Abstract
Lipid droplets (LDs) are highly conserved and dynamic intracellular organelles. Their functions are not limited to serving as neutral lipid reservoirs; they also participate in non-energy storage functions, such as cell lipid metabolism, protection from cell stresses, maintaining protein homeostasis, and regulating nuclear function. During a Zika virus (ZIKV) infection, the viruses hijack the LDs to provide energy and lipid sources for viral replication. The co-localization of ZIKV capsid (C) protein with LDs supports its role as a virus replication platform and a key compartment for promoting the generation of progeny virus particles. However, in view of the multiple functions of LDs, their role in ZIKV infection needs further elucidation. Here, we review the basic mechanism of LD biogenesis and biological functions and discuss how ZIKV infection utilizes these effects of LDs to facilitate virus replication, along with the future application strategy of developing new antiviral drugs based on the interaction of ZIKV with LDs.
1. Introduction
Zika virus (ZIKV) is a mosquito (Aedes)-borne human pathogen that spreads via human bodily fluids. The viral infection causes fever, rash, headache, joint and muscle pain, conjunctivitis, severe eye lesions, newborn microcephaly, and Guillain–Barré syndrome [,,,,,]. The virus has been transmitted and has spread rapidly in the Americas, especially in Brazil []; the World Health Organization (WHO) declared it a global public health concern for public health on 1 February 2016. The viral infection is also likely to spread to many uninfected countries and regions due to the widespread distribution of the Aedes that transmits it, the frequent international trade activities, and rising tourism []. In addition, no vaccines or specific antiviral therapy have been approved to prevent or treat its infection. Nevertheless, researchers have been actively developing effective prevention and control strategies in response to the severe threat of ZIKV infection, which is a vital research goal.
ZIKV belongs to the genus Flavivirus and the family Flaviviridae [], which also includes viruses such as the dengue virus (DENV) and West Nile virus (WNV). Its genome is a single-stranded positive RNA with a length of about 11 kb, which encodes a single open reading frame and translates into a multi-protein precursor. This polyprotein is usually processed by the virus and host protease into three structural proteins, including the capsid ©, the pre-membrane (prM), and the envelope (E). The polyprotein is also processed into seven non-structural (NS) proteins, namely, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5, which participates in viral replication, pathogenesis, and host antiviral reactions (Figure 1) [,]. Lipidomics and transcriptomics analysis on the different ZIKV-infected mosquito cells, small glial cells, primary retinal pigmented epithelial (RPE) cells, or serum from patients indicate that the viral infection significantly regulates lipid metabolism, causing changes in a large number of lipids in cells. The changes in lipids include those to glycerophospholipids, such as phosphatidylcholine (PC) and phosphatidylserine, sphingolipids, such as ceramide and sphingomyelin, plasmalogens, and lysophospholipids, such as lysophosphatidylcholine [,,,]. These lipids may participate in the critical processes of ZIKV infection, such as genome replication and virion biogenesis. The envelope of ZIKV has been reported from the membranes of the host cells, while the association of the virus with the remodeling of lipid metabolism demonstrates that its successful infection depends on the host lipid metabolism.
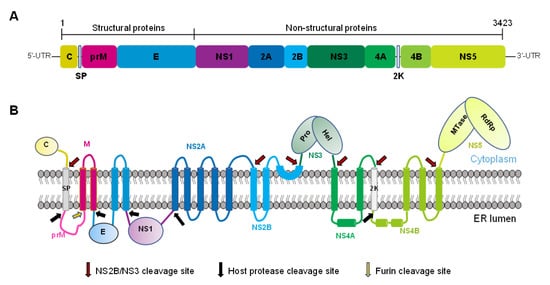
Figure 1.
Schematic diagram of ZIKV polyprotein organization and processing. (A) Linear organization of the ZIKV polyprotein precursor, including the three structural and seven nonstructural proteins. (B) Topology of the polyprotein in the ER membrane and its sites of splicing by the host or viral proteases.
Most of ZIKV’s structural and NS proteins possess transmembrane domains, allowing them to have specific locations on cellular organelles [,,,]. Other ZIKV proteins without transmembrane sequences, such as NS1, are also associated with the cell membrane [], indicating that viral proliferation during the infection is closely related to the membranes of plasma and other cellular organelles to which the viral proteins bind. For example, the endoplasmic reticulum (ER) plays an essential role in ZIKV infection by undergoing substantial rearrangement after viral infection in cells such as C6/36 in mosquitoes, Vero, human hepatic cells (Huh7), and human neural progenitor cells (hNPCs) in mammals. The ER remodeling is related to virus infection because viral replication sites, assembly, and budding occur in the vicinity of the ER region [,,]. The NS3 protease also needs the activation of the co-factor NS2B that anchors onto the ER membrane []. The NS2A protein is located in the peroxisome; its transient expression changes the organelle’s morphology and distribution, verifying the role of peroxisomes in ZIKV infection []. The analysis of the serum from patients infected with ZIKV showed that the levels of several types of phosphatidylethanolamine (PE) increased, and the synthesis of plasmalogens depended on functional peroxisome, which further supports the role of the lipids during ZIKV infection []. In addition, lipids are stored in cells as lipid droplets (LDs), in which the majority of the ZIKV C protein is found. This association is eliminated by substituting specific amino acids in the C-protein []. Although the consequences of this association have not yet been deciphered, it may be important for virion biogenesis, as described for hepatitis C virus (HCV) and DENV infections [,].
2. Basics of Lipid Droplets
Cellular LDs are highly conservative and dynamic intracellular organelles that are common in animals, plants, fungi, and bacteria [,] and fulfill a variety of functions, including storing neutral lipids in cells.
2.1. LDs Composition
The mature LDs mainly consist of a hydrophobic core composed of neutral lipids and a monolayer surface composed of phospholipids (Figure 2A). The hydrophobic core of LDs contains more than 100 different types of neutral lipids [,,,], which, in adipose and liver cells, are mainly composed of triacylglycerol (TAG) and cholesterol or sterol ester (SE). The phospholipid monolayer wrapping the hydrophobic core also has a unique composition of fatty acids (FAs) []. For LDs in mammalian cells, PC is the main component, accounting for approximately 60% of the phospholipid monolayer membrane. The changes in the ratios of phospholipids in the membrane affect the synthesis, maturity, and degradation of LDs [,].
The surface of LDs has several integration and peripheral proteins, which are divided into class I and class II proteins. Class II proteins are mainly from the PAT family, which is composed of five proteins in humans, namely, perilipin (also known as perilipin 1 or PLIN1), adipose-differentiation related protein (ADRP, also known as PLIN2), the 47 kDa tail interaction protein (TIP47, also named PLIN3), PLIN4. and PLIN5 [,,,]. These LD surface-bound proteins stabilize LDs and regulate the access of lipase and other enzymes to the neutral lipids in LDs. They also accumulate and control the size and interaction of LDs with other cellular organelles and the accessibility to lipolysis. In addition to PLINs, other LD proteins include enzymes participating in TAG and phospholipid synthesis or lipid transport.
LDs are mostly spherical and range from 50 to 200 µm in size. The specific LD size depends mainly on the state and type of the cells [,,]. The underlining mechanism of LD regulation mainly involves cellular TAG contents or LDs fusion pathways. The latter determines the size of the LDs in mammary epithelial cells (MECs), as shown through live-cell imaging using a confocal fluorescence microscope. The degree of LDs fusion depends on the composition of phospholipids in the cell membrane and is enhanced by increased PE content, which also increases the size of LDs []. However, whether the mechanism of LDs, as regulated by the fusion pathway, has broad adaptability in different cell types requires further investigation on a more experimental basis.
2.2. Lipid Droplet Biogenesis
Several studies have been conducted in the past few decades to elucidate LD biogenesis (Figure 2B) [,,,]. Traditionally, LDs were only present in the cytoplasm of eukaryotic cells and were known as cytoplasm LDs (cLDs). It was believed that LDs originated from the ER, where the enzymes for the generation of neutral lipids are located [,]. The generation of these neutral lipids in the ER and their behavior in the membrane drive LD biogenesis. The LD formation begins with the accumulation of neutral lipids, such as TAG and SE molecules, which are synthesized by diacylglycerol acyltransferase 1 (DGAT1) in the ER between two leaflets of its membrane [,]. Because the two layers of ER phospholipids can only accommodate a small number of neutral lipids, once the concentration of neutral lipids in the double layers exceeds the critical point, the formation of LDs will be triggered. In fact, when the nascent LDs were formed in yeast, a lens of about 50 nm was observed in the ER []. As the nascent LDs grow, they will emerge (or bud) from the ER and separate [], indicating that LDs seem to be formed spontaneously from the ER. This spontaneous bud mechanism does not require energy-consuming machinery, curvature induction reagents, or the intrinsic asymmetry of double layers []. Enzymes are necessary to synthesize neutral lipids in the process of LDs’ spontaneous budding, while proteins transform mature LDs, which process involves structural changes and the adjustment of LD biogenesis [,]. These proteins include PLIN3, glycerol-3-phosphate acyltransferase 4 (GPAT4), DGAT1, DGAT2, seipin, and fat storage-inducing transmembrane protein 2 (FIT2) [,,]. Seipins are located at the ER–LD contact site, which directly affects the occurrence of LD biogenesis or is involved in regulating lipid metabolism [,,,,]. The function of FIT is not completely clear, but the overexpression of FIT2 increases the size and number of LDs. The depletion of FIT2 causes the nascent LDs not to bud from ER but to still be embedded in the ER membrane, indicating that FIT regulates the budding of LDs from the ER. FITs may not directly mediate the budding but may affect the lipid homeostasis at the sites of LD biogenesis [,]. Molecular dynamic studies have shown that the protein recruited onto the ER membrane will be expelled precisely from the sites of LD formation, due to changes in the underlying membrane properties []. Lipids such as diacylglycerol (DAG) and phosphatidic acid also contribute to the formation of LDs by promoting shape changes in the same direction as the membrane curvature formation [,,,]. In the case of viral infection, the occurring process of LD biogenesis is observed and reflected by comparing their dynamics in virus-infected or uninfected cells. The data also demonstrate that LDs may be formed by peripheral TAG accumulation on the ER membrane and that HCV NS5A is responsible for interacting with those sites on the ER membrane that may form LDs [].
