Abstract
Cold stress usually causes the abscission of floral organs and a decline in fruit setting rate, seriously reducing tomato yield. Auxin is one of the key hormones that affects the abscission of plant floral organs; the YUCCA (YUC) family is a key gene in the auxin biosynthesis pathway, but there are few research reports on the abscission of tomato flower organs. This experiment found that, under low temperature stress, the expression of auxin synthesis genes increased in stamens but decreased in pistils. Low temperature treatment decreased pollen vigor and pollen germination rate. Low night temperature reduced the tomato fruit setting rate and led to parthenocarpy, and the treatment effect was most obvious in the early stage of tomato pollen development. The abscission rate of tomato pTRV-Slfzy3 and pTRV-Slfzy5 silenced plants was higher than that of the control, which is the key auxin synthesis gene affecting the abscission rate. The expression of Solyc07g043580 was down-regulated after low night temperature treatment. Solyc07g043580 encodes the bHLH-type transcription factor SlPIF4. It has been reported that PIF4 regulates the expression of auxin synthesis and synthesis genes, and is a key protein in the interaction between low temperature stress and light in regulating plant development.
1. Introduction
The plant hormone auxin plays a central role in shaping plant growth and development [1]. Auxin also plays a central role in control of organ abscission, and it is thought that changes in the auxin gradient across the abscission zone are the primary determinant of the onset of abscission [2]. After decades of genetic and biochemical studies, numerous core molecular components and their networks, underlying auxin biosynthesis, and transport and signaling have been identified [3,4]. The TAA/YUC pathway is the major endogenous auxin biosynthetic pathway involved in major biological processes mediated by auxin activity, and its conservation in the plant kingdom has been functionally examined in many plant species [5]. Many studies have shown that several YUC genes are highly expressed in seed tissues such as maize [6], rice [7], melon [8], and strawberry [9], suggesting that auxin biosynthesis via the TAA/YUC pathway may be dominant in flower and fruit development. AUXIN RESPONSE FACTOR 17 (AtARF17) directly binds to the promoters of CALS5 and MYB108 to regulate pollen wall formation [10], and overexpression of ARF17 in Arabidopsis leads to defects in tapetum development and male sterility [11]. OsPID regulates the transport of auxin by phosphorylating PIN1, changes the polar distribution of auxin, and regulates the production and development of rice floral organs; OsPID also participates in the regulation of rice floral organ development by interacting with transcription factors such as OsMADS16 and LAX1 [12]. OsYUC11-mediated auxin biosynthesis regulates rice endosperm development [13]. OsFTIP7 interacts with OsH1 and promotes its nuclear localization, directly represses the transcription of the auxin biosynthesis gene OsYUCCA4, and down-regulates auxin levels after stage 9, leading to up-regulation of JA levels required for anther dehiscence and pollen maturation [14]. Recent studies have excavated the homologous genes of YUCCA (YUC), a key rate-limiting enzyme for auxin synthesis in maize, and found that ZmYUC2 and ZmYUC4 are specifically expressed in the aerial root tips of maize and regulate the local synthesis of auxin in aerial root tips. This then affected the gravity and growth angle of aerial roots [15]. Elevated cytoplasmic levels of H2O2 caused a suppressed auxin signal in the early abscission stage and enhanced ETH production during abscission [16].
Tomatoes, an important fruit and vegetable in China, are often subjected to low-temperature stress during production, especially in protected cultivation. Low temperature can regulate the transport and signal transduction pathway of auxin [17], but there are few reports on the effect of low temperature on auxin biosynthesis [18]. Low temperature may reduce the auxin content in the ovary by causing stamen abortion, eventually leading to flower drop and fruit drop. Studies have shown that exogenous application of auxin can effectively rescue low-temperature-induced flower drop [19]. Studies have shown that low temperature affects auxin biosynthesis, transport, and signal transduction. The low temperature treatment of apple seedlings resulted in a significant decrease in auxin content in their roots [20]. Further transcriptome data confirmed that low temperatures suppress the expression of the auxin synthesis gene YUC2 [21]. However, there are also results of increased auxin content at low temperature. For example, after wheat was treated at 4 °C for 21 days, the IAA content on the spikes increased significantly [22]. The effect of low temperature on auxin synthesis may have different effects due to differences in species and organs. YUC-dependent IAA biosynthesis plays a role in Arabidopsis seedling cotyledon somatic embryogenesis [23]. Recent studies on apical hook formation and maintenance found that cotyledon and shoot apical meristems are protected from mechanical damage during seedling emergence from soil, revealing an alternative auxin signaling mechanism [24]. Relatively low auxin levels on the convex side of the apical hook promote cell elongation, whereas higher auxin levels on the concave side [25] inhibit this process [26]. Higher KNOTTED1-LIKE HOMEOBOX PROTEIN1 (SlKD1) and FRUITFULL (SlFUL2) expression in the abscission zone (AZ), thereby perturbing the auxin response gradient and causing increased ethylene production, eventually lead to the initiation of abscission [27]. There have been new findings indicating that SlHB15A mediates the antagonistic effect of auxin and JA-Ile during tomato pedicel abscission, while auxin inhibits abscission through the SlHB15A-SlJAR1 module [28]. However, the involvement of YUC-mediated auxin in low-temperature flower and fruit drop is still unclear.
Tomato flower drop is mainly caused by biotic and abiotic stresses, with low temperature being the most common factor. Thus, studying the effect of mitigating or preventing low temperature on tomato flower drop has great practical value. Clarifying the relationship between low temperature, auxin, and tomato flower drop will provide a theoretical foundation for preventing low temperature from affecting tomato flower drop. In our previous study, through bioinformatics and qRT-PCR analysis, members of the auxin synthesis gene family YUC were found in the tomato genome and SlFZY2, SlFZY3, SlFZY4-1, SlFZY5 were highly expressed in tomato floral organs; then, through field treatment experiments, the critical period of low temperature affecting tomato fruit setting rate was determined; virus-induced gene silencing (VIGS) technology was used to determine the effect of low night temperature on tomato flower organ abscission. The key genes of auxin synthesis, also through transcriptome sequencing, clearly show that low night temperature affects the expression of genes related to tomato stamen development. This study will lay the foundation for further analysis of the mechanism by which low temperature regulates auxin synthesis and affects tomato flower drop. Subsequently, it will provide a theoretical basis for preventing low temperature from affecting tomato flower drop.
2. Results
2.1. Effect of Day and Night Low Temperature Treatments on the Abscission Rate of Tomato Pedicel
The environment is an important factor leading to tomato flower and fruit drop, and low temperature is one of the environmental stresses that warm crops often suffer from. This study investigated the abscission rate of tomato flower stalks under low temperature stress day and night (Figure 1). The results are as follows: after being treated with low temperature 16/6 °C day and night, tomato flower stalks showed abscission at 4 h after flowering, and the abscission rate at 6 h was significantly higher than the control. The abscission rate at 16 h reached 100%; the tomato flower stalks in the control group fell off at 6 h and all fell off at 20 h. Research shows that low temperature treatment day and night promotes and accelerates the occurrence of tomato floral organ abscission.
