The Conserved and Specific Roles of the LUX ARRHYTHMO in Circadian Clock and Nodulation
Abstract
1. Introduction
2. Results
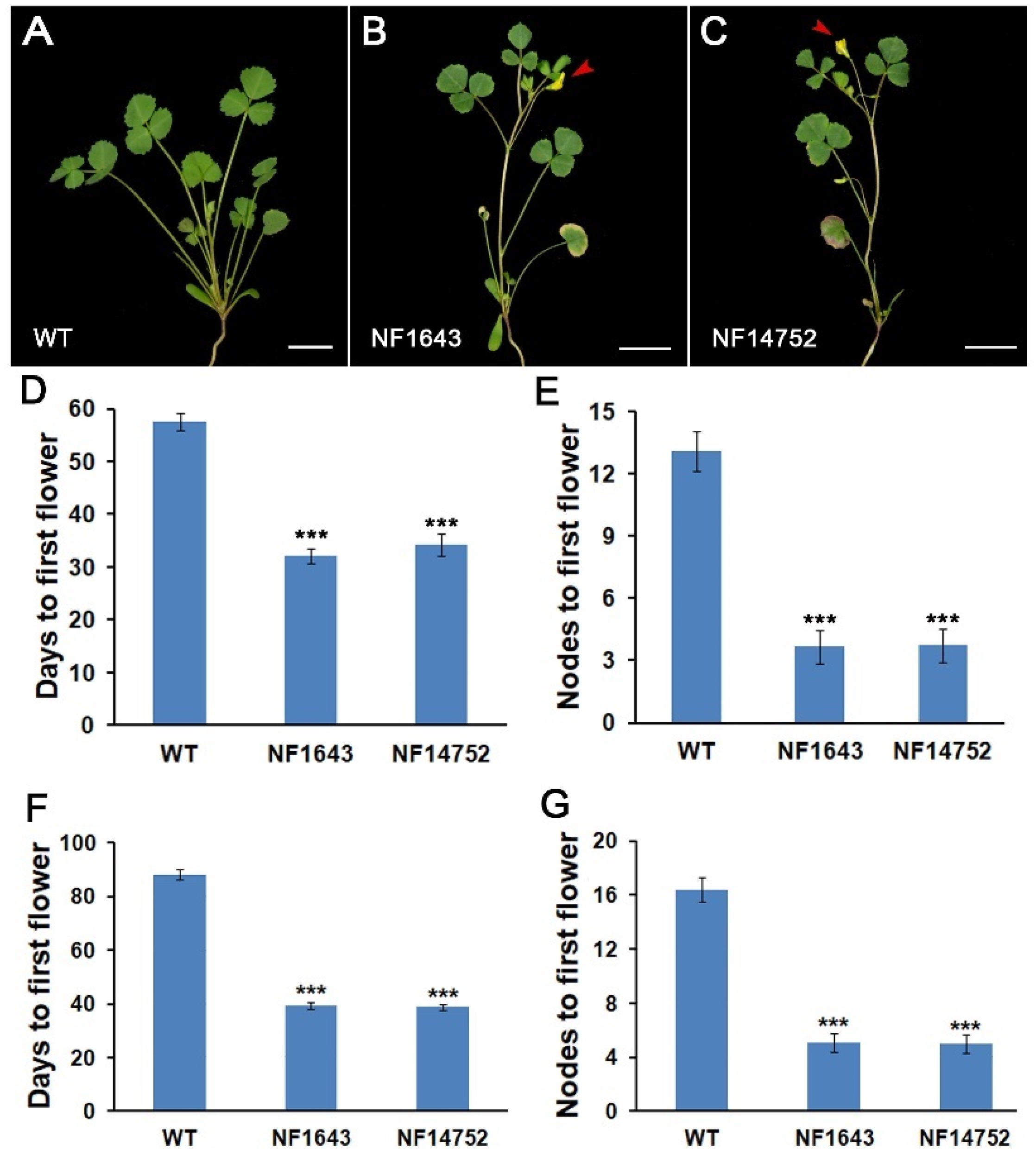
2.1. Identification and Phenotypic Characterization of the Early Flowering Mutants in M. truncatula
2.2. Molecular Cloning of MtLUX
2.3. MtLUX Expression Pattern and Subcellular Localization
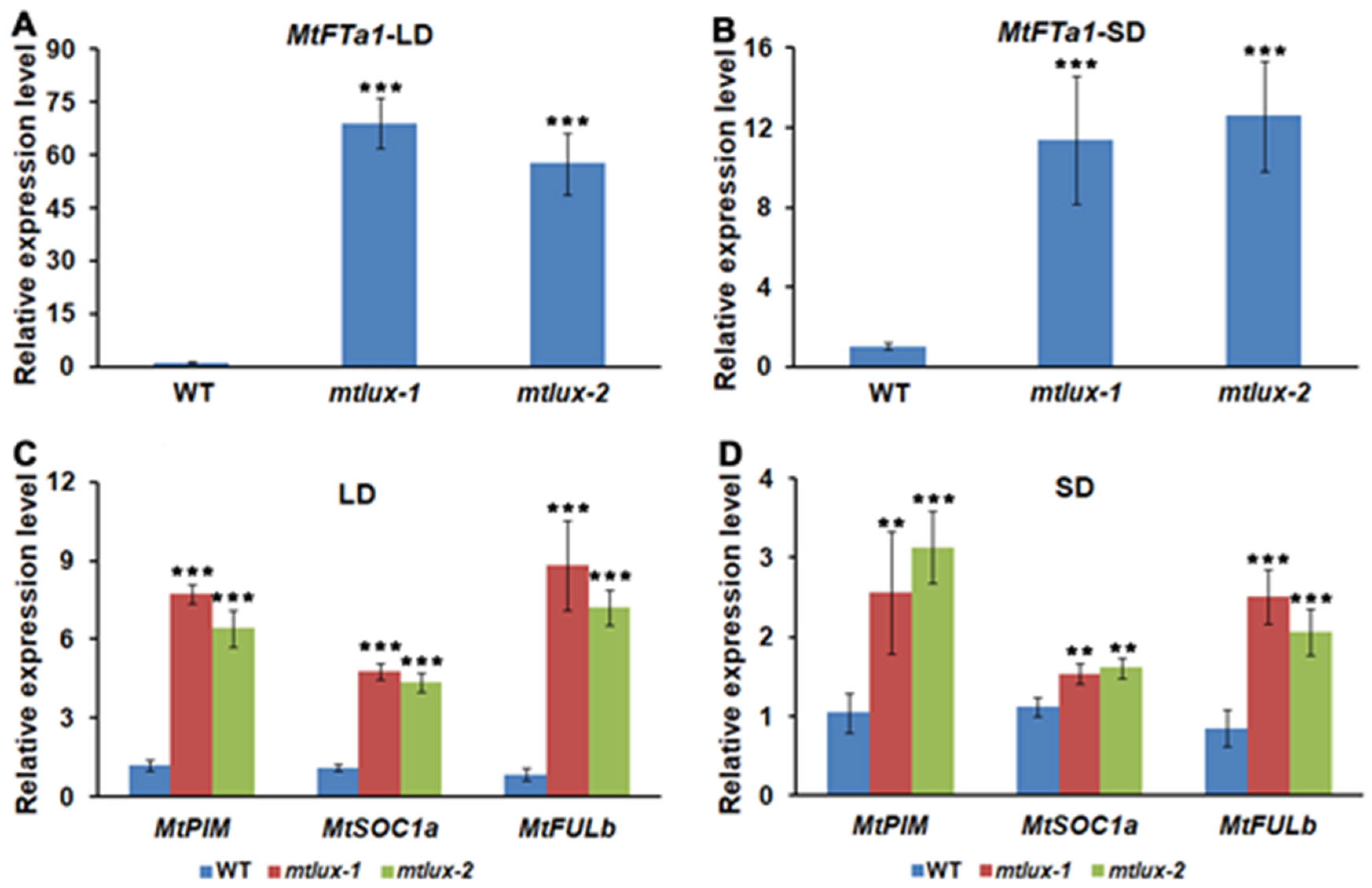
2.4. MtLUX Is Negatively Correlated with Transcript Levels of MtFTa1 Gene but Not MtCOL Genes
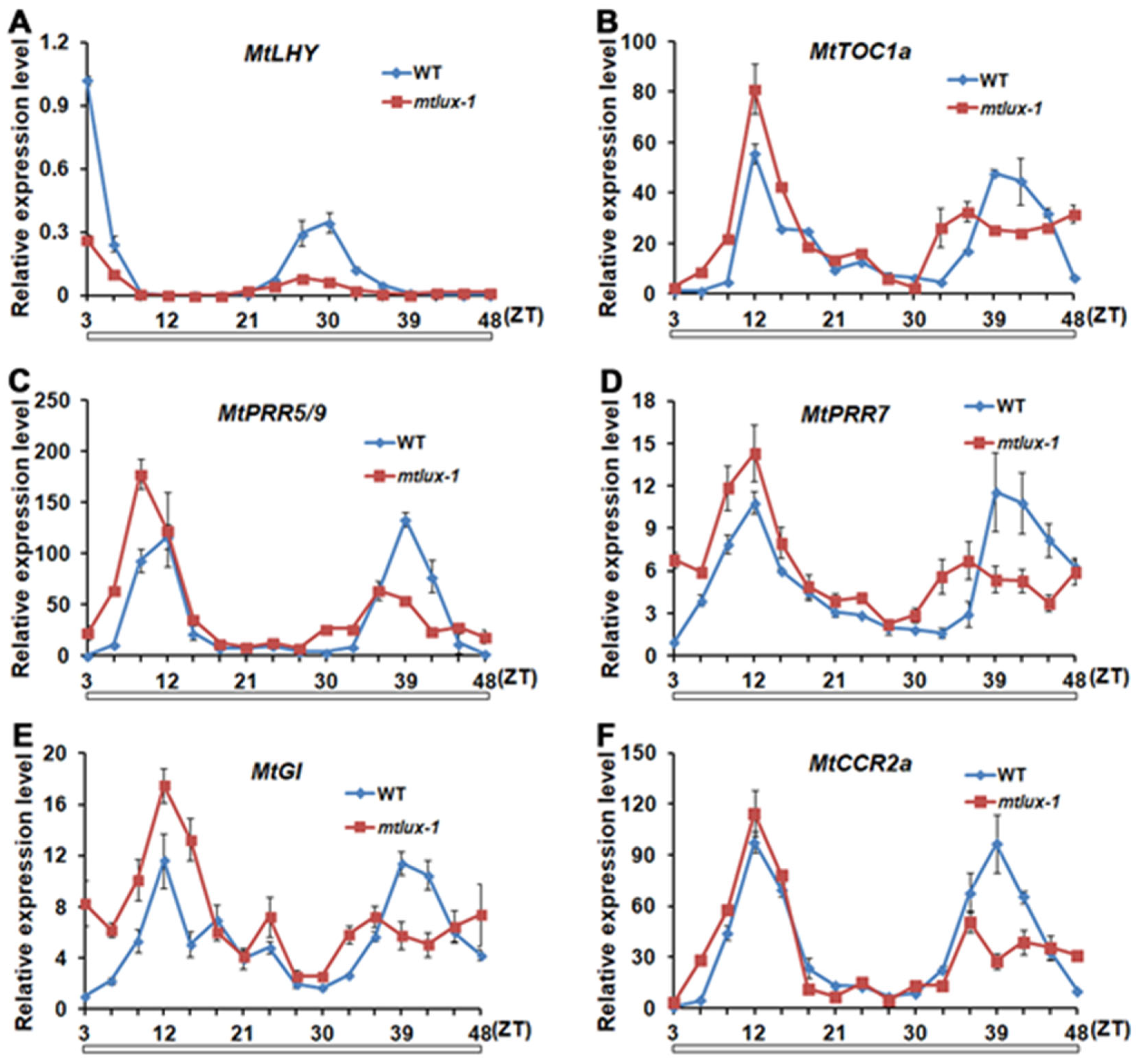
2.5. The Expression Pattern of Genes Associated with Circadian Clock Is Altered in Mtlux Mutants
2.6. The Transcriptomic Profiles of the Loss-of-Function of MtLUX
2.7. MtLUX Indirectly Regulates Nodulation and Nyctinastic Leaf Movement
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phylogenetic Analysis
4.3. Subcellular Localization of MtLUX
4.4. Root Nodule Induction
4.5. RNA Isolation and Quantitative Realtime PCR (qRT-PCR) Analysis
4.6. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yanovsky, M.J.; Kay, S.A. Living by the calendar: How plants know when to flower. Nat. Rev. Mol. Cell Biol. 2003, 4, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kévei, E.; Tóth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Hirata, Y.; Aihara, K.; Mas, P. A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell 2015, 163, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Kamioka, M.; Takao, S.; Suzuki, T.; Taki, K.; Higashiyama, T.; Kinoshita, T.; Nakamichi, N. Direct Repression of Evening Genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis Circadian Clock. Plant Cell 2016, 28, 696–711. [Google Scholar] [CrossRef]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef]
- Nagel, D.H.; Kay, S.A. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 2012, 22, R648–R657. [Google Scholar] [CrossRef]
- Hazen, S.P.; Schultz, T.F.; Pruneda-Paz, J.L.; Borevitz, J.O.; Ecker, J.R.; Kay, S.A. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 2005, 102, 10387–10392. [Google Scholar] [CrossRef]
- Onai, K.; Ishiura, M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells Devoted Mol. Cell. Mech. 2005, 10, 963–972. [Google Scholar] [CrossRef]