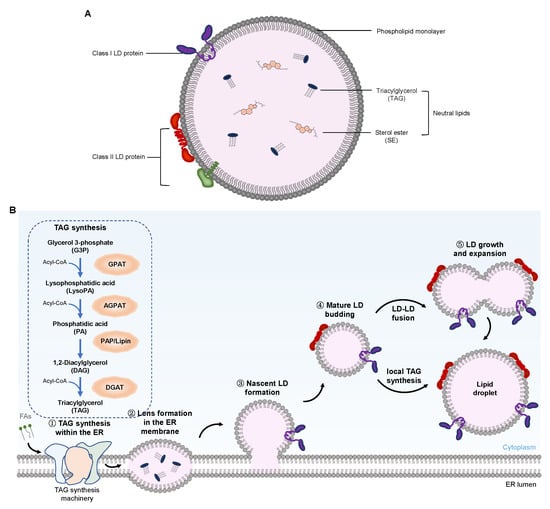
Figure 2.
Schematic representation of the LD structure and a model of the key steps in LD biogenesis. (A) LD is composed of a core of neutral lipids, such as TAGs and SEs, surrounded by a phospholipid monolayer. LD surface-bound proteins are classified into two groups. Class I LD proteins, such as GPAT2, mostly contain a hydrophobic hairpin that is associated with the lipid monolayer; Class II LD proteins, such as perilipins, are inserted into the LD from the cytosol via amphipathic helices or other short hydrophobic domains. (B) Five essential steps are involved in LD formation, growth, and expansion. Abbreviations: LD, lipid droplet; TAG, triacylglycerol; SE, sterol esters; FAs, fatty acids; Acyl-CoA, acyl-coenzyme A; ER, endoplasmic reticulum; GPAT, glycerol-3-phosphate acyltransferase; AGPAT, acyl-glycerol-3-phosphate acyltransferase; PAP, phosphatidate phosphatase; DGAT, diacylglycerol acyltransferase.
Extensive research on LD biogenesis, carried out over several years, showed the important role ER plays in the growth of nascent and mature LDs. However, the hypothesis that LDs can spontaneously form from a symmetric elongated lens of ER membrane remains controversial [], with some key questions, such as the location of the sites formed by LDs in ER, not yet having been answered. Other evidence suggests that LD formation in yeast and mammalian cells might occur at a unique ER site [,] with unknown characteristics, or it may be a process that occurs in random positions. Although the protein participating in TAG synthesis could be synthesized or enriched at the local discrete zones where LDs may be generated [], most of the enzymes participating in neutral lipids synthesis are distributed throughout the ER, indicating that certain lipids may not only be generated at the site of LD assembly. Alternatively, neutral lipid synthase may be activated at the sites of LD biogenesis, while the lipin homolog, known as phosphatidic acid phosphohydrolase 1 (Pah1p) in yeast, is a regulator of lipid synthesis and may be positioned at the location of LD biogenesis in the ER. Though seipin and FIT2 may play a role in the sites of LD biogenesis [,,], the way in which they regulate LD biogenesis in the ER remains to be clarified. When the nascent LDs transition to mature LDs, another question is whether they are separated from ER physically or are reconnected to the ER through the membrane bridge, whether they move from the cell periphery to the nucleus, and, if these bridges do exist, how can they form? In Saccharomyces cerevisiae, membrane bridge formation may not be required, given that the LD is still connected to the ER []. There also seems to be a unique mechanism in mammalian cells that uses the coat protein I (COPI) and adenosine diphosphate (ADP)–ribosylation factor 1 (ARF1) machinery to connect with the LDs separated from ER []. The answers to these questions will help us understand the machinery of LD biogenesis in the cell and elucidate the LDs’ interactions with other cellular organelles.
Recent evidence also shows the existence of nuclear LDs (nLDs), which originate from the inner nuclear membrane (INM) and are associated with INM via the predicted transmembrane protein, seipin [,]. Seipin forms a complex with LD assembly factor 1 (LDAF1) in the ER and plays a key role in cLD maturation []. The formation of nLDs is also related to promyelocytic leukemia (PML) nuclear bodies, which are dot structures in the nuclear matrix that regulates transcription and apoptosis in response to cell stress, including antiviral defense [,]. Since they were only found recently, their role has not yet been fully described, but, as with cLDs, nLDs may act as a lipid supplier for membrane expansion and as a bracket platform for proteins [,].
Besides nLDs, the secretory autophagosome released from DENV-infected cells also contains LDs. These extracellular LDs (eLDs) may involve the mechanism of cLD extracellular trafficking, which has not yet been thoroughly studied, but may be related to viral pathogenesis and could play an important role in the extracellular environment related to virus infection []. Whether the eLDs promote the early innate defense of DENV or other flaviviruses or are hijacked to spread the virus still needs further study.
Given the recent discovery of more types of LDs, their biogenesis should also depend on their positioning. In particular, the presence of nLDs indicates that LDs may also exist in the mitochondria derived from the endosymbionts. Therefore, it is necessary to determine whether they exist in other intracellular organelles or extracellular vesicles in eukaryotic cells. Theoretically, this speculation is reasonable because conservative LDs form a stable connection with ER and are also related to mitochondria, the inner nuclear envelope, lysosomes, vesicles, and endosomes [,].
2.3. Lipid Droplet Function and Its Regulation Mechanism
The LD is a highly complex organelle that stores energy and participates in the lipid metabolism of cells by acting as a hub for FA transport to the mitochondria. The LD also regulates nuclear functions through the availability of proteins and signaling lipids in the nucleus and protects cells from different forms of cellular stress, such as ER stress, and the lipotoxic effects of unesterified lipids []. In addition, LDs contribute to the maturity, degradation, storage, and turnover of many different proteins and maintain protein homeostasis [,,]. Here, the main features of LD function are briefly described in the following sections.
2.3.1. Participating in Cell Energy Storage and Lipid Metabolism
As an essential cellular organelle that regulates lipids and energy metabolism in cells, LDs are involved in many of the processes of cells, including lipid metabolism, membrane biogenesis, membrane transportation, and signal transduction. During nutritional stress, the lipids released from LDs are used either for hormone synthesis or as the primary repository of sterols, FAs, cholesterol, and the membrane phospholipid precursors used for cell membrane formation. They are used to maintain the homeostasis of ER and other membranes [,,,]. The lipids in the hydrophobic core of LDs are mostly TAG and SE, which play an important role in cell energy storage [].
In cell lipid metabolism, a key metabolic pivot involves the regulation of phospholipid acid (PA) in the ER membrane, which is used to synthesize membrane phospholipids or LD TAGs. In yeast, PA is converted into DAG, a precursor of TAG, by PAH1. When nutrition is lacking, the PAH1 pathway is activated through the Nem1-Spo7 complex, which transfers the fatty acyl chain from PA to membrane biogenesis and keeps it stored in the LDs [,]. This may be an important part of cell survival reaction and the lipid flux-balancing mechanism that causes the LDs to allow cellular regulation into membranes for growth or storage, and for starvation/stress survival.
Cellular lipid metabolism is a complex and highly regulated process. The neutral lipids stored in LDs can be catabolized by cytoplasm lipolysis or lysosome-mediated lipophagy, which is the selective autophagy of LDs (Figure 3). Lipolysis is mediated by LD-related lipases, such as adipose triglyceride lipase (ATGL), while lipophagy involves the key enzymes that hydrolyze TAG and cholesterol in the lysosome, namely, lysosomal acid lipase (LAL), which acts on the LDs delivered to autolysosomes via autophagy.
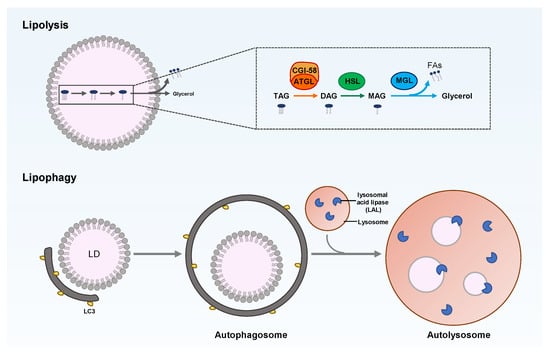
Figure 3.
