Abstract
Mitochondria are key organelles that combine features inherited from their bacterial endosymbiotic ancestor with traits that arose during eukaryote evolution. These energy producing organelles have retained a genome and fully functional gene expression machineries including specific ribosomes. Recent advances in cryo-electron microscopy have enabled the characterization of a fast-growing number of the low abundant membrane-bound mitochondrial ribosomes. Surprisingly, mitoribosomes were found to be extremely diverse both in terms of structure and composition. Still, all of them drastically increased their number of ribosomal proteins. Interestingly, among the more than 130 novel ribosomal proteins identified to date in mitochondria, most of them are composed of a-helices. Many of them belong to the nuclear encoded super family of helical repeat proteins. Here we review the diversity of functions and the mode of action held by the novel mitoribosome proteins and discuss why these proteins that share similar helical folds were independently recruited by mitoribosomes during evolution in independent eukaryote clades.
1. Introduction
Ribosomes are key molecular machines, fundamental to all life on Earth. They decode information carried by messenger RNAs (mRNAs) and translate it into proteins [1]. Ribosomes are universally composed of two subunits, the small one (SSU) binds mRNA and reads the genetic information, while the large subunit (LSU) catalyses the actual formation of peptidyl bonds between amino-acids that allows protein synthesis. Likewise, all ribosomes are ribonucleoprotein (RNP) complexes composed of numerous proteins and ribosomal RNAs (rRNAs), the LSU rRNA being a catalytic RNA, i.e., a ribozyme. Despite these common features, ribosomes diverged significantly during evolution. In bacteria and archaea, ribosomes both make 70S particles with about 54 proteins as well as 16S, 23S and 5S rRNAs [2]. However, archaeal ribosomes often evolved to adapt to harsh environmental conditions e.g., with the recruitment of specific r-proteins to respond to high temperature or halophilic conditions. Thus archaeal ribosomes often have a more rigid structure than their bacterial counterparts [3]. In contrast, ribosomes that occur in eukaryote cytosol are larger, forming 80S RNP complexes. They evolved from an ancestral archaeal ribosome but have more proteins and larger rRNAs, e.g., with 80 proteins and 18S, 28S, 5S and 5.8S rRNAs in humans [4]. Beyond these cytosolic ribosomes, which are often referred to as “eukaryote ribosomes”, other types of ribosomes occur in eukaryote cells. These additional ribosomes are found in genome-containing organelles: mitochondria, as well as chloroplasts for the Archaeplastida kingdom. Until the last decade, very little was known of these organellar ribosomes [5,6].
Mitochondria and chloroplasts are essential energy producing organelles of bacterial origin that have arisen through distinct endosymbiotic events. Mitochondria derive from a bacteria belonging to a sister group of α-proteobacteria that was engulfed by the archaeal-type ancestor of eukaryote cells about 1.5–2 billion years ago [7,8,9,10]. The second endosymbiotic event took place 500 million years later with the engulfment of a cyanobacteria by a fully ledged eukaryote that gave rise to the tripartite phylum (red, blue and green algae) Archaeplastida [11,12]. Both mitochondria and chloroplasts are considered as semi-autonomous organelles because they have retained a genome and fully functional gene expression machineries [5]. While the vast majority of bacterial genes coming from the ancestral endosymbionts were either lost or transferred to the host nuclei, some genes—most of them encoding essential proteins of the respiratory chain or the photosynthetic machinery—remain to this date in organellar genomes. It is notable that many of these genes encode hydrophobic core proteins of the respective energy producing systems. The difficulty to synthesize in the cytosol and import to the organelles such membrane proteins might have been an evolutionary indicator for the retention of their genes in organellar genomes [13]. Another hypothesis proposes that a colocation of genes and of their products is required for the redox regulation of gene expression [14]. Nonetheless, the retention of mRNA coding genes in organelles implied that comprehensive gene expression machineries, including in particular functional ribosomes, should be present in organelles. For their biogenesis, mitochondrial ribosomes thus rely on the coordinated synthesis of ribosomal proteins, most of whose mRNAs are coded in the nucleus, translated in the cytosol, and imported into mitochondria, together with the synthesis of moieties encoded by the mitochondrial genome, i.e., mitochondrial rRNAs and ribosomal proteins, in some eukaryotes. Few studies have investigated the mitoribosomes assembly, although the evidence suggests that, in human and yeast, this process resembles bacterial ribosome assembly [5]. Because of their bacterial origin, it was assumed for a long time that organellar ribosomes would be very similar to bacterial ones. This assumption turned out to be true for chloroplasts, where the chlororibosomes composition and three-dimensional structure strongly resemble those of bacterial ribosomes, as revealed by recent studies [15,16,17,18].
In marked contrast with chloroplasts, the recent characterization of mitochondrial ribosomes (mitoribosomes) in a diversity of species representing the major eukaryote groups has revealed how mitoribosomes strikingly diverged from prokaryotes and eukaryotes ribosomes. Even more stunning is their diversity between the different eukaryote clades [5,6]. While the 3D structures of bacterial and cytosolic ribosomes could be obtained by X-ray crystallography in 1999 and 2011, respectively [19,20], the characterisation and determination of 3D structures from very low abundant ribosomes such as mitoribosomes was only possible thanks to the cryo-electron microscopy (cryo-EM) revolution that started a decade ago. Since 2015, the compositions and structures of ribosomes from animals, fungi, kinetoplasts, land plants, a ciliate and a photosynthetic algae could be determined (Figure 1) [21,22,23,24,25,26,27,28], thus providing examples of mitoribosomes from Opisthokonta, Excavata, Archaeplastida and Stramenopiles, Alveolata, and Rhizaria (SAR), four out of the five major super-groups of eukaryotes. The overall architecture of all these mitoribosomes is extremely diverse, in particular that of the SSUs with numerous specific rRNA expansion segments and additional domains [5,29]. Mitochondrial ribosomal RNAs are extremely diverse. They can be larger than in bacteria, as seen in yeast and particularly in land plants, or highly reduced, as observed in animals and particularly in kinetoplasts. The most extreme example of rRNA divergence was observed in the algae Chlamydomonas, where rRNAs are fragmented into 13 pieces that are non-contiguously encoded as gene pieces in the Chlamydomonas mitochondrial genome, expressed as separate small RNAs and brought together to reconstitute a functional ribosome [27]. Beyond these divergences, a common feature of all mitoribosomes is the marked increase in the number of ribosomal proteins. To date, over 130 novel ribosomal proteins, specific to mitoribosomes, have been identified, although only about 30 novel proteins are common to at least two mitoribosomes [30]. This implies that beyond a set of common proteins that might have been present in the common ancestor of all mitoribosomes, the different eukaryote groups independently recruited specific sets of proteins to make specialised ribosomes, potentially adapted to different environmental niches. Still, despite this diversity, it is remarkable that most of the novel mitochondrial ribosomal proteins belong to the group of a-helical repeat proteins, including proteins such as pentatricopeptide repeat (PPR) proteins, octotricopeptide repeat (OPR) proteins or mitochondrial transcription termination factor (mTERF) proteins that share a similar type of tridimensional fold. The diversity of structures and functions of the novel mitochondrial ribosomal proteins, for rRNA stabilisation, for the recruitment of mRNA, to bind other ribosomal proteins, to anchor the ribosomes to the mitochondrial membrane and for the assembly of mitoribosomes will be discussed here, and insights will be provided to understand the diversity of their mode of action. This ensemble of functions is summarized in Figure 2.
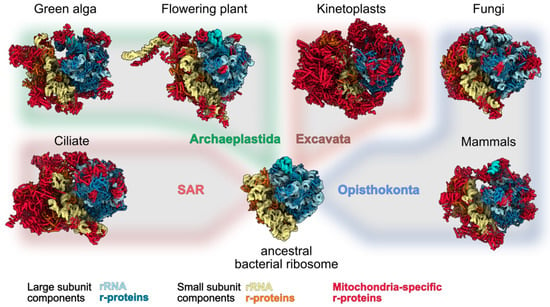
Figure 1.
Structural comparison between available mitoribosome structures. To highlight the divergence of mitoribosomes as compared to bacteria and their diversity in terms of size and shape across eukaryotes, high resolution structures of mitoribosomes are presented and overlaid on a schematic evolutionary tree of eukaryotes. From left to right, mitoribosomes structures are from: ciliate (T. thermophilus), green alga (C. reinhardtii), flowering plant (A. thaliana), kinetoplasts (T. brucei), fungi (N. crassa) and mammals (H. sapiens). They are compared with E. coli ribosome structure, representing the ancestral form of mitoribosomes. Large subunits components are shown in blue shades and small subunit components in yellow shades. Mitochondria-specific ribosomal proteins (which includes shared and species-specific proteins) are highlighted in red. No structures have been solved yet for the supergroup Amoebozoa (hence not shown on the figure). SAR corresponds to the supergroup which includes the Stramenopiles, Alveolates, and Rhizaria subgroups.
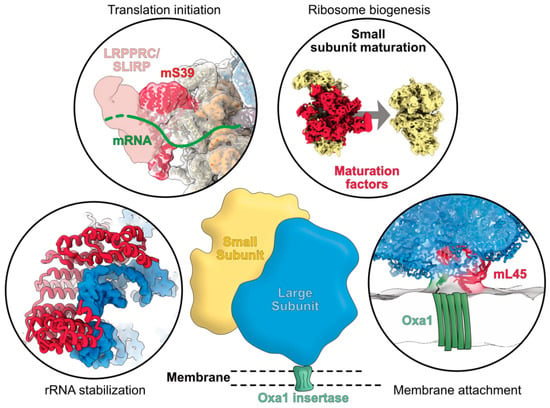
Figure 2.
Graphical summary of the functions of mitoribosome-specific ribosomal proteins. The recently identified proteins specifically occurring in mitochondrial ribosomes are shown in red. They can be involved in rRNA stabilization, in the recruitment of mRNA for translation initiation, in the assembly and maturation of mitoribosomes, and in its attachment to the mitochondrial inner membrane through the binding with the insertase Oxa1. The exemplary shown mS39 and mL45 specifically occur in mammalian mitoribosomes.
2. Technologies That Allowed to Identify Mitoribosome-Specific Proteins
As early as the late 1950s, it was speculated that complexes capable of protein synthesis would be present in mitochondria [31,32], only five years after the first description of ribosomes [33]. In the following 10 years, biochemical characterisation of mitoribosomes was achieved for fungal and animal species [34]. These early reports described the sedimentation coefficient and the probable rRNA content of these mitoribosomes, already linking them to bacteria. However, the clarification of their protein composition would also only come more than 10 years later, with the two-dimensional poly acrylamide gel electrophoresis (PAGE) analysis of purified bovine mitoribosomes, revealing that these mitoribosomes would be composed of more than 80 proteins, hence more than found in bacteria, yet without knowing exactly their nature [35]. The development of protein sequencing and mass-spectrometry in the 1990s allowed, in the early 2000s, for the first characterisation of the putative protein composition of the small and large subunit of the mammalian mitoribosome [36,37]. These studies already revealed the conservation and absence of bacterial homologs, as well as putative proteins shared with yeast mitoribosomes, and some being specific to animals. However, their role in the ribosome was unknown. In 2003, the first cryo-EM characterization of a mitoribosome was published [38]. This study reported the architecture of the bovine mitoribosome at about 10Å resolution, not enough for a clear assignation of the mitoribosome specific r-proteins. It confirmed, however, the largely proteic nature of this mitoribosome, looking significantly different from bacterial and cytosolic ribosomes.