- Helfer, A.; Nusinow, D.A.; Chow, B.Y.; Gehrke, A.R.; Bulyk, M.L.; Kay, S.A. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011, 21, 126–133. [Google Scholar] [CrossRef]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farré, E.M.; Kay, S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef]
- Huang, H.; Nusinow, D.A. Into the Evening: Complex Interactions in the Arabidopsis Circadian Clock. Trends Genet. 2016, 32, 674–686. [Google Scholar] [CrossRef]
- Zhang, C.; Gao, M.; Seitz, N.C.; Angel, W.; Hallworth, A.; Wiratan, L.; Darwish, O.; Alkharouf, N.; Dawit, T.; Lin, D.; et al. LUX ARRHYTHMO mediates crosstalk between the circadian clock and defense in Arabidopsis. Nat. Commun. 2019, 10, 2543. [Google Scholar] [CrossRef]
- Mizuno, T.; Nomoto, Y.; Oka, H.; Kitayama, M.; Takeuchi, A.; Tsubouchi, M.; Yamashino, T. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 958–976. [Google Scholar] [CrossRef] [PubMed]
- Campoli, C.; Pankin, A.; Drosse, B.; Casao, C.M.; Davis, S.J.; von Korff, M. HvLUX1 is a candidate gene underlying the early maturity 10 locus in barley: Phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 2013, 199, 1045–1059. [Google Scholar] [CrossRef] [PubMed]
- Liew, L.C.; Hecht, V.; Sussmilch, F.C.; Weller, J.L. The Pea Photoperiod Response Gene STERILE NODES Is an Ortholog of LUX ARRHYTHMO. Plant Physiol. 2014, 165, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Gawroński, P.; Ariyadasa, R.; Himmelbach, A.; Poursarebani, N.; Kilian, B.; Stein, N.; Steuernagel, B.; Hensel, G.; Kumlehn, J.; Sehgal, S.K.; et al. A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat. Genetics 2014, 196, 1253–1261. [Google Scholar] [CrossRef]
- Mizuno, N.; Nitta, M.; Sato, K.; Nasuda, S. A wheat homologue of PHYTOCLOCK 1 is a candidate gene conferring the early heading phenotype to einkorn wheat. Genes Genet. Syst. 2012, 87, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Liew, L.C.; Singh, M.B.; Bhalla, P.L. A novel role of the soybean clock gene LUX ARRHYTHMO in male reproductive development. Sci. Rep. 2017, 7, 10605. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Müller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef]
- Sanchez, S.E.; Kay, S.A. The Plant Circadian Clock: From a Simple Timekeeper to a Complex Developmental Manager. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef]
- Romera-Branchat, M.; Andrés, F.; Coupland, G. Flowering responses to seasonal cues: What’s new? Curr. Opin. Plant Biol. 2014, 21, 120–127. [Google Scholar] [CrossRef]