Overview of LD degradation through lipolysis or lipophagy. TAG breakdown in LD is catalyzed by cytoplasmic or lysosomal lipase. Lipolysis is composed of three sequential catalyzations of TAG, DAG, and MAG by adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL), and monoacylglycerol lipase (MGL), respectively. Lipophagy belongs to selective autophagy; that is, the LDs are engulfed into the autophagosome and are then fused with lysosomes to form autolysosomes, wherein the neutral lipids in LDs are hydrolyzed by lysosomal acid lipase (LAL).
Various proteins and lipases on the surfaces of LDs regulate LD catabolism. For example, perilipin is responsible for lipid homeostasis by controlling the availability of lipases to neutral lipids in LDs. At the basic level, perilipin combines with the comparative gene identification-58 (CGI-58) in an inactive state. Under starvation conditions, perilipin is phosphorylated by protein kinase A, releasing the combined CGI-58, which then combines with ATGL to drive the hydrolysis of TAG []. The phosphorylated perilipin can also add phosphate to the hormone-sensitive lipase (HSL) and transfer it to LDs. The HSL hydrolyzes DAG into monoacylglycerol (MAG) [], which is hydrolyzed into free FAs and glycerol by monoacylglycerol lipase (MGL) []. The free FAs released by neutral lipid breakdown undergo β-oxidation to generate energy via mitochondria or the peroxisome []. Therefore, LDs play a key role in maintaining cellular homeostasis by balancing lipogenesis and lipolysis. This homeostatic effect is essential for normal cellular and organismal functions, which are closely related to human health. LD disorders can also cause many human diseases, such as fatty liver disease, obesity, diabetes, cardiovascular disease, and cancer [,].
2.3.2. Preventing ER Stress
ER stress is caused by the activation of the unfolded protein response (UPR) by cells in response to conditions such as the aggregation of misfolded and unfolded proteins in the lumen of ER, the dysregulation of calcium homeostasis, and/or lipid composition imbalance, and the reaction process to re-establish ER homeostasis []. For example, three ER-integrated membrane proteins, also known as UPR sensors, are often activated in response to the accumulation of unfolded proteins. These proteins are the inositol-requiring protein 1 (IRE1), protein kinase RNA (PKR)-like ER kinase (PERK, also known as EIF2AK3), and the activated transcription factor 6 (ATF6). The activation of these three main pathways will trigger the cell signaling cascade, slowing the translation of related proteins and increasing the gene expression participating in ER protein-folding, degradation, and lipid biogenesis.
The disruption of LD biogenesis and TAGs synthesis often leads to UPR activation in yeast and mammalian cells. Studies on the mutant yeast strain (LDΔ) that is deficient in the enzymes required for TAG and SE biosynthesis with the loss of LDs showed an altered ER morphology. The alteration, however, could be reversed by inhibiting de novo FA synthesis or deleting the Opi1 inhibitor to induce ER phospholipid synthesis, suggesting that phospholipid synthesis could compensate for the loss of LD formation. Without LDs, the ER phospholipid composition changes, displaying an increased level of phosphatidylinositol, leading to impaired autophagosome biogenesis []. Although LDs function primarily by providing a lipid source to the autophagosome membrane [,,], mutant yeast studies suggest that LDs are more likely to regulate autophagy through ER quality-control functions. Yeast cells with PC biosynthesis inhibition activate the UPR to form abnormally fragmented ER aggregates []. Excessive saturated FA triggers UPR more effectively than unsaturated FA [,]. However, UPR triggering does not require the unfolded protein-sensing domains of IRE1 and PERK and may involve sensing perturbed ER membrane composition []. In adipocytes, the lipolysis of stored lipids is stimulated by a noradrenergic signaling cascade, releasing large amounts of FAs, followed by the production of large numbers of small LDs, which are not fragmented or in fission with LDs, as initially thought [,]. Instead, these FAs are re-esterified and packaged into DGAT1-dependent LDs []. Under these lipolytic conditions, DGAT1-dependent fatty acid re-esterification and LD biogenesis are critical for protection against ER stress, lipotoxicity, and UPR activation. Furthermore, in 3T3-L1 preadipocytes, the lack of RAB18 resulted in a substantial reduction in the number of mature LDs and UPR activation, indicating increased ER stress after oleate treatment [].
The molecular mechanism between impaired LD biogenesis and UPR activation is unclear. Because the abundance of LDs is usually found to increase when UPR is activated, an investigation needs to determine whether LDs improve or exacerbate this stress response. Studies also show that increased TAG storage in LDs mitigates ER stress in mammalian cells and tissues. The TAG storage of the liver LDs increases in ATGL-knockout mice, and the ER stress induced by the tunicamycin (TM) is prevented by reduced stress markers, such as the glucose-regulating protein 78 (GRP78) and the slicing X-box binding protein 1 (XBP1) []. Similarly, in human cardiomyocyte-derived cells, the overexpression of ATGL promotes the release of free FAs from the TAG in LDs, thereby increasing the ER stress markers. The ER stress induced by palmitate is also inhibited by increased TAG, through the overexpression of peroxisome proliferator-activated receptor γ (PPARγ) or acyl-CoA synthetase (ACSL1) []. The overexpression of autophagy-related 14 (ATG14) in Hela cells stimulates lipophagy, leading to increased free FAs and ER stress []. The constitutive hypoxia-inducible factor 2α (HIF-2α) upregulates the expression of the LD coat protein PLIN2 in clear-cell renal cell carcinoma (ccRCC). The knockdown of PLIN2, however, eliminates LD, expands the ER, and activates PERK, IRE-1α, and other UPR targets. Nevertheless, the supplies of the exogenous PLIN2 are sufficient to restore LDs and inhibit cell death [].
The inhibition of PC biosynthesis in yeast cells activates UPR and forms LDs at the abnormal fragmented ER aggregates. The polyubiquitinated proteins and the ER chaperone heat-shock protein (Hsp104p) contained in LDs appear to be degraded via the endosomal sorting complex required for transport (ESCRT)-dependent microlipophagy in the yeast vacuole cells []. Data also indicate that LDs formed by an ER sequester unfolded, misfolded, or aggregated ER proteins and further promoted their removal through microlipophagy. However, whether these ubiquitinated proteins are located on the surface of LDs or on LD-related ER subdomains is unclear. The specific substrates involved in the assumed degradation pathways have not yet been determined. In mammalian cells, we observed a connection between LDs and selected ER-associated degradation (ERAD) substrates. Hydroxymethylglutaryl (HMG)-CoA reductase enzyme and apolipoprotein B (ApoB) undergo ERAD under specific metabolic conditions [,]. Interestingly, both proteins bind with LDs or are related to the LD-associated ER subdomains before degradation. In addition, changes in the lipid composition of the ER can also directly activate the UPR without requiring the luminal sensing domains of yeast and mammalian IRE1s and PERKs. Consistent with this finding, the activation of PERK and IRE1 by saturated FAs in recombinant liposomes requires that their transmembrane domains should be independent of the luminal misfolded protein-sensing domains [,]. These studies suggest that abnormal FAs storage in LDs can either activate UPR directly by changing the lipid composition of the ER membrane or indirectly, by changing ER homeostasis, leading to impaired ER protein-folding or calcium storage. The LDs provide a vehicle for rebalancing ER lipid homeostasis and removing misfolded ER proteins. This LDs-mediated function for ER quality control is likely to play a role, together with the ERAD pathway [,,].
2.3.3. Participating in Autophagy
Intracellular LDs are degraded into free FAs and glycerol in the form of autophagy, called lipophagy. It is manifested as the co-localization of autophagic LC3 and LD coat proteins, such as PLIN2. This kind of selective autophagy is another form of lipolysis and is necessary for the clearance of LDs and TAG in hepatocytes []. In addition to acting as an energy source, lipophagy controls the quality of LD proteins and lipid homeostasis in cells [,].
The occurrence of lipophagy mainly starts from the recognition of cargoes by the autophagosome membrane via interaction with the microtubule-associated protein 1 light chain 3 (MAP1LC3) []. This usually involves the assistance of one or more cargo adapters, such as P62, Optineurin, NBR1, and NDP52, which connect the organelle membrane to LC3. This has been confirmed by Tatsumi’s study [] on the role of lipophagy in embryonic development. The study showed that the forced lipophagy system, through the fusion of the LD binding domain and p62, significantly reduced the number of LDs and TAG levels during mouse embryonic development, ultimately retarding the development. Meanwhile, lipophagy may also require the polyubiquitination of organelle surface proteins as a recruitment signal [].
The LDs’ surface proteins involved in lipophagy and promoting LD recognition are not fully understood. Before autophagy or cytosolic lipase degrades LDs, the perilipin on the surface of LDs needs to be removed, which mainly occurs through chaperone-mediated autophagy, regulated by adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK) signaling [,]. Studies also show that huntingtin is necessary for lipophagy under stress conditions since it connects p62 with LC3-II, releasing Unc-51-like autophagy-activating kinase 1 (ULK1), which is a pro-autophagic kinase inhibited by mammalian target of rapamycin (mTOR) []. The ancient ubiquitous protein 1 (AUP1) is a type-III membrane protein and is expressed most commonly in LDs and the ER. It recruits the E2 ubiquitin-binding enzyme G2, accumulating LDs and controlling ER protein quality []. AUP1 is used by DENV and ZIKV to trigger lipophagy and may be a specific factor of lipophagy, but the mechanism of the initiation of lipophagy has not been fully elucidated. DENV deubiquitinates and transports AUP1 from LDs to autophagosomes by binding the viral NS4A and NS4B proteins in infected cells. DENV also stimulates the acyltransferase activity of AUP1 to upregulate lipophagy and promote the generation of progeny viruses. The knockdown of AUP1 using the clustered, regularly interspaced short palindromic repeats (CRISPR)-Cas9 genome editing technique eliminates the generation of the infectious DENV and ZIKV virion [].
