Abstract
Warm temperatures induce plant bolting accompanied by flower initiation, where endogenous auxin is dynamically associated with accelerated growth. Auxin signaling is primarily regulated by a family of plant-specific transcription factors, AUXIN RESPONSE FACTORS (ARFs), which either activate or repress the expression of downstream genes in response to developmental and environmental cues. However, the relationship between ARFs and bolting has not been completely understood in lettuce yet. Here, we identified 24 LsARFs (Lactuca sativa ARFs) in the lettuce genome. The phylogenetic tree indicated that LsARFs could be classified into three clusters, which was well supported by the analysis of exon–intron structure, consensus motifs, and domain compositions. RNA-Seq analysis revealed that more than half of the LsARFs were ubiquitously expressed in all tissues examined, whereas a small number of LsARFs responded to UV or cadmium stresses. qRT-PCR analysis indicated that the expression of most LsARFs could be activated by more than one phytohormone, underling their key roles as integrative hubs of different phytohormone signaling pathways. Importantly, the majority of LsARFs displayed altered expression profiles under warm temperatures, implying that their functions were tightly associated with thermally accelerated bolting in lettuce. Importantly, we demonstrated that silencing of LsARF8a, expression of which was significantly increased by elevated temperatures, resulted in delayed bolting under warm temperatures, suggesting that LsARF8a might conduce to the thermally induced bolting. Together, our results provide molecular insights into the LsARF gene family in lettuce, which will facilitate the genetic improvement of the lettuce in an era of global warming.
1. Introduction
The phytohormone auxin (indole-3-acetic acid) is involved in a multitude of growth and developmental processes in land plants. In the past decade, auxin perception and signaling pathways that mediate the changes in gene expression have been explored intensively, leading to the identification of three dedicated protein families [1,2,3]. Theoretically, in the absence of auxin, the pathway is repressed by a group of AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) inhibitors. Aux/IAA proteins physically bind to the AUXIN RESPONSE FACTORs (ARFs) that allow the expression of auxin-dependent genes. To unlock the pathway, Aux/IAA proteins, in the presence of auxin, were targeted to the F-box-containing TRANSPORT INHIBITOR RESPONSE and AUXIN-SIGNALING F-BOX PROTEIN (TIR1/AFB) ubiquitin ligases, resulting in their degradation by the 26S proteasome [4]. Subsequently, de-repression of ARF transcription factors allows them to directly mediate the transcription of their downstream genes [5]. As ARF proteins couple the perception of the hormone auxin to gene expression programs, this represents the core of the auxin signaling pathway.
It is well established that ARFs play crucial roles in almost every developmental aspect such as flowering and bolting in Arabidopsis [6]. For instance, AtARF1 and AtARF2 were involved in the regulation of floral organ abscission, while AtARF3/ETTIN functions in gynoecium morphogenesis, self-incompatibility, and flower development [7,8,9]. Genetic analysis suggested that AtARF6 and AtARF8 redundantly regulate the flower maturation in Arabidopsis as well as tomato [10,11,12]. In addition to their developmental roles, recent studies found that some ARFs were involved in plant responses to abiotic stress. Overexpression of AtARF3 stimulated the expression of a set of drought-stress-responsive genes including RAP2.6, NAC019, ERD5, ZAT12, and RD26 in Arabidopsis; conversely, these ARF-activated genes remained at very lower expression levels in arf3 mutants in response to the drought stress [13]. Similarly, expression levels of the majority of SlARFs were attenuated under salinity stress, whereas several SlARF genes such as SlARF1, SlARF4, and SlARF19 exhibited significant upregulation upon salt stress [14]. Thus, these observations highlighted the neglected roles of ARFs that might also participate in the plant response to abiotic stress [15]. In our previous studies, we showed that the transcript accumulation of LsARF2 and LsARF5 in flower stalk kept increasing with the progression of lettuce bolting and revealed that tryptophan metabolism pathways were involved in the early bolting under high temperatures in lettuce [16,17]. These observations are consistent with the fact that the auxin content in stem tissues increased when flower stalks initiated their elongation [17]. As the major components of auxin signaling, LsARFs might integrate thermal and floral signals to coordinate floral development and plant growth in lettuce. Given that ARFs are tightly associated with multiple developmental and environmental cues, it is reasonable that they might be the potential intersections between two signaling pathways, wherein plant optimizes their developmental strategy to tackle the thermal challenges.
Lettuce (Lactuca sativa L.) is a globally important leaf vegetable in the family Asteraceae (Compositae) cultivated preliminarily for its fleshy leaves or succulent stems [18]. Lettuce growth is sensitive to heat stress, and exposure of lettuce to warm temperatures can reduce vegetative growth by hastening floral transition (premature bolting and accelerated flowering), ultimately resulting in severe economic loss to world producers, especially during summer productions [19,20]. Although ARFs probably link thermal response and floral transition signaling, the involvement of ARFs in lettuce flowering induction by warm temperatures remains largely unknown thus far.
As of 2020, the world production of lettuce (chicory included) reached 27.7 million tons, and China is ranked the highest in lettuce production, followed by USA and India [21]. Despite the economic importance of lettuce, a genome-wide, systematic study of the lettuce ARF gene family has not been reported yet. The first annotated genome of lettuce was released in 2017, and afterward, genome sequencing of 445 lettuce varieties showed the origins and breeding history of the crop [22,23]. The genome information facilitates the research and enables us to conduct genome-wide analysis of ARFs in lettuce. In the present study, a genome-wide, comprehensive analysis of lettuce ARF genes was performed, and a total of 24 LsARF genes were identified. The physical and chemical characteristics, genomic structures, chromosomal locations, evolutionary relationships, and expression profiles of the LsARF gene family were investigated in detail. Finally, the virus-induced gene silencing (VIGS) approach was employed to analyze the biological roles of LsARF8a under thermally induced bolting. These findings provide deep insights into the function of LsARF genes during the lettuce bolting upon warm temperatures.
2. Results
2.1. Identification of LsARFs from the Lettuce Genome
To identify lettuce ARF genes, protein sequences of functionally validated ARFs from Arabidopsis and rice were used as the queries to perform BLASTp searches against the lettuce genome. After removing the non-representative splicing forms of the same gene locus, 27 ARF-like genes were obtained from the genome sequences of L. sativa. Further, the presence of the B3 DNA-binding and AuxRE domains in these ARF-like proteins were scanned using the conserved domain search (CD-search) with an e-value <10−10. Due to the lack of an AuxRE motif, three genes (Lsat_1_v5_gn_5_62560, Lsat_1_v5_gn_6_46381, and Lsat_1_v5_gn_8_118440) were removed from our analysis, and ultimately, a total of 24 genes were identified as LsARFs. According to the homologies against Arabidopsis ARFs (Figure S1), the nomenclature of these LsARF genes are listed in Table 1. The predicted proteins encoded by LsARFs varied from 432 amino acids (LsARF3b) to 1025 amino acids (LsARF7a), with corresponding molecular weights from 47.53 to 115.76 kDa. Of these putative LsARF proteins, the theoretical isoelectric points ranged from 5.32 (LsARF5b) to 7.93 (LsARF16d), indicating that they could participate in biochemical processes under distinct in vivo environments.

Table 1.
List of putative the gene family of auxin response factors in the L. sativa genome.
2.2. Chromosomal Distribution and Duplication Events among LsARF Genes
The physical map position of LsARF genes on nine lettuce chromosomes was established. Chromosomes 2 and 7 contain the largest number of LsARFs, comprising six members; chromosome 5, 6, and 9 each contain two members; chromosome 6 has one member; and chromosome 1, 4, and 8 each contain a single LsARF. The number of LsARFs in the lettuce genome is similar to their counterparts in Arabidopsis. Pairwise sequence comparison of LsARF proteins suggests that the homology broadly ranged from 21.40% (LsARF3a and LsARF7b) to 79.52% (LsARF2a and LsARF2b). Strikingly, LsARF members in the ARF9 subclade comprising LsARF9a/9b/9c share high sequence identity (76.35–79.04%), suggesting that these LsARFs are likely to be originated from gene duplications while they are positioned to different chromosomes. A similar event was found in ARF10/16 subclade, which includes LsARF16a/16b/16c/16d/10 with identity from 57.72% to 70.85%. Duplication analysis revealed the expansion of the ARF gene family in lettuce (Figure 1B). Among 24 LsARFs, 10 were found to be segmentally duplicated, such as LsARF7a/7b, LsARF8a/8b, LsARF16a/16b/16/c/16d, and LsARF19a/19b. Accordingly, high sequence similarity between duplicated gene pairs was observed, suggesting they might participate in the regulation of similar biological processes. Interestingly, tandem duplication did not occur in LsARF genes, whereas it was common in other plant species such as AtARFs.
