 2021, 16, e0248042. [Google Scholar] [CrossRef]
- Moon, Y.-J.; Lee, S.-Y.; Oh, S.-W. A Review of Isothermal Amplification Methods and Food-Origin Inhibitors against Detecting Food-Borne Pathogens. Foods 2022, 11, 322. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol 2006, 4, e204. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase Polymerase Amplification: Basics, Applications and Recent Advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Song, Y.; Huang, P.; Yu, M.; Li, Y.; Jin, H.; Qiu, J.; Li, Y.; Gao, Y.; Zhang, H.; Wang, H. Rapid and Visual Detection of SARS-CoV-2 RNA Based on Reverse Transcription-Recombinase Polymerase Amplification with Closed Vertical Flow Visualization Strip Assay. Microbiol. Spectr. 2023, 11, e02966-22. [Google Scholar] [CrossRef]
- Farrera-Soler, L.; Gonse, A.; Kim, K.T.; Barluenga, S.; Winssinger, N. Combining Recombinase Polymerase Amplification and DNA-templated Reaction for SARS-CoV-2 Sensing with Dual Fluorescence and Lateral Flow Assay Output. Biopolymers 2022, 113, 5307–5315. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A Comprehensive Summary of a Decade Development of the Recombinase Polymerase Amplification. Analyst 2019, 144, 31–67. [Google Scholar] [CrossRef]
- Oliveira, B.B.; Veigas, B.; Baptista, P.V. Isothermal Amplification of Nucleic Acids: The Race for the Next “Gold Standard”. Front. Sens. 2021, 2, 752600. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science (1979) 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science (1979) 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Gostimskaya, I. CRISPR—Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing. Biochemistry 2022, 87, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Dong, H.; Cui, Y.; Cong, L.; Zhang, D. Application of Different Types of CRISPR/Cas-Based Systems in Bacteria. Microb. Cell Factories 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Shebanova, R.; Nikitchina, N.; Shebanov, N.; Mekler, V.; Kuznedelov, K.; Ulashchik, E.; Vasilev, R.; Sharko, O.; Shmanai, V.; Tarassov, I.; et al. Efficient Target Cleavage by Type V Cas12a Effectors Programmed with Split CRISPR RNA. Nucleic Acids Res. 2022, 50, 1162–1173. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, M.I.; Makhawi, A.M. SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases. J. Clin. Microbiol. 2021, 59, e00745-20. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic Acid Detection with CRISPR Nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Macaluso, N.C.; Pizzano, B.L.M.; Cash, M.N.; Spacek, J.; Karasek, J.; Miller, M.R.; Lednicky, J.A.; Dinglasan, R.R.; Salemi, M.; et al. A Thermostable Cas12b from Brevibacillus Leverages One-Pot Discrimination of SARS-CoV-2 Variants of Concern. EBioMedicine 2022, 77, 103926. [Google Scholar] [CrossRef]
- Azmi, I.; Faizan, M.I.; Kumar, R.; Raj Yadav, S.; Chaudhary, N.; Kumar Singh, D.; Butola, R.; Ganotra, A.; Datt Joshi, G.; Deep Jhingan, G.; et al. A Saliva-Based RNA Extraction-Free Workflow Integrated with Cas13a for SARS-CoV-2 Detection. Front. Cell. Infect. Microbiol. 2021, 11, 632646. [Google Scholar] [CrossRef]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.-G.; Woolley, A.E.; Segel, M.; Barretto, R.P.J.; Ranu, A.; Macrae, R.K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively Multiplexed Nucleic Acid Detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-Based Diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Portable CRISPR-Based Diagnostics. Nat. Biotechnol. 2019, 37, 832. [CrossRef]
- Jolany, V.S.; Katalani, C.; Boone, H.A.; Hajizade, A.; Sijercic, A.; Ahmadian, G. CRISPR-Based Diagnosis of Infectious and Noninfectious Diseases. Biol. Proced. Online 2020, 22, 22. [Google Scholar] [CrossRef]
- Harvard Wyss Institute. INSPECTRTM. Available online: https://wyss.harvard.edu/technology/inspectr-a-direct-to-consumer-molecular-diagnostic/ (accessed on 16 February 2023).
- Sherlock Biosciences. Available online: https://sherlock.bio/platforms/synthetic-biology/ (accessed on 16 February 2023).