Proteins of the Rab molecular switch family may also be involved in the process of lipophagy. Many Rab molecules have been identified on LDs [], some of which are related to autophagy regulation, especially Rab7, Rab10, and Rab25. They have been proven to be essential for lipophagy of hepatocytes under certain conditions. For example, Rab7 is activated in hepatocytes under nutrient deficiency; however, during lipophagy, it promotes the recruitment of multivesicular bodies and lysosomes to the LD surface. The deletion of Rab7 causes morphological changes in multivesicular bodies, lysosomes, and autophagosomes, and leads to reduced lipophagy []. In addition, chronic alcohol exposure inhibits the activation of Rab7, leading to the damage of lysosomes that degrade LDs and ultimately block the lipophagy of hepatocytes [,]. Rab10 forms a complex with the adaptor protein EH domain-binding protein 1 (EHBP1), while the membrane-modified adenosine triphosphatase EH-Domain Containing 2 (EHD2) to promote the migration of LC3-positive autophagic membrane to the LD surface. However, the loss of Rab10 function leads to LD accumulation []. The disappearance of LDs often accompanies the activation of hepatic stellate cells (HSCs). The activation of HSC stimulates the expression of Rab25 and promotes the formation of complexes with PI3KCIII, thereby guiding autophagy to recognize, encapsulate, and degrade LDs. The knockdown of Rab25 expression with specific siRNA, however, prevents lipophagy and inhibits the disappearance of LDs [].
Lipase recognizes LDs and directly promotes the formation of autophagosomes by inducing the recruitment of TAG and SEs, which help to initiate lipophagy [,]. The cytoplasmic ATGL (also known as PNPLA2) regulates lipolysis and lipophagy, but the way in which it coordinates the regulation of both processes is unclear. ATGL is a necessary and sufficient positive regulator for inducing lipophagy in mouse liver. It interacts with LC3 via its LC3-interacting region (LIR) motif to promote its movement to LDs, upon which it causes lipophagy []. ATGL also promotes lipophagy by enhancing sirtuin 1 (SIRT1) activity, which regulates the catabolism of liver LDs []. The patatin-like phospholipase-domain-containing enzyme 5 (PNPLA5) that is present in LDs contributes to lipophagy and autophagic proteolysis []. PNPLA8 mediates the lipophagy driven by sterol-regulatory element binding protein 2 (SREBP-2) through its dynamic interaction with LC3 in the hepatocytes of high-fat diet (HFD)-fed mice and regulates lipid homeostasis in patients with nonalcoholic fatty liver disease (NAFLD) []. Through mutation studies, PNPLA3 was confirmed as a key player in the autophagosome formation stage of the lipophagy process in starved human hepatocyte HepG2 []. In lysosomes, LDs can also be degraded by acidic lipases. Under nutrient deficiency, the expression of lysosomal lipases in Caenorhabditis elegans and mouse hepatocytes is regulated by the lysosomal biogenesis transcription factor EB (TFEB) []. TFE3 regulates the steatosis of hepatocytes by inducing lipophagy []. Under fasting conditions, the forkhead homeobox transcription factor (FoxO1) triggers the lipophagy of adipocytes by inducing lysosomal acid lipase [].
Similar to general autophagy, lipophagy is also regulated by cellular nutritional state-sensing signals and response systems, to ensure that the level of free FAs in cells matches their energy requirements. These sensory systems include AMPK, mTOR, the nuclear receptor farnesoid X receptor (FXR), PPARα, and transcription activator cAMP response element-binding protein (CREB) [,,,]. If the cells are in a nutrient-rich state and do not need free FAs as an energy source, lipophagy will be inhibited. However, in the case of insufficient nutrition, lipophagy is triggered, leading to the decomposition of TAG in LDs. The SIRT3 protein activates lipophagy by stimulating the AMPK-ULK1 pathway, inducing smaller-sized LDs, and reducing lipid accumulation in mature adipocytes []. For example, DENV induces AMPK kinase activity as well as AMPK-independent mTORC1 suppression to cause the lipophagy that promotes viral infection []. In addition, treatment with AMPK and an autophagy activator (AICAR and rapamycin) or mTOR inhibitor (Torin-1) significantly alleviates the symptoms in patients with NAFLD [,,], indicating that restoring the function of lipophagy may be an important treatment for improving fatty liver disease.
3. Roles of LDs in the ZIKV Life Cycle
3.1. ZIKV Infection Causes Changes in LDs
To confirm that LDs participate in ZIKV infection, LDs are stained with specific dyes, such as BODIPY, Oil Red O, LipidTox Deep Red, and Nile Red, and then observed under a microscope to detect their intracellular changes after viral infection. To ensure that the effects of LD measurement occur in a single cycle of ZIKV replication, a low multiplicity of infection (MOI), i.e., infection with ZIKV at an MOI of 0.1 and a limited virus replication cycle (i.e., 24 h) are used for LDs analysis. Confocal microscopy showed that compared with uninfected cells, the number, content, and size of LDs in ZIKV-infected cells were significantly reduced []. This result is consistent with the previous findings that flavivirus infection induces pro-viral selective autophagy against LDs [,].
However, some studies have also reported increased LDs during the early stages of ZIKV infection. For example, two hours after ZIKV infection, both in vivo and in vitro, the induced LDs were transiently upregulated and controlled by the epidermal growth factor receptor (EGFR). The inhibition of EGFR suppresses the expression of LDs during infection, while the production of types I and III IFN, which reflects the ability to mount an effective immune response in infected cells, is significantly reduced. This ultimately leads to increased ZIKV replication, indicating that LDs can also serve as an important cellular organelle in the innate antiviral immune response [,]. In the model of infected astrocytes, cells were infected with ZIKV or stimulated with poly I:C, a dsRNA virus mimic. The LD displacement and average velocity in cells were significantly enhanced 2 h after infection or stimulation, indicating that the dynamics of LDs had changed in the early stage of pathogen infection, further supporting the emerging role of LDs in the innate host response []. Another study showed that hydroxysteroid (17β) dehydrogenase (HSD17B)12 promotes the replication of ZIKV and the production of infectious particles by increasing LD biosynthesis, which plays a key role in virus assembly. This study confirms the indispensability of LDs in ZIKV infection [].
No explanation exists for the differences seen in LD changes following ZIKV infection. However, the differences may be due to the different cell systems or ratios of the virus to cells adapted to infect the monolayer cells [,]. Other reasons may be the research purposes used to determine the various analysis and measurement targets under corresponding experimental conditions. For example, some authors may pay more attention to the changes in a single LD or the number or total area of LDs, while others may only focus on the number of LDs. Given that the size of LDs may vary, the observed phenotype of LD changes may correspond to the overall activation of different cellular processes. The differences come from the various cell lines and the time of infection used. In various cell systems, viruses may adopt different LD hijacking paradigms. For example, ZIKV hijacks LDs by manipulating the SREBP pathway, which is the main regulator of LD biogenesis, or via secreted autophagosomes containing LDs [,]. There is also the possibility that the virus adopts different LD utilization modes at the various stages of infection. Therefore, in the early stages of viral infection, the virus may induce LD biogenesis to stimulate the initial replication and may then trigger lipophagy to reduce the number of LDs, to release free FAs from these lipid structures.
3.2. Interaction of LDs with ZIKV Proteins
Similar to other flaviviruses, the ZIKV-C protein is also one of the viral structural proteins assembled with the viral genome to form the nucleocapsid. In addition to the classical structural function that accommodates and protects the virus genome, the C-protein also has multi-functional characteristics. It plays an important role in viral replication and infection by interacting with cellular proteins and regulating cell metabolism, apoptosis, and immune response.
Previous studies have shown that the ZIKV-C protein is localized in the LDs of the cells in infected HEK293, BHK21, and Vero [,,], representing the primary location of the C-protein in host cells. This is also consistent with DENV and HCV-C protein localization in the LDs [,]. The interaction between the C-protein and host LDs is important for viral genome encapsulation, replication, and assembly (Figure 4). In addition, the accumulation of C-protein around the LD structure is the key prerequisite for the efficient production of viral particles [,].
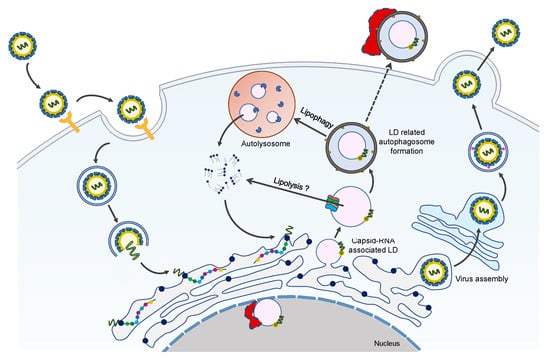
Figure 4.