The release of complete organisms genomes combined with basic local alignment search tool (BLAST) and mass-spectrometry analyses allowed to investigate the composition of mitoribosomes in new organisms, revealing conserved and specific proteins [39,40,41,42]. Still, small proteins can escape detection by mass-spectrometry, and really pure samples are required to draw a clear picture of the protein composition, otherwise leading to an overestimation of the protein content, and the mis-assignation of proteins as r-proteins. The combination of different purification techniques and analyses, such as immunoprecipitation and classical sucrose density gradient separation, or large pore Blue Native PAGE and complexome profiling, or grad-seq, provides more accurate protein composition determination [26,43,44,45] but is still not 100% reliable and does not provide functional and spatial localization of the proteins in the complex of interest. These experiments can be complemented by crosslinking mass-spectrometry which allows for the determination of what proteins or domains are close to each other in the complex [46].
More than 50 years have been necessary to achieve a clear idea of the complete composition of a mitoribosome. Structural biology elucidates both the composition and function of the studied complex. Thanks to the rapid technical advancement of cryo-EM in the 2010s, notably with the development of new direct electron detectors [47], larger and more challenging to purify complexes could be resolved, such as mitoribosomes [21,22,23,48,49], to resolutions that allowed unambiguous assignation of protein densities, thus allowing for the direct determination of the complexes’ composition.
Today, thanks to the technical progress in proteomics, metagenomic and structural biology, the combination of these techniques allows for the investigation of the composition of mitoribosomes and their evolution in ways that were unimaginable before.
3. Prominence of Helical Repeat Proteins in Mitochondrial Gene Expression
As mentioned above, recent technological advances, in particular cutting edge single particle cryo-EM [47], has enabled the identification of a large number of ribosomal proteins that specifically occur in mitochondria. Many of these factors belong to the protein super-family of helical repeat proteins that are composed of repeated modules of helices. The occurrence of helical repeat proteins as ribosomal proteins might not be surprising because, for two decades, a fast growing number of genomic and functional investigations have identified that many organelle specific post-transcriptional processes are performed by helical repeat proteins belonging to recently recognized gene families such as pentatricopeptide repeat (PPR), octotricopeptide repeat (OPR), mTERF, half-a-tetratricopeptide repeat (HAT) and heptatricopeptide repeat (HPR) proteins [50].
For instance, PPR proteins that occur in all eukaryotes and are particularly prevalent in plants with hundreds of PPR genes were found to be involved in the maturation of RNA ends, RNA editing, splicing, RNA stabilization and translation [51]. Interestingly a mode of action has been described for PPR proteins where specific amino-acids at given positions in individual PPR motifs give specificity for a particular ribonucleotide. Thus, a succession of PPR motifs recognizes single stranded RNA in a sequence specific manner. This connection between amino-acids of PPR motifs and nucleotides is referred to as the “PPR code” [52]. This code was validated experimentally for many proteins, i.e., for RNA editing factors [51]. However, this mode of action is not applicable to all PPR proteins, as, for example, recently recognized PPR-NYN proteins holding RNase P activity, that recognize folded pre-tRNAs in a non-canonical way [53,54,55].
HAT proteins make a smaller family of proteins, conserved in eukaryotes, where they were found to be involved in a variety of organellar but also nucleo-cytoplasmic processes [56]. For instance, the HAT protein HCF107 was found to bind and most probably stabilize the 5′ ends of chloroplast mRNAs [57].
Likewise, the mTERF protein family that occurs in plants and animals appears to be specific to organelles [58]. mTERF proteins were found to bind DNA but also RNA with, fodr example, MTERF4 that was found to bind rRNA to regulate ribosome biogenesis in humans [59], and mTERF9 that interacts with the 16S rRNA to promote chloroplast ribosome biogenesis in Arabidopsis [60].
In contrast, OPR proteins appear to be restricted to the plant lineage as well as to Alveolata, and are particularly prevalent in Chlorophyta such as Chlamydomonas. OPR proteins are related to a variety of RNA related processes in the chloroplast, for example, [61,62], and the OPR protein RAP was found to be required for chloroplast 16S rRNA maturation [63].
Finally, HPR proteins were recently recognized to be a family of putative organellar proteins which are mostly found in Alveolata. They were experimentally shown to occur in mitochondria of the malaria parasite Plasmodium, where they were proposed to be involved in RNA processing and/or stabilization [64], possibly for the assembly of the fragmented rRNAs that occur in apicomplexa mitoribosomes, similar to Chlamydomonas.
Beyond OPR and HPR that appear to be related [64], all these gene families do not seem to have a common evolutionary origin [50]. In particular, consensus motifs derived from the respective protein families do not appear to hold conserved sequence features [50]. Nonetheless, all these proteins have a similar modular organization, with tandem arrays of individual repeats that all contain a similar secondary structure relying on antiparallel a-helices. In all these different protein families, the succession of repeats folds into a solenoid structure. It is this conserved fold that assigns these organelle proteins to the super-group of helical repeat proteins that also includes PUF and TALE proteins [65,66]. Interestingly, the comparison of structure models for representative factors from the different families had suggested that many helical repeat proteins contain nucleic acid binding platforms composed of positively charged residues in the concave surface of the super-helices [50]. This feature that was proposed for PPR and mTERF proteins [59,67] has been shown experimentally for PUF proteins [68] and also observed recently for many of the ribosomal helical repeat proteins by the recent cryo-EM studies of mitoribosomes. In this light, it is tempting to speculate that helical repeat proteins might share at least some features used for RNA recognition.
To date, among helical repeat proteins, the PPR family has clearly attracted the most attention, for instance with the description of a PPR code. Interestingly, as described in details below, none of the PPR proteins identified as ribosomal proteins seem to recognize RNAs according to the PPR code. At this stage, it is unclear whether the occurrence of PPR proteins as ribosomal proteins constituted an ancestral function for these proteins and the PPR code was acquired later for specific processes or whether the mode of action using the PPR code was ancestral and subsequently lost for some functions, such as the association to ribosomes in different eukaryote groups. Future investigations will contribute to decipher the diversity of the helical repeat proteins mode of action to try to understand why this type of structure was independently selected for nucleic acid binding several times in evolution, in particular in the context of mitoribosomes.
4. Functions and Modes of Action of Mitoribosome-Specific Proteins
4.1. rRNA Stabilization Mediated by Mitoribosome-Specific Proteins
Ribosomes are complex ribonucleoprotein particles composed of rRNAs and r-proteins. Most of the r-proteins are located at the periphery of the ribosomes. These proteins are usually composed of a globular domain facing the solvent, and long domains extending into the ribosome, between rRNA helices, to anchor the protein and/or stabilize the rRNA. Even though prokaryotic and cytosolic ribosomes have now been characterized in detail, it is unclear what actual function these r-proteins possess. Given that the majority of them are largely positively charged, it is suggested that the general function of r-proteins is to stabilize and counteract the negatively charged rRNAs which perform the catalytic activity [69]. Hence, their main role is to stabilize rRNAs to promote efficient protein synthesis.
In mitoribosomes, most of the additional r-proteins that were acquired during eukaryote evolution also appear to serve this function (additional functions are described in the following sections). However, due to the diversity of rRNA composition across mitoribosomes, many of these proteins have been recruited to fit a certain need, e.g., stabilization of an rRNA extension, or compensation of rRNA loss.
Some of these proteins are conserved between mitoribosomes and were surely acquired early in their evolution. This is, for example, the case of mS29, a GTPase located on the head of the small subunit, first described in humans [21,22]. Resolution was sufficient in human, yeast and kinetoplastids to show that the GTPase do contain GDP and might be active, however its role in the mitoribosome is unclear [23,24,29]. In all cases, it binds to rRNA of the small subunit, and might be involved in subunit association and translocation by making specific bridges between the SSU and the LSU.
One interesting case is the systematic acquisition of PPR proteins at the foot of the small subunit (Figure 3D). In bacteria, the foot of the SSU is formed by the tip of the rRNA helix h44 and the helix h6—the only protein present in that area is bS20, which probably contributes to h44 stabilization. However, bS20 was universally lost in the mitochondria [5]. The presence of a PPR protein in the that area was first described in humans, where the protein mS27 compensates for the strong reduction and unwinding of h44 and the loss of h6 [21,22]. It was later discovered that mS27 is also present in yeast, however the protein here carries an additional function, as the rRNA is more conserved with bacteria in yeast compared to humans and is even expanded [70]. There, it stabilizes h44, h6 and the fungi expansion segment h44-ES1. In land plants, a PPR protein is positioned similar to the yeast mS27, and helps stabilize the extended h44 tip [25]. In the ciliate Tetrahymena, the rRNA in this area is significantly expanded, and two proteins were recruited to stabilize it [28]. mS93 is an mTERF protein involved in the stabilization of the expanded h44, and mS90, which resembles a PPR protein, is positioned similar to the yeast mS27 and the PPR protein found in land plants. Finally, in the green algae Chlamydomonas, where the rRNAs are reduced—h6 is noticeably absent —a PPR protein, mS106, is found, and is positioned similar to mS27 in humans [27]. Interestingly, in kinetoplastids, two PPR proteins, mS51 and mS63, are located in that area, which no longer contains RNA due to the drastic rRNA reduction in these organisms [24]. mS51 and mS63 are positioned similar to mS90 and mS93, respectively, in Tetrahymena. It is remarkable to systematically find these very similar proteins at that position in all mitoribosomes investigated to date. One explanation could be that similar proteins would have been independently recruited to perform a similar function in the different eukaryote groups. Alternatively, a PPR protein might have been recruited very early during mitoribosome evolution to compensate for bS20 loss, and have drifted significantly during evolution. This remains to be properly investigated.
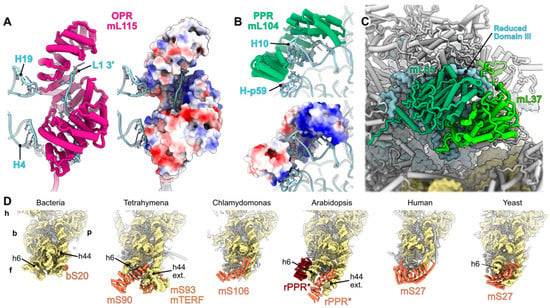
Figure 3.