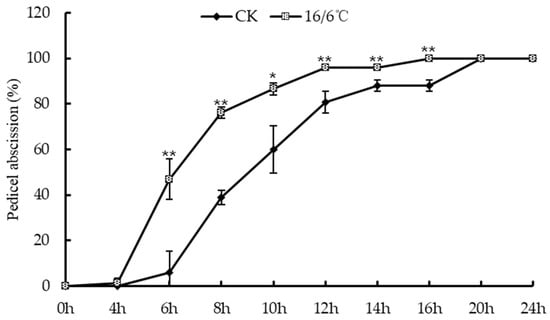
Figure 1.
Peeling off rate of tomato petiole under low temperature treatment. * Indicates significant difference (p < 0.05, Student’s t-test), ** indicates extremely significant difference (p < 0.01, Student’s t-test).
2.2. Effect of Day and Night Low Temperature Treatments on the Vitality and Germination Rate of Tomato Pollen
In this study, tomato pollen vitality and germination were observed under a laser confocal microscope using flowers that were fully opened under both day and night low temperature treatment and control (Figure 2A,B,D,E). I2-KI staining was used to observe the vitality of the tomato pollen. The pollen with deeper staining had higher vitality, while the pollen with lighter staining had lower vitality. After low temperature treatment at 16/6 °C, the pollen vitality was significantly lower than that of the control. Under low temperature, the pollen vitality was 51.84%, while the control was 99.67% (Figure 2C). Stamens were taken from low temperature and control flowering periods at day and night, the pollen was shaken out and added to the germination solution. After 0.5 h, the germination was observed under a microscope. The results showed that the pollen germination rate under low temperature was significantly lower than the control, with a pollen germination rate of only 31.34% under low temperature and 89.39% under the control (Figure 2F).
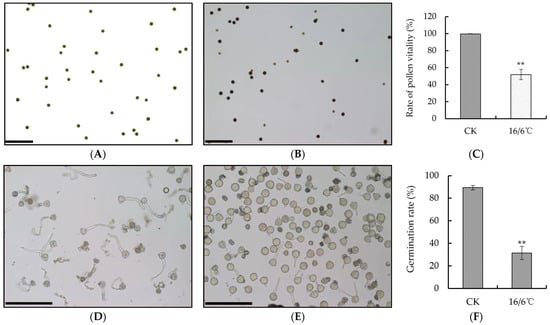
Figure 2.
(A,B) The dyeing situation of tomato pollen vigor at 25/15 °C and 16/6 °C respectively, scale bar = 100 μm; (C) statistics of pollen vitality; (D,E) the pollen germination of tomato of 25/15 °C and 16/6 °C number, scale bar = 100 μm; (F) statistics of pollen germination rate. ** Indicates extremely significant difference (p < 0.01, Student’s t-test).
2.3. Effects of Day and Night Low Temperature on the Expression of Tomato Auxin Synthesis Genes
Auxin is one of the important hormones regulating organ shedding. In this study, in order to understand the mechanism of tomato pedicel shedding under diurnal low temperature, the expression level of the auxin synthesis gene SlFZY in pistils (Figure 3A) and stamens (Figure 3B) was analyzed after the tomato seedlings were subjected to low temperature. After diurnal low temperature treatment, the expression trends of different genes in different reproductive organs of tomato were not consistent. The expression levels of SlFZY3 and SlFZY4-1, which were highly expressed in stamens, were up-regulated after low temperature treatment, but the expression levels in pistils were significantly lower than those of the control. SlFZY1 and SlFZY2 were opposite to these two genes, that is, they were lower in tomato stamens after 16/6 °C treatment, but higher in pistils. SlFZY5 was significantly higher in tomato stamens and pistils than the control. The expression levels of SlFZY4-2 and SlFZY6 in tomato stamens had no significant difference between diurnal low temperature treatment and control, but in tomato pistil, the expression levels of these two genes were significantly lower than those of control after diurnal low temperature treatment.
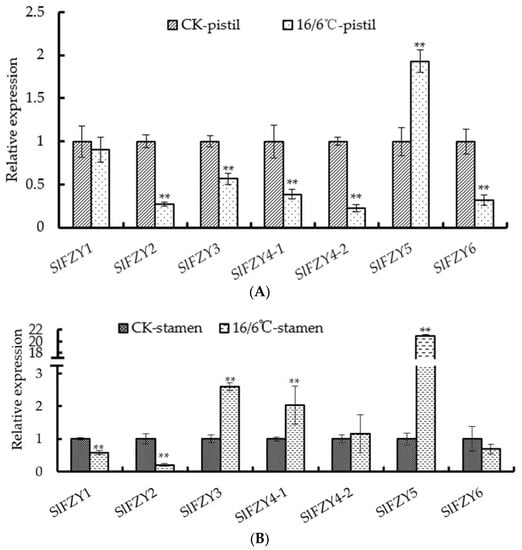
Figure 3.
Analysis of expression levels of SlFZY in pistils (A) and stamens (B) in tomatoes under low temperature treatment. ** Indicates extremely significant difference (p < 0.01, Student’s t-test).
2.4. Effect of Low Night Temperature on Fruit Setting Rate of Tomato
In this study, ‘AC’ tomato was used as the test material, and the low night temperature treatment of 25/6 °C was carried out in the artificial climate chamber at the early stage of flower bud differentiation, the stage of stamen primordium differentiation, the stage of carpel formation, the early stage of pollen development, and the flowering stage until the fruit setting stage. The statistical fruit setting rate is shown in Figure 4A. After investigation, it was found that the tomato fruit setting rate in the first four periods was significantly lower than that of the control, and the fruit setting rate in the fifth period was not significantly different from that of the control (Figure 4B). After the tomato fruit was cut crosswise, it was sent down. The low night temperature treatment led to parthenocarpy. The plants in the first four stages were all under low night temperature treatment from the early stage of pollen development to fruit setting, and there was no significant difference in fruit setting rate. Therefore, the early stage of selective pollen development is a critical period that affects the fruit setting rate after low temperature stress.
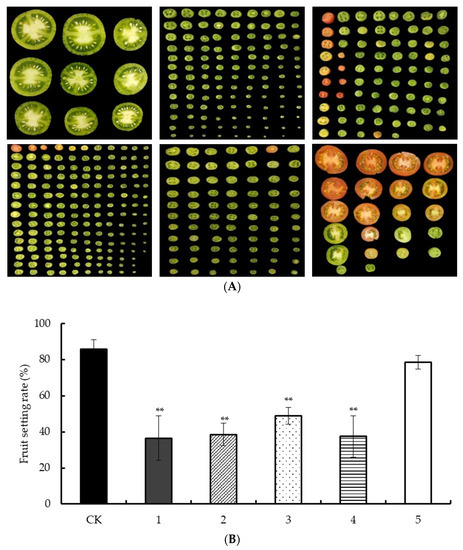
Figure 4.
Investigation of tomato fruit set rate under low temperature treatment in different periods: in (A) from top to bottom, from left to right, they are CK, flower bud differentiation stage, stamen primordium differentiation stage, carpel formation stage, pollen development stage, and flowering stage, respectively; (B) uses CK, 1, 2, 3, 4, and 5 to represent, respectively. ** Indicates extremely significant difference (p < 0.01, Student’s t-test).