- Ietswaart, R.; Wu, Z.; Dean, C. Flowering time control: Another window to the connection between antisense RNA and chromatin. Trends Genet. 2012, 28, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Fudge, J.B.; Lee, R.H.; Laurie, R.E.; Mysore, K.S.; Wen, J.; Weller, J.L.; Macknight, R.C. Medicago truncatula SOC1 Genes Are Up-regulated by Environmental Cues That Promote Flowering. Front. Plant Sci. 2018, 9, 496. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R.; Lou, P.; Hermand, V.; Kim, J.A. The Importance of Ambient Temperature to Growth and the Induction of Flowering. Front. Plant Sci. 2016, 7, 1266. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Laurie, R.; Macknight, R. It’s time to flower: The genetic control of flowering time. BioEssays News Rev. Mol. Cell. Dev. Biol. 2004, 26, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation tagging of the floral inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.K.; Chung, K.S.; Kim, J.; Lee, J.H.; Hong, S.M.; Yoo, S.J.; Yoo, S.Y.; Lee, J.S.; Ahn, J.H. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 2005, 139, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, V.; Fornara, F. Molecular control of flowering in response to day length in rice. J. Integr. Plant Biol. 2013, 55, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Oikawa, T.; Sugiyama, N.; Tanisaka, T.; Yano, M.; Shimamoto, K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 2002, 16, 2006–2020. [Google Scholar] [CrossRef]
- González-Schain, N.D.; Díaz-Mendoza, M.; Zurczak, M.; Suárez-López, P. Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. Cell Mol. Biol. 2012, 70, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Chia, T.Y.; Müller, A.; Jung, C.; Mutasa-Göttgens, E.S. Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Exp. Bot. 2008, 59, 2735–2748. [Google Scholar] [CrossRef]
- Hicks, K.A.; Millar, A.J.; Carré, I.A.; Somers, D.E.; Straume, M.; Meeks-Wagner, D.R.; Kay, S.A. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 1996, 274, 790–792. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Tobin, E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93, 1207–1217. [Google Scholar] [CrossRef]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Más, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, T.; Wright, L.; Fujiwara, S.; Cremer, F.; Lee, K.; Onouchi, H.; Mouradov, A.; Fowler, S.; Kamada, H.; Putterill, J.; et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 2005, 17, 2255–2270. [Google Scholar] [CrossRef] [PubMed]