- Hernández, M.; Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M. Aplicación de La Secuenciación Masiva y La Bioinformática al Diagnóstico Microbiológico Clínico. Rev. Argent. De Microbiol. 2020, 52, 150–161. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The Spike Glycoprotein of the New Coronavirus 2019-NCoV Contains a Furin-like Cleavage Site Absent in CoV of the Same Clade. Antiviral Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Chen, J.; Wang, R.; Wang, M.; Wei, G.-W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020, 432, 5212–5226. [Google Scholar] [CrossRef]
- Yu, W.-B.; Tang, G.-D.; Zhang, L.T.; Corlett, R. Decoding the Evolution and Transmissions of the Novel Pneumonia Coronavirus (SARS-CoV-2/HCoV-19) Using Whole Genomic Data. Zool. Res. 2020, 41, 247–257. [Google Scholar] [CrossRef]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-Cell RNA-Seq Data Analysis on the Receptor ACE2 Expression Reveals the Potential Risk of Different Human Organs Vulnerable to 2019-NCoV Infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef]
- World Health Organization. Guidance for Surveillance of SARS-CoV-2 Variants: Interim Guidance; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Yager, P.; Edwards, T.; Fu, E.; Helton, K.; Nelson, K.; Tam, M.R.; Weigl, B.H. Microfluidic Diagnostic Technologies for Global Public Health. Nature 2006, 442, 412–418. [Google Scholar] [CrossRef]
- Yang, S.M.; Lv, S.; Zhang, W.; Cui, Y. Microfluidic Point-of-Care (POC) Devices in Early Diagnosis: A Review of Opportunities and Challenges. Sensors 2022, 22. [Google Scholar] [CrossRef]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic Lab-on-a-Chip Platforms: Requirements, Characteristics and Applications. Chem. Soc. Rev. 2010, 39, 1153. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi-Aghdam, A.; Majidi, M.R.; Omidi, Y. Microfluidic Paper-Based Analytical Devices (ΜPADs) for Fast and Ultrasensitive Sensing of Biomarkers and Monitoring of Diseases. BioImpacts 2018, 8, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.Y.; Theek, B.; Storm, G.; Kiessling, F.; Lammers, T. Recent Progress in Nanomedicine: Therapeutic, Diagnostic and Theranostic Applications. Curr. Opin. Biotechnol. 2013, 24, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Misra, R.; Acharya, S.; Sushmitha, N. Nanobiosensor-Based Diagnostic Tools in Viral Infections: Special Emphasis on COVID-19. Rev. Med. Virol. 2022, 32. [Google Scholar] [CrossRef]
- A Matter of Scale. Nat. Nanotechnol. 2016, 11, 733. [CrossRef]
- Shubhika, K. Nanotechnology and Medicine-The Upside and the Downside. Int. J. Drug Dev. Res. 2012, 5, 1–10. [Google Scholar]
- Khaitov, M.; Nikonova, A.; Shilovskiy, I.; Kozhikhova, K.; Kofiadi, I.; Vishnyakova, L.; Nikolskii, A.; Gattinger, P.; Kovchina, V.; Barvinskaia, E.; et al. Silencing of SARS-CoV-2 with Modified SiRNA-peptide Dendrimer Formulation. Allergy 2021, 76, 2840–2854. [Google Scholar] [CrossRef]
- Volpatti, L.R.; Wallace, R.P.; Cao, S.; Raczy, M.M.; Wang, R.; Gray, L.T.; Alpar, A.T.; Briquez, P.S.; Mitrousis, N.; Marchell, T.M.; et al. Polymersomes Decorated with the SARS-CoV-2 Spike Protein Receptor-Binding Domain Elicit Robust Humoral and Cellular Immunity. ACS Cent. Sci. 2021, 7, 1368–1380. [Google Scholar] [CrossRef]
- Thorn, C.R.; Sharma, D.; Combs, R.; Bhujbal, S.; Romine, J.; Zheng, X.; Sunasara, K.; Badkar, A. The Journey of a Lifetime—Development of Pfizer’s COVID-19 Vaccine. Curr. Opin. Biotechnol. 2022, 78, 102803. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, J.; Li, Y.; Tan, G.; Sun, M.; Shan, Y.; Zhang, Y.; Wang, X.; Song, K.; Shi, R.; et al. SARS-CoV-2 Detection Using Quantum Dot Fluorescence Immunochromatography Combined with Isothermal Amplification and CRISPR/Cas13a. Biosens. Bioelectron. 2022, 202, 113978. [Google Scholar] [CrossRef]
- Agha, A.; Waheed, W.; Stiharu, I.; Nerguizian, V.; Destgeer, G.; Abu-Nada, E.; Alazzam, A. A Review on Microfluidic-Assisted Nanoparticle Synthesis, and Their Applications Using Multiscale Simulation Methods. Discov. Nano 2023, 18, 18. [Google Scholar] [CrossRef]
- Unser, S.; Bruzas, I.; He, J.; Sagle, L. Localized Surface Plasmon Resonance Biosensing: Current Challenges and Approaches. Sensors 2015, 15, 15684–15716. [Google Scholar] [CrossRef]
- Piliarik, M.; Kvasnička, P.; Galler, N.; Krenn, J.R.; Homola, J. Local Refractive Index Sensitivity of Plasmonic Nanoparticles. Opt. Express 2011, 19, 9213. [Google Scholar] [CrossRef]
- Huang, L.; Ding, L.; Zhou, J.; Chen, S.; Chen, F.; Zhao, C.; Xu, J.; Hu, W.; Ji, J.; Xu, H.; et al. One-Step Rapid Quantification of SARS-CoV-2 Virus Particles via Low-Cost Nanoplasmonic Sensors in Generic Microplate Reader and Point-of-Care Device. Biosens. Bioelectron. 2021, 171, 112685. [Google Scholar] [CrossRef]