2.5. Effect of Low Night Temperature on Pollen Vitality of Tomato
The pollen was taken from the tomato at flowering stage, and the pollen vigor and germination rate were counted under low night temperature treatment and control respectively (Figure 5A–F). The staining of tomato pollen I2-KI observed under the microscope after low temperature treatment was lighter (Figure 5A,B,D,E), and the statistics found that its viability was 46.24%, which was significantly lower than that of the control 95.74% (Figure 5C). The pollen germination rate of ‘AC’ tomato after low night temperature treatment was 72.28%, which was significantly lower than that of the control 87.32% (Figure 5F).
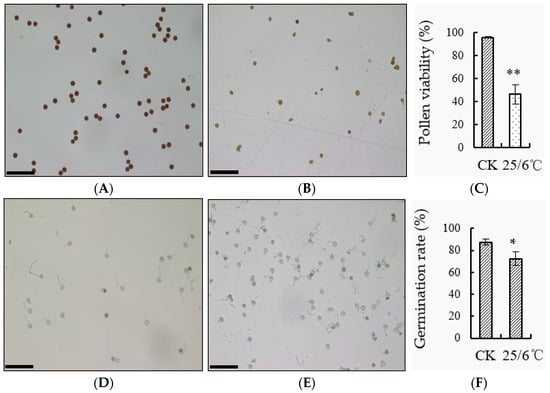
Figure 5.
Effect of low night temperature on pollen vitality of tomato. (A,B) The dyeing situation of tomato pollen vigor under low temperature treatment at 25/15 °C and 25/6 °C, respectively, scale bar = 100 μm; (C) statistics of pollen viability; (D,E) the pollen germination of tomato under low temperature treatment at 25/15 °C and 25/6 °C, respectively, scale bar = 100 μm; (F) statistics of pollen germination rate. * Indicates significant difference (p < 0.05, Student’s t-test), ** indicates extremely significant difference (p < 0.01, Student’s t-test).
2.6. Investigation on Pedicel Abscission Rate of Silenced Tomato Plants with pTRV-SlFZY
In order to preliminarily study the functions of the SlFZY family of genes found in tomato, this study used VIGS technology to silence four genes with high expression levels in floral organs and investigated tomato flower stalk abscission (Figure 6). The results showed that the flower organ abscission rate of pTRV-Slfzy2 and pTRV-Slfzy4 tomato silenced plants was not significantly different from that of the control. The abscission rate of floral organs of pTRV-Slfzy3 and pTRV-Slfzy5 silenced plants was extremely significantly higher than that of the control at 8 h after anthesis, significantly higher at 10 h, and higher than that of the control at 12 h to complete abscission. The tomato floral organ of pTRV-Slfzy2fzy3 was significantly higher than that of the control at 10 h and 12 h after deflowering, and completely abscised after 14 h pTRV-Slfzy2fzy4 tomato floral organs abscised at 4 h and completely abscised at 14 h, and the abscission rate at 8 h was 58.33%, which was significantly higher than that of the control (30.95%). The shedding rate of pTRV-Slfzy2fzy5 at 10 h was 88.19%, which was extremely significantly higher than that of the control, at which point the shedding rate of the control was 68.81%.
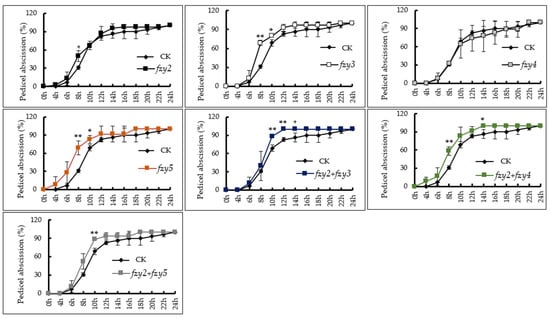
Figure 6.
Petiole shedding rate of pTRV-Slfzy silent plants that were highly expressed in tomato flower organs. * Indicates significant difference (p < 0.05, Student’s t-test), ** indicates extremely significant difference (p < 0.01, Student’s t-test).
2.7. Investigation on Pollen Viability of Silenced Tomato Plants with pTRV-SlFZY
I2-KI staining was used to observe the pollen viability of tomato pTRV-Slfzy silenced plants (Figure 7A–H). According to staining and statistics, only the pollen viability of pTRV-Slfzy2fzy5 silenced plants was significantly lower than that of the control, and there was no significant difference between the other silenced plants and the control (Figure 7I).
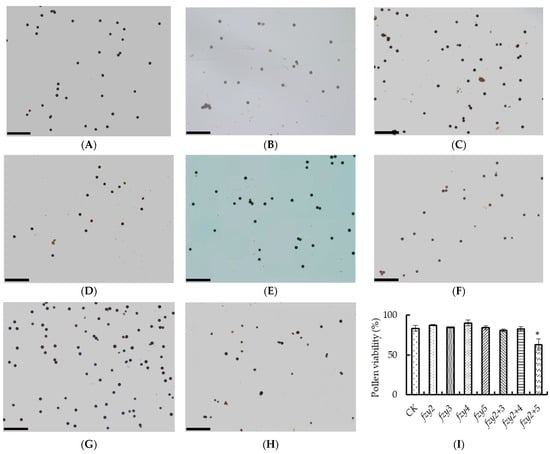
Figure 7.
(A–H) pTRV-Slfzy silent plant tomato pollen vitality dyeing was observed by I2-KI staining. (A) Represents CK; (B–H) represent pTRV-Slfzy2, pTRV-Slfzy3, pTRV-Slfzy4, pTRV-Slfzy5, pTRV-Slfzy2+3, pTRV-Slfzy2+4, pTRV-Slfzy2+5, respectively, scale bar = 100 μm; (I) statistics of pollen vitality. * Indicates significant difference (p < 0.05, Student’s t-test).
2.8. High-Throughput Transcriptome Sequencing Analysis of Tomato Stamens under Low Night Temperature Treatment
Selecting an appropriate reference genome is an important step in information analysis. The sequence data clean data are compared with the reference genome, and the data comparison rate reflects the similarity between the sequenced sample and the selected reference genome. As shown in Table 1, the total mapped percentages of the three samples (CK1, CK2, CK3) under the control and the three samples (LT-1, LT-2, LT-3) under the low night temperature treatment are greater than 70%, multiple mapped is less than 10%, all of which meet the follow-up analysis requirements.

Table 1.
The match rate of RNA-seq data and reference genome.
In the volcano map of differential genes in tomato stamens under low night temperature treatment, when the q value of a gene is less than or equal to 0.05, and the expression changes by more than 2 times, the gene is considered to be a differential gene. If log2 (Fold Change) is less than −1, it means that the differential gene is significantly down-regulated, and if it is greater than 1, it means that the significant differential gene is up-regulated. Under the low night temperature treatment, there were 92 differential genes in tomato flowering stamens, of which 84 genes were down-regulated and 8 genes were up-regulated (Figure 8A).