Examples of RNA-binding proteins recruited to mitoribosomes. (A,B) present close-up views of large alpha-helical proteins which were largely recruited to mitoribosomes. (A) The OPR mL115 involved in the stabilization of fragment L1 of the fragmented rRNAs of C. reinhardtii is shown. The OPR enlaces the 3′ single-stranded extremity of the L1 fragment which is stabilized via charge interaction. It also interacts with H19 and H4. The electrostatic potential of mL115 is shown with blue surfaces showing positive charges and red surfaces showing negative charges. (B) The PPR mL104 of the flowering plant mitoribosome is shown. It englobes helix H10, interacting with the rRNA backbone which is a different mode of action compared to non-ribosomal PPR proteins. It also interacts with H-p59, a plant specific RNA helix. As in (A), the electrostatic potential of mL115 is shown, revealing that the rRNA interaction is mostly mediated by positively charged residues. (C) The pseudo-dimer composed of mL65 and mL37, probably repurposed endonucleases, are shown as an example of non-alpha-helical mitoribosome-specific r-proteins. The r-proteins interact with the reduced domain III of the large subunit of the mammalian mitoribosome. (D) Structural comparison of the foot of the small subunit. This comparison highlights the conserved helical proteins involved in the stabilization of the foot of the small subunit—where the tip of h44 and h6 are either extended, reduced, or deleted—most likely due to the loss of the bacterial r-protein bS20 in mitoribosomes. All mitoribosomes share a PPR protein at this position (mS27, mS90, rPPR* and mS106), which may be the product of a single ancestral PPR protein. In trypanosoma, PPR protein mS63 is also present at this position but no longer interacts via RNA. The different parts of the small subunit are indicated on the bacterial model; h: head, b: body, f: foot, p: platform.
rPPR proteins seem to have a totally distinct RNA binding process than the canonical PPRs, where a “PPR code” was described. In particular, in plants, no RNA targets could be predicted using the PPR code when looking at positions 5 and 35 of each rPPR protein motif, suggesting that RNA interactions are performed differently. This was confirmed by the cryo-EM structure of a plant mitoribosome. It revealed the contact sites between rPPR proteins and rRNAs [25]. It appears that most rPPRs contact double stranded RNA helices of the rRNA instead of single stranded RNA (Figure 3B). This interaction is mediated by positively charged residues of the rPPRs and the negatively charged phosphate backbone of the rRNAs. How this process achieves the specific binding of individual rPPR proteins to their interaction site on the mitoribosome is still undetermined.
In the case of specific proteins being recruited, in Chlamydomonas, where rRNAs are reduced and fragmented, several OPR proteins were recruited to the ribosome, and help stabilize the extremities of the rRNA fragments [27]. They mainly interact with single stranded rRNAs by enlacing the rRNA ends, but can also interact with rRNA helices via their solvent exposed side. This is the case for mL115, which stabilizes the L1 rRNA 3′ end, and also interacts with H4 and H19 of the same rRNA fragment (Figure 3A). A similar case is found in the ciliate Tetrahymena, where the mL106 protein is also involved in stabilizing the 3′ end of one of the two rRNAs of the large subunit [28].
These RNA/protein interactions are reminiscent of the canonical ssRNA/PPR interactions, although no recognition code between OPR proteins and single stranded RNA could be recognized at this stage. In contrast, mS107, an OPR from the SSU of Chlamydomonas, binds the double stranded RNA of the S2 rRNA, similar to the land plant mitoribosome rPPRs. mL113, an OPR protein, binds the tip of a helix from L3b thanks to a domain inserted between OPR repeats; in that case OPR repeats are not at all involved in RNA binding.
In animals, specific proteins, a pseudo-dimer made by mL65 and mL37, repurposed endonucleases, interacts with the reduced domain III of the large subunit. There, it probably stabilizes the rRNA and compensates for its loss (Figure 3C) [21,22]. In Chlamydomonas, the strong reduction of domain III is compensated for by the acquisition of the OPR mL113 and the mTERF mL114 [27]. As opposed to these cases, rRNAs are expanded in flowering plants. In the large subunit, the rPPR proteins mL101 and mL104 are involved in the stabilisation of the refolded and extended domain III and domain I. mL104, for example, englobes the tip of H10 from domain I and interacts with H-p59, a helix from the refolded domain III (Figure 3B) [25].
Altogether, mitoribosomes have recruited a wide variety of binders that bind their RNA targets through very diverse processes.
4.2. Mitochondrial Specific mRNA Recruitment Processes
During translation initiation, the mRNA is recruited to the SSU, where its start codon is aligned with the ribosomal P site and recognized by the initiator tRNA. This first step of the translation cycle exhibits a great variation across lineages and has attracted a lot of interest in the last decades. In bacteria, the mRNA binding to the ribosome is often directed by an interaction between a conserved sequence motif, the Shine-Dalgarno (SD), and a complementary sequence, the anti Shine-Dalgarno, located at the 16S rRNA [71,72,73]. The SD motif is positioned upstream of the start codon and aids in aligning the ribosome to select and decode the open reading frame (Figure 4A). This Shine-Dalgarno dependent mechanism is the most widely used for prokaryotic mRNAs, but other mechanisms involve, for example, the S1 protein [74] and some mRNAs that are completely devoid of SD sequence, or even 5′UTR [75]. For the later, it remains elusive how they are recruited to the ribosome [71]. Thus, mRNAs with discrete characteristics require distinct mechanisms for their recruitment.
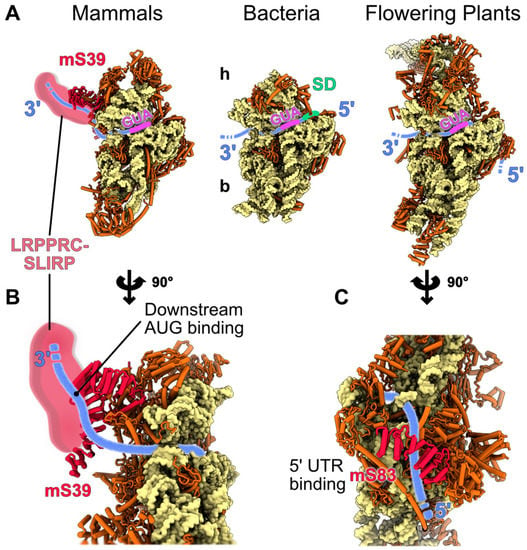
Figure 4.
Specific proteins involved in the translation initiation process in mitochondria. Proposed mechanisms of mitochondria-specific translation initiation events involving mitochondria-specific r-proteins are shown. (A) The proposed positions and orientations of the mRNA on the small subunits of mammalian, plant and bacteria during translation initiation are indicated on the figure. The mRNA is shown in blue, the initiation codon (AUG) is in purple and the Shine-Dalgarno (SD) consensus, only present in bacteria, is in green. (B,C) present close-up views of mammalian and plant mRNA binders. (B) The mammalian mRNA stabilization by the PPR protein mS39 and probably the LRPPRC-SLIRP complex is shown. mS39 and LRPPRC-SLIRP interact with the ORF, downstream of the AUG. (C) In flowering plants, a proposed mechanism of mRNA stabilization would involve the PPR protein mS83, which would bind a consensus sequence located in the 5′ UTR of the mitochondrial mRNAs. A similar process was also proposed in yeast, involving specific proteins for each yeast mRNA.
In mitochondria, despite the plethora of proteins that partake in their proteome, only a few are encoded by the mitochondrial expression machinery, i.e., 13 in human, eight in yeast [6], and around 30 in land plants [76,77], where the majority are components of the oxidative phosphorylation complexes. In most eukaryotes, mitochondria-encoded mRNAs lack the SD sequence and the 5′ m7G cap, features that aid their translation in bacteria and cytosol, respectively, the exception being some Jakobid species such as Reclinomonas, which retained SD-like sequences [78]. Moreover, the 5′ untranslated region (5′UTR), which contains cis-elements essential for expression, varies in length across lineages and in some cases is missing. The heterogeneity that mitochondrial mRNAs present, coupled with the profound divergence of mitoribosomes structure and composition across various phyla, have resulted in specialized recruitment mechanisms. For instance in mammals, most mitochondrial mRNAs do not possess a 5′ UTR and when they do, it is up to 3 nt in length [79,80,81], leaving little, if any space for a 5′ UTR-dependent regulation. In compliance with this notion, a specific ribosomal protein has been proposed to direct mRNA binding in humans. The PPR protein mS39 is located at the entry site of the mRNA channel on the ribosomal small subunit [21,22]. Allegedly, mS39 PPR motifs interact with a U-rich region placed downstream of the start codon in the vicinity of the mRNA’s 7th codon, facilitating the recruitment of the mRNA to the ribosome [82]. Moreover, a recent study revealed an additional moiety, the leucine-rich PPR motif-containing protein (LRPPRC), forming a complex with the Stem-Loop Interacting RNA binding Protein (SLIRP), that interacts with mS39 in translationally active mitoribosomes, i.e., bound mRNA and tRNA present in A or P site [83]. The LRPPRC-SLIRP complex shows a strong affinity for RNAs via several PPR motifs [84], promotes mRNA polyadenylation in mitochondria [85], and acts as a chaperon to mediate the relaxation of mRNA secondary structures [86]. Also, SLIRP has been linked with the presence of mRNAs in active mitoribosomes [87]. Collectively, mS39 may serve as a platform for the docking of the LRPPRC-SLIRP complex which delivers mitochondrial mRNAs to the ribosome [83]. Then, mS39 facilitates the threading of mRNA through the SSU, and the start codon is selected for the initiation of translation (Figure 4).
LRPPRC-SLIRP complex constitutes a linking bond between transcription and translation and seems to act as a universal activator of translation in mammals. To date, only one translational activator has been found in mammals, that being TACO1, which is involved in the translation of COX1 [88]. In contrast, other eukaryotes are heavily dependent on translational activators for mitochondrial translation. These are regulatory proteins that promote the protein synthesis of specific mitochondrial mRNAs. In yeast, the limited number of protein-encoding mRNAs has allowed the evolution of a specialized mRNA recruitment mechanism where particular protein factors promote the translation of distinct mRNAs according to cellular needs. These translational activators utilize the 5′ UTR to bind and deliver the transcripts to the ribosome [89,90]. Interestingly, yeast’s mRNA exit channel has been remodeled, possibly following the alteration of the SSU rRNA 3′ end which no longer facilitates the start codon selection via the Shine-Dalgarno sequence. Instead, the alignment of the yeast mRNAs might be coordinated by the action of translation activators. The species-specific mS42 and mS43 form a heterodimer located at the edge of the remodeled mRNA exit channel, and along with a series of specific protein elements placed on this site, could act as a platform for translation activators [23].
In plants, the mRNA recruitment mechanism is yet to be determined. However, recent findings point to another ribosomal-specific PPR protein, mS83, which may have a role similar to the protein mammalian mS39. mS83 is positioned at the SSU near the mRNA exit channel and could possibly bind to an AxAAA motif, present in nearly half of the plant’s mitochondrial mRNAs via its PPR moieties [25]. Thus, mS83 could potentially aid the mRNA binding to the mitoribosome, align the mRNA with the SSU and assist in the selection of the start codon from the initiation complex (Figure 4). Nonetheless, although mS83 may constitute a potential recruitment mechanism for a number of the plant’s mRNAs, it is unlikely to be unique, since not all mRNAs contain the AxAAA motif [25]. Rather, it is more likely that a combination of distinct mechanisms is involved in mRNA recruitment in plant mitoribosomes [25]. In compliance with this notion, the existence of translational activators in plant mitochondria is expected, especially if we take into consideration the long 5′ UTRs of plant mitochondrial mRNAs, which can act as a binding site similar to yeast. For instance, proteins such as the recently described RFL8 likely act similar to yeast translation activators in plant mitochondria [91]. In the green algae Chlamydomonas, the initiation mechanism is unknown; however, like in humans, the mRNAs lack 5′ UTRs. Additionally, they are post-transcriptionally polycytidylated at their 3′ UTRs [92]. It is possible that the polycytidylation is important to have translatable mRNA, either by creating a binding platform for trans-acting factors—RNA binding proteins could recognize this cytidine stretch and act as translation activators—or may contribute to mRNA circulation to also promote translation [93].