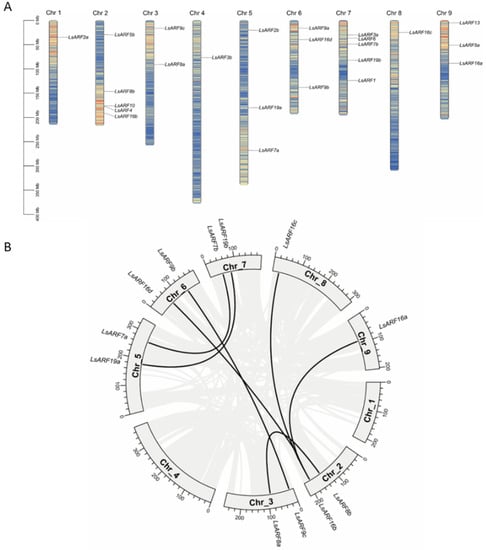
Figure 1.
Chromosomal distribution and duplication events of LsARF genes. (A) Chromosomal location of members of the LsARF gene family. The chromosome numbers and size are indicated at the top and bottom of each bar, respectively. The number on the right side of the bars designated the approximate physical position of the first exon of corresponding LsARF genes on lettuce chromosomes. (B) Mapping of duplicated LsARF genes on the lettuce genome. Grey ribbons indicate a collinear relationship among the blocks in the whole genome, and black ribbons show LsARF paralogs.
2.3. Structural Analysis and Conserved Motifs of LsARF Genes
To better understand the structure of the exon and intron, the full-length cDNA sequences of LsARF genes were aligned against the corresponding genome DNA sequences (Figure 2). Gene structure analysis suggested that the number of exons of LsARFs ranged from 2 to 14. It is noteworthy that LsARF members in the ARF8 and ARF9 subfamily share identical intron–exon structure, suggesting they are highly conserved within the same subclade. Three members of the LsARF10/16 subclade also exhibited similar gene structures, despite the fact that LsARF16a is a truncated gene. Although the exon–intron structure of LsARFs varies between subclades, it is similar within subclades, which was supported by the phylogenetic tree constructed with the cDNA sequences of LsARFs.
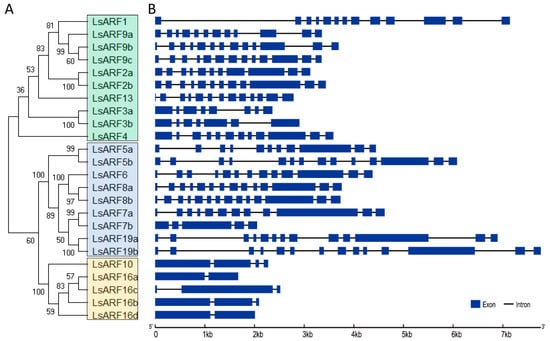
Figure 2.
Classification of L. sativa ARF proteins. (A). Neighbor-joining tree were generated using MEGA to determine the phylogenetic relationship between L. sativa ARF proteins (left). According to classification proposed by Finet et al. (2013), LsARFs were divided into three subgroups: A (top), B (middle) and C (bottom), and shadow colors were used to distinguish different LsARF subgroups. (B). The intron–exon organization of LsARF genes was plotted using Gene Structure Display Server (Version 2.0). Blue boxes represent exons, and grey lines represent introns.
Furthermore, we performed multiple sequence alignment using the deduced amino acid sequences of LsARF genes. As shown in Figure 3A, most LsARF proteins contain highly conserved DBD domains at N-terminal and PB1 domain at C-terminal. LsARF7b had a truncated DBD domain in which the B3 region was deleted, indicating that its DNA-binding ability could be impaired despite the remaining AuxRE motif. The PB1 domain of LsARF3a, LsARF3b, and LsARF16a was completely lost, suggesting that it might be independent of IAA/AUX-induced degradation. Although the DBD and PB1 domains are highly conserved, the MRs showed the highest divergence in the amino acid composition of ARF proteins. On the basis of the enrichment of specific amino acids in the MR, as well as on the ability of some tested AtARFs in transient experiments, it is predictable whether ARFs act as activators or repressors. Composition analysis of LsARF proteins suggest that the MRs of LsARF7a/7b, LsARF6, and LsARF19a/19b are abundant in Q residues (Figure 3B), thereby suggesting they might be transcriptional activators.
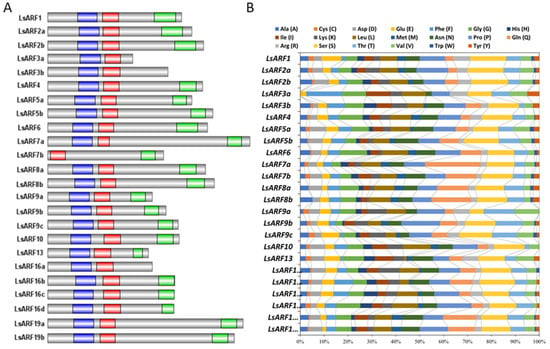
Figure 3.
Analysis of conserved domains in LsARF porteins. (A) Schematic organization of conserved domains in LsARF proteins. The B3 DNA-binding domain, Aux_Resp domain, and PB1 domain are shown in blue, red, and green, respectively. (B) Amino acid composition of MR domains in LsARF proteins. Bars represent the percentage of different amino acid residues in MR domains of LsARFs, and each color represents one kind of amino acid.
To identify the novel conserved motif, the MEME online program was used to analyze the motif distribution of LsARF proteins (Figure S2). In our analysis, eight motifs were identified from LsARF proteins. Among them, motifs 1, 2, 3, and 6 correspond to the B3 domain (PF02362); motifs 5, 7, and 8 correspond to the Auxin_Resp (Pf06507); and motif 4 represents the PB1 domain (PF00564). Motifs 3 and 5 are conserved among LsARFs, suggesting that they might be novel motifs essential to ARF proteins. Furthermore, LsARFs in the same subfamily shared similar patterns of motif composition, indicating their functional similarities. Thus, the distribution of the motifs also revealed that LsARFs were likely conserved during evolution.
2.4. Analysis of Cis-Elements in the Promoter Regions and Gene Ontology
In order to identify the potential regulatory regime, we analyzed the cis-elements in the 2 kb upstream of all identified LsARF genes by using the PlantCare database (Figure 4A). The promoter regions of LsARF3b, LsARF7b, and LsARF9c fall in the sequencing gap that contains artificially filled ambiguous N nucleotides, and thus removal of these regions resulted in reduced size of these promoter sequences. Our analysis identified 305 major cis-acting elements in these putative LsARF promoter regions (Figure 4B), and the five most highly represented cis-elements were the ABA-responsive element (ABRE, 54), GT1-motif (GT1, 36), G-box (32), ethylene-responsive element (ERE, 30), and low-temperature-responsive element (LTR, 24). As the promoter sequence of LsARF7b is very short, no motif was identified in these regions, and LsARF7b was excluded from the analysis. Surprisingly, ABRE was found in 17 out of 23 LsARF members, and 11 of them contain more than one such element. The auxin-responsive elements (AuxRE) were only identified in three LsARF members, while TGACG-motif, found to be associated with auxin response, was presented in the promoter of many LsARF genes. Gibberellin-responsive element (GARE) was found in eight members of the LsARF gene family. A total of 46 elements involved in MeJA-responsiveness (CGTAC and TGACG) were found in the promoter regions of almost all LsARF members, and similarly, 33 cis-elements (as-1 and TCA motif) were identified as being involved in the salicylic-acid-responsiveness elements. The results reveal that these lettuce ARFs might be involved in the hormonal crosstalk due to the presence of such hormone-responsive motifs.
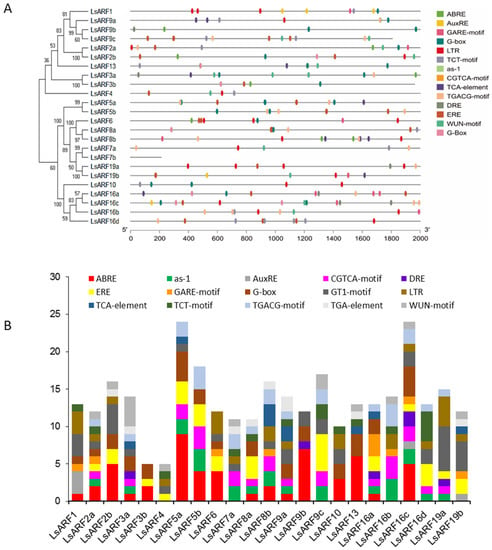
Figure 4.