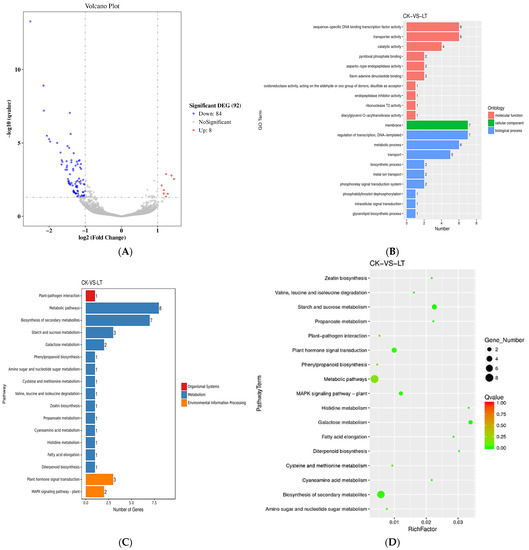
Figure 8.
Transcriptome analysis of tomato stamens at low night temperature. (A) Differential gene volcano map; (B) GO enrichment histogram; (C) annotated histogram of significantly enriched KEGG annotation; (D) scattered graph of KEGG enrichment of differential genes.
The GO enrichment histogram of differential genes intuitively reflects the number distribution of molecular functions, cellular components, and biological processes. As shown in Figure 8B, in the GO enrichment of differential genes in tomato stamens under low night temperature treatment, 26 differential genes were enriched in molecular functions, 7 differential genes were enriched in cellular components, and 27 differential genes were enriched in set in biological processes.
In organisms, different genes participate in plant metabolism and signaling pathways through mutual cooperation, and through pathway significance enrichment analysis, the most important biochemical metabolic pathways and signal transduction pathways in which differentially expressed genes participate can be determined. KEGG enrichment analysis of differential genes in tomato stamens under low night temperature treatment found that there were five differential genes in environmental signaling pathways, of which three were involved in plant hormone signal transduction pathways and two were involved in plant MAPK pathways (Figure 8C,D).
Through KEGG analysis, we found that there were four down-regulated genes enriched in the plant hormone signal transduction pathway, namely Solyc07g043580 encoding the bHLH-type transcription factor SlPIF4, the expression of SlPIF4 was significantly down-regulated at low night temperature; the gene encoding SlRRA7 protein Solyc06g048600; Solyc06g048930 encoding TRR16-17 protein, SlRRA7 and TRR16-17 are induced by cytokinin in tomato root; Solyc12g008900 encoding SlCKX6 protein, SlCKX6 is induced by cytokinin in tomato root (Table 2). Solyc09g009110 encodes GA20 oxidase, which can catalyze the production of gibberellins with biological activity. MAPK3, encoded by Solyc06g005170, is a member of the tomato protein kinase gene family that plays an important role in mediating responses to biotic and abiotic stresses. SlWRKY33 encoded by Solyc09g014990 is a member of the WRKY transcription factor gene family and has been related to multiple biological processes in plants. It has a C2H2 zinc finger domain and its expression level is down-regulated under low temperature treatment. Solyc10g083290 encoded lin6, which is an extracellular invertase that catalyzes the cleavage of sucrose in the apoplast and its supply from the site of synthesis to the ‘sink’. Furthermore, lin6 is expressed in floral organs and roots, induced by glucose and zeatin. The 3-methyl-2-oxobutyrate dehydrogenase encoded by Solyc06g05985 is a component of E1, and the expression of it and Solyc09g009110 genes were up-regulated at low temperature (Table 2).

Table 2.
Differential genes.
3. Discussion
Auxin is almost accompanied by the whole process of plant growth and development, and its locality is crucial to plant growth and development [29]. Low temperature stress is an important environmental stress that affects plant development, so it is speculated that there may be a regulatory relationship between low temperature stress and auxin. The transportation and signal transduction of auxin in tomato have been extensively studied; however, the research on its biosynthesis is still limited [30]. Under low temperature stress, the expression of auxin synthesis genes increased in stamens, but decreased in pistils. Low temperature treatment decreased pollen vigor and pollen germination rate. Low night temperature reduced the tomato fruit setting rate and led to parthenocarpy, and the treatment effect was most obvious in the early stage of tomato pollen development. The abscission rate of tomato pTRV-Slfzy3 and pTRV-Slfzy5 silenced plants was higher than that of the control, which is the key auxin synthesis gene affecting the abscission rate. The expression of Solyc07g043580 was down-regulated after low night temperature treatment. Solyc07g043580 encodes the bHLH-type transcription factor SlPIF4.
3.1. Low Temperature Promotes Abscission of Tomato Flower Organs
Auxin is locally synthesized [31,32,33], and analyzing the expression patterns of SlFZYs genes in different tissues in tomato is an important part of studying the function of this family of genes [34]. In our previous study, through qRT-PCR analysis, it was found that SlFZY2, SlFZY3, SlFZY4-1, SlFZY5 were highly expressed in tomato floral organs, and it was preliminarily speculated that these four genes regulate the development of tomato floral organs. The expression level of SlFZY3 was the highest in the underground part of tomato, the expression level of SlFZY5 was the highest in the stem and leaf, and the expression level of SlFZY4-2 was the highest in the young fruit.
Temperature stress includes high temperature and low temperature, which may be of reference to a certain extent [35]. At high temperature, HISTONE DEACETYLASE 9 (HDA9) accumulates, facilitating the H2A.Z removal from the YUC8 locus and providing a looser chromatin environment that allows PIF4-mediated activation of YUC8 transcription [36]. High temperatures additionally promote auxin biosynthesis through the temperature-specific recruitment of PIF4 to the promoters of the IPyA glycosylase UGT76F1 and the IAOx-pathway-related CYP79B2 gene to repress and promote their transcription, respectively, by unknown epigenetic mechanisms [37]. After the tomato was treated with diurnal low temperature during the flower bud stage, it was found that the diurnal low temperature promoted the abscission of tomato flower organs. Under diurnal low temperature stress, the expression of auxin synthesis genes increased in stamens, but decreased in pistils (Figure 3). The auxin in the tomato pedicel mainly comes from the base of the ovary, and the decrease of the auxin content in the ovary promotes the abscission of the pedicel. Diurnal low temperature treatment decreased pollen vigor and pollen germination rate (Figure 2). Stamen abortion is one of the important factors affecting flower and fruit drop. The increase of auxin content in rice stamens inhibits the synthesis of JA, which can lead to non-dehiscence of anthers and affect pollination [14]. The reduction of auxin content in pistil may be the result of poor pollination and fertilization.