- Dietzek, B.; Cialla, D.; Schmitt, M.; Popp, J. Introduction to the Fundamentals of Raman Spectroscopy. In Confocal Raman Microscopy; Springer: Berlin/Heidelberg, Germany, 2018; pp. 47–68. [Google Scholar]
- Yin, G.; Li, L.; Lu, S.; Yin, Y.; Su, Y.; Zeng, Y.; Luo, M.; Ma, M.; Zhou, H.; Orlandini, L.; et al. An Efficient Primary Screening of COVID-19 by Serum Raman Spectroscopy. J. Raman Spectrosc. 2021, 52, 949–958. [Google Scholar] [CrossRef]
- Li, X.; Qin, Z.; Fu, H.; Li, T.; Peng, R.; Li, Z.; Rini, J.M.; Liu, X. Enhancing the Performance of Paper-Based Electrochemical Impedance Spectroscopy Nanobiosensors: An Experimental Approach. Biosens. Bioelectron. 2021, 177, 112672. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Li, Z.; Sfeir, M.M.; Ballesteros, E.; Liu, C. Autonomous Lab-on-Paper for Multiplexed, CRISPR-Based Diagnostics of SARS-CoV-2. Lab A Chip 2021, 21, 2730–2737. [Google Scholar] [CrossRef]
- Wang, H.; Xu, J.; Li, S.; Wang, X.; Liu, G.; Yang, S.; Zhao, F.; Liu, Q.; Chen, X.; He, C.; et al. An Integrated Dual-Layer Microfluidic Platform for Multiple Respiratory Viruses Screening. Anal. Chim. Acta 2023, 1242, 340812. [Google Scholar] [CrossRef]
- Ganguli, A.; Mostafa, A.; Berger, J.; Aydin, M.Y.; Sun, F.; de Ramirez, S.A.S.; Valera, E.; Cunningham, B.T.; King, W.P.; Bashir, R. Rapid Isothermal Amplification and Portable Detection System for SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 22727–22735. [Google Scholar] [CrossRef]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef]
- Park, Y.; Ryu, B.; Ki, S.J.; Chen, M.; Liang, X.; Kurabayashi, K. Bioinspired Plasmo-Virus for Point-of-Care SARS-CoV-2 Detection. Nano Lett. 2023, 23, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Robin, P.; Barnabei, L.; Marocco, S.; Pagnoncelli, J.; Nicolis, D.; Tarantelli, C.; Tavilla, A.C.; Robortella, R.; Cascione, L.; Mayoraz, L.; et al. A DNA Biosensors-Based Microfluidic Platform for Attomolar Real-Time Detection of Unamplified SARS-CoV-2 Virus. Biosens. Bioelectron. X 2023, 13, 100302. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, C.; Kang, X.; Li, Y. A Visual Detection Strategy for SARS-CoV-2 Based on Dual Targets-Triggering DNA Walker. Sens. Actuators B Chem. 2023, 379, 133252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, X.; Pan, J.; Zhang, Y.; Zhang, L.; Wang, C.; Yan, X.; Liu, X.; Lu, G. Ultrasensitive Detection of SARS-CoV-2 Spike Protein in Untreated Saliva Using SERS-Based Biosensor. Biosens. Bioelectron. 2021, 190, 113421. [Google Scholar] [CrossRef]
- Liu, J.; Chen, P.; Hu, X.; Huang, L.; Geng, Z.; Xu, H.; Hu, W.; Wang, L.; Wu, P.; Liu, G.L. An Ultra-Sensitive and Specific Nanoplasmonic-Enhanced Isothermal Amplification Platform for the Ultrafast Point-of-Care Testing of SARS-CoV-2. Chem. Eng. J. 2023, 451, 138822. [Google Scholar] [CrossRef]
- Chen, Y.; Zong, N.; Ye, F.; Mei, Y.; Qu, J.; Jiang, X. Dual-CRISPR/Cas12a-Assisted RT-RAA for Ultrasensitive SARS-CoV-2 Detection on Automated Centrifugal Microfluidics. Anal. Chem. 2022, 94, 9603–9609. [Google Scholar] [CrossRef]
- Mahari, S.; Roberts, A.; Shahdeo, D.; Gandhi, S. ECovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of NCovid-19 Antigen, a Spike Protein Domain 1 of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric Field-Driven Microfluidics for Rapid CRISPR-Based Diagnostics and Its Application to Detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef]
- Xing, W.; Li, Q.; Han, C.; Sun, D.; Zhang, Z.; Fang, X.; Guo, Y.; Ge, F.; Ding, W.; Luo, Z.; et al. Customization of Aptamer to Develop CRISPR/Cas12a-Derived Ultrasensitive Biosensor. Talanta 2023, 256, 124312. [Google Scholar] [CrossRef]
- Idili, A.; Parolo, C.; Alvarez-Diduk, R.; Merkoçi, A. Rapid and Efficient Detection of the SARS-CoV-2 Spike Protein Using an Electrochemical Aptamer-Based Sensor. ACS Sens. 2021, 6, 3093–3101. [Google Scholar] [CrossRef]
- Sampaio, I.; Takeuti, N.N.K.; Gusson, B.; Machado, T.R.; Zucolotto, V. Capacitive Immunosensor for COVID-19 Diagnosis. Microelectron. Eng. 2023, 267, 111912. [Google Scholar] [CrossRef]
- Nguyen, P.Q.; Soenksen, L.R.; Donghia, N.M.; Angenent-Mari, N.M.; de Puig, H.; Huang, A.; Lee, R.; Slomovic, S.; Galbersanini, T.; Lansberry, G.; et al. Wearable Materials with Embedded Synthetic Biology Sensors for Biomolecule Detection. Nat. Biotechnol. 2021, 39, 1366–1374. [Google Scholar] [CrossRef]
- Cepheid OS. Xpert Xpress SARS-CoV-2. 2020. Available online: https://www.cepheid.com/content/dam/www-cepheid-com/documents/package-insert-files/Xpert%20Xpress%20SARS-CoV-2%20CE-IVD%20GeneXpert%20System%20With%20Touchscreen%20302-8405-NL%20Rev%20B.pdf (accessed on 13 June 2023).