Altogether, the vast divergence of mitoribosomes across eukaryotes, the presence of novel ribosomal specific proteins, the specialized nature of mitoribosome to decode membrane proteins coupled with the distinct characteristics of mitochondrial mRNAs, had an immense influence in mRNA recruitment, resulting in elegant and highly adapted mechanisms.
4.3. Involvement of Mitoribosome Specific Proteins for Protein Binding
Mitoribosomes have followed separate evolutionary routes regarding their RNA contents from being expanded in plants [27] and fungi [23,70], reduced in metazoa [21,22], and kinetoplastids [24], or even fragmented in the green algae Chlamydomonas and in members of the Apicomplexa clade [27,94], to being mostly unchanged in jakobids [30] compared to their bacterial counterparts. In contrast, their protein composition has universally increased across the eukaryotic lineages as a result of various expansions in bacterial proteins as well as the presence of novel species-specific ribosomal proteins, shifting the RNA-protein ratio from the bacterial 2:1 to 1:2 in mammals and 1:6 in kinetoplastids [6].
The novel ribosomal proteins hold a central role in ribosome architecture, assembly, and translation through interactions either with rRNAs (discussed above) or with other proteins. In mammals, for instance, two novel ribosomal proteins mS29 and mS27 form mitochondria-specific bridges between the two subunits. As discussed in 4.1, mS29 is a guanosine 5′-triphosphate (GTP) binding protein located at the head of the small subunit. It interacts with the central protuberance in the LSU creating two bridges with mL46 and mL48 [22]. Another inter-subunit bridge is formed by the novel RP mS27 and the N-terminal extension of bL19m, connecting the bottom of the SSU with the LSU.
Inter-subunit bridges coordinate the relative movements of the subunits, crucial for the active translation. In yeast, the specific connection of mS44 and bL19m contributes to the multiple contact sites between the small and the large subunit of the species. The increased number of inter-subunit bridges present in yeast mitoribosomes compared to humans allows for a more stiff ribosome with the movement of the subunits to be more firm [23].
The presence of novel ribosomal proteins is even more profound in kinetoplastids. Trypanosomal mitoribosomes render an extreme case of divergence, with severely reduced rRNAs and the enrolment of over 120 proteins, half of them being species-specific [24,95]. Notably, the two largest ribosomal proteins mS48 and mS49 (1788 and 1181 aa, respectively) are specific to kinetoplastids. mS48 expands from the lower body of the SSU up to the head interacting with a series of ribosomal proteins (mS51, mS22, mS47, mS60, uS5m, mS59, and mS49), displaying an intrinsic role in the assembly of the SSU and its protein network. mS49 exhibits a similar role through interactions with most of the proteins located at the head of the SSU (uS10m, mS53, mS35, mS55, mS52, and mS50) [24]. A repercussion of the rRNA contraction is the presence of functionally important regions, like the L1 stalk and the central protuberance (CP), which is totally depleted of rRNA. The L1 stalk, through conformational changes, regulates the tRNA positioning and release from the E-site [96,97]. The novel ribosomal proteins mL70, mL74, m91 and the bacterial bL9m that form the L1 stalk compensate for the missing rRNA. Similarly, the CP is solely formed by proteins including the specific mL73, mL82 and mL96, and probably assembles as a module which docks to the LSU at a late intermediate state [95,98,99]. The kinetoplastid’s mitoribosomal exit tunnel is also highly remodeled. The unique r-proteins mL72 and mL109 interact with the conserved uL4m, uL22m, uL23m and uL29m to shape the mature peptide exit channel [95,99].
Another notable example, where novel ribosomal proteins have reshaped the mitoribosome’s architecture, is the evolutionarily distant Tetrahymena thermophila, a ciliate protist from the phylum Alveolata. The SSU is characterized by its two protein rich regions, the body extension and the back protuberance [28]. Both regions have a limited rRNA presence, and subsequently they are shaped by multiple protein-protein interactions, many of them attributed to mitoribosomes or proteins only found in ciliate. The body extension is formed by eight proteins (four unique) uS11, mS23, mS26, mS37, mS76, mS77, mS78-NTD and mS79, whereas the back protuberance consists of eleven proteins mS45, mS47, mS78-CTD, mS83–88, mS91 and mS92 (one mitoribosome specific, 10 unique). The two moieties are connected with mS78, the largest ribosomal protein present in ciliate mitoribosomes. In addition, both regions are interconnected with the SSU head, with the body extension to interact with the head through mS23, mS26, mS29, and mS37 and the back protuberance through mS31, mS45, mS47, mS85, mS87, mS88, mS89, mS92, and uS9m [28]. This extensive protein network formed between the head and the body could affect and may limit the movement of the head upon the conformational changes of the SSU during translation elongation [96,100]. Interestingly, half of the proteins synthesized in ciliate mitoribosomes are soluble [28]; however, how mitoribosomes can adapt their mode of synthesis from membrane to matrix proteins is yet to be determined. A notable unique ribosomal protein, mL105, is located near the exit channel and interacts with the uL22m and the uL24m at the solvent side, whereas the mL105-CTD enters the exit tunnel, interacting with bL23m and possibly with the nascent chain. In addition, mL105 possesses a ‘signal peptide-binding domain’ which may constitute some early evidence regarding a putative protein targeting the system in ciliate mitochondria [28]. Finally, an interesting feature of the mRNA entry channel is the reconstitution of uS3 ribosomal proteins out of three separate proteins: mS31m, mS92, and uS3m. The two first, which are nucleus-encoded, interact with the mitochondria-encoded uS3m to form the complete uS3 ribosomal protein, showcasing the significance of the crosstalk between the two genomes as a repercussion of the genetic drift throughout mitochondrial evolution [28].
Collectively, unique mitoribosomal proteins have been recruited, following several evolutionary pressures, to vastly reshape the architecture of eukaryote’s mitoribosomes and take over many elegant mechanisms of the mitoribosomes by establishing a wide protein network.
4.4. Specific Processes for Mitoribosome Attachment to Membranes
The majority of proteins still encoded in the mitochondrial genome are hydrophobic membrane proteins, components of the respiratory chain complexes. Due to their hydrophobic nature, these proteins would be challenging to import from the cytosol. This specificity seems to have caused a specialization of the mitochondrial protein translation machinery for the synthesis of hydrophobic membrane proteins [101]. As a consequence of this specialization, it appears that protein synthesis occurs in the mitochondrial matrix at or near the inner membrane. This localization allows for simultaneous membrane insertion of the neo-synthetized peptide during translation [83]. In order to bring the mitochondrial ribosome close to the inner membrane and close to the protein insertion machinery, specific proteins allow the tethering of ribosomes directly to the main component of the Oxidative Assembly (OXA) translocase, the complex responsible for most of the membrane protein insertion of both nuclear and mitochondrially encoded proteins in the mitochondrial inner membrane [102].
The main component of the OXA complex is the Oxa1 protein insertase. It is part of the Oxa1/YidC/Alb3 conserved protein family, which is an evolutionarily conserved protein machinery for the insertion of hydrophobic proteins into bacterial and organellar membranes [103,104,105]. All proteins from this family have five transmembrane domains; YidC proteins can be found in prokaryotes, whereas Alb3 proteins are present in chloroplasts [106,107]. Oxa1 is found in mitochondria, in an orientation with its N-terminal part in the membrane and its C-terminal part located in the mitochondrial matrix. This C-terminal region plays the role of a “Ribosome Binding Domain” (RBD) [108,109]. The RBD is positively charged, but the precise mechanism of interaction between this RBD and mitochondrial ribosome is still unclear. In yeast, it has been shown that cells expressing C-terminal truncated versions of Oxa1 have impaired respiration functions and difficulties to use non-fermentable carbon sources [109]. In addition, yeasts lacking the C-terminal region accumulate improperly inserted mitochondrial proteins in the matrix which should have been inserted in the inner membrane [110]. Oxa1 knock-out mutation leads to a critical deficiency in respiratory complexes III, IV and V, especially for the two subunits of complex IV (cytochrome oxidase c), Cox1 and Cox2 proteins, both mitochondrially encoded. Insertion of critical membrane proteins in the inner membrane, such as the cytochrome b protein, part of the complex III (cytochrome bc1), is also severely reduced.
Mechanisms and proteins involved in mitochondrial ribosome binding with Oxa1 seem to strongly vary between eukaryotes. Nevertheless, these mechanisms are well described for some organisms [111,112].
The link between Oxa1 and the mitochondrial ribosome in yeast is mediated by a non-ribosomal linker protein named Mba1. This protein is able to bind mitochondrial ribosomes near the peptide exit channel, and can then bind the C-terminal domain of Oxa1 [113]. Structural studies using Cryo-EM and Cryo-ET techniques showed that this interaction is stabilized with the help of the rRNA expansion segment 96-ES1 of the large subunit, making a second contact site directly with the inner membrane of mitochondria [114]. This rRNA stabilizing expansion is not found in any other organism for which the mitochondrial ribosome structure has already been resolved.
Unlike in yeast, the structural homolog of Mba1 in mammals, mL45, was identified as an integral part of the large subunit of the mitochondrial ribosome. This protein directly binds on the Oxa1 homolog, the Oxa1L insertase, and positions the peptide exit channel just above it [115]. Recent structural studies show that the mL45 protein is not the only ribosomal protein making contact with Oxa1L. It seems that the C-terminal region of Oxa1L containing the ribosome binding domain makes two additional contact sites, one with uL24m protein, and another with bL28m, uL29m and the large subunit rRNA [112]. These proteins are located near the peptide channel exit and contact the C-terminal extension of Oxa1. The last contact site with rRNA is mediated by a small extension of the C-terminal region of Oxa1L only found in mammals.
In the green algae Chlamydomonas, in situ imaging using cryo-electron tomography suggests that mitoribosomes are all bound to the mitochondrial inner membrane. Moreover, the determination of the Chlamydomonas mitoribosom’s high resolution structure combined with tomography suggests that a specific protein, mL119, located at the peptide channel exit appears to be involved in the interaction between the mitoribosome and Oxa1 to mediate membrane attachment. This protein is not related to the mammals mL45 or the yeast Mba1 [27]. In contrast with green algae, little is known about membrane-tethered mitochondrial ribosome in land plants, but high concentrations of non-ionic detergents are necessary to solubilise ribosomes from Arabidopsis mitochondrial extracts, suggesting that at least part of these ribosomes are tightly bound to the mitochondrial membranes [26]. Still, no homolog proteins of the mammals mL45, the yeast Mba1 or Chlamydomonas mL119 was found encoded in the Arabidopsis genome, implying that land plants might use yet another specific process for membrane attachment [25]. Nonetheless, non-dissociative immunoprecipitation of plant mitochondrial ribosomes co-precipitate Oxa1a [26], and expression of Oxa1a in Oxa1 knock-out yeast cells leads to a partial complementation of the respiratory defects normally present in the mutant [116]. These clues tend to suggest that plant Oxa1a performs similar functions and might have the possibility to bind the ribosome despite the low amino acid identity between the two homolog proteins.