Identification of the cis-acting elements in the putative promoter region of LsARF genes. (A) The distribution of 15 major cis-elements in the putative promoter of LsARF genes. Fifteen cis-elements including auxin-responsive elements (AuxRE), as-1, ABA-responsive element (ABRE), CGTGCA motif, dehydration-responsive element (DRE), ethylene-responsive element (ERE), gibberellin-responsive motif (GARE), G-box, GT1-motif, low-temperature-responsive element (LRE), TCA-element, TCT-motif, TGACG motif, TGA element, and wound-responsive element (WUN) are designated with different colors. (B) Number of different cis-elements in the LsARF promoter regions.
2.5. Phylogenetic Analysis of LsARFs
On the basis of ARF protein sequences from liverwort (Marchantia polymorpha), moss (Physcomitrella patens), tomato, potato, rice, and Arabidopsis, a phylogenetic tree was constructed to explore the evolutionary scenario of ARF genes (Figure 5). One B3 domain-containing sequence (Cre13g562400) from green algae (Chlamydomonas reinhardtii) was used as an outgroup. Consistent with analyses by Mutte et al. (2018), 131 ARFs were generally classified into 3 major clades: clade A (52 members) includes homologs of AtARF5, AtARF6/8, and AtARF7/19; clade B (33 members) represents the homologs to AtARF1, AtARF2, AtARF3/4, and AtARF13; and the remaining 27 ARFs belonged to clade C, which consists of AtARF16/10 and AtARF17 homologs. According to the phylogenetic tree, members of LsARFs were detected from almost all subclades, despite the fact that lettuce ARFs homologous to AtAARF17 were not identified in our analysis. Generally, our phylogenetic tree revealed that the ARF proteins from the lettuce and Solanaceae (tomato and potato) usually were clustered into the same subclades, indicating the closer relationship between the Solanaceae and Asteraceae families compared to Arabidopsis or rice.
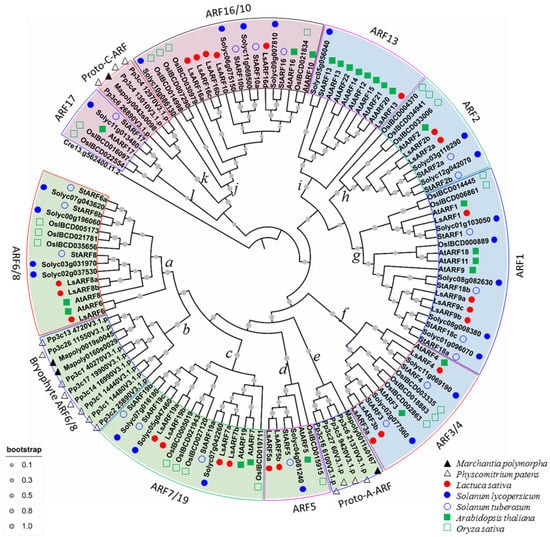
Figure 5.
Phylogenetic relationship among ARF proteins from liverwort, moss, tomato, potato, Arabidopsis, rice, and lettuce. The deduced full-length amino acid sequences of ARFs from liverwort, moss, tomato, potato, Arabidopsis, rice, and lettuce were aligned by the MUSCLE method, and the neighbor-joining tree was generated using MEGA X to determine the phylogenetic relationship between those ARF proteins. According to the classification proposed by Mutte et al. (2017), LsARFs were divided into three subgroups: A (purple), B (blue), and C (green). Shadow colors are used to distinguish different LsARF subgroups.
The phylogeny of the ARF family was investigated in detail according to the previous proposed standard [24]. As illustrated in Figure 5, clade A contained five subclades represented by nodes a, b, c, d, and e, and the phylogenetic relationships within the subclades of clade A were well supported by bootstrap percentage. Node a (ARF6/8 subclade) subclade was placed as a twin-sister to node b (bryophytes ARF6/8 subclade). Node c was represented by ARF7/19, while node d included AtARF5 and its homologs. Notably, the node e subclade, containing class A-ARFs of bryophytes, was basal to the other four subclades, suggesting they might represent precursors of class A-ARFs. Clade B was further divided into four subclades, represented by node f (ARF3/4 subclade), g (ARF1/9 subclade), h (ARF2 subclade), and i (ARF13 subclade). Notably, class B-ARFs consisted of members only from land plants, suggesting that these higher plant-specific ARFs. Clade C included node j (ARF10/16 subclade), k (proto-C subclade), and l (ARF17 subclade). It was noted that the basal node k subclade was only represented by members from bryophytes, implying they might be a group of proto-C-ARFs.
2.6. Expression Profiles of LsARFs among Various Tissues and Organs
To determine the tissue-specific expression patterns of LsARF genes, we utilized the transcriptome data derived from the illumine RNA-Seq reads stored in the NCBI. The RNA-seq data provide the expression of 40,341 lettuce genes in five tissues (root, stem, leaf, flower, and seed). According to the RNA-Seq libraries, 22 members of LsARFs were detected in various organs and exhibited tissue-specific expression patterns, while LsARF9a and LsARF13 were absent, probably due to their low transcriptional levels or spatiotemporal expression patterns. As shown in Figure 6A, LsARF1/2a/2b/4/6/8a/8b/9b/9c/19a were ubiquitously and highly expressed in tissues, suggesting that these ARFs might be engaged in the auxin-regulated plant growth and development in various organs. Of 24 expressed lettuce ARFs, LsARF8a/8b were the most abundant transcripts in stem and flower tissues, implying that they might be implicated in floral development such as bolting and flowering. On the contrary, some LsARFs such as LsARF7a and LsARF16b/16c/16d were mildly expressed (Figure 6A), suggesting these LsARFs might be less required in those organs. Despite low transcriptional abundance in several tissues, LsARF7a, as well as LsARF3a, was preferentially expressed in the flower tissues compared to other organs (Figure 6A), indicating that they might serve as transcriptional regulators of auxin-signaling cascade mainly in the flower development. Thus, the results suggest that these LsARF genes have specific functions in different organs.
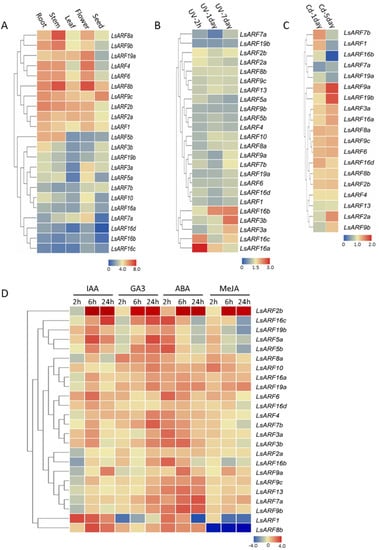
Figure 6.
Expression profiles of LsARF genes. (A) Expression profiles of LsARFs with hierarchical clustering in different organs. (B,C) Heatmap representation and hierarchical clustering of LsARFs under UV treatments and cadmium stresses. (D) Heatmap showing expression patterns of the LsARFs under different phytohormone treatments. The FPKM value or normalized qRT-PCR data of representative transcripts of LsARFs was used to generate heatmap with hierarchical clustering on the basis of the Manhattan correlation with average linkage using the TBtools software package. The color scales below heatmap show the expression levels. Red indicates high transcript abundance, while blue indicates low abundance.
2.7. Expression Responses of LsARFs to UV and Cd Stresses
As the auxin signaling pathway is associated with abiotic stress, we examined its expression response to abiotic stress with RNA-Seq data available in the NCBI database. As shown in Figure 6B, the transcript levels of most LsARF genes remained unchanged or slightly upregulated under UV treatments, suggesting that their expressions might be independent of UV stress. However, expression of LsARF16a and LsARF16c increased by 2.3- and 3.1-fold after 4 h UV treatment and remained at a relatively high level at 7-day UV treatment, suggesting they might be involved in the early response of lettuce plants to the UV stress. Transcript accumulations of LsARF16b, LsARF3a, and LsARF3b reached their highest levels after 7-day UV treatment, indicating that they were late-responsive genes.