3.2. The Early Stage of Pollen Development Is the Key Period When Low Night Temperature Affects the Fruit Setting Rate of Tomato
The traditional view is that auxin is mainly produced in vigorously divided apical meristems, young shoots, leaves, and germinating seeds, and is transported between cells through polar transport [18,38,39]. However, recent research advances have elucidated the molecular mechanism of auxin biosynthesis, revealing that its biosynthesis is not ubiquitous but localized [32,33]. Site-specific accumulation of auxin is regulated by the balance of cellular auxin efflux and local auxin biosynthesis [40]. The low temperature may affect the distribution of local auxin in the flower stalk, resulting in flower and fruit drop [41]. Low temperature is one of the main environmental factors affecting tomato yield, so this study screened the critical period when low night temperature affects tomato fruit set. Low night temperature reduced the tomato fruit setting rate and led to parthenocarpy, and the treatment effect was most obvious in the early stage of tomato pollen development, and the effect was less after tomato flowering, that is, after the pollen matured [42]. Through the detection of pollen viability and germination rate, it was found that low night temperature significantly reduced pollen viability and pollen germination rate. Low temperature at night leads to a decrease in fruit setting rate by affecting the development of pollen (Figure 4). Previous studies have shown that Arabidopsis yuc2yuc6 mutants have reduced pollen vigor [43] and a lower seed setting rate than the wild type, indicating that auxin can regulate pollen development. It is preliminarily speculated that the increase of auxin content in stamens leads to decreased stamen fertility, developmental defects in unfertilized pistils, and the down-regulation of auxin synthesis gene expression levels, resulting in reduced transport of auxin to flower stalks, and eventually flower and fruit drop and parthenogenesis solid physiology.
3.3. SlFZY3, SlFZY5 Are the Main Auxin Synthesis Genes Affecting Flower Organ Abscission in Tomato
In this study, VIGS technology was used to verify the effect of auxin synthesis genes in tomato on abscission. The results found that pTRV-Slfzy2 and pTRV-Slfzy4 silenced plants had no significant difference in flower stalk abscission compared with wild type. After 8 h, the tomato pedicel shedding rate was significantly higher than that of the control, indicating that the gene family in tomato is functionally redundant. It is known that this family has been confirmed to have functional redundancy in Arabidopsis [31]. The conclusion of this experiment is consistent with that of the control group. The abscission rate of tomato pTRV-Slfzy3 and pTRV-Slfzy5 silenced plants was higher than that of the control. I2-KI staining showed that only when pTRV-Slfzy2fzy5 was simultaneously silenced, the pollen viability of tomato was significantly lower than that of the control, which indicated that auxin synthesis deficiency in tomato affects pollen development (Figure 6).
3.4. Solyc07g043580 May Be a Key Transcription Factor Regulating Auxin Synthesis Genes in Tomato Stamens
High-throughput transcriptome sequencing was performed on the stamens of tomato at flowering stage after low night temperature treatment. A total of 94 differential genes were found under low night temperature treatment, of which 8 were up-regulated and 84 were down-regulated. GO enrichment analysis found that 26 differential genes were enriched in molecular functions, 7 differential genes were enriched in cellular components, and 27 genes were enriched in biological processes. Through KEGG analysis, it was found that three genes were enriched in the plant hormone signal transduction pathway, namely Solyc07g043580, Solyc06g048600, and Solyc06g048930, and these three genes were all down-regulated after low night temperature treatment. Among them, Solyc07g043580 encodes the bHLH-type transcription factor SlPIF4. CBFs directly interact with PIF3, stabilizing the PIF3-phyB structure; on the one hand, inhibiting the expression of PIF1, PIF4, PIF5, and on the other hand, promoting the binding of ubiquitin ligase E3 to PIF1, PIF4, PIF5 protein and degrading it [44]. It is known that AtPIF4 can activate the expression of YUC4 in Arabidopsis [45], indicating that nighttime low temperature treatment in tomato stamens may affect auxin synthesis through the transcription factor encoded by Solyc07g043580, and low temperature stress and light play a role in regulating plant development. There are interactions between them. GO analysis showed that 2 genes were enriched in the MAPK signaling pathway, namely Solyc06g005170 and Solyc09g014990. These 2 genes encoded SlMAPK3 and SlWRKY33 respectively, and their expression levels were both down-regulated. The MAPK family is involved in plant response to abiotic stress, and it is speculated that low night temperature can affect auxin synthesis by regulating SlMAPK. MPK14, another auxin-activatable MPK family member, was reported to phosphorylate and enhance the stability of non-canonical Aux/IAA Aux/IAA33 in root tips with high auxin concentrations [46]. MPK1/2/14 were recently found to be activated by 1-naphthylacetic acid (NAA), suggesting a role for MPK family members in auxin signaling [47].
Low temperature may affect the expression of auxin synthesis genes through one or more transcription factors regulated by it, but it does not rule out the regulation of the expression of the FZYs family through epigenetics. It is speculated that there is functional redundancy in the tomato FZY family through VIGS, and further experiments are needed to verify the gene function. The effect of low temperature on the auxin content in tomato pistil may be through two ways: one is that low temperature regulates one or more transcription factors, which affects the expression of auxin synthesis genes in pistil, the other is that fertilization fails and auxin synthesis is reduced in the ovary. Low temperature and failure of pollination and fertilization may regulate auxin synthesis in tomato pistil through two different signaling pathways. Previous studies have shown that low temperature inhibits the transport of auxin, and low temperature may regulate the abscission of tomato floral organs by affecting the transport and signal transduction of auxin. Therefore, the research on auxin synthesis in tomato still needs to be further in-depth.
4. Materials and Methods
4.1. Plants and Growth Conditions
The tomato varieties tested in this experiment were ‘MicroTom’ and ‘Alisa Craig’ (AC). The ‘MicroTom’ was sown in the hole tray of the energy-saving solar greenhouse of the Facility Vegetable Research Base of Shenyang Agricultural University. When the seedlings sprouted flower buds, they were moved to the light incubator for treatment. The day and night low temperature treatment was 16 °C during the day and 6 °C at night; 25 °C during the day and 15 °C at night. The photoperiod was 12/12 h; the humidity was 65%; the light intensity was 400 μmol·m−2·s−1 [48]. The ‘AC’ was sown in the plug trays of the energy-saving solar greenhouse of Shenyang Agricultural University Facility Vegetable Research Base, and the flower bud differentiation stage, stamen primordium differentiation stage, carpel formation stage, pollen development stage, and flowering stage were respectively moved into artificial light source climate chambers. Then, night temperature treatment was carried out; night low temperature treatment temperature was 6 °C, control night temperature was 15 °C, and daytime temperature was 25 °C. The photoperiod was 12/12 h; the humidity was 65%; the light intensity was 400 μmol·m−2·s−1.
4.2. Extraction and Detection of Total RNA from Different Tomato Tissues
Take tomato roots, stems, leaves, flower buds, fully open flowers, stamens, pistils, and young fruits under normal growth conditions, and use Kangwei Century RNA Extraction Kit (CWBIO, Cambridge, MA, USA) to extract total RNA for analysis of the expression of the tomato FZY gene family in different tissues model. Tomato stamens and pistils under day and night low temperature treatment and low night temperature treatment were taken to prepare for analyzing the effects of different low temperature treatments on the expression of tomato FZY gene. The total RNA concentration was measured using a microplate reader (Thermo Fisher Scientific Inc., Waltham, MA, USA) and its integrity was checked by agarose gel electrophoresis [49].
4.3. Preparation of cDNA from Different Tissues of Tomato
The extracted total RNA was reverse transcribed using a TAKARA reverse transcription kit (Takara, Kusatsu, Japan). Reaction system: mix 4 μL; RNA + ddH2O 16 μL. After the reverse transcription procedure, the cDNA template was stored in a −20 °C low-temperature refrigerator.