4.5. Functions of Mitoribosome Specific Proteins for Ribosome Assembly
Ribosome biogenesis is the crucial process where all the ribosomes components, r-proteins, and rRNAs come together to form the functional translation machinery. This process involves a large number of proteins (maturation factors) and RNAs (such as snoRNAs in cytosolic ribosome maturation) factors that will aid to properly fold, assemble and modify the different elements of the ribosome [117]. Even if some of these maturation factors are conserved from prokaryotes to eukaryotes—these are usually factors involved in the maturation of conserved catalytic areas—the process of ribosome maturation in eukaryotes involves many additional factors, and is much more complex. In eukaryotes, the maturation process is restricted in the nucleus [118], and recent cryo-EM structures have highlighted the intricacy of this process [119,120,121,122,123]. In prokaryotes, biogenesis appears to be quicker and is thus more challenging to capture by structural means, even if some studies do exist [124,125].
In mitochondria, the biogenesis of mitoribosomes is even more complex, as it has to be coordinated between two gene expression compartments [126]. Indeed, the rRNAs, and part of the r-proteins, depending of the organisms, are encoded by the mitochondrial genome and are thus transcribed and processed inside the mitochondria. The rest of the r-proteins are encoded by the nuclear genome and have to be imported into the mitochondria. This is also the case for all the maturation factors involved in mitoribosome biogenesis. Recently, several studies managed to capture mitoribosome assembly intermediates from humans [127,128,129,130,131] and kinetoplastids [95,98,99,132,133], describing different steps and factors involved in the maturation of the large and small mitoribosomal subunits. A portion of these factors were previously described by non-structural means [59,134,135,136], but most of them were described for the first time with these studies, and most importantly, directly observed in action.
Collectively, the studies on the human LSU maturation structurally describe the mode of action of 13 maturation factors mainly involved in the maturation of the PTC. A large portion of these factors are GTPases and helicases involved in the correct folding of the rRNA, or methylases involved in its modification. Most are conserved and have bacterial or eukaryotic homologs; this is the case for GTPB5 and GTBP10 (homologs to ObgE) or are conserved in the same process with other eukaryotes. Thus, DDX28 is mt-LAF2 in kinetoplastids, and most likely Mss116 in yeast [136] and MRM3 corresponds to mt-LAF5-6. One complex that might be conversed across eukaryotes is the ACP block, composed of MALSU1–L0R8F8–mt-ACP in human and mt-Rsfs–L0R8F8–mt-SAF32 in kinetoplastids. It is always present on the large subunit during its maturation and prevents LSU-SSU association, it however does not bind RNA. The complex is composed of MALSU1/mt-Rsfs, a conserved bacterial factor, involved in ribosome hibernation and not in maturation, complemented by the two factors L0R8F8 and mt-ACP, which are specific to mitochondria. Moreover, the redundancy of assembly factors between LSU and SSU, notably mt-ACP and LYR motif proteins, with components of complex I in kinetoplastids, might point towards potential crosstalks between the assembly of ribosomal subunits and respiratory complexes [95,132,137]. On the contrary, the mTERF4 protein involved rRNA stabilization during the PTC maturation appears to be specific to humans.
In kinetoplastids, the detailed structures of maturation complexes of the large subunit present over 30 different maturation factors [95,98,99]. Given that kinetoplastid mitoribosomes are probably among the most divergent mitoribosomes described to date, it is not surprising that so many additional maturation factors were recruited in kinetoplastids (which is also the case for the SSU maturation described later) [24]. Many of the specific maturation factors are RNA binders, but also repurposed mitochondrial proteins, similar to the additional mitochondria-specific r-proteins that were recruited to mitoribosomes [5]. The strong complexity of this mitoribosome, and the extensive number of maturation factors involved, probably makes the biogenesis of this mitoribosome much slower compared to classical ribosomes, which may well have played an important role in the capture of these maturation states. A tryptic of GTPases, mt-EngA, mt-EngB, and mt-Rgb1 (also found in the human LSU intermediates, named MTG1), notably plays an important role in the maturation of the PTC by interacting with the rRNA and locking it in a specific conformation.
Finally, several maturation states were also resolved for the small subunit of Trypanosoma brucei, involving in total 39 maturation factors [132,133]. During that process, the SSU goes through drastic remodelling, and many of the assembly factors complement the huge protein shell formed by the already in place r-proteins. Similar to the LSU, a large portion of these factors are involved in the correct folding of the rRNA, notably for the decoding centre. A portion of these maturation factors are KRIPP proteins, kinetoplastids-specific proteins PPR proteins that were previously identified as potential r-proteins [138] that directly interact with single stranded rRNA. For example, KRIPP2 interacts with the 5′ end of the immature rRNA, very similar to OPR mL115 in Chlamydomonas or mL106 in Tetrahymena interactions with mature rRNA.
In summary, mitoribosome assembly involves protein factors conserved with bacteria but also new factors that are similar to mitoribosome r-proteins, either shared between eukaryotes, or specific to certain clades. Many of those factors interact with RNA, notably to promote its correct folding.
5. Concluding Remarks
The extraordinary and unexpected diversity of mitochondrial ribosomes began to be recognised less than a decade ago, in particular due to the development of revolutionary cryo-electron microscopy techniques that allowed for the obtaining of high resolution structures of mitoribosomes from a number of distantly related eukaryotes. Initial biochemical analyses suggested that mitoribosomes are protein-rich [36,39,78]. This was corroborated by the first cryo-EM structures obtained with animal models and led to the initial assumption that the recruitment of many novel proteins to mitoribosomes had been driven by the necessity to spatially replace rRNA segments or sub-domains that were lost during evolution. This “structural patching” hypothesis was however contradicted by the observation that yeast and particularly plant mitoribosomes do not have reduced rRNAs, contrary to animals or kinetoplastids, but still have a widely expanded set of ribosomal proteins. Likewise, the bioinformatic and biochemical analysis of mitoribosomes in jakobids, a group of protists with the most bacterial-like mitochondrial genomes and rRNAs, also found that jakobid mitoribosomes are protein rich as compared to their bacterial ancestors [30]. This led to the hypothesis that the last eukaryote common ancestor (LECA) mitoribosome might have already been protein-rich. Indeed, while tens of specific r-proteins were acquired in independent eukaryote clades, phylogeny analyses suggest that LECA might have contained as many as 33 SSU and 46 LSU proteins, thus with a 50% increase of protein numbers as compared to bacterial ribosomes [30]. This raises the question of the nature of the evolutionary forces that might have driven the very early recruitment of many novel proteins to mitoribosomes. One possible explanation might reside in the biochemical environment constituted by the mitochondrial matrix. This compartment is much more basic, with a pH close to 8, than the mitochondrial inter membrane space and the cytosol that has a pH close to 7 [139]. This pH gradient generated by the oxidative phosphorylation (OXPHOS) machinery and required for energy production through the ATP synthase might be deleterious to rRNA stability. Thus, the recruitment of many proteins to the mitoribosome might have been favoured to decrease contacts between rRNAs and a high pH solvent that could result in rRNA hydrolysis. In turn, the occurrence of many additional proteins might have been the evolutionary hint to allow the drift of mitochondrial rRNA genes through either losses or expansions.
Beyond the evolutionary history of mitoribosomes composition, many open questions remain with regard to the diversity of mitoribosomes within the same species, on the regulation of their function, and on their interaction with other gene expression complexes, as well as the assembly machineries of OXPHOS complexes. For instance, it is still unclear if different subpopulations of mitoribosomes, with different protein contents, occur within the same mitochondria. In some eukaryotes, i.e., in plants, hydrophilic proteins are encoded in the mitochondrial genome in addition to the common set of hydrophobic OXPHOS proteins. Does this imply that a subpopulation of mitoribosomes is attached to the inner membrane to synthesize hydrophobic proteins, while another subset of mitoribosomes would be soluble in the matrix for the synthesis of hydrophilic proteins? Answers to such questions are likely to be brought by in situ approaches such as cryo-electron tomography. These analyses are also expected to reveal if some of the additional ribosomal proteins described here play novel or additional functions, e.g., for the interaction of ribosomes with other complexes or for other yet unassigned regulatory processes related or not to mitochondrial translation. Likewise, since mitoribosome protein functions were already related to diseases in humans, e.g., tumorigenesis [140], future studies will also reveal whether dysfunctions of the novel mitochondrial-specific ribosomal proteins described here may result in disorders ranging from decreased agriculture yield to human diseases.