Likewise, the expression of almost all LsARF genes could respond to the cadmium stress, except LsARF3b, LsARF5a, LsARF10, and LsARF16b/16c (Figure 6C). Of the 19 LsARFs, most could be induced by 3-day or 5-day cadmium stress. In particular, LsARF9a and LsARF19b exhibited a relatively higher expression in Cd-treated lettuce seedlings, which pointed to their crucial roles in plant adaption to heavy metal stress. Conversely, expressions of LsARF7a, LsARF16b, and LsARF19a were downregulated at 5-day cadmium treatment, indicating they might be negative regulators of auxin-related signaling pathways under heavy metal stress. Thus, the results suggest that the LsARF-mediated auxin cascade might contribute to cadmium stress.
2.8. Expression of LsARFs in Response to Various Phytohormone Treatments
The expression changes of LsARFs under different phytohormone or chemical analog treatments by the qRT-PCR analysis were evaluated in the leaf tissues of one-month-old seedlings. In general, compared to the controls, the majority of lettuce ARFs considered in the study were upregulated by ABA, GA3, or IAA treatments (Figure 6D). In auxin treatments, LsARF1 expression was activated at 2h-IAA treatment, but its expression reduced gradually, suggesting it might be early responsive genes; expression of most LsARF genes peaked at 6h- or 24hIAA treatments, indicating that they could be involved in the plant late responses to auxin; by contrast, LsARF8a expression was repressed by auxin treatments, implying its negative roles in auxin signaling pathways. Moreover, expression levels of several LsARFs were kept unchanged in auxin-treated leaves, suggesting that they were insensitive to auxin. It is worth noting that ABA could stimulate the expression of many LsARF genes, and their expression peaks were reached mainly at 2h-ABA treatments. The results are consistent with the findings that ABRE cis-elements were identified in the promoter regions of most LsARFs. Almost all LsARFs were gibberellin-responsive genes, while their expressions were elevated to the maximum probably after 24-h GA3 treatments. In contrast, methyl jasmonate had minor effects on the expression of LsARF genes, despite the fact that a few LsARFs including LsARF2b, LsARF10, and LsARF19 could be triggered upon the MeJA treatments. Together, these results provide experimental evidence that ARFs play central roles during the interplay between diverse phytohormone signaling pathways.
2.9. Gene Expression Analysis of LsARFs under Thermal Treatments
To explore their association with thermally induced bolting, we examined the expression levels of LsARF genes in the stem tissues of lettuce plants exposed to heat treatment for 0, 2, 4, 8, 16, and 24 days (Figure 7). As predicted, transcripts of most LsARF genes were accumulated to high levels during the bolting of lettuce plants. Among them, expression levels of LsARF1, LsARF2a/2b, LsARF8a, and LsARF13 were dramatically increased with the elongation of the stem within 24 days, while other LsARF genes, including LsARF3a/3b, LsARF6, LsARF9a, and LsARF16b, displayed a mild increase in transcript accumulations compared with their controls, which implies that they could be actively involved in the floral transition such as bolting. In contrast, several LsARFs such as LsARF5a/5b, LsARF7a, LsARF9b/9c, and LsARF16c/16d did not show any significant changes in expression levels, indicating that they might not be required for bolting under normal growth conditions.
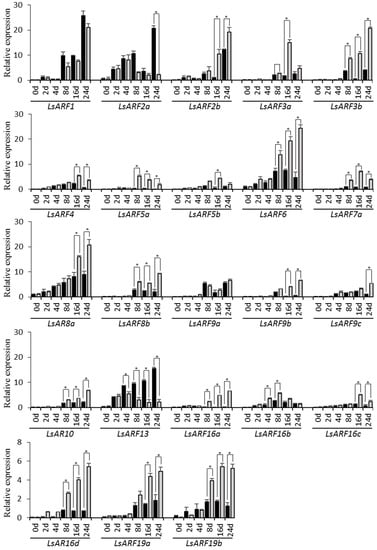
Figure 7.
qRT-PCR analysis of LsARF genes in response to heat treatments. LsARF transcript levels measured by real-time RT-qPCR using cDNAs from the shoot apex at indicated time points. Data are means of three biological replicates (8 pooled plants each), and error bars denote SE. Lettuce eIF4ɑ gene was used as an internal control. Stars above the error bars indicate significant differences between treatments and the mocks (according to Student’s t-test, p < 0.05). qRT-PCR primers for each LsARF gene are provided in Table S1.
High temperature has a profound impact on the expression patterns of LsARF genes (Figure 7). Transcript levels of the majority of LsARF genes were induced by thermal stress, including LsARF2b, LsARF3a, LsARF6, LsARF8a, and LsARF16a. Conversely, high temperature significantly decreased the expression of many LsARFs such as LsARF2a and LsARF13. Thus, those LsARFs with up- or downregulated expressions in response to high temperatures might mediate the expression of their downstream genes in the auxin signaling pathways, ultimately leading to developmental alternation upon warm temperatures. In addition, the expression profiles of LsARF1, as well as LsARF9a, were almost identical between heat-treated and control samples, suggesting that they might be insensitive to thermal stimuli.
2.10. LsARF8a Encodes an ARF Transcription Factor
According to the gene expression analysis, LsARF8a exhibited much higher expression in stem tissues, and importantly, its expression was strongly repressed by high temperatures during the floral transition stage. Therefore, LsARF8a might be tightly associated with the elongation of stem stalk in lettuce, and its possible role during thermally induced bolting was further investigated. Sequence analysis identified a nuclear localization signal in the N-terminal region of the putative LsAR8a protein, consistent with its predicted role as a transcription factor. To test the possibility, we transiently expressed LsARF8a-GFP in N. benthamiana leaf epidermal cells by infiltration with A. tumefaciens harboring a construct designed to express the LsARF8a ORF N-terminally fused to GFP. As revealed by the microscopy, GFP-labelled LsARF8a proteins were observed only in the nucleus, while free GFPs were distributed throughout the nucleus and the cytoplasm (Figure 8A). The results indicated that LsARF8a could redirect the GFP from the cytosol to the nucleus, supporting its function as a transcription factor.
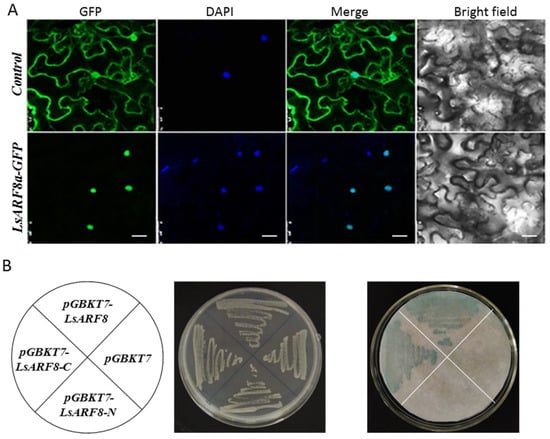
Figure 8.
LsARF8a encodes a transcriptional activator. (A) Subcellular localization of LsARF8a-GFP fusion proteins. Indicated constructs were agro-infiltrated into N. benthamiana leaves, and the subcellular localization of GFP alone (upper panel) or LsARF8a-GFP fusion proteins (lower panel) was analyzed by confocal laser-scanning microscopy. Images of representative leaf epidermal cells were taken 48 h after agro-infiltration. The photographs (left to right): bright field, GFP fluorescence, DAPI fluorescence, and merge. Bar = 20 μm. (B) Transcriptional activity analysis of LsARF8a in yeast cells. pGBKT7-LsARF8a, pGBKT7-LsARF8a-N (1–385 AA), pGBKT7-LsARF8a-C (386–799 AA), and pGBK&7 vector were transformed into yeast strain AH109 (left); the transformants grew well on SD/-Trp plate (middle), and colony-lift filter assay showed their β-galactosidase activities (right).
To test the transcriptional activity of LsARF8a, the fusion plasmids pGBKT7-LsARF8a, pGBKT7-LsARF8a-N, and pGBKT7-LsARF8a-C were separately transformed into yeast strain AH109, and pGBKT7 vector was used as the negative control. The transformed yeast strains grew well on the synthetic dextrose (SD) medium lacking tryptophan (SD/-Trp) but containing 20 mM 3-AT. The β-galactosidase activity assay suggested that LacZ expression in the yeast cells harboring pGBKT7-LsARF8a or pGBKT7-LsARF8a-C was transactivated by LsARF8a or its C-terminal alone, yet no significant expression could be detected in those yeast strains carrying pGBKT7-LsARF8a_C or pGBKT7 empty vector (Figure 8B). Thus, the results demonstrated that N-terminal polypeptide confers LsARF8a in the transcriptional activation activity in yeast cells. Hence, the results suggest that LsARF8 might be a functional transcriptional factor in lettuce.