- Suárez-López, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Lasswell, J.; Rogg, L.E.; Cohen, M.A.; Bartel, B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 2000, 101, 331–340. [Google Scholar] [CrossRef]
- Fowler, S.; Lee, K.; Onouchi, H.; Samach, A.; Richardson, K.; Morris, B.; Coupland, G.; Putterill, J. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999, 18, 4679–4688. [Google Scholar] [CrossRef]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef]
- Niwa, Y.; Ito, S.; Nakamichi, N.; Mizoguchi, T.; Niinuma, K.; Yamashino, T.; Mizuno, T. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 2007, 48, 925–937. [Google Scholar] [CrossRef]
- Weller, J.L.; Ortega, R. Genetic control of flowering time in legumes. Front. Plant Sci. 2015, 6, 207. [Google Scholar] [CrossRef]
- Hecht, V.; Foucher, F.; Ferrándiz, C.; Macknight, R.; Navarro, C.; Morin, J.; Vardy, M.E.; Ellis, N.; Beltrán, J.P.; Rameau, C.; et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005, 137, 1420–1434. [Google Scholar] [CrossRef]
- Cao, D.; Li, Y.; Lu, S.; Wang, J.; Nan, H.; Li, X.; Shi, D.; Fang, C.; Zhai, H.; Yuan, X.; et al. GmCOL1a and GmCOL1b Function as Flowering Repressors in Soybean Under Long-Day Conditions. Plant Cell Physiol. 2015, 56, 2409–2422. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.C.; Hecht, V.F.; Picard, K.; Diwadkar, P.; Laurie, R.E.; Wen, J.; Mysore, K.; Macknight, R.C.; Weller, J.L. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 2014, 5, 486. [Google Scholar] [CrossRef] [PubMed]
- Laurie, R.E.; Diwadkar, P.; Jaudal, M.; Zhang, L.; Hecht, V.; Wen, J.; Tadege, M.; Mysore, K.S.; Putterill, J.; Weller, J.L.; et al. The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 2011, 156, 2207–2224. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Laurie, R.E.; Vander Schoor, J.K.; Ridge, S.; Knowles, C.L.; Liew, L.C.; Sussmilch, F.C.; Murfet, I.C.; Macknight, R.C.; Weller, J.L. The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 2011, 23, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Liu, B.; Xia, Z.; Sato, S.; Kim, B.M.; Watanabe, S.; Yamada, T.; Tabata, S.; Kanazawa, A.; Harada, K.; et al. Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 2010, 154, 1220–1231. [Google Scholar] [CrossRef]