Author Contributions
Writing—original draft preparation, V.S., N.C., F.W. and P.G.; Figures preparation, F.W.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by ANR grants “MITRA, ANR 16 CE11-0024” and “DAMIA, ANR 20 CE11-0021” to P.G.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors apologize to those whose contributions were not cited because of length restrictions. This work was supported by the French “Centre National de la Recherche Scientifique”, by the University of Strasbourg, by the LabEx consortium “MitoCross” and by an Alexander von Humboldt postdoctoral grant to FW.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Melnikov, S.; Ben-Shem, A.; de Loubresse, N.G.; Jenner, L.; Yusupova, G.; Yusupov, M. One core, two shells: Bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 2012, 19, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Schmeing, T.M.; Ramakrishnan, V. What recent ribosome structures have revealed about the mechanism of translation. Nature 2009, 461, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Londei, P.; Ferreira-Cerca, S. Ribosome Biogenesis in Archaea. Front. Microbiol. 2021, 12, 686977. [Google Scholar] [CrossRef] [PubMed]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef]
- Waltz, F.; Giegé, P. Striking Diversity of Mitochondria-Specific Translation Processes across Eukaryotes. Trends Biochem. Sci. 2020, 45, 149–162. [Google Scholar] [CrossRef]
- Kummer, E.; Ban, N. Mechanisms and regulation of protein synthesis in mitochondria. Nat. Rev. Mol. Cell Biol. 2021, 22, 307–325. [Google Scholar] [CrossRef]
- Eme, L.; Spang, A.; Lombard, J.; Stairs, C.W.; Ettema, T.J.G. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 2017, 15, 711–723. [Google Scholar] [CrossRef]
- Martijn, J.; Vosseberg, J.; Guy, L.; Offre, P.; Ettema, T.J.G. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 2018, 557, 101–105. [Google Scholar] [CrossRef]
- Muñoz-Gómez, S.A.; Susko, E.; Williamson, K.; Eme, L.; Slamovits, C.H.; Moreira, D.; López-García, P.; Roger, A.J. Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known Alphaproteobacteria. Nat. Ecol. Evol. 2022, 6, 253–262. [Google Scholar] [CrossRef]
- Roger, A.J.; Muñoz-Gómez, S.A.; Kamikawa, R. The Origin and Diversification of Mitochondria. Curr. Biol. 2017, 27, R1177–R1192. [Google Scholar] [CrossRef]
- Dorrell, R.G.; Howe, C.J. What makes a chloroplast? Reconstructing the establishment of photosynthetic symbioses. J. Cell Sci. 2012, 125, 1865–1875. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef] [PubMed]
- Björkholm, P.; Harish, A.; Hagström, E.; Ernst, A.M.; Andersson, S.G.E. Mitochondrial genomes are retained by selective constraints on protein targeting. Proc. Natl. Acad. Sci. USA 2015, 112, 10154–10161. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.F. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, 10231–10238. [Google Scholar] [CrossRef]
- Zoschke, R.; Bock, R. Chloroplast Translation: Structural and Functional Organization, Operational Control, and Regulation. Plant Cell 2018, 30, 745–770. [Google Scholar] [CrossRef]
- Bieri, P.; Leibundgut, M.; Saurer, M.; Boehringer, D.; Ban, N. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2017, 36, 475–486. [Google Scholar] [CrossRef]
- Boerema, A.P.; Aibara, S.; Paul, B.; Tobiasson, V.; Kimanius, D.; Forsberg, B.O.; Wallden, K.; Lindahl, E.; Amunts, A. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants 2018, 4, 212–217. [Google Scholar] [CrossRef]
- Graf, M.; Arenz, S.; Huter, P.; Dönhöfer, A.; Nováček, J.; Wilson, D.N. Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 2016, 45, gkw1272. [Google Scholar] [CrossRef]
- Clemons, W.M.; May, J.L.C.; Wimberly, B.T.; McCutcheon, J.P.; Capel, M.S.; Ramakrishnan, V. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature 1999, 400, 833–840. [Google Scholar] [CrossRef]
- Ben-Shem, A.; de Loubresse, N.G.; Melnikov, S.; Jenner, L.; Yusupova, G.; Yusupov, M. The Structure of the Eukaryotic Ribosome at 3.0 A Resolution. Science 2011, 334, 1524–1529. [Google Scholar] [CrossRef]
- Greber, B.J.; Bieri, P.; Leibundgut, M.; Leitner, A.; Aebersold, R.; Boehringer, D.; Ban, N. The complete structure of the 55S mammalian mitochondrial ribosome. Science 2015, 348, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Amunts, A.; Brown, A.; Toots, J.; Scheres, S.H.W.; Ramakrishnan, V. The structure of the human mitochondrial ribosome. Science 2015, 348, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Desai, N.; Brown, A.; Amunts, A.; Ramakrishnan, V. The structure of the yeast mitochondrial ribosome. Science 2017, 355, 528–531. [Google Scholar] [CrossRef] [PubMed]
- Ramrath, D.J.F.; Niemann, M.; Leibundgut, M.; Bieri, P.; Prange, C.; Horn, E.K.; Leitner, A.; Boehringer, D.; Schneider, A.; Ban, N. Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes. Science 2018, 362, eaau7735. [Google Scholar] [CrossRef]
- Waltz, F.; Soufari, H.; Bochler, A.; Giegé, P.; Hashem, Y. Cryo-EM structure of the RNA-rich plant mitochondrial ribosome. Nat. Plants 2020, 6, 377–383. [Google Scholar] [CrossRef]
- Waltz, F.; Nguyen, T.; Arrivé, M.; Bochler, A.; Chicher, J.; Hammann, P.; Kuhn, L.; Quadrado, M.; Mireau, H.; Hashem, Y.; et al. Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants 2019, 5, 106–117. [Google Scholar] [CrossRef]
- Waltz, F.; Salinas-Giegé, T.; Englmeier, R.; Meichel, H.; Soufari, H.; Kuhn, L.; Pfeffer, S.; Förster, F.; Engel, B.D.; Giegé, P.; et al. How to build a ribosome from RNA fragments in Chlamydomonas mitochondria. Nat. Commun. 2021, 12, 7176. [Google Scholar] [CrossRef]
- Tobiasson, V.; Amunts, A. Ciliate mitoribosome illuminates evolutionary steps of mitochondrial translation. eLife 2020, 9, e59264. [Google Scholar] [CrossRef]
- Bieri, P.; Greber, B.J.; Ban, N. High-resolution structures of mitochondrial ribosomes and their functional implications. Curr. Opin. Struct. Biol. 2018, 49, 44–53. [Google Scholar] [CrossRef]
- Valach, M.; Alcazar, J.A.G.; Sarrasin, M.; Lang, B.F.; Gray, M.W.; Burger, G. An Unexpectedly Complex Mitoribosome in Andalucia godoyi, a Protist with the Most Bacteria-like Mitochondrial Genome. Mol. Biol. Evol. 2021, 38, 788–804. [Google Scholar] [CrossRef]
- Mager, J. Chloramphenicol and chlortetracycline inhibition of amino acid incorporation into proteins in a cell-free system from Tetrahymena pyriformis. Biochim. Biophys. Acta 1960, 38, 150–152. [Google Scholar] [CrossRef]
- Rendi, R. On the occurrence of intramitochondrial ribonucleoprotein particles. Exp. Cell Res. 1959, 17, 585–587. [Google Scholar] [CrossRef]
- Palade, G.E. A small particulate component of the cytoplasm. J. Cell Biol. 1955, 1, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Borst, P.; Grivell, L.A. Mitochondrial ribosomes. FEBS Lett. 1971, 13, 73–88. [Google Scholar] [CrossRef]
- Matthews, D.E.; Hessler, R.A.; Denslow, N.D.; Edwards, J.S.; O’Brien, T.W. Protein composition of the bovine mitochondrial ribosome. J. Biol. Chem. 1982, 257, 8788–8794. [Google Scholar] [CrossRef]
- Koc, E.C.; Burkhart, W.; Blackburn, K.; Moyer, M.B.; Schlatzer, D.M.; Moseley, A.; Spremulli, L.L. The Large Subunit of the Mammalian Mitochondrial Ribosome. J. Biol. Chem. 2001, 276, 43958–43969. [Google Scholar] [CrossRef] [PubMed]
- Koc, E.C.; Burkhart, W.; Blackburn, K.; Moseley, A.; Koc, H.; Spremulli, L.L. A Proteomics Approach to the Identification of Mammalian Mitochondrial Small Subunit Ribosomal Proteins. J. Biol. Chem. 2000, 275, 32585–32591. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.R.; Koc, E.C.; Datta, P.P.; Booth, T.M.; Spremulli, L.L.; Agrawal, R.K. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 2003, 115, 97–108. [Google Scholar] [CrossRef]
- Bonen, L.; Calixte, S. Comparative Analysis of Bacterial-Origin Genes for Plant Mitochondrial Ribosomal Proteins. Mol. Biol. Evol. 2006, 23, 701–712. [Google Scholar] [CrossRef]
- Gan, X.; Arita, K.; Isono, S.; Kitakawa, M.; Yoshino, K.; Yonezawa, K.; Kato, A.; Inoue, H.; Isono, K. Identification and comparative analysis of the large subunit mitochondrial ribosomal proteins of Neurospora crassa. FEMS Microbiol. Lett. 2006, 254, 157–164. [Google Scholar] [CrossRef]
- Sharma, M.R.; Booth, T.M.; Simpson, L.; Maslov, D.A.; Agrawal, R.K. Structure of a mitochondrial ribosome with minimal RNA. Proc. Natl. Acad. Sci. USA 2009, 106, 9637–9642. [Google Scholar] [CrossRef] [PubMed]
- Maslov, D.A.; Sharma, M.R.; Butler, E.; Falick, A.M.; Gingery, M.; Agrawal, R.K.; Spremulli, L.L.; Simpson, L. Isolation and characterization of mitochondrial ribosomes and ribosomal subunits from Leishmania tarentolae. Mol. Biochem. Parasitol. 2006, 148, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Rugen, N.; Straube, H.; Franken, L.E.; Braun, H.-P.; Eubel, H. Complexome profiling reveals association of PPR proteins with ribosomes in the mitochondria of plants. Mol. Cell. Proteom. 2019, 18, 1345–1362. [Google Scholar] [CrossRef]
- Smirnov, A.; Förstner, K.U.; Holmqvist, E.; Otto, A.; Günster, R.; Becher, D.; Reinhardt, R.; Vogel, J. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. USA 2016, 113, 11591–11596. [Google Scholar] [CrossRef]
- Gerovac, M.; Vogel, J.; Smirnov, A. The World of Stable Ribonucleoproteins and Its Mapping with Grad-Seq and Related Approaches. Front. Mol. Biosci. 2021, 8, 186. [Google Scholar] [CrossRef]
- O’Reilly, F.J.; Rappsilber, J. Cross-linking mass spectrometry: Methods and applications in structural, molecular and systems biology. Nat. Struct. Mol. Biol. 2018, 25, 1000–1008. [Google Scholar] [CrossRef]
- Kühlbrandt, W. The Resolution Revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef]
- Amunts, A.; Brown, A.; Bai, X.C.; Llacer, J.L.; Hussain, T.; Emsley, P.; Long, F.; Murshudov, G.; Scheres, S.H.W.; Ramakrishnan, V. Structure of the Yeast Mitochondrial Large Ribosomal Subunit. Science 2014, 343, 1485–1489. [Google Scholar] [CrossRef]