2.11. Silencing of LsARF8a Resulted in Delayed Bolting under Thermal Treatments
To investigate the roles of LsARFs during lettuce bolting, we selected LsARF8a for functional analysis by the virus-induced gene silencing (VIGS) method. A 250 bp fragment targeting the LsARF8a 5′-region was chosen for the insertion into the viral vector pTRV2 vector, and the resulting plasmid pTRV2-LsARF8a was introduced into Agrobacterium. The Agrobacterium lines harboring pTRV2-PDS and empty pTRV2 vector served as controls. Lettuce seedlings with four true leaves were transformed by leaf injection with the aforementioned Agrobacterium as well as the prepared pTRV1 agrobacterium lines. After a three-week recovery under normal growth conditions, all silencing lines were verified by the qRT-PCR method. We found that leaves of pTRV2-PDS lines exhibited photo-bleaching phenotypes, which agreed with the reduction of PDS genes. qRT-PCR analysis with leaf tissues suggested that transcript accumulation of LsARF8a, compared to the control plants, significantly decreased in LsARF8a-sliciencing lines, whereas expression of LsARF6 and LsARF8b was not affected (Figure S3), suggesting that LsARF8a was specifically silenced. Subsequently, half LsARF8-knockdown (TRV:LsARF8a) plants were transferred to another growth chamber for heat treatments, and lettuce plants transformed with pRTV2 empty vectors (TRV:00) and WT plants were used as controls.
After growth in the thermal chamber for about one month, we assessed the responses of these lettuce lines upon heat treatments. After heat treatments, the stems of the LsARF8a-silencing plants were still immersed among the rosette leaves, whereas the elongated inflorescence stems emerged in the control groups (Figure 9A). Moreover, the stem length of LsARF8a knockdown lines was significantly lower than that of WT, whereas there was no difference between WT and TRV::00 lines, suggesting that silencing of LsARF8a could suppress stem elongation under thermal conditions (Figure S4). Moreover, the anatomical comparisons of the stem apex (inflorescence meristem) were conducted using these one-month heat-treated plants. As illustrated in Figure 9B, the stem apex of LsARF8a-silenced plants became much larger and more flattened, indicating the initial stage of floral differentiation; on the contrary, flower bud differentiation was observed in these non-silenced lines (TRV:00 or WT plants), suggesting that their floral transitions were completed after one-month heat treatments. In addition, it is noticeable that, under control temperatures, the bolting time of LsARF8a silencing lettuce plants was delayed significantly compared to the control groups, indicating that absence of LsARF8a led to the late bolting under normal conditions. Together, the results suggest that downregulation of LsARF8a resulted in the delayed bolting, thus supporting the fact that LsARF8a might positively regulate plant bolting in lettuce.
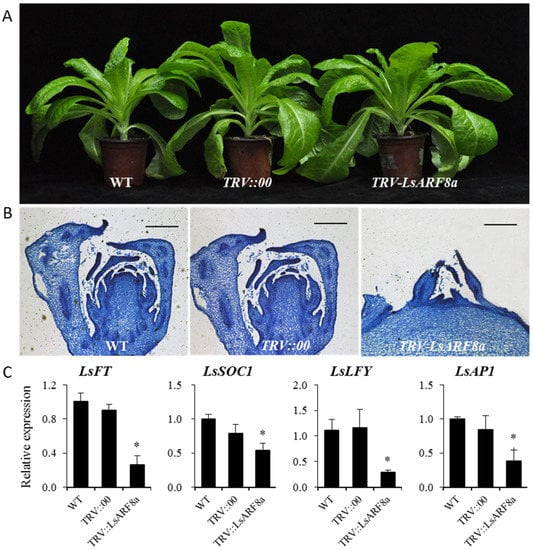
Figure 9.
Functional characterization of LsARF8a during the thermally induced bolting in lettuce. (A) Phenotypic characterization of TRV2-LsARF8a, TRV:00, and WT lettuce plants after one-month heat treatments. (B) Microscopic observation of shoot apices from TRV:LsARF8a, TRV:00, and WT lettuce plants after one-month thermal stress. Bar = 200 μm. (C) qRT-PCR analysis of LsFT, LsSOC1, LsAP1, and LsLFY in LsARF8a-silencing and control plants. Error bars indicate SE from three biological replicates, and asterisks indicate statistically significant differences between WT and treatments, as determined by Student’s t-test (* p < 0.05).
Next, we still needed to fill up the lacuna on the identification of downstream genes of LsARF8a in floral transition signaling pathways. As some genes such as LsFT, LsLFY, LsAP1, and LsSOC1 have been reported to be the inducers that are involved in the lettuce bolting [25,26,27], their expression profiles were investigated in the LsARF8a-silencing lines through qRT-PCR analysis. As shown in Figure 9C, we found that the expression levels of LsFT, LsLFY, and LsSOC1 were downregulated in the LsARF8a-silencing lettuce. The results indicate that LsARF8a might be a positive regulator to activate these floral activator genes directly or indirectly, thereby accelerating plant bolting in lettuce.
3. Discussion
3.1. Expansion of Lettuce ARF Gene Family
As the major effector of numerous aspects of auxin responses, ARF transcription factors translate the chemical signal into the transcriptional regulation of a defined set of genes. Due to their functional importance in the auxin cascades, the ARF gene family has been investigated intensively in the model plant Arabidopsis and to varying degrees in non-model plant species. Previous studies revealed that the Arabidopsis genome contains 23 ARF genes, while 25, 51, and 36 ARF members were identified in rice, soybean, and corn genomes [28,29,30], respectively. The genome size of lettuce is 2.5 Gb, which is one of the largest plant genomes assembled thus far [22]. Considering that lettuce has such a large genome, it was assumed that the lettuce genome could encode a larger ARF gene family as previously reported in soybean and corn [29,30]. Intriguingly, only 24 ARFs were finally identified from the lettuce genome, indicating that ARF transcription factors might be encoded by a moderate-sized gene family. Although the lettuce genome is nearly 17 times larger than the Arabidopsis counterpart, the numbers of the ARF gene family in the two genomes are still comparable, which contradicted the genome complexity between the two species. It is noteworthy that Arabidopsis has seven members of the ARF13 subclade while only one belongs to the subclade. It is plausible that several independent, small-scale, segmental duplication events and chromosome rearrangements that occurred at ARF13 loci resulted in multiple members of the AtARF13 subclade, which led to the expansion of the ARF gene family in Arabidopsis.
Furthermore, we investigated the distribution of lettuce and Arabidopsis ARF members in different subclades. With respect to the class A-ARFs, five LsARFs belong to the ARF10/16 subclade, while only two AtARF genes were grouped into this subclade; surprisingly, lettuce ortholog to LsARF17 was absent from the ARF17 subclade, whereas its counterparts could be found in rice, tomato, and potato. Despite the lack of ARF17 ortholog in lettuce, lettuce ARF16/10 subclade expanded in class A-ARFs due to the duplications of LsARF16s. Similarly, duplication events in the lettuce genome were observed in class B- and class C-ARFs, except in the ARF13 subclade. Hence, we conclude that the total number of AtARF genes is smaller than that of LsARFs. There is a whole-genome triplication event in lettuce since the divergence from the grape lineage and the triplicated regions cover at least 26% of the lettuce genome [22,31]. Consequently, genes encoding some but not all transcription factors and DNA-binding proteins were enriched in the triplicated regions of the lettuce genome [22]. Therefore, it is reasonable that some LsARFs might be present in these duplicated regions and that their duplicated copies contribute to the expansion of the ARF gene family in the Asteraceae.
With the expansion of the lettuce ARF family, functional redundancy and divergence could occur among different LsARFs. Phylogenetic analysis suggested that LsARF5a and LsARF5b form sister pairs within the ARF5 subclade, and thereby they might execute similar functions in auxin-regulated pathways. Despite the lack of experimental evidence, the expression profiles of two LsARF5s were almost identical in the respective hormone treatments, supporting the assumption that they might play overlapping roles in these signaling pathways. However, this is not always the case. It seems that subfunctionalization is common for LsARFs in the same subclade. For example, LsARF2a and LsARF2b genes displayed contrasting expression behaviors in response to various phytohormone applications. The qRT-PCR analysis revealed that LsARF2a was insensitive to all phytohormones, whereas LsARF2b was highly inducible genes upon ABA, auxin, gibberellin, or MeJA. Given that the expression profiles differ across individual members of the LsARF16 subclade, their functions might have diverged considerably. Similar assumptions could be applied to other LsARF members that were grouped in the same subclade. Together, our findings reflected that, in most cases, LsARF members from the same subclade might have acquired their unique roles.