Overview of LDs-C protein interaction during the ZIKV life cycle. ZIKV infection promotes LDs formation at the early stages of infection. With the increasing interaction of LDs with ZIKV-C protein, lipophagy is induced to generate sufficient free fatty acids for robust viral replication and/or assembly. During this process, ZIKV NS4A/4B proteins may also contribute to the occurrence of lipophagy. We hypothesize that the eLDs and nLDs are involved in the ZIKV life cycle, and their formation and function possibly need the help of some unknown host factors, which are shown as red irregular polygons with a border of dashed lines.
Shang et al. [] resolved the crystal structure of the ZIKV C protein with a resolution of 1.9 Å. The structure contains four α helices with a unique long pre-α1 loop, contributing to C-C dimer formation. Compared with the known forms of WNV and DENV-C proteins, the ZIKV-C protein has different hydrophobic characteristics at the lipid bilayer interface. The interaction between the ZIKV-C protein and LDs was confirmed by confocal microscopy analysis. The substitution mutation of key amino acids (F27S/K31S/R32S) in the pre-α1 loop of ZIKV-C protein disrupted the interaction with LDs, indicating that this loop is critical for membrane association [].
The dynamic N-terminal domain of the C-protein is related to its functional diversity. A specific peptide (pep14–23) in the DENV-C protein mediates the binding of the C-protein to LDs through conformational transition []. The characteristics of the structural dynamics of the N-terminal domain of the ZIKV-C protein were analyzed by dividing the domain into three truncated fragments. The circular dichroism, dynamic light scattering, Zeta potential, and molecular dynamic simulation confirmed that the truncated fragments, 5–26 and 1–30, are prone to adopt an α-helical conformation, but the conserved fragment, 14–23, is unstructured. This fragment does not undergo conformational conversion, which is different from the DENV-C protein, suggesting that this conserved region in the ZIKV-C protein may not be involved in the association with LDs []. The molecular mechanism of the interaction between ZIKV-C protein and LDs needs further study.
In addition to the interaction of ZIKV-C protein with LDs, ZIKV NS4A and NS4B proteins are also associated with the selective autophagy of LDs. DENV NS4A and NS4B proteins induce lipophagy to improve the production of infectious particles by interacting with AUP1 to hijack its acyltransferase function. This mechanism seems also to play a role in ZIKV infection and is likely to be a universal mechanism adopted by flavivirus infection [].
4. Conclusions
Lipid droplets serve as a platform for ZIKV replication and a lipid reservoir for cells and provide energy and lipid sources for virus replication. By comparing the size, location, accumulation, and dynamic changes of LDs through real-time visualization techniques in uninfected and infected cells, we reveal the important details of how ZIKV hijacks cellular LDs for its successful replication and understand in what stages of the virus life cycle the LDs may play a role. However, the molecular mechanism of LD involvement in ZIKV infection still needs further elucidation. How ZIKV establishes contact, interacts, and communicates with cellular LDs needs to be answered. Identifying and characterizing viral factors (such as structural and NS proteins) and host factors, especially the specific LD-related factors involved in the interaction between ZIKV and LDs, is also important. Identifying and characterizing these interaction factors can provide basic information for developing effective therapeutic drugs that inhibit ZIKV replication. In recent years, with the improvement of the isolation and purification of LDs from cultured cells, the morphology, lipidomics, proteomics, and even “ZIKV-LDs interaction omics” after viral infection will be characterized in depth. The data will provide important information for further understanding the role of LDs in the pathogenesis of flaviviruses.
Since there are no effective antiviral drugs for ZIKV, developing antiviral drugs is still challenging. In this sense, positioning lipid metabolism regulators as antiviral drugs is a promising strategy for the future. Existing studies also support the possibility of developing antiviral treatment strategies based on LDs, a basic and conserved cellular organelle. For example, the use of SREBP pathway inhibitors (such as PF-429242) to control LD abundance and change LD size and proteome has been proven to be an attractive option for controlling flavivirus infection [,,]. However, this lipid metabolism antiviral drug has a long way to go before it can be applied in clinical practice.
Therefore, a comprehensive understanding of the important role of LDs in the life cycle of ZIKV helps to understand the pathogenic mechanism of viruses and may make them possible targets for developing new antiviral therapies.
Author Contributions
Conceptualization, Z.-L.Q.; writing—original draft preparation, Z.-L.Q. and Q.-F.Y.; writing—review and editing, Z.-L.Q., Q.-F.Y., H.R., P.Z. and Z.-T.Q.; visualization, Z.-L.Q. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Petersen, E.; Wilson, M.E.; Touch, S.; McCloskey, B.; Mwaba, P.; Bates, M.; Dar, O.; Mattes, F.; Kidd, M.; Ippolito, G.; et al. Rapid Spread of Zika Virus in The Americas—Implications for Public Health Preparedness for Mass Gatherings at the 2016 Brazil Olympic Games. Int. J. Infect. Dis. 2016, 44, 11–15. [Google Scholar] [CrossRef]
- Araujo, A.Q.; Silva, M.T.; Araujo, A.P. Zika virus-associated neurological disorders: A review. Brain 2016, 139, 2122–2130. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Gubler, D.J.; Schaub, B.; Lanteri, M.C.; Musso, D. An update on Zika virus infection. Lancet 2017, 390, 2099–2109. [Google Scholar] [CrossRef]
- Watrin, L.; Ghawche, F.; Larre, P.; Neau, J.P.; Mathis, S.; Fournier, E. Guillain-Barre Syndrome (42 Cases) Occurring During a Zika Virus Outbreak in French Polynesia. Medicine 2016, 95, e3257. [Google Scholar] [CrossRef]
- Malkki, H. CNS infections: Zika virus infection could trigger Guillain-Barre syndrome. Nat. Rev. Neurol 2016, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Ferraris, P.; Yssel, H.; Misse, D. Zika virus infection: An update. Microbes Infect. 2019, 21, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Bogoch, I.I.; Brady, O.J.; Kraemer, M.U.G.; German, M.; Creatore, M.I.; Kulkarni, M.A.; Brownstein, J.S.; Mekaru, S.R.; Hay, S.I.; Groot, E.; et al. Anticipating the international spread of Zika virus from Brazil. Lancet 2016, 387, 335–336. [Google Scholar] [CrossRef]
- Lessler, J.; Chaisson, L.H.; Kucirka, L.M.; Bi, Q.; Grantz, K.; Salje, H.; Carcelen, A.C.; Ott, C.T.; Sheffield, J.S.; Ferguson, N.M.; et al. Assessing the global threat from Zika virus. Science 2016, 353, aaf8160. [Google Scholar] [CrossRef]
- Barrows, N.J.; Campos, R.K.; Liao, K.C.; Prasanth, K.R.; Soto-Acosta, R.; Yeh, S.C.; Schott-Lerner, G.; Pompon, J.; Sessions, O.M.; Bradrick, S.S.; et al. Biochemistry and Molecular Biology of Flaviviruses. Chem. Rev. 2018, 118, 4448–4482. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.C.; Vazquez, C.A.; Giovannoni, F.; Russo, C.A.; Cordo, S.M.; Alaimo, A.; Damonte, E.B. Cellular Organelles Reorganization During Zika Virus Infection of Human Cells. Front. Microbiol. 2020, 11, 1558. [Google Scholar] [CrossRef]
- Melo, C.F.; de Oliveira, D.N.; Lima, E.O.; Guerreiro, T.M.; Esteves, C.Z.; Beck, R.M.; Padilla, M.A.; Milanez, G.P.; Arns, C.W.; Proenca-Modena, J.L.; et al. A Lipidomics Approach in the Characterization of Zika-Infected Mosquito Cells: Potential Targets for Breaking the Transmission Cycle. PLoS ONE 2016, 11, e0164377. [Google Scholar] [CrossRef] [PubMed]