- Greber, B.J.; Boehringer, D.; Leitner, A.; Bieri, P.; Voigts-Hoffmann, F.; Erzberger, J.P.; Leibundgut, M.; Aebersold, R.; Ban, N. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 2013, 505, 515–519. [Google Scholar] [CrossRef]
- Hammani, K.; Bonnard, G.; Bouchoucha, A.; Gobert, A.; Pinker, F.; Salinas, T.; Giegé, P. Helical repeats modular proteins are major players for organelle gene expression. Biochimie 2014, 100, 141–150. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Rojas, M.; Fujii, S.; Yap, A.; Chong, Y.S.; Bond, C.S.; Small, I. A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins. PLoS Genet. 2012, 8, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.; Pinker, F.; Fuchsbauer, O.; Gutmann, B.; Boutin, R.; Roblin, P.; Sauter, C.; Giegé, P. Structural insights into protein-only RNase P complexed with tRNA. Nat. Commun. 2013, 4, 1353. [Google Scholar] [CrossRef] [PubMed]
- Pinker, F.; Schelcher, C.; Fernandez-Millan, P.; Gobert, A.; Birck, C.; Thureau, A.; Roblin, P.; Giegé, P.; Sauter, C. Biophysical analysis of Arabidopsis protein-only RNase P alone and in complex with tRNA provides a refined model of tRNA binding. J. Biol. Chem. 2017, 292, 13904–13913. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, T.; Kaitany, K.J.; Kakuta, Y.; Kimura, M.; Fierke, C.A.; Hall, T.M.T. Pentatricopeptide repeats of protein-only RNase P use a distinct mode to recognize conserved bases and structural elements of pre-tRNA. Nucleic Acids Res. 2020, 48, 11815–11826. [Google Scholar] [CrossRef]
- Preker, P.J.; Keller, W. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci. 1998, 23, 15–16. [Google Scholar] [CrossRef]
- Hammani, K.; Cook, W.B.; Barkan, A. RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc. Natl. Acad. Sci. USA 2012, 109, 5651–5656. [Google Scholar] [CrossRef]
- Roberti, M.; Polosa, P.L.; Bruni, F.; Manzari, C.; Deceglie, S.; Gadaleta, M.N.; Cantatore, P. The MTERF family proteins: Mitochondrial transcription regulators and beyond. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 303–311. [Google Scholar] [CrossRef]
- Spåhr, H.; Habermann, B.; Gustafsson, C.M.; Larsson, N.G.; Hallberg, B.M. Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc. Natl. Acad. Sci. USA 2012, 109, 15253–15258. [Google Scholar] [CrossRef]
- Méteignier, L.-V.; Ghandour, R.; Zimmerman, A.; Kuhn, L.; Meurer, J.; Zoschke, R.; Hammani, K. Arabidopsis mTERF9 protein promotes chloroplast ribosomal assembly and translation by establishing ribonucleoprotein interactions in vivo. Nucleic Acids Res. 2021, 49, 1114–1132. [Google Scholar] [CrossRef]
- Ozawa, S.-I.; Cavaiuolo, M.; Jarrige, D.; Kuras, R.; Rutgers, M.; Eberhard, S.; Drapier, D.; Wollman, F.-A.; Choquet, Y. The OPR Protein MTHI1 Controls the Expression of Two Different Subunits of ATP Synthase CFo in Chlamydomonas reinhardtii. Plant Cell 2020, 32, 1179–1203. [Google Scholar] [CrossRef] [PubMed]
- Viola, S.; Cavaiuolo, M.; Drapier, D.; Eberhard, S.; Vallon, O.; Wollman, F.; Choquet, Y. MDA 1, a nucleus-encoded factor involved in the stabilization and processing of the atpA transcript in the chloroplast of Chlamydomonas. Plant J. 2019, 98, tpj.14300. [Google Scholar] [CrossRef]
- Kleinknecht, L.; Wang, F.; Stübe, R.; Philippar, K.; Nickelsen, J.; Bohne, A.-V. RAP, the Sole Octotricopeptide Repeat Protein in Arabidopsis, Is Required for Chloroplast 16S rRNA Maturation. Plant Cell 2014, 26, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, A.; Matz, J.M.; Almendinger, M.; Müller, K.; Matuschewski, K.; Schmitz-Linneweber, C. Identification of clustered organellar short (cos) RNAs and of a conserved family of organellar RNA-binding proteins, the heptatricopeptide repeat proteins, in the malaria parasite. Nucleic Acids Res. 2018, 46, 10417–10431. [Google Scholar] [CrossRef]
- Filipovska, A.; Rackham, O. Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol. Biosyst. 2012, 8, 699–708. [Google Scholar] [CrossRef]
- Rubinson, E.H.; Eichman, B.F. Nucleic acid recognition by tandem helical repeats. Curr. Opin. Struct. Biol. 2012, 22, 101–109. [Google Scholar] [CrossRef]
- Delannoy, E.; Stanley, W.A.; Bond, C.S.; Small, I.D. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 2007, 35, 1643–1647. [Google Scholar] [CrossRef]
- Edwards, T.A.; Pyle, S.E.; Wharton, R.P.; Aggarwal, A.K. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 2001, 105, 281–289. [Google Scholar] [CrossRef]
- Nikolay, R.; van den Bruck, D.; Achenbach, J.; Nierhaus, K.H. Ribosomal Proteins: Role in Ribosomal Functions. In eLS; Wiley: Hoboken, NJ, USA, 2015; pp. 1–12. [Google Scholar]
- Itoh, Y.; Naschberger, A.; Mortezaei, N.; Herrmann, J.M.; Amunts, A. Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi. Nat. Commun. 2020, 11, 5187. [Google Scholar] [CrossRef]
- Rodnina, M.V. Translation in Prokaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032664. [Google Scholar] [CrossRef]
- Shine, J.; Dalgarno, L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 1974, 71, 1342–1346. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, T.; Marzi, S.; Romby, P. The role of mRNA structure in translational control in bacteria. RNA Biol. 2009, 6, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Duval, M.; Korepanov, A.; Fuchsbauer, O.; Fechter, P.; Haller, A.; Fabbretti, A.; Choulier, L.; Micura, R.; Klaholz, B.P.; Romby, P.; et al. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol. 2013, 11, e1001731. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Green, R.; Buskirk, A.R. Translational initiation in E. Coli occurs at the correct sites genome-wide in the absence of mrna-rrna base-pairing. eLife 2020, 9, e55002. [Google Scholar] [CrossRef] [PubMed]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef]
- Waltz, F.; Corre, N.; Hashem, Y.; Giegé, P. Specificities of the plant mitochondrial translation apparatus. Mitochondrion 2020, 53, 30–37. [Google Scholar] [CrossRef]
- Burger, G.; Gray, M.W.; Forget, L.; Lang, B.F. Strikingly Bacteria-Like and Gene-Rich Mitochondrial Genomes throughout Jakobid Protists. Genome Biol. Evol. 2013, 5, 418–438. [Google Scholar] [CrossRef]
- Temperley, R.J.; Wydro, M.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M. Human mitochondrial mRNAs—Like members of all families, similar but different. Biochim. Biophys. Acta Bioenerg. 2010, 1797, 1081–1085. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Montoya, J.; Ojala, D.; Attardi, G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature 1981, 290, 465–470. [Google Scholar] [CrossRef]
- Kummer, E.; Leibundgut, M.; Rackham, O.; Lee, R.G.; Boehringer, D.; Filipovska, A.; Ban, N. Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 2018, 560, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Aibara, S.; Singh, V.; Modelska, A.; Amunts, A. Structural basis of mitochondrial translation. eLife 2020, 9, e58362. [Google Scholar] [CrossRef] [PubMed]
- Baggio, F.; Bratic, A.; Mourier, A.; Kauppila, T.E.S.; Tain, L.S.; Kukat, C.; Habermann, B.; Partridge, L.; Larsson, N.-G. Drosophila melanogaster LRPPRC2 is involved in coordination of mitochondrial translation. Nucleic Acids Res. 2014, 42, 13920–13938. [Google Scholar] [CrossRef] [PubMed]
- Ruzzenente, B.; Metodiev, M.D.; Wredenberg, A.; Bratic, A.; Park, C.B.; Cámara, Y.; Milenkovic, D.; Zickermann, V.; Wibom, R.; Hultenby, K.; et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012, 31, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Siira, S.J.; Spåhr, H.; Shearwood, A.-M.J.; Ruzzenente, B.; Larsson, N.-G.; Rackham, O.; Filipovska, A. LRPPRC-mediated folding of the mitochondrial transcriptome. Nat. Commun. 2017, 8, 1532. [Google Scholar] [CrossRef]
- Lagouge, M.; Mourier, A.; Lee, H.J.; Spåhr, H.; Wai, T.; Kukat, C.; Ramos, E.S.; Motori, E.; Busch, J.D.; Siira, S.; et al. SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from Degradation. PLoS Genet. 2015, 11, e1005423. [Google Scholar] [CrossRef]
- Weraarpachai, W.; Antonicka, H.; Sasarman, F.; Seeger, J.; Schrank, B.; Kolesar, J.E.; Lochmüller, H.; Chevrette, M.; Kaufman, B.A.; Horvath, R.; et al. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 2009, 41, 833–837. [Google Scholar] [CrossRef]
- Herrmann, J.M.; Woellhaf, M.W.; Bonnefoy, N. Control of protein synthesis in yeast mitochondria: The concept of translational activators. Biochim. Biophys. Acta 2013, 1833, 286–294. [Google Scholar] [CrossRef]
- Derbikova, K.S.; Levitsky, S.A.; Chicherin, I.V.; Vinogradova, E.N.; Kamenski, P.A. Activation of Yeast Mitochondrial Translation: Who Is in Charge? Biochemistry 2018, 83, 87–97. [Google Scholar] [CrossRef]
- Nguyen, T.-T.; Planchard, N.; Dahan, J.; Arnal, N.; Balzergue, S.; Benamar, A.; Bertin, P.; Brunaud, V.; Dargel-Graffin, C.; Macherel, D.; et al. A Case of Gene Fragmentation in Plant Mitochondria Fixed by the Selection of a Compensatory Restorer of Fertility-Like PPR Gene. Mol. Biol. Evol. 2021, 38, 3445–3458. [Google Scholar] [CrossRef]
- Salinas-Giegé, T.; Cavaiuolo, M.; Cognat, V.; Ubrig, E.; Remacle, C.; Duchêne, A.-M.; Vallon, O.; Maréchal-Drouard, L. Polycytidylation of mitochondrial mRNAs in Chlamydomonas reinhardtii. Nucleic Acids Res. 2017, 45, 12963–12973. [Google Scholar] [CrossRef] [PubMed]
- Cahoon, A.B.; Qureshi, A.A. Leaderless mRNAs are circularized in Chlamydomonas reinhardtii mitochondria. Curr. Genet. 2018, 64, 1321–1333. [Google Scholar] [CrossRef] [PubMed]
- Feagin, J.E.; Harrell, M.I.; Lee, J.C.; Coe, K.J.; Sands, B.H.; Cannone, J.J.; Tami, G.; Schnare, M.N.; Gutell, R.R. The Fragmented Mitochondrial Ribosomal RNAs of Plasmodium falciparum. PLoS ONE 2012, 7, e38320. [Google Scholar] [CrossRef]
- Soufari, H.; Waltz, F.; Parrot, C.; Durrieu-Gaillard, S.; Bochler, A.; Kuhn, L.; Sissler, M.; Hashem, Y. Structure of the mature kinetoplastids mitoribosome and insights into its large subunit biogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 29851–29861. [Google Scholar] [CrossRef] [PubMed]