4.4. Investigation on Abscission Rate of Tomato Floral Organs
Configuration of 1% agar medium: weigh 5 g of agar, add it to 500 g of distilled water, heat it twice in a microwave oven for 2 min each time, and stir it with a glass rod until it is completely dissolved. Pour the culture medium into a disposable plastic petri dish, cool and solidify at room temperature, and seal it with parafilm for later use [50,51]. The flower stalks of ‘MicroTom’ tomato at the flowering stage under the control and day and night low temperature treatment were taken, and the pistil base was removed with a double-sided knife, and then the proximal end was inserted into 1% agar medium, and the tomato flower organ abscission rate was measured in vitro. Taking the moment of inserting into the medium after deflowering as 0 h, the abscission rate was measured after 4 h, 6 h, 8 h, 10 h, 12 h, 16 h, 20 h, and 24 h, respectively. The number of pedicels that finally fell off was taken as the total number, that is, the shedding rate at this time was 100%; the shedding rate at other times was the percentage of the number of pedicels that fell off at this time to the total number of shedding [52].
4.5. Tomato Pollen Viability Assay
Configuration of I2-KI staining solution: weigh 25.4 g of I2 and 16.6 g of KI into a volumetric flask, add 900 mL of distilled water, dissolve completely, and dilute to 1 L [53]. Then pour into a brown threaded bottle and store in darkened light. At 9:00 in the morning, the flowers that fully opened under the control and low temperature treatment were taken, and the stamens were cut out with tweezers, cut into three sections with a double-sided blade, put into a 1.5 mL centrifuge tube, vortexed for five minutes, and the stamens were taken out with tweezers. Use a pipette gun to transfer 1 mL of staining solution into a centrifuge tube, then transfer the staining solution containing pollen into a cell culture dish and observe the staining situation with a laser microscope after 15 min [54].
4.6. Investigation on Pollen Germination Rate of Tomato
Weigh 2.62 g of MES into a volumetric flask, add 950 mL of sterilized water, and adjust its pH to 6.00 with NaOH. Take the tomato stamens under the control and low temperature treatment, shake out the pollen, add 1 mL pollen germination solution into the centrifuge tube with a pipette gun, then transfer the pollen germination solution containing pollen into the cell culture dish, and observe it with the laser microscope. After 30 min observe the germination [55].
4.7. Investigation on Tomato Fruit Setting Rate
‘Alisa Craig’ (AC) tomatoes treated with low temperature at night at different stages, when growing and developing to the fruiting stage, calculate the fruit setting rate of the first inflorescence.
4.8. RNA-Seq and Data Analysis
After low night temperature (25/6 °C) treatment at the early stage of tomato pollen development, the stamens at flowering stage were selected for high-throughput transcriptome sequencing. Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and mRNA was enriched with magnetic beads of Oligo (dT) for PCR amplification. Illumina Hiseq 4000 was used, followed by sequencing and bioinformation analysis in Suzhou Jinweizhi Biotechnology Co., LTD. After the library construction was completed, the library was diluted to 1.5 ng·μL−1 using Qubit2.0 Fluorometer for initial quantification, and the insert size of the library was tested using Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). qRT-PCR was used to detect the library with a concentration higher than 2 nM, and Illumina sequencing was performed after qualified library detection.
The Gene properties were analyzed by the enrichment analysis method of Gene Ontology (GO), including molecular functions, cellular locations, and biological processes involved in the differential genes. Differentially expressed genes were selected with p < 0.05 correction value and significantly enriched on the GO pathway. KEGG was used to build the main public database for screening pathways. According to the data analysis of LT vs. CK, Rich factor was mainly included in the expression of enrichment degree.
4.9. Statistics Statement
Excel software was used for statistical analysis and mapping of differential genes and other data. SPSS24.0 software was used to calculate the mean and standard errors, and the significant differences were analyzed.
5. Conclusions
Tomato is one of the most widely cultivated heat-loving vegetables. However, in the north of China, low temperature stress often occurs during winter, particularly in protected cultivation. In this study, low temperature treatment was found to decrease pollen vigor and pollen germination rate. Low night temperature reduced the tomato fruit setting rate and led to parthenocarpy, and the treatment effect was most obvious in the early stage of tomato pollen development. The abscission rate of tomato pTRV-Slfzy3 and pTRV-Slfzy5 silenced plants was higher than that of the control, which is the key auxin synthesis gene affecting the abscission rate. The expression of Solyc07g043580 was down-regulated after low night temperature treatment.
Author Contributions
Data curation, S.M. and H.X.; investigation, X.Y. and Y.Y.; methodology, T.X., Y.L., F.W. and C.T.; project administration, M.Q. and T.L.; validation, X.Y. and L.H.; writing—original draft, S.M. and H.X.; writing—review & editing, S.M., H.X., X.Y., M.Q. and T.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, grant number 32102460 and 31972397, 2021 Scientific Research Funding Project of Liaoning Provincial Department of Education, grant number LJKZ0639.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The authors will supply the relevant date in response to reasonable requests.
Acknowledgments
The authors are grateful to National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Modern Protected Horticulture Engineering & Technology Center (Shenyang Agricultural University) and Key Laboratory of Protected Horticulture (Shenyang Agricultural University) institute for supporting the project.
Conflicts of Interest
All authors have no competing financial interest to declare.