- Tu, Y.-P.; Iqbal, J.; O’Leary, T. Sensitivity of ID NOW and RT–PCR for Detection of SARS-CoV-2 in an Ambulatory Population. eLife 2021, 10, e65726. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef]
- Aguiar, E.R.G.R.; Navas, J.; Pacheco, L.G.C. The COVID-19 Diagnostic Technology Landscape: Efficient Data Sharing Drives Diagnostic Development. Front. Public Health 2020, 8, 309. [Google Scholar] [CrossRef]
- Center for Disease Control and Prevention (CDC). CDC’s Role in Tracking Variants. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/cdc-role-surveillance.html (accessed on 20 February 2020).
- Ashcroft, P.; Lehtinen, S.; Bonhoeffer, S. Test-Trace-Isolate-Quarantine (TTIQ) Intervention Strategies after Symptomatic COVID-19 Case Identification. PLoS ONE 2022, 17, e0263597. [Google Scholar] [CrossRef]
- Duma, Z.; Chuturgoon, A.A.; Ramsuran, V.; Edward, V.; Naidoo, P.; Mpaka-Mbatha, M.N.; Bhengu, K.N.; Nembe, N.; Pillay, R.; Singh, R.; et al. The Challenges of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Testing in Low-Middle Income Countries and Possible Cost-Effective Measures in Resource-Limited Settings. Glob. Health 2022, 18, 5. [Google Scholar] [CrossRef]
- Contreras, S.; Dehning, J.; Loidolt, M.; Zierenberg, J.; Spitzner, F.P.; Urrea-Quintero, J.H.; Mohr, S.B.; Wilczek, M.; Wibral, M.; Priesemann, V. The Challenges of Containing SARS-CoV-2 via Test-Trace-and-Isolate. Nat. Commun. 2021, 12, 378. [Google Scholar] [CrossRef]
- Rando, H.M.; Bennett, T.D.; Byrd, J.B.; Bramante, C.; Callahan, T.J.; Chute, C.G.; Davis, H.E.; Deer, R.; Gagnier, J.; Koraishy, F.M.; et al. Challenges in Defining Long COVID: Striking Differences across Literature, Electronic Health Records, and Patient-Reported Information. MedRxiv 2021. [Google Scholar] [CrossRef]