- Diop, F.; Vial, T.; Ferraris, P.; Wichit, S.; Bengue, M.; Hamel, R.; Talignani, L.; Liegeois, F.; Pompon, J.; Yssel, H.; et al. Zika virus infection modulates the metabolomic profile of microglial cells. PLoS ONE 2018, 13, e0206093. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, A.; Pinto, I.F.D.; Lima, M.; Giovanetti, M.; de Jesus, J.G.; Xavier, J.; Barreto, F.K.; Canuto, G.A.B.; do Amaral, H.R.; de Filippis, A.M.B.; et al. Lipidomic Analysis Reveals Serum Alteration of Plasmalogens in Patients Infected With ZIKA Virus. Front. Microbiol. 2019, 10, 753. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Khatri, I.; Jha, A.; Pretto, C.D.; Spindler, K.R.; Arumugaswami, V.; Giri, S.; Kumar, A.; Bhasin, M.K. Determination of system level alterations in host transcriptome due to Zika virus (ZIKV) Infection in retinal pigment epithelium. Sci. Rep. 2018, 8, 11209. [Google Scholar] [CrossRef]
- Hou, W.; Cruz-Cosme, R.; Armstrong, N.; Obwolo, L.A.; Wen, F.; Hu, W.; Luo, M.H.; Tang, Q. Molecular cloning and characterization of the genes encoding the proteins of Zika virus. Gene 2017, 628, 117–128. [Google Scholar] [CrossRef]
- Coyaud, E.; Ranadheera, C.; Cheng, D.; Goncalves, J.; Dyakov, B.J.A.; Laurent, E.M.N.; St-Germain, J.; Pelletier, L.; Gingras, A.C.; Brumell, J.H.; et al. Global Interactomics Uncovers Extensive Organellar Targeting by Zika Virus. Mol. Cell. Proteom. 2018, 17, 2242–2255. [Google Scholar] [CrossRef]
- Brown, W.C.; Akey, D.L.; Konwerski, J.R.; Tarrasch, J.T.; Skiniotis, G.; Kuhn, R.J.; Smith, J.L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 2016, 23, 865–867. [Google Scholar] [CrossRef]
- Barreto-Vieira, D.F.; Jacome, F.C.; da Silva, M.A.N.; Caldas, G.C.; de Filippis, A.M.B.; de Sequeira, P.C.; de Souza, E.M.; Andrade, A.A.; Manso, P.P.A.; Trindade, G.F.; et al. Structural investigation of C6/36 and Vero cell cultures infected with a Brazilian Zika virus. PLoS ONE 2017, 12, e0184397. [Google Scholar] [CrossRef]
- Cortese, M.; Goellner, S.; Acosta, E.G.; Neufeldt, C.J.; Oleksiuk, O.; Lampe, M.; Haselmann, U.; Funaya, C.; Schieber, N.; Ronchi, P.; et al. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017, 18, 2113–2123. [Google Scholar] [CrossRef]
- Rossignol, E.D.; Peters, K.N.; Connor, J.H.; Bullitt, E. Zika virus induced cellular remodelling. Cell Microbiol. 2017, 19, e12740. [Google Scholar] [CrossRef]
- Majerova, T.; Novotny, P.; Krysova, E.; Konvalinka, J. Exploiting the unique features of Zika and Dengue proteases for inhibitor design. Biochimie 2019, 166, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Song, H.; Shi, Y.; Qi, J.; Gao, G.F. Crystal Structure of the Capsid Protein from Zika Virus. J. Mol. Biol. 2018, 430, 948–962. [Google Scholar] [CrossRef] [PubMed]
- Samsa, M.M.; Mondotte, J.A.; Iglesias, N.G.; Assuncao-Miranda, I.; Barbosa-Lima, G.; Da Poian, A.T.; Bozza, P.T.; Gamarnik, A.V. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009, 5, e1000632. [Google Scholar] [CrossRef] [PubMed]
- Ambroggio, E.E.; Costa Navarro, G.S.; Perez Socas, L.B.; Bagatolli, L.A.; Gamarnik, A.V. Dengue and Zika virus capsid proteins bind to membranes and self-assemble into liquid droplets with nucleic acids. J. Biol. Chem. 2021, 297, 101059. [Google Scholar] [CrossRef]
- Hashemi, H.F.; Goodman, J.M. The life cycle of lipid droplets. Curr. Opin. Cell. Biol. 2015, 33, 119–124. [Google Scholar] [CrossRef]
- Walther, T.C.; Farese, R.V., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012, 81, 687–714. [Google Scholar] [CrossRef]
- Grillitsch, K.; Connerth, M.; Kofeler, H.; Arrey, T.N.; Rietschel, B.; Wagner, B.; Karas, M.; Daum, G. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: Lipidome meets proteome. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2011, 1811, 1165–1176. [Google Scholar] [CrossRef]
- Chitraju, C.; Trotzmuller, M.; Hartler, J.; Wolinski, H.; Thallinger, G.G.; Lass, A.; Zechner, R.; Zimmermann, R.; Kofeler, H.C.; Spener, F. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J. Lipid. Res. 2012, 53, 2141–2152. [Google Scholar] [CrossRef]
- Schmidt, C.; Ploier, B.; Koch, B.; Daum, G. Analysis of yeast lipid droplet proteome and lipidome. Methods Cell Biol. 2013, 116, 15–37. [Google Scholar]
- Vrablik, T.L.; Petyuk, V.A.; Larson, E.M.; Smith, R.D.; Watts, J.L. Lipidomic and proteomic analysis of Caenorhabditis elegans lipid droplets and identification of ACS-4 as a lipid droplet-associated protein. Biochim. Biophys. Acta 2015, 1851, 1337–1345. [Google Scholar] [CrossRef]
- Tauchi-Sato, K.; Ozeki, S.; Houjou, T.; Taguchi, R.; Fujimoto, T. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J. Biol. Chem. 2002, 277, 44507–44512. [Google Scholar] [CrossRef] [PubMed]
- Wilfling, F.; Haas, J.T.; Walther, T.C.; Farese, R.V., Jr. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 2014, 29, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Czaja, M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013, 20, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Chen, X.; Moktan, H.; Arrese, E.L.; Duan, L.; Wang, L.; Soulages, J.L.; Zhou, D.H. Membrane attachment and structure models of lipid storage droplet protein 1. Biochim. Biophys. Acta 2014, 1838, 874–881. [Google Scholar] [CrossRef]
- Bersuker, K.; Olzmann, J.A. Establishing the lipid droplet proteome: Mechanisms of lipid droplet protein targeting and degradation. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 1166–1177. [Google Scholar] [CrossRef]
- Krahmer, N.; Guo, Y.; Farese, R.V., Jr.; Walther, T.C. SnapShot: Lipid Droplets. Cell 2009, 139, 1024. [Google Scholar] [CrossRef]
- Sztalryd, C.; Brasaemle, D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar]
- Kimmel, A.R.; Sztalryd, C. The Perilipins: Major Cytosolic Lipid Droplet-Associated Proteins and Their Roles in Cellular Lipid Storage, Mobilization, and Systemic Homeostasis. Annu. Rev. Nutr. 2016, 36, 471–509. [Google Scholar] [CrossRef]
- Reue, K. A thematic review series: Lipid droplet storage and metabolism: From yeast to man. J. Lipid Res. 2011, 52, 1865–1868. [Google Scholar] [CrossRef]
- Cohen, B.C.; Raz, C.; Shamay, A.; Argov-Argaman, N. Lipid Droplet Fusion in Mammary Epithelial Cells is Regulated by Phosphatidylethanolamine Metabolism. J. Mamm. Gland. Biol. Neoplasia 2017, 22, 235–249. [Google Scholar] [CrossRef]
- Pol, A.; Gross, S.P.; Parton, R.G. Review: Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 2014, 204, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, V.; Golani, G.; Joshi, A.S.; Cottier, S.; Schneiter, R.; Prinz, W.A.; Kozlov, M.M. Architecture of Lipid Droplets in Endoplasmic Reticulum Is Determined by Phospholipid Intrinsic Curvature. Curr. Biol. 2018, 28, 915–926. [Google Scholar] [CrossRef]
- Jackson, C.L. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 2019, 59, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Deslandes, F.; Thiam, A.R.; Foret, L. Lipid Droplets Can Spontaneously Bud Off from a Symmetric Bilayer. Biophys. J. 2017, 113, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Buhman, K.K.; Chen, H.C.; Farese, R.V., Jr. The enzymes of neutral lipid synthesis. J. Biol. Chem. 2001, 276, 40369–40372. [Google Scholar] [CrossRef]
- Joshi, A.S.; Zhang, H.; Prinz, W.A. Organelle biogenesis in the endoplasmic reticulum. Nat. Cell Biol. 2017, 19, 876–882. [Google Scholar] [CrossRef]
- Thiam, A.R.; Farese, R.V., Jr.; Walther, T.C. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 2013, 14, 775–786. [Google Scholar] [CrossRef]
- Vanni, S. Intracellular Lipid Droplets: From Structure to Function. Lipid Insights 2017, 10, 1178635317745518. [Google Scholar] [CrossRef]
- Szymanski, K.M.; Binns, D.; Bartz, R.; Grishin, N.V.; Li, W.P.; Agarwal, A.K.; Garg, A.; Anderson, R.G.; Goodman, J.M. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA 2007, 104, 20890–20895. [Google Scholar] [CrossRef]
- Grippa, A.; Buxo, L.; Mora, G.; Funaya, C.; Idrissi, F.Z.; Mancuso, F.; Gomez, R.; Muntanya, J.; Sabido, E.; Carvalho, P. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 2015, 211, 829–844. [Google Scholar] [CrossRef]
- Wang, H.; Becuwe, M.; Housden, B.E.; Chitraju, C.; Porras, A.J.; Graham, M.M.; Liu, X.N.; Thiam, A.R.; Savage, D.B.; Agarwal, A.K.; et al. Seipin is required for converting nascent to mature lipid droplets. Elife 2016, 5, e16582. [Google Scholar] [CrossRef] [PubMed]