- Tadege, M.; Wen, J.; He, J.; Tu, H.; Kwak, Y.; Eschstruth, A.; Cayrel, A.; Endre, G.; Zhao, P.X.; Chabaud, M.; et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. Cell Mol. Biol. 2008, 54, 335–347. [Google Scholar] [CrossRef]
- Yeoh, C.C.; Balcerowicz, M.; Zhang, L.; Jaudal, M.; Brocard, L.; Ratet, P.; Putterill, J. Fine mapping links the FTa1 flowering time regulator to the dominant spring1 locus in Medicago. PLoS ONE 2013, 8, e53467. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Jiang, A.; Thomson, G.; Kerr-Phillips, M.; Phan, C.; Krueger, T.; Jaudal, M.; Wen, J.; Mysore, K.S.; Putterill, J. Overexpression of Medicago MtCDFd1_1 Causes Delayed Flowering in Medicago via Repression of MtFTa1 but Not MtCO-Like Genes. Front. Plant Sci. 2019, 10, 1148. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wei, H.; Li, N.; Tian, W.; Chong, K.; Wang, L. Circadian Evening Complex Represses Jasmonate-Induced Leaf Senescence in Arabidopsis. Mol. Plant 2018, 11, 326–337. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, X.; Hu, Y.; Liu, S.; Nan, H.; Li, X.; Fang, C.; Cao, D.; Shi, X.; Kong, L.; et al. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 2017, 49, 773–779. [Google Scholar] [CrossRef]
- Gutiérrez, R.A.; Stokes, T.L.; Thum, K.; Xu, X.; Obertello, M.; Katari, M.S.; Tanurdzic, M.; Dean, A.; Nero, D.C.; McClung, C.R.; et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc. Natl. Acad. Sci. USA 2008, 105, 4939–4944. [Google Scholar] [CrossRef] [PubMed]
- Dornbusch, T.; Michaud, O.; Xenarios, I.; Fankhauser, C. Differentially phased leaf growth and movements in Arabidopsis depend on coordinated circadian and light regulation. Plant Cell 2014, 26, 3911–3921. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352. [Google Scholar] [CrossRef]
- Yon, F.; Kessler, D.; Joo, Y.; Cortés Llorca, L.; Kim, S.G.; Baldwin, I.T. Fitness consequences of altering floral circadian oscillations for Nicotiana attenuata. J. Integr. Plant Biol. 2017, 59, 180–189. [Google Scholar] [CrossRef]
- Joo, Y.; Fragoso, V.; Yon, F.; Baldwin, I.T.; Kim, S.G. Circadian clock component, LHY, tells a plant when to respond photosynthetically to light in nature. J. Integr. Plant Biol. 2017, 59, 572–587. [Google Scholar] [CrossRef]