- Noller, H.F.; Lancaster, L.; Zhou, J.; Mohan, S. The ribosome moves: RNA mechanics and translocation. Nat. Struct. Mol. Biol. 2017, 24, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Trabuco, L.G.; Schreiner, E.; Eargle, J.; Cornish, P.; Ha, T.; Luthey-Schulten, Z.; Schulten, K. The Role of L1 Stalk–tRNA Interaction in the Ribosome Elongation Cycle. J. Mol. Biol. 2010, 402, 741–760. [Google Scholar] [CrossRef] [PubMed]
- Jaskolowski, M.; Ramrath, D.J.F.; Bieri, P.; Niemann, M.; Mattei, S.; Calderaro, S.; Leibundgut, M.; Horn, E.K.; Boehringer, D.; Schneider, A.; et al. Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis. Mol. Cell 2020, 79, 629–644. [Google Scholar] [CrossRef]
- Tobiasson, V.; Rej Gahura, O.; Aibara, S.; Baradaran, R.; Ikov, A.; Amunts, A. Interconnected assembly factors regulate the biogenesis of mitoribosomal large subunit. EMBO J. 2021, 40, e106292. [Google Scholar] [CrossRef]
- Ratje, A.H.; Loerke, J.; Mikolajka, A.; Brünner, M.; Hildebrand, P.W.; Starosta, A.L.; Dönhöfer, A.; Connell, S.R.; Fucini, P.; Mielke, T.; et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 2010, 468, 713–716. [Google Scholar] [CrossRef]
- Ott, M.; Amunts, A.; Brown, A. Organization and Regulation of Mitochondrial Protein Synthesis. Annu. Rev. Biochem. 2016, 85, 77–101. [Google Scholar] [CrossRef]
- Keil, M.; Bareth, B.; Woellhaf, M.W.; Peleh, V.; Prestele, M.; Rehling, P.; Herrmann, J.M. Oxa1-Ribosome Complexes Coordinate the Assembly of Cytochrome c Oxidase in Mitochondria. J. Biol. Chem. 2012, 287, 34484–34493. [Google Scholar] [CrossRef] [PubMed]
- Stuart, R.A. Insertion of proteins into the inner membrane of mitochondria: The role of the Oxa1 complex. Biochim. Biophys. Acta—Mol. Cell Res. 2002, 1592, 79–87. [Google Scholar] [CrossRef]
- Krüger, V.; Deckers, M.; Hildenbeutel, M.; van der Laan, M.; Hellmers, M.; Dreker, C.; Preuss, M.; Herrmann, J.M.; Rehling, P.; Wagner, R.; et al. The Mitochondrial Oxidase Assembly Protein1 (Oxa1) Insertase Forms a Membrane Pore in Lipid Bilayers. J. Biol. Chem. 2012, 287, 33314–33326. [Google Scholar] [CrossRef] [PubMed]
- Stiller, S.B.; Höpker, J.; Oeljeklaus, S.; Schütze, C.; Schrempp, S.G.; Vent-Schmidt, J.; Horvath, S.E.; Frazier, A.E.; Gebert, N.; van der Laan, M.; et al. Mitochondrial OXA Translocase Plays a Major Role in Biogenesis of Inner-Membrane Proteins. Cell Metab. 2016, 23, 901–908. [Google Scholar] [CrossRef]
- Wang, P.; Dalbey, R.E. Inserting membrane proteins: The YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. Biophys. Acta—Biomembr. 2011, 1808, 866–875. [Google Scholar] [CrossRef]
- Hennon, S.W.; Soman, R.; Zhu, L.; Dalbey, R.E. YidC/Alb3/Oxa1 Family of Insertases. J. Biol. Chem. 2015, 290, 14866–14874. [Google Scholar] [CrossRef]
- Haque, M.E.; Elmore, K.B.; Tripathy, A.; Koc, H.; Koc, E.C.; Spremulli, L.L. Properties of the C-terminal Tail of Human Mitochondrial Inner Membrane Protein Oxa1L and Its Interactions with Mammalian Mitochondrial Ribosomes. J. Biol. Chem. 2010, 285, 28353–28362. [Google Scholar] [CrossRef]
- Jia, L. Yeast Oxa1 interacts with mitochondrial ribosomes: The importance of the C-terminal region of Oxa1. EMBO J. 2003, 22, 6438–6447. [Google Scholar] [CrossRef]
- Szyrach, G.; Ott, M.; Bonnefoy, N.; Neupert, W.; Herrmann, J.M. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003, 22, 6448–6457. [Google Scholar] [CrossRef]
- Hildenbeutel, M.; Theis, M.; Geier, M.; Haferkamp, I.; Neuhaus, H.E.; Herrmann, J.M.; Ott, M. The Membrane Insertase Oxa1 Is Required for Efficient Import of Carrier Proteins into Mitochondria. J. Mol. Biol. 2012, 423, 590–599. [Google Scholar] [CrossRef]
- Itoh, Y.; Andréll, J.; Choi, A.; Richter, U.; Maiti, P.; Best, R.B.; Barrientos, A.; Battersby, B.J.; Amunts, A. Mechanism of membrane-tethered mitochondrial protein synthesis. Science 2021, 371, 846–849. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.; Prestele, M.; Bauerschmitt, H.; Funes, S.; Bonnefoy, N.; Herrmann, J.M. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 2006, 25, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Woellhaf, M.W.; Herrmann, J.M.; Förster, F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat. Commun. 2015, 6, 6019. [Google Scholar] [CrossRef] [PubMed]
- Englmeier, R.; Pfeffer, S.; Förster, F. Structure of the Human Mitochondrial Ribosome Studied In Situ by Cryoelectron Tomography. Structure 2017, 25, 1574–1581.e2. [Google Scholar] [CrossRef]
- Benz, M.; Soll, J.; Ankele, E. Arabidopsis thaliana Oxa proteins locate to mitochondria and fulfill essential roles during embryo development. Planta 2013, 237, 573–588. [Google Scholar] [CrossRef]
- Kressler, D.; Hurt, E.; Baßler, J. A Puzzle of Life: Crafting Ribosomal Subunits. Trends Biochem. Sci. 2017, 42, 640–654. [Google Scholar] [CrossRef]
- Erdmann, P.S.; Hou, Z.; Klumpe, S.; Khavnekar, S.; Beck, F.; Wilfling, F.; Plitzko, J.M.; Baumeister, W. In situ cryo-electron tomography reveals gradient organization of ribosome biogenesis in intact nucleoli. Nat. Commun. 2021, 12, 5364. [Google Scholar] [CrossRef]
- Klinge, S.; Woolford, J.L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef]
- Kargas, V.; Castro-Hartmann, P.; Escudero-Urquijo, N.; Dent, K.; Hilcenko, C.; Sailer, C.; Zisser, G.; Marques-Carvalho, M.J.; Pellegrino, S.; Wawiórka, L.; et al. Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. eLife 2019, 8, e44904. [Google Scholar] [CrossRef]
- Ameismeier, M.; Zemp, I.; van den Heuvel, J.; Thoms, M.; Berninghausen, O.; Kutay, U.; Beckmann, R. Structural basis for the final steps of human 40S ribosome maturation. Nature 2020, 587, 683–687. [Google Scholar] [CrossRef]
- Cheng, J.; Lau, B.; La Venuta, G.; Ameismeier, M.; Berninghausen, O.; Hurt, E.; Beckmann, R. 90S pre-ribosome transformation into the primordial 40S subunit. Science 2020, 369, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Lau, B.; Cheng, J.; Flemming, D.; La Venuta, G.; Berninghausen, O.; Beckmann, R.; Hurt, E. Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome. Mol. Cell 2021, 81, 293–303.e4. [Google Scholar] [CrossRef] [PubMed]
- Nikolay, R.; Hilal, T.; Qin, B.; Mielke, T.; Bürger, J.; Loerke, J.; Textoris-Taube, K.; Nierhaus, K.H.; Spahn, C.M.T. Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution. Mol. Cell 2018, 70, 881–893.e3. [Google Scholar] [CrossRef] [PubMed]
- Nikolay, R.; Hilal, T.; Schmidt, S.; Qin, B.; Schwefel, D.; Vieira-Vieira, C.H.; Mielke, T.; Bürger, J.; Loerke, J.; Amikura, K.; et al. Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization. Mol. Cell 2021, 81, 1200–1215.e9. [Google Scholar] [CrossRef]
- Sissler, M.; Hashem, Y. Mitoribosome assembly comes into view. Nat. Struct. Mol. Biol. 2021, 28, 631–633. [Google Scholar] [CrossRef]
- Cipullo, M.; Gesé, G.V.; Khawaja, A.; Hällberg, B.M.; Rorbach, J. Structural basis for late maturation steps of the human mitoribosomal large subunit. Nat. Commun. 2021, 12, 3673. [Google Scholar] [CrossRef]
- Hillen, H.S.; Lavdovskaia, E.; Nadler, F.; Hanitsch, E.; Linden, A.; Bohnsack, K.E.; Urlaub, H.; Richter-Dennerlein, R. Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling. Nat. Commun. 2021, 12, 3672. [Google Scholar] [CrossRef]
- Cheng, J.; Berninghausen, O.; Beckmann, R. A distinct assembly pathway of the human 39S late pre-mitoribosome. Nat. Commun. 2021, 12, 4544. [Google Scholar] [CrossRef]
- Lenarčič, T.; Jaskolowski, M.; Leibundgut, M.; Scaiola, A.; Schönhut, T.; Saurer, M.; Lee, R.G.; Rackham, O.; Filipovska, A.; Ban, N. Stepwise maturation of the peptidyl transferase region of human mitoribosomes. Nat. Commun. 2021, 12, 3671. [Google Scholar] [CrossRef]
- Chandrasekaran, V.; Desai, N.; Burton, N.O.; Yang, H.; Price, J.; Miska, E.A.; Ramakrishnan, V. Visualizing formation of the active site in the mitochondrial ribosome. eLife 2021, 10, e68806. [Google Scholar] [CrossRef]
- Saurer, M.; Ramrath, D.J.F.; Niemann, M.; Calderaro, S.; Prange, C.; Mattei, S.; Scaiola, A.; Leitner, A.; Bieri, P.; Horn, E.K.; et al. Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery. Science 2019, 365, 1144–1149. [Google Scholar] [CrossRef] [PubMed]
- Lenarčič, T.; Niemann, M.; Ramrath, D.J.F.; Calderaro, S.; Flügel, T.; Saurer, M.; Leibundgut, M.; Boehringer, D.; Prange, C.; Horn, E.K.; et al. Mitoribosomal small subunit maturation involves formation of initiation-like complexes. Proc. Natl. Acad. Sci. USA 2022, 119, e2114710118. [Google Scholar] [CrossRef] [PubMed]
- Rorbach, J.; Boesch, P.; Gammage, P.A.; Nicholls, T.J.J.; Pearce, S.F.; Patel, D.; Hauser, A.; Perocchi, F.; Minczuk, M. MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol. Biol. Cell 2014, 25, 2542–2555. [Google Scholar] [CrossRef]
- De Silva, D.; Tu, Y.-T.; Amunts, A.; Fontanesi, F.; Barrientos, A. Mitochondrial ribosome assembly in health and disease. Cell Cycle 2015, 14, 2226–2250. [Google Scholar] [CrossRef]
- Zeng, R.; Smith, E.; Barrientos, A. Yeast Mitoribosome Large Subunit Assembly Proceeds by Hierarchical Incorporation of Protein Clusters and Modules on the Inner Membrane. Cell Metab. 2018, 27, 645–656.e7. [Google Scholar] [CrossRef]
- Van Vranken, J.G.; Nowinski, S.M.; Clowers, K.J.; Jeong, M.-Y.; Ouyang, Y.; Berg, J.A.; Gygi, J.P.; Gygi, S.P.; Winge, D.R.; Rutter, J. ACP Acylation Is an Acetyl-CoA-Dependent Modification Required for Electron Transport Chain Assembly. Mol. Cell 2018, 71, 567–580.e4. [Google Scholar] [CrossRef]
- Aphasizheva, I.; Maslov, D.; Wang, X.; Huang, L.; Aphasizhev, R. Pentatricopeptide Repeat Proteins Stimulate mRNA Adenylation/Uridylation to Activate Mitochondrial Translation in Trypanosomes. Mol. Cell 2011, 42, 106–117. [Google Scholar] [CrossRef]
- Porcelli, A.M.; Ghelli, A.; Zanna, C.; Pinton, P.; Rizzuto, R.; Rugolo, M. pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant. Biochem. Biophys. Res. Commun. 2005, 326, 799–804. [Google Scholar] [CrossRef]
- Pecoraro, A.; Pagano, M.; Russo, G.; Russo, A. Ribosome Biogenesis and Cancer: Overview on Ribosomal Proteins. Int. J. Mol. Sci. 2021, 22, 5496. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).