- Wolinski, H.; Hofbauer, H.F.; Hellauer, K.; Cristobal-Sarramian, A.; Kolb, D.; Radulovic, M.; Knittelfelder, O.L.; Rechberger, G.N.; Kohlwein, S.D. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta 2015, 1851, 1450–1464. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Wang, W.; Liu, Z.; Huang, X.; Jiang, Q.; Liu, G.; Wang, Y.; Huang, X. Seipin promotes adipose tissue fat storage through the ER Ca2+-ATPase SERCA. Cell Metab. 2014, 19, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, V.; Ojha, N.; Golden, A.; Prinz, W.A. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 2015, 211, 261–271. [Google Scholar] [CrossRef]
- Gross, D.A.; Zhan, C.; Silver, D.L. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl. Acad. Sci. USA 2011, 108, 19581–19586. [Google Scholar] [CrossRef]
- Skinner, J.R.; Shew, T.M.; Schwartz, D.M.; Tzekov, A.; Lepus, C.M.; Abumrad, N.A.; Wolins, N.E. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 2009, 284, 30941–30948. [Google Scholar] [CrossRef]
- Martins, A.S.; Martins, I.C.; Santos, N.C. Methods for Lipid Droplet Biophysical Characterization in Flaviviridae Infections. Front. Microbiol. 2018, 9, 1951. [Google Scholar] [CrossRef]
- Adeyo, O.; Horn, P.J.; Lee, S.; Binns, D.D.; Chandrahas, A.; Chapman, K.D.; Goodman, J.M. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 2011, 192, 1043–1055. [Google Scholar] [CrossRef]
- Nevo-Yassaf, I.; Lovelle, M.; Nahmias, Y.; Hirschberg, K.; Sklan, E.H. Live cell imaging and analysis of lipid droplets biogenesis in hepatatis C virus infected cells. Methods 2017, 127, 30–36. [Google Scholar] [CrossRef]
- Kassan, A.; Herms, A.; Fernandez-Vidal, A.; Bosch, M.; Schieber, N.L.; Reddy, B.J.; Fajardo, A.; Gelabert-Baldrich, M.; Tebar, F.; Enrich, C.; et al. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 2013, 203, 985–1001. [Google Scholar] [CrossRef]
- Jacquier, N.; Mishra, S.; Choudhary, V.; Schneiter, R. Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. J. Cell Sci. 2013, 126, 5198–5209. [Google Scholar] [PubMed]
- Shockey, J.M.; Gidda, S.K.; Chapital, D.C.; Kuan, J.C.; Dhanoa, P.K.; Bland, J.M.; Rothstein, S.J.; Mullen, R.T.; Dyer, J.M. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant. Cell 2006, 18, 2294–2313. [Google Scholar] [CrossRef]
- Jacquier, N.; Choudhary, V.; Mari, M.; Toulmay, A.; Reggiori, F.; Schneiter, R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 2011, 124, 2424–2437. [Google Scholar] [CrossRef]
- Wilfling, F.; Thiam, A.R.; Olarte, M.J.; Wang, J.; Beck, R.; Gould, T.J.; Allgeyer, E.S.; Pincet, F.; Bewersdorf, J.; Farese, R.V., Jr.; et al. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife 2014, 3, e01607. [Google Scholar] [CrossRef]
- Romanauska, A.; Kohler, A. The Inner Nuclear Membrane Is a Metabolically Active Territory that Generates Nuclear Lipid Droplets. Cell 2018, 174, 700–715. [Google Scholar] [CrossRef] [PubMed]
- Hariri, H.; Rogers, S.; Ugrankar, R.; Liu, Y.L.; Feathers, J.R.; Henne, W.M. Lipid droplet biogenesis is spatially coordinated at ER-vacuole contacts under nutritional stress. EMBO Rep. 2018, 19, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Wu, X.; Lambert, T.J.; Lai, Z.W.; Walther, T.C.; Farese, R.V., Jr. LDAF1 and Seipin Form a Lipid Droplet Assembly Complex. Dev. Cell 2019, 51, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Ohsaki, Y.; Kawai, T.; Yoshikawa, Y.; Cheng, J.; Jokitalo, E.; Fujimoto, T. PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 2016, 212, 29–38. [Google Scholar] [CrossRef]
- Everett, R.D.; Chelbi-Alix, M.K. PML and PML nuclear bodies: Implications in antiviral defence. Biochimie 2007, 89, 819–830. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef]
- Layerenza, J.P.; Gonzalez, P.; de Bravo, M.M.G.; Polo, M.P.; Sisti, M.S.; Ves-Losada, A. Nuclear lipid droplets: A novel nuclear domain. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2013, 1831, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.W.; Mettling, C.; Wu, S.R.; Yu, C.Y.; Perng, G.C.; Lin, Y.S.; Lin, Y.L. Autophagy-associated dengue vesicles promote viral transmission avoiding antibody neutralization. Sci. Rep. 2016, 6, 32243. [Google Scholar] [CrossRef] [PubMed]
- Welte, M.A. Expanding roles for lipid droplets. Curr. Biol. 2015, 25, R470–R481. [Google Scholar] [CrossRef]
- Schuldiner, M.; Bohnert, M. A different kind of love—lipid droplet contact sites. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 1188–1196. [Google Scholar] [CrossRef]
- Welte, M.A.; Gould, A.P. Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2017, 1862, 1260–1272. [Google Scholar] [CrossRef] [PubMed]
- Shpilka, T.; Welter, E.; Borovsky, N.; Amar, N.; Mari, M.; Reggiori, F.; Elazar, Z. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015, 34, 2117–2131. [Google Scholar] [CrossRef]
- Velazquez, A.P.; Graef, M. Autophagy regulation depends on ER homeostasis controlled by lipid droplets. Autophagy 2016, 12, 1409–1410. [Google Scholar] [CrossRef]
- Velazquez, A.P.; Tatsuta, T.; Ghillebert, R.; Drescher, I.; Graef, M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J. Cell Biol. 2016, 212, 621–631. [Google Scholar] [CrossRef]
- Barbosa, A.D.; Sembongi, H.; Su, W.M.; Abreu, S.; Reggiori, F.; Carman, G.M.; Siniossoglou, S. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol. Biol. Cell 2015, 26, 3641–3657. [Google Scholar] [CrossRef]
- Hansen, J.S.; de Mare, S.; Jones, H.A.; Goransson, O.; Lindkvist-Petersson, K. Visualization of lipid directed dynamics of perilipin 1 in human primary adipocytes. Sci. Rep. 2017, 7, 15011. [Google Scholar] [CrossRef]
- Ducharme, N.A.; Bickel, P.E. Lipid droplets in lipogenesis and lipolysis. Endocrinology 2008, 149, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Fredrikson, G.; Tornqvist, H.; Belfrage, P. Hormone-sensitive lipase and monoacylglycerol lipase are both required for complete degradation of adipocyte triacylglycerol. Biochim. Biophys. Acta 1986, 876, 288–293. [Google Scholar] [CrossRef]
- Eaton, S. Control of mitochondrial beta-oxidation flu.ux. Prog. Lipid Res. 2002, 41, 197–239. [Google Scholar] [CrossRef]
- Chen, Z.; Shrestha, R.; Yang, X.; Wu, X.; Jia, J.; Chiba, H.; Hui, S.P. Oxidative Stress and Lipid Dysregulation in Lipid Droplets: A Connection to Chronic Kidney Disease Revealed in Human Kidney Cells. Antioxidants 2022, 11, 1387. [Google Scholar] [CrossRef]
- Krahmer, N.; Farese, R.V., Jr.; Walther, T.C. Balancing the fat: Lipid droplets and human disease. EMBO Mol. Med. 2013, 5, 973–983. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef]
- Dupont, N.; Chauhan, S.; Arko-Mensah, J.; Castillo, E.F.; Masedunskas, A.; Weigert, R.; Robenek, H.; Proikas-Cezanne, T.; Deretic, V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr. Biol. 2014, 24, 609–620. [Google Scholar] [CrossRef]
- Vevea, J.D.; Garcia, E.J.; Chan, R.B.; Zhou, B.; Schultz, M.; Di Paolo, G.; McCaffery, J.M.; Pon, L.A. Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast. Dev. Cell 2015, 35, 584–599. [Google Scholar] [CrossRef]
- Volmer, R.; Ron, D. Lipid-dependent regulation of the unfolded protein response. Curr. Opin. Cell Biol. 2015, 33, 67–73. [Google Scholar] [CrossRef]
- Han, J.; Kaufman, R.J. The role of ER stress in lipid metabolism and lipotoxicity. J. Lipid Res. 2016, 57, 1329–1338. [Google Scholar] [CrossRef]
- Volmer, R.; van der Ploeg, K.; Ron, D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. USA 2013, 110, 4628–4633. [Google Scholar] [CrossRef] [PubMed]
- Brasaemle, D.L.; Dolios, G.; Shapiro, L.; Wang, R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 2004, 279, 46835–46842. [Google Scholar] [CrossRef] [PubMed]
- Marcinkiewicz, A.; Gauthier, D.; Garcia, A.; Brasaemle, D.L. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 2006, 281, 11901–11909. [Google Scholar] [CrossRef] [PubMed]
- Chitraju, C.; Mejhert, N.; Haas, J.T.; Diaz-Ramirez, L.G.; Grueter, C.A.; Imbriglio, J.E.; Pinto, S.; Koliwad, S.K.; Walther, T.C.; Farese, R.V., Jr. Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell Metab. 2017, 26, 407–418. [Google Scholar] [CrossRef]
- Xu, D.; Li, Y.; Wu, L.; Li, Y.; Zhao, D.; Yu, J.; Huang, T.; Ferguson, C.; Parton, R.G.; Yang, H.; et al. Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 2018, 217, 975–995. [Google Scholar] [CrossRef]