References
- Sharif, R.; Su, L.; Chen, X.; Qi, X. Hormonal interactions underlying parthenocarpic fruit formation in horticultural crops. Hortic. Res. 2022, 9, uhab024. [Google Scholar] [CrossRef]
- Dong, X.F.; Ma, C.; Xu, T.; Reid, M.S.; Jiang, C.-Z.; Li, T.L. Auxin response and transport during induction of pedicel abscission in tomato. Hortic. Res. 2021, 8, 192. [Google Scholar] [CrossRef]
- Tan, S.; Luschnig, C.; Friml, J. Pho-view of Auxin: Reversible Protein Phosphorylation in Auxin Biosynthesis, Transport and Signaling. Mol. Plant 2021, 14, 151–165. [Google Scholar] [CrossRef]
- Qu, G.; Peng, D.; Yu, Z.; Chen, X.; Cheng, X.; Yang, Y.; Ye, T.; Lv, Q.; Ji, W.; Deng, X.; et al. Advances in the role of auxin for transcriptional regulation of lignin biosynthesis. Funct. Plant Biol. 2021, 48, 743–754. [Google Scholar] [CrossRef]
- Matthes, M.S.; Best, N.B.; Robil, J.M.; Malcomber, S.; Gallavotti, A.; McSteen, P. Auxin EvoDevo: Conservation and Diversification of Genes Regulating Auxin Biosynthesis, Transport, and Signaling. Mol. Plant 2018, 12, 298–320. [Google Scholar] [CrossRef]
- Gallavotti, A.; Barazesh, S.; Malcomber, S.; Hall, D.; Jackson, D.; Schmidt, R.J.; McSteen, P. sparse inflorescence1 encodes a monocot-specific YUCCA—Like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 2008, 105, 15196–15201. [Google Scholar] [CrossRef]
- Abu-Zaitoon, Y.M.; Bennett, K.; Normanly, J.; Nonhebel, H.M. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol. Plant. 2012, 146, 487–499. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, L.; Duan, K.; Zhu, Z.-P.; Ye, Z.-W.; Gao, Q.-H. YUCCA type auxin biosynthesis genes encoding flavin monooxygenases in melon: Genome-wide identification and developmental expression analysis. S. Afr. J. Bot. 2016, 102, 142–152. [Google Scholar] [CrossRef]
- Feng, J.; Dai, C.; Luo, H.; Han, Y.; Liu, Z.; Kang, C. Reporter gene expression reveals precise auxin synthesis sites during fruit and root development in wild strawberry. J. Exp. Bot. 2018, 70, 563–574. [Google Scholar] [CrossRef]
- Yang, J.; Tian, L.; Sun, M.-X.; Huang, X.-Y.; Zhu, J.; Guan, Y.; Jia, Q.-S.; Yang, Z.-N. AUXIN RESPONSE FACTOR17 Is Essential for Pollen Wall Pattern Formation in Arabidopsis. Plant Physiol. 2013, 162, 720–731. [Google Scholar] [CrossRef]
- Wang, B.; Xue, J.-S.; Yu, Y.-H.; Liu, S.-Q.; Zhang, J.-X.; Yao, X.-Z.; Liu, Z.-X.; Xu, X.-F.; Yang, Z.-N. Fine regulation of ARF17 for anther development and pollen formation. BMC Plant Biol. 2017, 17, 243. [Google Scholar] [CrossRef]
- Wu, H.; Xie, D.; Tang, Z.; Shi, D.; Yang, W. PINOID regulates floral organ development by modulating auxin transport and interacts with MADS16 in rice. Plant Biotechnol. J. 2020, 18, 1778–1795. [Google Scholar] [CrossRef]
- Xu, X.E.Z.; Zhang, D.; Yun, Q.; Zhou, Y.; Niu, B.; Chen, C. OsYUC11-mediated auxin biosynthesis is essential for endosperm development of rice. Plant Physiol. 2020, 185, 934–950. [Google Scholar] [CrossRef]
- Song, S.; Chen, Y.; Liu, L.; See, Y.H.B.; Mao, C.; Gan, Y.; Yu, H. OsFTIP7 determines auxin-mediated anther dehiscence in rice. Nat. Plants 2018, 4, 495–504. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, B.; Zhuo, C.; Xie, Y.; Zhang, X.; Liu, Y.; Zhang, G.; Ding, H.; Zhao, B.; Tian, M.; et al. Local auxin biosynthesis regulates brace root angle and lodging resistance in maize. New Phytol. 2023, 238, 142–154. [Google Scholar] [CrossRef]
- Wang, R.; Li, R.Z.; Cheng, L.N.; Wang, X.Y.; Fu, X.; Dong, X.F.; Qi, M.F.; Jiang, C.-Z.; Xu, T.; Li, T.L. SlERF52 regulates SlTIP1;1 expression to accelerate tomato pedicel abscission. Plant Physiol. 2021, 185, 1829–1846. [Google Scholar] [CrossRef]
- Tiwari, M.; Kumar, R.; Subramanian, S.; Doherty, C.J.; Jagadish, S.K. Auxin–cytokinin interplay shapes root functionality under low-temperature stress. Trends Plant Sci. 2023, 28, 447–459. [Google Scholar] [CrossRef]
- Guo, T.; Chen, K.; Dong, N.; Ye, W.; Shan, J.; Lin, H. Tillering and small grain 1 dominates the tryptophan aminotransferase family required for local auxin biosynthesis in rice. J. Integr. Plant Biol. 2019, 62, 581–600. [Google Scholar] [CrossRef]
- Meir, S.; Philosoph-Hadas, S.; Sundaresan, S.; Selvaraj, V.; Burd, S.; Ophir, R.; Kochanek, B.; Reid, M.S.; Jiang, C.-Z.; Lers, A. Microarray Analysis of the Abscission-Related Transcriptome in the Tomato Flower Abscission Zone in Response to Auxin Depletion. Plant Physiol. 2010, 154, 1929–1956. [Google Scholar] [CrossRef]
- Rietveld, P.L.; Wilkinson, C.; Franssen, H.M.; Balk, P.A.; van der Plas, L.H.; Weisbeek, P.J.; de Boer, A.D. Low temperature sensing in tulip (Tulipa gesneriana L.) is mediated through an increased response to auxin. J. Exp. Bot. 2000, 51, 587–594. [Google Scholar] [CrossRef]
- Yuan, H.; Zhao, K.; Lei, H.; Shen, X.; Liu, Y.; Liao, X.; Li, T. Genome-wide analysis of the GH3 family in apple (Malus × domestica). BMC Genom. 2013, 14, 297. [Google Scholar] [CrossRef]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef]
- Li, M.; Wrobel-Marek, J.; Heidmann, I.; Horstman, A.; Chen, B.; Reis, R.; Angenent, G.C.; Boutilier, K. Auxin biosynthesis maintains embryo identity and growth during BABY BOOM-induced somatic embryogenesis. Plant Physiol. 2021, 188, 1095–1110. [Google Scholar] [CrossRef]
- Huang, R.; Zheng, R.; He, J.; Zhou, Z.; Wang, J.; Xiong, Y.; Xu, T. Noncanonical auxin signaling regulates cell division pattern during lateral root development. Proc. Natl. Acad. Sci. USA 2019, 116, 21285–21290. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, D.; Qiu, Y.; Xiao, Z.; Ji, Y.; Li, W.; Xia, Y.; Wang, Y.; Guo, H. Growth asymmetry precedes differential auxin response during apical hook initiation in Arabidopsis. J. Integr. Plant Biol. 2021, 64, 5–22. [Google Scholar] [CrossRef]
- McLaughlin, H.M.; Ang, A.C.H.; Østergaard, L. Noncanonical Auxin Signaling. Cold Spring Harb. Perspect. Biol. 2021, 13, a039917. [Google Scholar] [CrossRef]
- Cheng, L.N.; Li, R.Z.; Wang, X.Y.; Ge, S.-Q.; Wang, S.; Liu, X.F.; He, J.; Jiang, C.-Z.; Qi, M.F.; Xu, T.; et al. A SlCLV3-SlWUS module regulates auxin and ethylene homeostasis in low light-induced tomato flower abscission. Plant Cell 2022, 34, 4388–4408. [Google Scholar] [CrossRef]