3.2. LsARFs Are Involved in the Heat-Induced Bolting
Plant exposure to high temperatures causes diverse developmental, physiological, and morphological responses, including early flowering, accelerated shoot and root growth, and changes in stomatal differentiation [32,33,34,35]. Nevertheless, the signaling pathways of how plants sense and respond to elevated temperatures are incompletely understood. Recent studies have demonstrated that PHYTOCHROME B (PHYB) perceives the thermal signals and therefore connects plant growth and ambient temperatures [36,37]. PHYB exists in two relatively stable forms: a red-light-absorbing inactive Pr form and the far-red-light-absorbing active Pfr form, and the Pfr form can be converted into Pr form through a temperature-dependent process called thermos-reversion. In Arabidopsis seedlings, the phytochrome null mutants (phyABCDE) lacking phytochrome activity exhibit constitutive high-temperature responses such as elongated hypocotyl, which resemble the plant behavior under heat treatments [36]. Interestingly, it is worth noting that a small number of genes associated with auxin signaling and elongation growth including YUCCA8, YUCCA9, LIKE AUXIN RESISTNAT1, TRANSPORT INHIBITOR1, ARF7, and ARF9 are upregulated in the mutant background, which argues in favor of crucial contributions of auxin signaling during plant heat responses [36]. In previous experiments, we have shown that exogenous application of auxin could promote early bolting, albeit blocking of auxin transport with TIBA (2,3,5-triiodobenzoic acid) led to the delayed bolting in lettuce, which is in agreement with the hypothesis that auxin signaling is needed in lettuce bolting [16,38]. Additionally, we demonstrated that “loss-of-function” of LsARF3 led to the delayed bolting in lettuce under heat-treated or control conditions, further indicating that auxin signaling allows for lettuce bolting [35]. Consistent with the aforementioned findings, we further suggested that most LsARFs, the central components of the auxin signaling pathways, displayed altered expression profiles upon heat treatments, implying their contribution to the warm temperature-induced bolting. Therefore, our results further showed that ARF-mediated auxin signaling is responsible, at least partially, for these altered phenotypes upon heat treatments.
3.3. LsARF8a Postively Regulates Lettuce Bolting under Thermal Growth Conditions
Accelerated bolting is regarded as the hallmark of lettuce responses to thermal conditions [19], being regulated by an elaborate network of genetic pathways responsive to endogenous and external signals. The ambient temperatures accelerate the thermos-reversion rate of the PHYB, leading to a reduction of the steady-state level of the active Pfr. Because the active Pfr can repress the transcription of a group of bHLH transcriptional factors such as PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and PIF5, thermal treatment leads to the induction of PIF4 at the transcriptional levels in Arabidopsis [39]. PIF4 directly regulates the expression of a defined set of growth-relevant genes, particularly those genes in auxin biosynthesis and signaling [40,41]. In our study, promoter analysis of LsARF8a identified two G-box (5′-CACGTG-3′) motifs that could be recognized by bHLH proteins [42], suggesting LsARF8 might be the direct target of PIF4. Intriguingly, G-box motifs were also found in the promoter regions of other 15 LsARFs, expression profiles of which were largely altered upon warm temperatures. For example, the promoter regions of LsARF3a, LsARF3b, and LsARF13 contain two, two, and three G-box motifs, respectively. Thus, the results suggest that more than half LsARFs could be dynamically regulated by PIFs in a temperature-dependent manner.
In addition, it is unusual that there are so many ABRE cis-elements in the promoter regions of LsARF8a and other 16 LsARFs, implying that their expressions were possibly influenced by the ABRE-binding protein (AREB)/ABRE-binding factor (ABF) transcription factors. It was revealed that AREB1/ABF2, AREB2/ABF4, and ABF3 redundantly control a major part of ABA-mediated transcriptional changes when plants encounter dehydration, cold, heat stresses, or other abiotic stresses [43,44]. qRT-PCR demonstrated that ABA could induce the expression of LsARF8a, which is in line with the cis-element analysis of its promoter regions. Taken together, PIF4, as well as ABRE/ARFs, could play equally important roles in stimulating the transcription of LsARFs such as LsARF8a under higher temperatures. The possible scenario is that the transcription of LsARF8a might be modulated either by the ABREs solely or by the PHYB-PIF4 node upon heat treatments.
The silencing of LsARF8a resulted in late flowering in warmer conditions, thereby suggesting that LsARF8a contributed to lettuce bolting. Furthermore, LsARF8a was preferentially expressed in stem tissues, supporting its potential role in stem development. On the basis of these observations, we propose that LsARF8a might positively regulate the floral transition process in lettuce. In particular, LsARF8a transcripts in the shoot apex accumulated with the transition from the vegetative to the floral stage, which indicated that LsARF8a is required for lettuce bolting and flowering under normal growth conditions. Given that LsARF8a is a transcription activator, these results imply that LsARF8a could promote plant bolting by stimulating the expression of the floral gene(s) during the early stage of floral transition. Several floral activators such as LsFT, LsMADS55, LsLFY, LsAP1, and LsSOC1 have been reported in lettuce [25,26,27,45]. Consistent with this hypothesis, we found an associated reduction in LsFT, LsSOC1, and LsLFY transcript levels in the LsARF8a knockdown lines. It is possible that the downregulation of these floral inducers due to the LsARF8a silencing is responsible for the delayed bolting and flowering. Therefore, our observations from LsARF8a might indicate the existence of a previously unknown mechanism whereby thermal stress regulates the timing of plant bolting via heat-induced, ARF-mediated floral signaling pathways. However, it is still unknown as to how LsARF8a controls the floral genes to coordinate the timing of lettuce bolting, especially during thermal conditions. Hence, our future work will be focused on the systematic identification of direct targets of LsARF8a through comparative transcriptome analysis of LsARF8a overexpressing and knockout lines. We are generating these stable LsARF8a transgenic lines that will be an impressive feat to better understand the LsARF8a-mediated bolting in lettuce.
4. Materials and Methods
4.1. Plant Material and Treatments
Experiments were performed in a greenhouse located at the National Engineering Research Center for Vegetables (Beijing, China). The seeds of leafy lettuce (Lactuca sativa L.) varieties GB-30 were provided by the Beijing University of Agriculture. Seeds were sown in the plug trays, using Jeffery perlite as the substrate, in a growth chamber. One-week-old seedlings with uniform height were transplanted to the greenhouse, enabling only one plant per pot, and the growth conditions are as follows: 16 h day, 20 °C/8 h night, 13 °C. The lettuce seedlings were grown until the fourth leaf was fully expanded. For heat treatment, lettuce plants were transferred to another greenhouse, and the growth conditions were changed to 16 h day, 33 °C/8 h night, 25 °C. The remaining plants grown in normal conditions were used as controls. For phytohormone treatments, lettuce plants with four true leaves were subjected to IAA (100 μM), ABA (50 μM), GA3 (10 μM), or MeJA (10 μM). After phytohormone treatments, the leaf tissues were collected at designated points and immediately frozen in liquid nitrogen for RNA extractions. Sample collections were performed on separate days for the replicates.
4.2. Identification of LsARF Genes from the L. sativa Genome
To investigate the ARF gene family in the lettuce genome, all members of ARF sequences from Arabidopsis thaliana and rice were used as querie sequences for a BLAST search against Phytozome (https://phytozome.jgi.doe.gov/, accessed on 19 July 2022) with default parameters [46]. These candidates were verified the presence of the B3 (PF02362), AuxRE (PF06507), and PB1 (PF00564) domain using SMART (http://smart.embl-heidelberg.de/smart/batch.pl, accessed on 25 July 2022) and CDD-search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 25 July 2022). The redundant LsARF sequences were manually removed by multiple sequence comparisons. Finally, the non-redundant LsARF sequences were used as queries to blast against the Phytozome database. The representing gene model for each LsARF locus was identified, and their corresponding information on chromosomal location, locus ID, and transcript ID were obtained simultaneously.