- Kong, Y.; Han, L.; Liu, X.; Wang, H.; Wen, L.; Yu, X.; Xu, X.; Kong, F.; Fu, C.; Mysore, K.S.; et al. The nodulation and nyctinastic leaf movement is orchestrated by clock gene LHY in Medicago truncatula. J. Integr. Plant Biol. 2020, 62, 1880–1895. [Google Scholar] [CrossRef]
- Earley, K.W.; Haag, J.R.; Pontes, O.; Opper, K.; Juehne, T.; Song, K.; Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. Cell Mol. Biol. 2006, 45, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Boivin, C.; Camut, S.; Malpica, C.A.; Truchet, G.; Rosenberg, C. Rhizobium meliloti Genes Encoding Catabolism of Trigonelline Are Induced under Symbiotic Conditions. Plant Cell 1990, 2, 1157–1170. [Google Scholar] [CrossRef]



| Gene ID | Description | Gene Name | log2(mtlux-1/WT) | FDR |
|---|---|---|---|---|
| Medtr1g098140 | chalcone and stilbene synthase family protein | - | 11.62570884 | 2.24 × 10−7 |
| Medtr7g016720 | chalcone and stilbene synthase family protein | - | 11.09011242 | 6.06 × 10−5 |
| Medtr3g082630 | B-box type zinc finger protein | - | 3.240051088 | 7.12× 10−9 |
| Medtr4g061360 | PRR response regulator | MtPRR3 | 2.841248051 | 3.09 × 10−121 |
| Medtr3g037390 | timing of cab expression 1/PRR response regulator | MtTOC1b | 2.182937767 | 2.54 × 10−16 |
| Medtr8g105590 | flavin-binding kelch repeat F-box protein, putative | MtFKF1 | 1.913744267 | 4.36 × 10−6 |
| Medtr3g091340 | CCT motif protein | - | 1.428077186 | 7.93 × 10−16 |
| Medtr4g108880 | two-component response regulator-like APRR7 protein | MtTOC1a | 1.397727614 | 9.32 × 10−12 |
| Medtr1g098160 | gigantea protein 1B | MtGI | 1.326863054 | 2.20 × 10−124 |
| Medtr3g092780 | PRR response regulator | MtPRR5/9 | 1.315561529 | 1.15 × 10−19 |
| Medtr3g103970 | early flowering protein | MtELF3 | 1.212819095 | 4.07 × 10−6 |
| Medtr7g083540 | zinc finger constans-like protein | - | 1.155346537 | 0.0002833 |
| Medtr4g046640 | B-box type zinc finger protein | - | −12.52160044 | 0.0001545 |
| Medtr4g008050 | B-box type zinc finger protein, putative | - | −2.731240606 | 1.29 × 10−32 |
| Medtr7g110810 | helix loop helix DNA-binding domain protein | MtPIF4like | −2.351726642 | 2.37 × 10−12 |
| Medtr1g067000 | myb transcription factor | - | −2.182338147 | 5.41 × 10−233 |
| Medtr2g089310 | B-box type zinc finger protein | - | −1.935687612 | 9.49 × 10−58 |