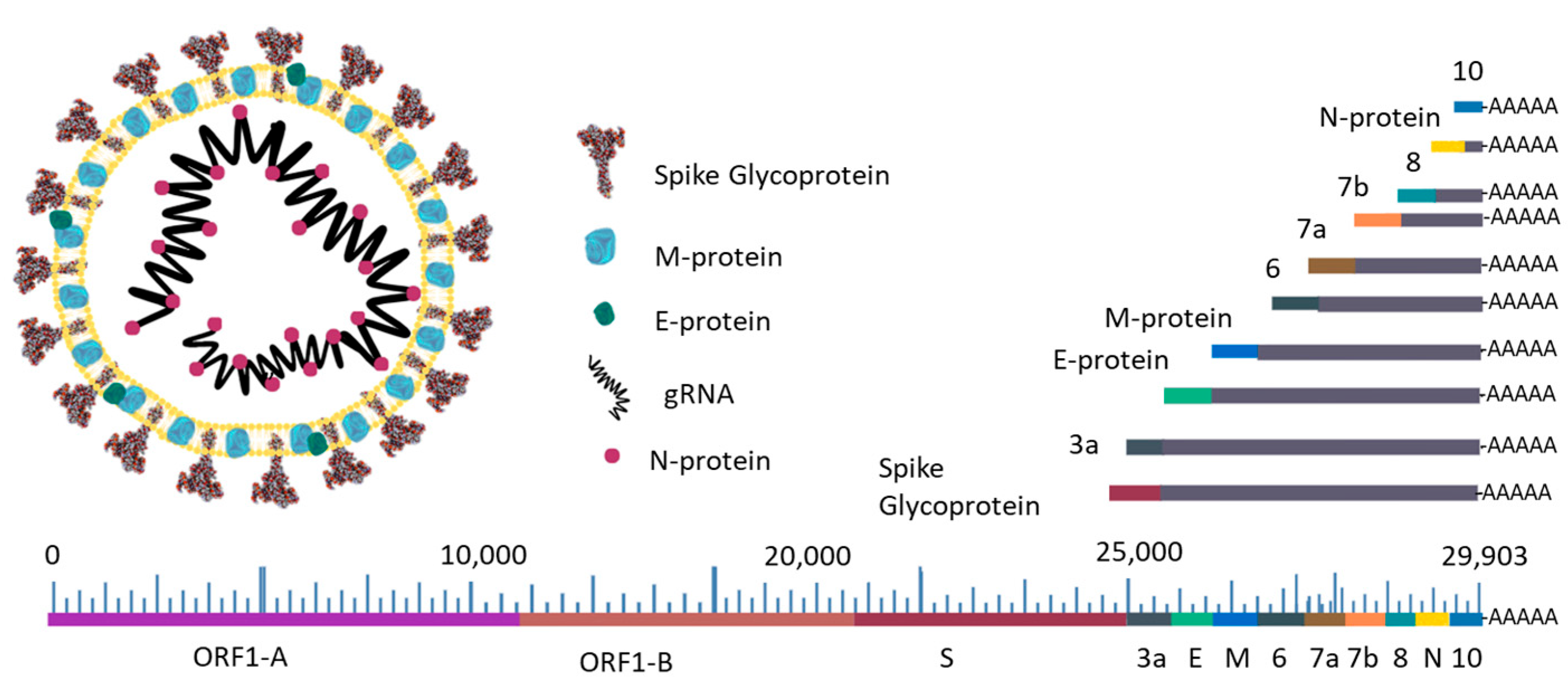
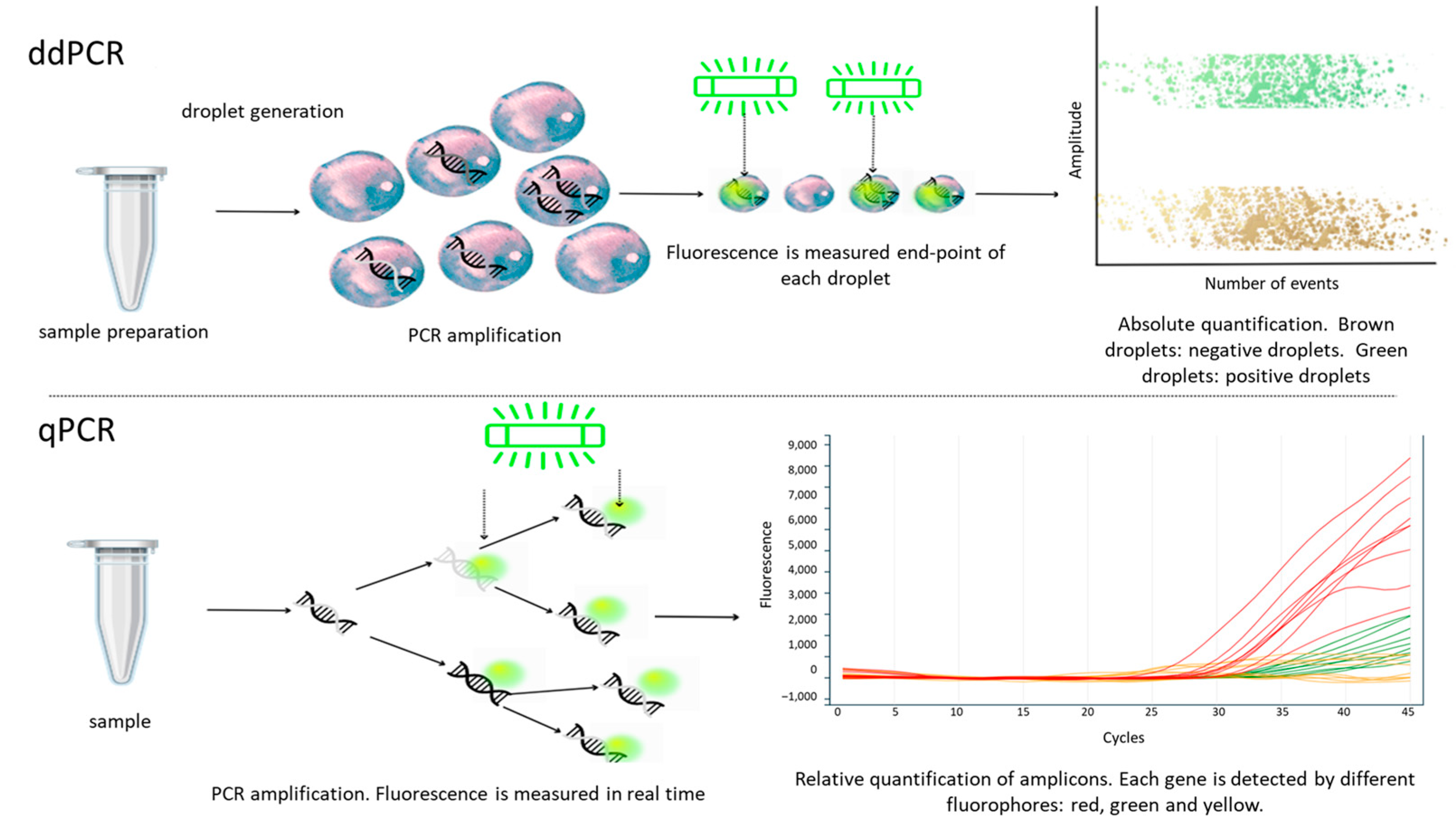
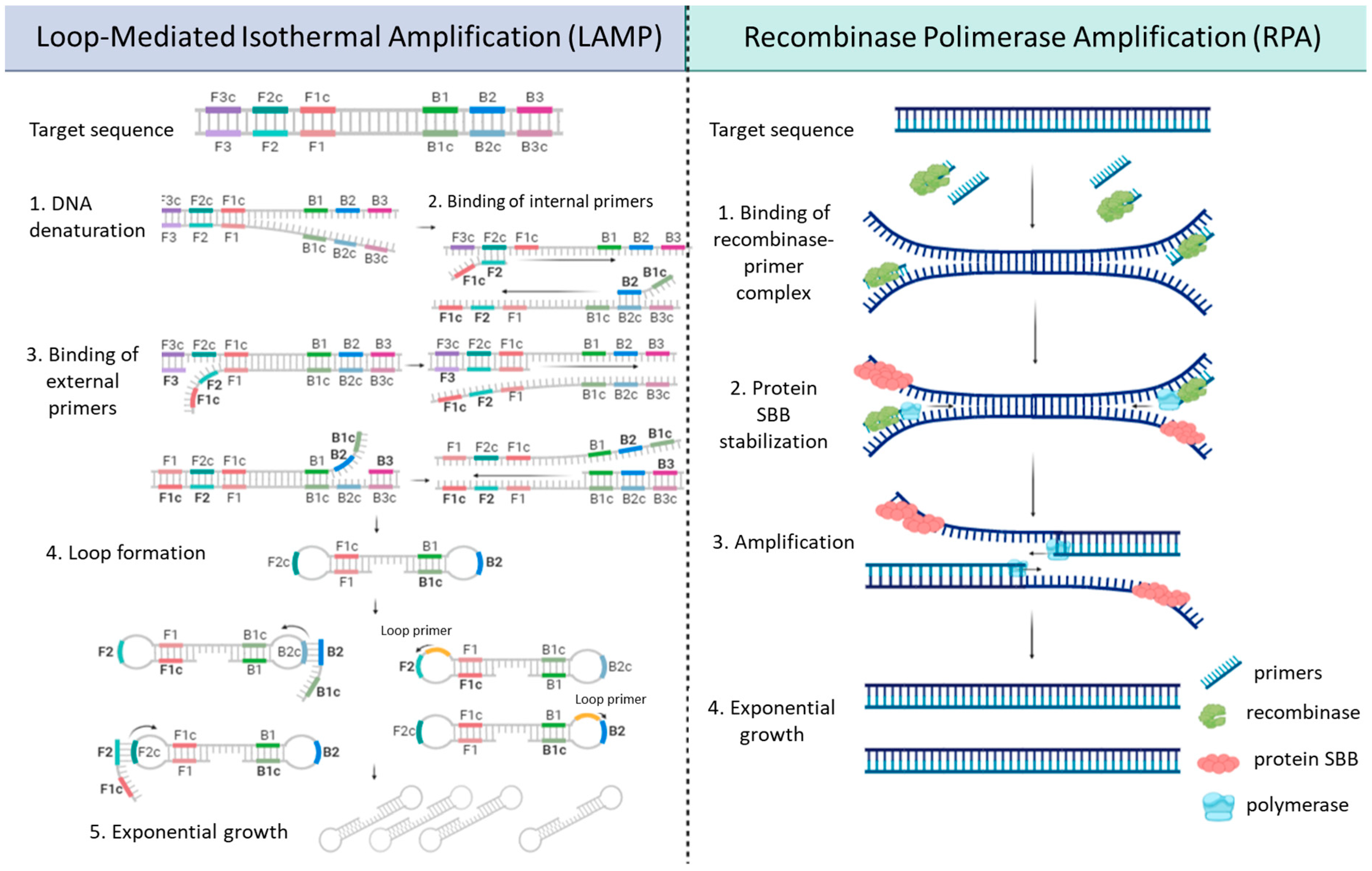
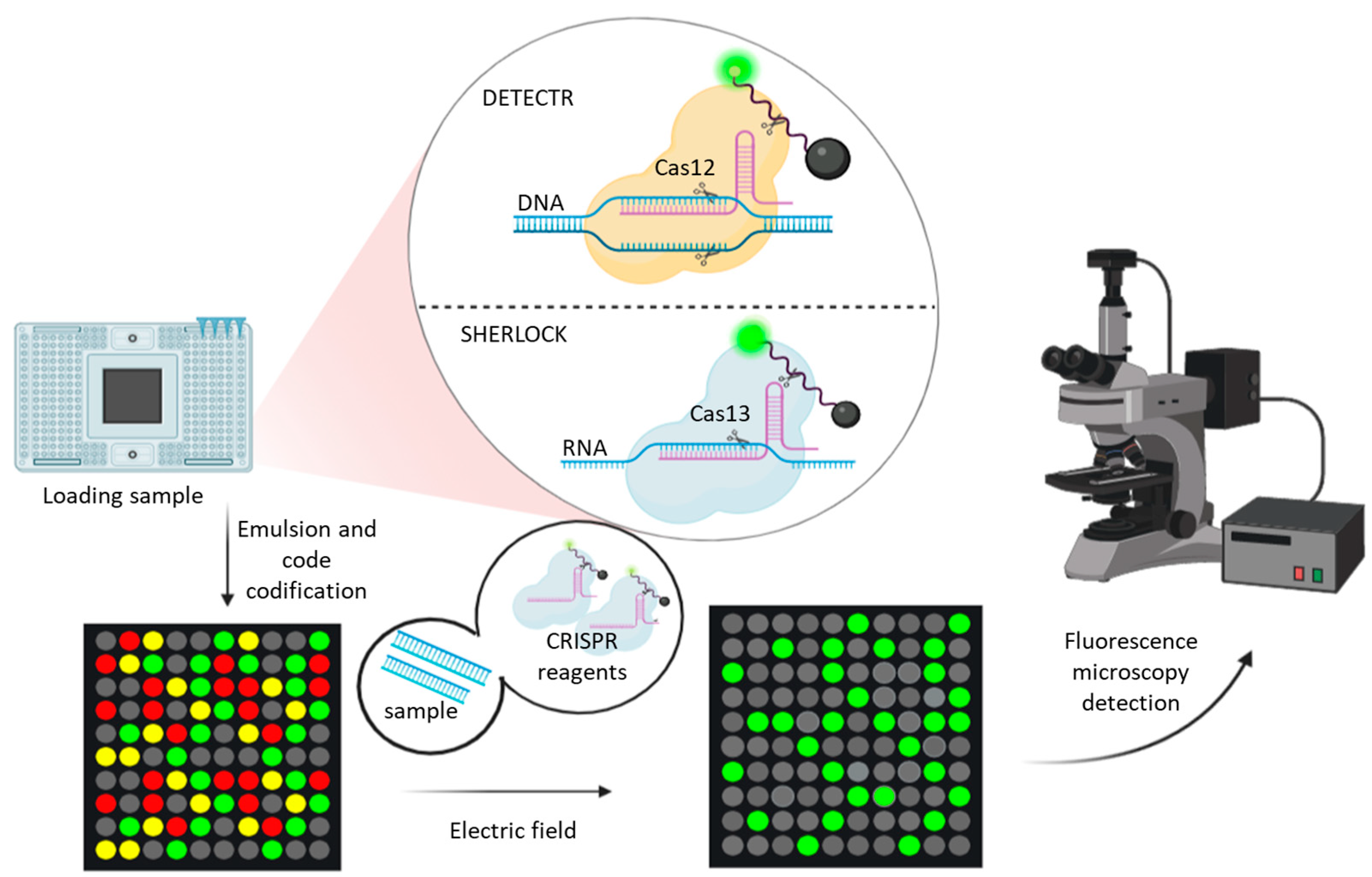
| VOIs | |||
|---|---|---|---|
| Pango Lineage | Nexstrain Clade | Genetic Features | Date of Designation and Risk Assessment |
| XBB.1.5 | 23A | Recombinant of BA.2.10.1 and BA.2.75 sublineages, namely BJ1 and BM.1.1.1, with a breakpoint in S1 XBB.1 + S:F486P (similar spike genetic profile as XBB.1.9.1) | 24 February 2023 |
| XBB.1.16 | 23B | Recombinant of BA.2.10.1 and BA.2.75 sublineages, i.e., BJ1 and BM.1.1.1 XBB.1 + S:E180V, S:K478R and S:F486P | 17 April 2023 |
| VUMs | |||
| BA.2.75 | 22D | BA.2 + S:K147E, S:W152R, S:F157L, S:I210V, S:G257S, S:D339H, S:G446S, S:N460K, S:Q493R reversion | 6 July 2022 |
| CH.1.1 | 22D | BA.2.75 + S:L452R, S:F486S | 8 February 2023 |
| BQ.1 | 22E | BA.5 + S:R346T, S:K444T, S:N460K | 21 September 2022 |
| XBB* | 22F | BA.2 + S:V83A, S:Y144-, S:H146Q, S:Q183E, S:V213E, S:G252V, S:G339H, S:R346T, S:L368I, S:V445P, S:G446S, S:N460K, S:F486S, S:F490S | 12 October 2022 |
| XBB.1.9.1 | Not assigned | Recombinant of BA.2.10.1 and BA.2.75 sublineages, i.e., BJ1 and BM.1.1.1 XBB.1 + S:F486P (similar spike genetic profile as XBB.1.5) | 30 March 2023 |
| XBB.1.9.2 | Not assigned | Recombinant of BA.2.10.1 and BA.2.75 sublineages, i.e., BJ1 and BM.1.1.1, XBB.1 + S:F486P, S:Q613H | 26 April 2023 |
| Type of Test | Name/Manufacturer | Target | Limit of Detection | Time-to-Result | Extraction of RNA/Pretreatment | Reference |
|---|---|---|---|---|---|---|
| RT-qPCR | ||||||