- Xiu, X.F.; Cheng, L.N.; Li, R.Z.; Cai, Y.; Wang, X.Y.; Fu, X.; Dong, X.F.; Qi, M.F.; Jiang, C.-Z.; Xu, T.; et al. The HD-Zip transcription factor SlHB15A regulates abscission by modulating jasmonoyl-isoleucine biosynthesis. Plant Physiol. 2022, 189, 2396–2412. [Google Scholar] [CrossRef]
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef]
- Goren, S.; Lugassi, N.; Stein, O.; Yeselson, Y.; Schaffer, A.A.; David-Schwartz, R.; Granot, D. Suppression of sucrose synthase affects auxin signaling and leaf morphology in tomato. PLoS ONE 2017, 12, e0182334. [Google Scholar] [CrossRef]
- Zhao, Y. Essential Roles of Local Auxin Biosynthesis in Plant Development and in Adaptation to Environmental Changes. Annu. Rev. Plant Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef]
- Liu, H.; Luo, Q.; Tan, C.; Song, J.; Zhang, T.; Men, S. Biosynthesis- and transport-mediated dynamic auxin distribution during seed development controls seed size in Arabidopsis. Plant J. 2023, 113, 1259–1277. [Google Scholar] [CrossRef]
- Nam, H.; Han, S.; Lee, S.; Nam, H.; Lim, H.; Lee, G.; Cho, H.S.; Dang, T.V.T.; Choi, S.; Lee, M.M.; et al. CPR5-mediated nucleo-cytoplasmic localization of IAA12 and IAA19 controls lateral root development during abiotic stress. Proc. Natl. Acad. Sci. USA 2023, 120, e2209781120. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef]
- Shapulatov, U.; van Zanten, M.; van Hoogdalem, M.; Meisenburg, M.; van Hall, A.; Kappers, I.; Fasano, C.; Facella, P.; Loh, C.C.; Perrella, G.; et al. The Mediator complex subunit MED25 interacts with HDA9 and PIF4 to regulate thermomorphogenesis. Plant Physiol. 2022, 192, 582–600. [Google Scholar] [CrossRef]
- van der Woude, L.C.; Perrella, G.; Snoek, B.L.; van Hoogdalem, M.; Novák, O.; van Verk, M.C.; van Kooten, H.N.; Zorn, L.E.; Tonckens, R.; Dongus, J.A.; et al. HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proc. Natl. Acad. Sci. USA 2019, 116, 25343–25354. [Google Scholar] [CrossRef]
- Chen, L.; Huang, X.-X.; Li, Y.-J.; Hou, B.-K. Glycosyltransferase UGT76F1 is involved in the temperature-mediated petiole elongation and the BR-mediated hypocotyl growth in Arabidopsis. Plant Signal. Behav. 2020, 15, 1777377. [Google Scholar] [CrossRef]
- Chen, Q.; Dai, X.; De Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin Overproduction in Shoots Cannot Rescue Auxin Deficiencies in Arabidopsis Roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Kneuper, I.; Teale, W.; Dawson, J.E.; Tsugeki, R.; Katifori, E.; Palme, K.; Ditengou, F.A. Auxin biosynthesis and cellular efflux act together to regulate leaf vein patterning. J. Exp. Bot. 2020, 72, 1151–1165. [Google Scholar] [CrossRef]
- Qin, H.; Wang, J.; Zhou, J.; Qiao, J.; Li, Y.; Quan, R.; Huang, R. Abscisic acid promotes auxin biosynthesis to inhibit primary root elongation in rice. Plant Physiol. 2022, 191, 1953–1967. [Google Scholar] [CrossRef]
- Mei, S.; Zhang, M.; Ye, J.; Du, J.; Jiang, Y.; Hu, Y. Auxin contributes to jasmonate-mediated regulation of abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2022, 35, 1110–1133. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.-Y.; Doležal, K.; Schlereth, A.; Jürgens, G.; Alonso, J.M. TAA1-Mediated Auxin Biosynthesis Is Essential for Hormone Crosstalk and Plant Development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-Induced CBF–PIF3 Interaction Enhances Freezing Tolerance by Stabilizing the phyB Thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef]
- Pucciariello, O.; Legris, M.; Rojas, C.C.; Iglesias, M.J.; Hernando, C.E.; Dezar, C.; Vazquez, M.; Yanovsky, M.J.; Finlayson, S.A.; Prat, S.; et al. Rewiring of auxin signaling under persistent shade. Proc. Natl. Acad. Sci. USA 2018, 115, 5612–5617. [Google Scholar] [CrossRef]
- Lv, B.; Yu, Q.; Liu, J.; Wen, X.; Yan, Z.; Hu, K.; Li, H.; Kong, X.; Li, C.; Tian, H.; et al. Non-canonical AUX/IAA protein IAA 33 competes with canonical AUX/IAA repressor IAA 5 to negatively regulate auxin signaling. EMBO J. 2019, 39, e101515. [Google Scholar] [CrossRef]
- Lv, B.; Wei, K.; Hu, K.; Tian, T.; Zhang, F.; Yu, Z.; Zhang, D.; Su, Y.; Sang, Y.; Zhang, X.; et al. MPK14-mediated auxin signaling controls lateral root development via ERF13-regulated very-long-chain fatty acid biosynthesis. Mol. Plant 2020, 14, 285–297. [Google Scholar] [CrossRef]
- Yu, J.; Lu, J.; Cui, X.; Guo, L.; Wang, Z.; Liu, Y.; Wang, F.; Qi, M.; Li, T. Melatonin mediates reactive oxygen species homeostasis via Sl CV to regulate leaf senescence in tomato plants. J. Pineal Res. 2022, 73, e12810. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, K.-X.; Wang, W.-S.; Gong, W.; Liu, W.-C.; Chen, H.-G.; Xu, H.-H.; Lu, Y.-T. Low Temperature Inhibits Root Growth by Reducing Auxin Accumulation via ARR1/12. Plant Cell Physiol. 2014, 56, 727–736. [Google Scholar] [CrossRef]
- Qi, X.; Hu, S.; Zhou, H.; Liu, X.; Wang, L.; Zhao, B.; Huang, X.; Zhang, S. A MADS-box transcription factor of ‘Kuerlexiangli’ (Pyrus sinkiangensis Yu) PsJOINTLESS gene functions in floral organ abscission. Gene 2017, 642, 163–171. [Google Scholar] [CrossRef]
- Li, Q.; Chai, L.; Tong, N.; Yu, H.; Jiang, W. Potential Carbohydrate Regulation Mechanism Underlying Starvation-Induced Abscission of Tomato Flower. Int. J. Mol. Sci. 2022, 23, 1952. [Google Scholar] [CrossRef]
- Han, H.; Adamowski, M.; Qi, L.; Alotaibi, S.S.; Friml, J. PIN-mediated polar auxin transport regulations in plant tropic responses. New Phytol. 2021, 232, 510–522. [Google Scholar] [CrossRef]
- Bao, H.; Ding, Y.; Yang, F.; Zhang, J.; Xie, J.; Zhao, C.; Du, K.; Zeng, Y.; Zhao, K.; Li, Z.; et al. Gene silencing, knockout and over-expression of a transcription factor ABORTED MICROSPORES (SlAMS) strongly affects pollen viability in tomato (Solanum lycopersicum). BMC Genom. 2022, 23, 346. [Google Scholar] [CrossRef]
- Song, J.; Tachibana, S. Loss of viability of tomato pollen during long-term dry storage is associated with reduced capacity for translating polyamine biosynthetic enzyme genes after rehydration. J. Exp. Bot. 2007, 58, 4235–4244. [Google Scholar] [CrossRef]
- Kormuťák, A.; Bolecek, P.; Galgóci, M.; Gömöry, D. Longevity and germination of Juniperus communis L. pollen after storage. Sci. Rep. 2021, 11, 12755. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).