| Medtr7g118330 | late elongated hypocotyl-like protein | MtLHY | −1.689747281 | 0 |
| Medtr5g021580 | salt tolerance-like protein | - | −1.379673589 | 3.91 × 10−48 |
| Medtr1g013450 | zinc finger constans-like protein | MtCOLb | −1.361552093 | 6.45 × 10−145 |
| Medtr7g018170 | zinc finger constans-like protein | MtCOLa | −1.324077697 | 1.87 × 10−102 |
| Medtr5g058920 | phosphatidylinositol-4-phosphate 5-kinase | - | −1.156725504 | 6.44 × 10−6 |
| Medtr1g016920 | early flowering protein, putative | MtELF3-like | −1.063984456 | 4.27 × 10−5 |
| Medtr3g449770 | transcription factor | MtPIF3like | −1.060477166 | 2.44 × 10−60 |
| Medtr1g084980 | phytochrome-interacting factor 3.1 | MtPIF5like | −1.020982564 | 1.78 × 10−26 |
| Medtr6g053620 | ubiquitin-protein ligase, putative | - | −1.411551885 | 0.0007789 |
| Medtr1g067110 | two-component response regulator-like APRR7 protein | MtPRR7 | 0.79438414 | 9.02 × 10−39 |
| Medtr4g070080 | RNA-binding (RRM/RBD/RNP motif) family protein | MtCCR2a | 1.794137127 | 3.88 × 10−167 |
| Medtr4g070190 | RNA-binding (RRM/RBD/RNP motif) family protein | MtCCR2b | 1.692850978 | 1.572 × 10−32 |
| Medtr4g070140 | RNA-binding (RRM/RBD/RNP motif) family protein | MtCCR2c | 2.237543475 | 9.1 × 10−103 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, Y.; Zhang, Y.; Liu, X.; Meng, Z.; Yu, X.; Zhou, C.; Han, L. The Conserved and Specific Roles of the LUX ARRHYTHMO in Circadian Clock and Nodulation. Int. J. Mol. Sci. 2022, 23, 3473. https://doi.org/10.3390/ijms23073473
Kong Y, Zhang Y, Liu X, Meng Z, Yu X, Zhou C, Han L. The Conserved and Specific Roles of the LUX ARRHYTHMO in Circadian Clock and Nodulation. International Journal of Molecular Sciences. 2022; 23(7):3473. https://doi.org/10.3390/ijms23073473
Chicago/Turabian StyleKong, Yiming, Yuxue Zhang, Xiu Liu, Zhe Meng, Xiaolin Yu, Chuanen Zhou, and Lu Han. 2022. "The Conserved and Specific Roles of the LUX ARRHYTHMO in Circadian Clock and Nodulation" International Journal of Molecular Sciences 23, no. 7: 3473. https://doi.org/10.3390/ijms23073473
APA StyleKong, Y., Zhang, Y., Liu, X., Meng, Z., Yu, X., Zhou, C., & Han, L. (2022). The Conserved and Specific Roles of the LUX ARRHYTHMO in Circadian Clock and Nodulation. International Journal of Molecular Sciences, 23(7), 3473. https://doi.org/10.3390/ijms23073473