| RT-qPCR | Cepheid Xpress® GenXpert | gene N, gene E | 0.005 and 0.02 pfu/mL, respectively | 45 min | Sample is mixed and transferred to the cartridge and loaded onto the system | [116] |
| Isothermal amplification | ||||||
| RT-LAMP | Abbott ID Now™ | gene RdRp | 125 GE/mL with variations in different studies | ≤13 min | Sample is transferred to the cartridge to the test base, initiating target amplification | [117] |
| RT-RPA/LF * | RT-RPA/LF | gene N | 1 copy/µL | 25 min | Without RNA (extraction infected samples with RNA) | [53] |
| CRISPR-Cas based systems | ||||||
| CRISPR-Cas13/LF * | CASSPIT | genes S and N | ~100 copies | ≤1 h | Without RNA extraction (untreated samples) | [66] |
| Isothermal amplification with CRISPR-Cas | ||||||
| RT-LAMP or RT-RPA/CRISPR-Cas13 * | SHERLOCK | DNA/RNA | 2 aM | 50 min | Without RNA extraction (heating samples) | [64] |
| RT-LAMP/CRISPR-Cas12b | CRISPR-SPADE | gene N of each VOC | 15 copies/µL α 25 copies/µL β 50 copies/µL γ 12 copies/µL δ | 30 min | Without RNA extraction (in vitro RNA transcripts chemically synthetized) | [65] |
| RT-LAMP/CRISPR-Cas13 * | STOP-COVID | gene N | 100 copies of the viral genome | 40–70 min depending on LF or fluorescence, respectively | RNA extraction using magnetic beads | [67] |
| NAATs-Microfluidics | ||||||
| RT-RPA/CRISPR-Cas12a | µPAD CRISPR | genes S and N | 100 copies of the viral genome | 1 h | RNA extraction of 15 min | [100] |
| RT-LAMP/CRISPR-Cas | Dµchip | SARS-CoV-2, influenza A H1N1, H3N2 e influenza B RNA | 10 copies | 55 min | RNA extraction separated from the chip | [101] |
| RT-LAMP/cartridge and smartphone | RT-LAMP 3D cartridge | gene N | 50 copies RNA (VTM), 5 × 104 copies in nasal solution | ≤40 min | Without RNA extraction (lysis 1 min using heat, 95 ºC) | [102] |