4.3. Analysis of Gene Structure and Conserved Domains
On the basis of the genome annotation available in Phytozome, the intron–exon structure of individual LsARF genes was predicated, and its genomic organization was visualized using the Gene Structure Display Server [47]. Conserved domains in protein sequences were verified using ScanProsite (http://pro-site.expasy.org/scanprosite/, accessed on 30 July 2022), which provides information about the positions of different domains in the protein sequence. The MEME program (https://meme-suite.org/meme/tools/meme, accessed on 19 July 2022) was used to predict the novel motifs with the default settings. This information was visualized with TBtools to show the representation of the distribution of domains in the deduced amino acid sequences of LsARF proteins [48].
4.4. Sequence Alignment and Phylogenetic Construction
Multiple alignments of LsARF protein sequences from Marchantia polymorpha, Physcomitrium patens, Solanum lycopersicum, Solanum tuberosum, Arabidopsis thaliana, Oryza sativa, and Lactuca sativa were conducted using ClustalW [49]. The neighbor-joining method was used to conduct a phylogenetic analysis in MEGA X, with 500 bootstrap replicates [50].
4.5. Expression Profiling of LsARF Genes in Different Tissues or under Various Stresses
The lettuce RNA-Seq data were downloaded from the Gene Expression Omnibus (GEO) in the NCBI (GSE105821, GSE143675) or Genome Sequence Archive in National Genomics Data Center (PRJCA007442) databases, and the corresponding FPKM (fragments per kilobase per million reads) values for LsARFs were obtained for five organs representing major organs including root, stem, leaf, flower, and seed. As described, abiotic-stress-treated samples include lettuce plants exposed to UV (2h and 4h) or cadmium stress (3 days and 5 days). Similarly, FPKM values for abiotic-stress-treated lettuce plants were calibrated by calculating the fold change of expression levels between treatments and the corresponding controls. The normalized expression data were used to generate heatmaps by using the TBtools software package [48].
4.6. RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted with an EasyPure Plant RNA Kit (Transgene, Beijing, China). RNA quantity and quality were assessed using a NanoDrop8000 (Thermo Scientific, Waltham, MA, USA). Total RNA isolation and reverse transcription with oligo (dT)18 were performed as described previously [51,52]. The amounts of individual genes were quantified with gene-specific primers by real-time PCR analysis with a LightCycler 480 System (Roche, Rotkreuz, Switzerland) and SYBR Green mixture (Toyobo, Shanghai, Japan). The relative expression of specific genes was quantitated with the 2−∆∆Ct calculation method, where ΔΔCt is the difference in the threshold cycles and the reference housekeeping gene [53]. Lettuce eIF4A was used as the reference gene for normalizations, and the sequences of specific primers are shown in Table S1.
4.7. Subcellular Location of LsARF8a
The cDNA fragment containing the coding region of LsARF8a was fused to the 5′-end of the GFP coding region, and the fused gene was subcloned into pRI101 under the controls of the 35S promoter through homologous recombination. The resulting plasmid pRI101-LsARF3 and the pRI101 vector were transformed into Agrobacterium tumefaciens strain GV3101 and infiltrated into tobacco (Nicotiana benthaminana) leaves. The plants were incubated in the dark at 24 °C for one day and then transferred to the greenhouse with a 16 h light/8 h dark photoperiod for an additional day. Nucleus DNA was counterstained with DAPI, and cells were visualized with excitation at 405 nm (DAPI) and 488 nm (GFP) under a confocal microscope.
4.8. In Vivo Transcriptional Activity Assay
The full-length coding region, 5′-fragment (ATG-1155), and 3′-fragments (1156–2397) of LsARF8a were fused to the GFP under the control of the 35S promoter, and the resulting plasmids pGBKT7-LsARF8a-N and pGBKT7-LsARF8a-C were transformed into yeast strain AH109, as described previously [54]. The pGBKT7 vector was used as a control. These yeast transformants were screened on the synthetic dextrose (SD) medium without histidine, leucine, and uracil. β-Galactosidase activities of these yeast transformants were examined according to the guidance in the Yeast Protocols Handbook (Clontech, Mountain View, CA, USA).
4.9. Virus-Induced Gene Silencing (VIGS)
pTRV2-LsARF8a was generated by cloning a PCR-fragment-amplified lettuce leaf cDNA template using specific oligonucleotide primers incorporating Sal I and Xba I restriction sites, respectively, at the 5′- and 3′-ends for cloning into virus vector pTRV2. The Agrobacterium tumefaciens strain GV3101 harboring the recombinant plasmids pTRV2-LsARF8a and pTRV1 were used in vitro agroinoculation by leaf-injecting of one-month-old lettuce plants. The Agrobacterium lines carrying the pTRV2 empty vector or pTRV2-PDS were used as negative and positive controls, respectively. Primers used for vector constructions are listed in Table S1.
5. Conclusions
In this study, 24 LsARF genes were identified from the lettuce genome. The phylogenetic relationship, chromosomal distribution, exon–intron structure, consensus motifs, domain compositions, and promoter analysis provide valuable insights into the evolutionary aspects of the LsARF gene family. Expression analysis suggested that LsARF is dynamically involved in diverse biological processes, including organ development, phytohormone, and responses to abiotic stimuli. Subcellular location and transcriptional activity analysis suggested that LsARF8a might encode an ARF transcriptional factor. VIGS analysis indicated that LsARF8a might be a positive regulator of accelerated bolting under thermal conditions. In addition, G-box and ABRE cis-elements were identified in the promoter regions of LsARF8a and other LsARFs, implying that PhyB and ABRE might control the expression of LsARF genes, respectively. On the basis of the previous findings in Arabidopsis and our results, we propose a novel molecular mechanism explaining the thermally accelerated bolting: Warm temperatures stimulate the conversion to inactive Pr, which releases the depression of PIF4 and other PIFs; the transcript accumulations of PIFs as well as heat-inducible ABREs/ABFs facilitate the activation of LsARF8a, resulting in the activation of floral activator genes including LsFT, LsLFY, and LsSOC1 and thus promoting stem elongation in lettuce. These findings expand our knowledge of the LsARF gene family, and importantly, identified the positive role of LsARF8a in the thermally induced bolting in lettuce, which will facilitate the genetic improvement of heat tolerance in horticultural crops.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232113509/s1.
Author Contributions
N.L. and J.H. conceived and designed the study; M.H., Z.Q. and Z.R. conducted the in vitro experiments; M.H., B.W. and J.T. were involved in bioinformatics analysis including transcriptome sequencing, assembly, and annotations; N.L. and J.H. analyzed the data and supervised the work; Z.W. was responsible for project administration; N.L. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This project was financially supported by the “Young Talent Supporting Program (YCXTD00002-10, N.L.)”, “Special Financial Funds (CZZJ202209, Z.W.)”, and “Youth funding (QNJJ202248, J.T.)” of the Beijing Agricultural and Forestry Sciences, and the “Science Innovation Program (KYCX202001-09 and KYCX202001-03, N.L. and J.T.)” of National Research Engineering Center for Vegetables.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Wei Liu (Beijing Academy of Agricultural and Forestry Sciences) for fruitful discussions on this manuscript.
Conflicts of Interest
The authors declare that they have no competing interest.