| RT-RAA/CRISPR-Cas | Centrifugal microfluidics | gene E | 1 copy/µL | 30 min | RNA extraction separated from the chip | [109] |
| Isochatophoresis-RT-LAMP/CRISPR-Cas | ITP/-RT-LAMP | gene E, gene N and human RNAse P | 10 copies/µL | 35 min | Without RNA extraction (untreated samples) | [111] |
| Nanomedicine | ||||||
| gold nanoparticle layer with antibodies | SERS-biosensor | protein S | 6.07 fg per mL | Not specified | Without RNA extraction (untreated samples) | [107] |
| AuNPs/plasmonic sensor and smartphone | Nanoplasmonic sensor | protein S | 370 viral particles/mL | 15 min | Without RNA extraction (SARS-CoV-2 pseudovirus) | [96] |
| Aptamers/CRISPR-Cas12a and potentiostat | Aptamers | S1 domain Spike | 1.5 pg/mL | 30 min | Without RNA extraction (untreated samples) | [112] |
| Nanomedicine-microfluidics | ||||||
| Hybridization DNA walker with a functionalized glass slide | DNA walker/glass slide | two parts of RdRp gene | 1.19 pM | 30 min | RNA extraction with commercial kit | [106] |
| Hybridization with a functionalized glass slide | Biosensor glass slide | RNA/DNA | 10 aM | 15 min | Fast RNA extraction automated | [105] |
| RT-RPA in gold-layer functionalized chip | NanoPEIA | gene E and orfab1 | 28.5 and 23.3 copies per mL | 6 min in Ct ≤ 25 | Samples lysed 95 °C 5 min | [108] |
| FTO electrode functionalized with antibodies | eCovSens | antigen protein spike | 90 fM | 10–30s | Without RNA extraction (buffer samples and spiked saliva samples) | [110] |
| capacitive biosensor | Capacitive biosensor | protein Spike | ≈760 pg/mL–76 ng/mL | 15–20 min | Without RNA extraction (spiked sample in phosphate-buffered saline) | [113,114] |
| SHERLOCK in a face mask-integrated sensor | FDCF Wearable face mask | gene S | 500 copies/17 aM | ~1.5 h | Viral lysis with lyophilized compounds | [115] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorta-Gorrín, A.; Navas-Méndez, J.; Gozalo-Margüello, M.; Miralles, L.; García-Hevia, L. Detection of SARS-CoV-2 Based on Nucleic Acid Amplification Tests (NAATs) and Its Integration into Nanomedicine and Microfluidic Devices as Point-of-Care Testing (POCT). Int. J. Mol. Sci. 2023, 24, 10233. https://doi.org/10.3390/ijms241210233
Dorta-Gorrín A, Navas-Méndez J, Gozalo-Margüello M, Miralles L, García-Hevia L. Detection of SARS-CoV-2 Based on Nucleic Acid Amplification Tests (NAATs) and Its Integration into Nanomedicine and Microfluidic Devices as Point-of-Care Testing (POCT). International Journal of Molecular Sciences. 2023; 24(12):10233. https://doi.org/10.3390/ijms241210233
Chicago/Turabian StyleDorta-Gorrín, Alexis, Jesús Navas-Méndez, Mónica Gozalo-Margüello, Laura Miralles, and Lorena García-Hevia. 2023. "Detection of SARS-CoV-2 Based on Nucleic Acid Amplification Tests (NAATs) and Its Integration into Nanomedicine and Microfluidic Devices as Point-of-Care Testing (POCT)" International Journal of Molecular Sciences 24, no. 12: 10233. https://doi.org/10.3390/ijms241210233
APA StyleDorta-Gorrín, A., Navas-Méndez, J., Gozalo-Margüello, M., Miralles, L., & García-Hevia, L. (2023). Detection of SARS-CoV-2 Based on Nucleic Acid Amplification Tests (NAATs) and Its Integration into Nanomedicine and Microfluidic Devices as Point-of-Care Testing (POCT). International Journal of Molecular Sciences, 24(12), 10233. https://doi.org/10.3390/ijms241210233