References
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Greenham, K.; Zhang, Y.; Santner, A.; Castillejo, C.; Mutka, A.M.; O’Malley, R.C.; Ecker, J.R.; Kunkel, B.N.; Estelle, M. The Arabidopsis auxin receptor F-box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 2016, 6, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Cancé, C.; Martin-Arevalillo, R.; Boubekeur, K.; Dumas, R. Auxin response factors are keys to the many auxin doors. New Phytol. 2022, 235, 402–419. [Google Scholar] [CrossRef]
- Cucinotta, M.; Cavalleri, A.; Chandler, J.W.; Colombo, L. Auxin and Flower Development: A Blossoming Field. Cold Spring Harb. Perspect. Biol. 2021, 13, a039974. [Google Scholar] [CrossRef]
- Cheng, Z.J.; Wang, L.; Sun, W.; Zhang, Y.; Zhou, C.; Su, Y.H.; Li, W.; Sun, T.T.; Zhao, X.Y.; Li, X.G.; et al. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 2013, 161, 240–251. [Google Scholar] [CrossRef]
- Fahlgren, N.; Montgomery, T.A.; Howell, M.D.; Allen, E.; Dvorak, S.K.; Alexander, A.L.; Carrington, J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006, 16, 939–944. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, R.; Zi, H.; Li, Y.; Cao, X.; Li, D.; Guo, L.; Tong, J.; Pan, Y.; Jiao, Y.; et al. AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 2018, 30, 324–346. [Google Scholar] [CrossRef]
- Reeves, P.H.; Ellis, C.M.; Ploense, S.E.; Wu, M.F.; Yadav, V.; Tholl, D.; Chételat, A.; Haupt, I.; Kennerley, B.J.; Hodgens, C.; et al. A regulatory network for coordinated flower maturation. PLoS Genet. 2012, 8, e1002506. [Google Scholar] [CrossRef]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Wu, S.; Van Houten, J.; Wang, Y.; Ding, B.; Fei, Z.; Clarke, T.H.; Reed, J.W.; van der Knaap, E. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 2014, 65, 2507–2520. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, K.; Guo, L.; Liu, X.; Zhang, Z. AUXIN RESPONSE FACTOR3 plays distinct role during early flower development. Plant Signal. Behav. 2018, 13, e1467690. [Google Scholar] [CrossRef] [PubMed]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef]
- Verma, S.; Negi, N.P.; Pareek, S.; Mudgal, G.; Kumar, D. Auxin response factors in plant adaptation to drought and salinity stress. Physiol. Plant. 2022, 174, e13714. [Google Scholar] [CrossRef]
- Wang, Y.; Li, B.; Li, Y.; Du, W.; Zhang, Y.; Han, Y.; Liu, C.; Fan, S.; Hao, J. Application of exogenous auxin and gibberellin regulates the bolting of lettuce (Lactuca sativa L.). Open Life Sci. 2022, 17, 438–446. [Google Scholar] [CrossRef]
- Hao, J.-H.; Zhang, L.-L.; Li, P.-P.; Sun, Y.-C.; Li, J.-K.; Qin, X.-X.; Wang, L.; Qi, Z.-Y.; Xiao, S.; Han, Y.-Y.; et al. Quantitative proteomics analysis of Lettuce (Lactuca sativa L.) reveals molecular basis-associated auxin and photosynthesis with bolting induced by high temperature. Int. J. Mol. Sci. 2018, 19, 2967. [Google Scholar] [CrossRef]
- Lindqvist, K. On the origin of cultivated lettuce. Hereditas 1960, 46, 319–350. [Google Scholar] [CrossRef]
- Thompson, H.C.; Knott, J.K. The effect of temperature and photoperiod on the growth of lettuce. Proc. Am. Soc. Hort. Sci. 1933, 30, 507–509. [Google Scholar]
- Fukuda, M.; Matsuo, S.; Kikuchi, K.; Mitsuhashi, W.; Toyomasu, T.; Honda, I. The endogenous level of GA(1) is upregulated by high temperature during stem elongation in lettuce through LsGA3ox1 expression. J. Plant Physiol. 2009, 166, 2077–2084. [Google Scholar] [CrossRef]
- FAO. FAOSTAT: Crops. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 27 July 2022).
- Reyes-Chin-Wo, S.; Wang, Z.; Yang, X.; Kozik, A.; Arikit, S.; Song, C.; Xia, L.; Froenicke, L.; Lavelle, D.O.; Truco, M.J.; et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017, 8, 14953. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; van Treuren, R.; Liu, X.; Zhang, Z.; Chen, J.; Liu, Y.; Dong, S.; Sun, P.; Yang, T.; Lan, T.; et al. Whole-genome resequencing of 445 Lactuca accessions reveals the domestication history of cultivated lettuce. Nat. Genet. 2021, 53, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Finet, C.; Berne-Dedieu, A.; Scutt, C.P.; Marletaz, F. Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol. Biol. Evol. 2013, 30, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Matsuo, S.; Kikuchi, K.; Kawazu, Y.; Fujiyama, R.; Honda, I. Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. J. Plant Physiol. 2011, 168, 1602–1607. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, W.; Ge, D.; Han, Y.; Ning, K.; Luo, C.; Wang, S.; Liu, R.; Zhang, X.; Wang, Q. LCM-seq reveals the crucial role of LsSOC1 in heat-promoted bolting of lettuce (Lactuca sativa L.). Plant J. 2018, 95, 516–528. [Google Scholar] [CrossRef]
- Chen, Z.; Han, Y.; Ning, K.; Ding, Y.; Zhao, W.; Yan, S.; Luo, C.; Jiang, X.; Ge, D.; Liu, R.; et al. Inflorescence Development and the role of LsFT in regulating bolting in lettuce (Lactuca sativa L.). Front. Plant Sci. 2017, 8, 2248. [Google Scholar] [CrossRef]
- Sato, Y.; Nishimura, A.; Ito, M.; Ashikari, M.; Hirano, H.Y.; Matsuoka, M. Auxin response factor family in rice. Genes Genet. Syst. 2001, 76, 373–380. [Google Scholar] [CrossRef][Green Version]
- Ha, C.V.; Le, D.T.; Nishiyama, R.; Watanabe, Y.; Sulieman, S.; Tran, U.T.; Mochida, K.; Dong, N.V.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; et al. The auxin response factor transcription factor family in soybean: Genome-wide identification and expression analyses during development and water stress. DNA Res. 2013, 20, 511–524. [Google Scholar]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef]
- Chapman, M.A.; Leebens-Mack, J.H.; Burke, J.M. Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol. Biol. Evol. 2008, 25, 1260–1273. [Google Scholar] [CrossRef]
- Quint, M.; Delker, C.; Franklin, K.A.; Wigge, P.A.; Halliday, K.J.; van Zanten, M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2016, 2, 15190. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J.; Balasubramanian, S. Thermomorphogenesis. Annu. Rev. Plant Biol. 2019, 70, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Wang, W.; Huq, E. Spatial regulation of thermomorphogenesis by HY5 and PIF4 in Arabidopsis. Nat. Commun. 2021, 12, 3656. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, J.; Feng, Y.; Li, Z.; Ren, Z.; Liu, N.; Liu, C.; Hao, J.; Han, Y. LsARF3 mediates thermally induced bolting through promoting the expression of LsCO in lettuce (Lactuca sativa L.). Front. Plant Sci. 2022, 13, 958833. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef]
- Legris, M.; Klose, C.; Burgie, E.S.; Rojas, C.C.; Neme, M.; Hiltbrunner, A.; Wigge, P.A.; Schäfer, E.; Vierstra, R.D.; Casal, J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 2016, 354, 897–900. [Google Scholar] [CrossRef]
- Qi, Z.-Y. Expression of ARFs (Auxin Response Factors) Gene Family and Functional Analysis of LsARF8 in Leaf Lettuce (Lactuca sativa L.) during Bolting Induced by High Temperature. Master’s Thesis, Beijing University of Agriculture, Beijing, China, 2019. [Google Scholar]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alós, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, G.; Kim, S.; Thi, T.N.; Kim, H.; Jeong, J.; Kim, J.; Kim, J.; Choi, G.; Oh, E. The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat. Commun. 2020, 11, 1053. [Google Scholar] [CrossRef]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef]
- Ezer, D.; Shepherd, S.J.K.; Brestovitsky, A.; Dickinson, P.; Cortijo, S.; Charoensawan, V.; Box, M.S.; Biswas, S.; Jaeger, K.E.; Wigge, P.A. The G-box transcriptional regulatory code in Arabidopsis. Plant Physiol. 2017, 175, 628–640. [Google Scholar] [CrossRef]
- Kim, J.-B.; Kang, J.-Y.; Kim, S.Y. Over-expression of a transcription factor regulating ABA-responsive gene expression confers multiple stress tolerance. Plant Biotech. J. 2004, 2, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Han, Y.; Chen, Z.; Luo, C.; Wang, S.; Zhang, W.; Li, L.; Zhang, X.; Fan, S.; Wang, Q. Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 2019, 42, 1868–1881. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Liu, N.; Staswick, P.E.; Avramova, Z. Memory responses of jasmonic acid-associated Arabidopsis genes to a repeated dehydration stress. Plant Cell Environ. 2016, 39, 2515–2529. [Google Scholar] [CrossRef]
- Song, S.; Hao, L.; Zhao, P.; Xu, Y.; Zhong, N.; Zhang, H.; Liu, N. Genome-wide identification, expression profiling and evolutionary analysis of auxin response factor gene family in potato (Solanum tuberosum Group Phureja). Sci. Rep. 2019, 9, 1755. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta Ct) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhong, N.Q.; Wang, G.L.; Li, L.J.; Liu, X.L.; He, Y.K.; Xia, G.X. Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens. Planta 2007, 226, 827–838. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).