- Fuchs, C.D.; Claudel, T.; Kumari, P.; Haemmerle, G.; Pollheimer, M.J.; Stojakovic, T.; Scharnagl, H.; Halilbasic, E.; Gumhold, J.; Silbert, D.; et al. Absence of adipose triglyceride lipase protects from hepatic endoplasmic reticulum stress in mice. Hepatology 2012, 56, 270–280. [Google Scholar] [CrossRef]
- Bosma, M.; Dapito, D.H.; Drosatos-Tampakaki, Z.; Huiping-Son, N.; Huang, L.S.; Kersten, S.; Drosatos, K.; Goldberg, I.J. Sequestration of fatty acids in triglycerides prevents endoplasmic reticulum stress in an in vitro model of cardiomyocyte lipotoxicity. Biochim. Biophys. Acta 2014, 1841, 1648–1655. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Schlaepfer, I.R.; Bergman, B.C.; Panda, P.K.; Praharaj, P.P.; Naik, P.P.; Agarwal, R.; Bhutia, S.K. ATG14 facilitated lipophagy in cancer cells induce ER stress mediated mitoptosis through a ROS dependent pathway. Free Radic. Biol. Med. 2017, 104, 199–213. [Google Scholar] [CrossRef]
- Qiu, B.; Ackerman, D.; Sanchez, D.J.; Li, B.; Ochocki, J.D.; Grazioli, A.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Keith, B.; Simon, M.C. HIF2alpha-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2015, 5, 652–667. [Google Scholar] [CrossRef]
- Jo, Y.; Hartman, I.Z.; DeBose-Boyd, R.A. Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol. Biol. Cell 2013, 24, 169–183. [Google Scholar] [CrossRef]
- Suzuki, M.; Otsuka, T.; Ohsaki, Y.; Cheng, J.; Taniguchi, T.; Hashimoto, H.; Taniguchi, H.; Fujimoto, T. Derlin-1 and UBXD8 are engaged in dislocation and degradation of lipidated ApoB-100 at lipid droplets. Mol. Biol. Cell 2012, 23, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Promlek, T.; Ishiwata-Kimata, Y.; Shido, M.; Sakuramoto, M.; Kohno, K.; Kimata, Y. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell 2011, 22, 3520–3532. [Google Scholar] [CrossRef] [PubMed]
- Olzmann, J.A.; Kopito, R.R.; Christianson, J.C. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 2013, 5, a013185. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.C.; Ye, Y. Cleaning up in the endoplasmic reticulum: Ubiquitin in charge. Nat. Struct. Mol. Biol. 2014, 21, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Liu, Y.; Li, X.; Zhang, H.; Zhang, S.; Mak, H.Y.; Liu, P. Dietary S. maltophilia induces supersized lipid droplets by enhancing lipogenesis and ER-LD contacts in C. elegans. Gut Microbes 2022, 14, 2013762. [Google Scholar] [CrossRef]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef]
- Garcia, E.J.; Vevea, J.D.; Pon, L.A. Lipid droplet autophagy during energy mobilization, lipid homeostasis and protein quality control. Front. Biosci. 2018, 23, 1552–1563. [Google Scholar]
- Kounakis, K.; Chaniotakis, M.; Markaki, M.; Tavernarakis, N. Emerging Roles of Lipophagy in Health and Disease. Front. Cell Dev. Biol. 2019, 7, 185. [Google Scholar] [CrossRef]
- Maus, M.; Cuk, M.; Patel, B.; Lian, J.; Ouimet, M.; Kaufmann, U.; Yang, J.; Horvath, R.; Hornig-Do, H.T.; Chrzanowska-Lightowlers, Z.M.; et al. Store-Operated Ca(2+) Entry Controls Induction of Lipolysis and the Transcriptional Reprogramming to Lipid Metabolism. Cell Metab. 2017, 25, 698–712. [Google Scholar] [CrossRef]
- Tatsumi, T.; Takayama, K.; Ishii, S.; Yamamoto, A.; Hara, T.; Minami, N.; Miyasaka, N.; Kubota, T.; Matsuura, A.; Itakura, E.; et al. Forced lipophagy reveals that lipid droplets are required for early embryonic development in mouse. Development 2018, 145, dev161893. [Google Scholar] [CrossRef]
- Rogov, V.; Dotsch, V.; Johansen, T.; Kirkin, V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 2014, 53, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Cuervo, A.M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 2015, 17, 759–770. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy 2016, 12, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.N.; Xu, Z.; Patel, B.; Chen, Z.; Chen, D.; Tito, A.; David, G.; Sun, Y.; Stimming, E.F.; Bellen, H.J.; et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015, 17, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Spandl, J.; Lohmann, D.; Kuerschner, L.; Moessinger, C.; Thiele, C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J. Biol. Chem. 2011, 286, 5599–5606. [Google Scholar] [CrossRef]
- Zhang, J.; Lan, Y.; Li, M.Y.; Lamers, M.M.; Fusade-Boyer, M.; Klemm, E.; Thiele, C.; Ashour, J.; Sanyal, S. Flaviviruses Exploit the Lipid Droplet Protein AUP1 to Trigger Lipophagy and Drive Virus Production. Cell Host. Microbe. 2018, 23, 819–831. [Google Scholar] [CrossRef]
- Kiss, R.S.; Nilsson, T. Rab proteins implicated in lipid storage and mobilization. J. Biomed. Res. 2014, 28, 169–177. [Google Scholar]
- Schroeder, B.; Schulze, R.J.; Weller, S.G.; Sletten, A.C.; Casey, C.A.; McNiven, M.A. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 2015, 61, 1896–1907. [Google Scholar] [CrossRef]
- Schulze, R.J.; Rasineni, K.; Weller, S.G.; Schott, M.B.; Schroeder, B.; Casey, C.A.; McNiven, M.A. Ethanol exposure inhibits hepatocyte lipophagy by inactivating the small guanosine triphosphatase Rab7. Hepatol. Commun. 2017, 1, 140–152. [Google Scholar] [CrossRef]
- Shin, D.W. Lipophagy: Molecular Mechanisms and Implications in Metabolic Disorders. Mol. Cells 2020, 43, 686–693. [Google Scholar]
- Li, Z.; Schulze, R.J.; Weller, S.G.; Krueger, E.W.; Schott, M.B.; Zhang, X.; Casey, C.A.; Liu, J.; Stockli, J.; James, D.E.; et al. A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci Adv. 2016, 2, e1601470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhao, S.; Yao, Z.; Wang, L.; Shao, J.; Chen, A.; Zhang, F.; Zheng, S. Autophagy regulates turnover of lipid droplets via ROS-dependent Rab25 activation in hepatic stellate cell. Redox. Biol. 2017, 11, 322–334. [Google Scholar] [CrossRef]
- Ward, C.; Martinez-Lopez, N.; Otten, E.G.; Carroll, B.; Maetzel, D.; Singh, R.; Sarkar, S.; Korolchuk, V.I. Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim. Biophys. Acta 2016, 1861, 269–284. [Google Scholar] [CrossRef]
- Martinez-Lopez, N.; Garcia-Macia, M.; Sahu, S.; Athonvarangkul, D.; Liebling, E.; Merlo, P.; Cecconi, F.; Schwartz, G.J.; Singh, R. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab. 2016, 23, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Sathyanarayan, A.; Mashek, M.T.; Mashek, D.G. ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Rep. 2017, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Jang, H.J.; Yang, Y.R.; Park, K.I.; Seo, J.; Shin, I.W.; Jeon, T.I.; Ahn, S.C.; Suh, P.G.; Osborne, T.F.; et al. SREBP-2/PNPLA8 axis improves non-alcoholic fatty liver disease through activation of autophagy. Sci. Rep. 2016, 6, 35732. [Google Scholar] [CrossRef]
- Negoita, F.; Blomdahl, J.; Wasserstrom, S.; Winberg, M.E.; Osmark, P.; Larsson, S.; Stenkula, K.G.; Ekstedt, M.; Kechagias, S.; Holm, C.; et al. PNPLA3 variant M148 causes resistance to starvation-mediated lipid droplet autophagy in human hepatocytes. J. Cell Biochem. 2019, 120, 343–356. [Google Scholar] [CrossRef]
- Settembre, C.; De Cegli, R.; Mansueto, G.; Saha, P.K.; Vetrini, F.; Visvikis, O.; Huynh, T.; Carissimo, A.; Palmer, D.; Klisch, T.J.; et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013, 15, 647–658. [Google Scholar] [CrossRef]
- Xiong, J.; Wang, K.; He, J.; Zhang, G.; Zhang, D.; Chen, F. TFE3 Alleviates Hepatic Steatosis through Autophagy-Induced Lipophagy and PGC1alpha-Mediated Fatty Acid beta-Oxidation. Int. J. Mol. Sci. 2016, 17, 387. [Google Scholar] [CrossRef]
- Barbato, D.L.; Tatulli, G.; Aquilano, K.; Ciriolo, M.R. FoxO1 controls lysosomal acid lipase in adipocytes: Implication of lipophagy during nutrient restriction and metformin treatment. Cell Death Dis. 2013, 4, e861. [Google Scholar] [CrossRef]
- Li, Y.; Yang, P.; Zhao, L.; Chen, Y.; Zhang, X.; Zeng, S.; Wei, L.; Varghese, Z.; Moorhead, J.F.; Chen, Y.; et al. CD36 plays a negative role in the regulation of lipophagy in hepatocytes through an AMPK-dependent pathway. J. Lipid Res. 2019, 60, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yan, S.; Khambu, B.; Ma, F.; Li, Y.; Chen, X.; Martina, J.A.; Puertollano, R.; Li, Y.; Chalasani, N.; et al. Dynamic MTORC1-TFEB feedback signaling regulates hepatic autophagy, steatosis and liver injury in long-term nutrient oversupply. Autophagy 2018, 14, 1779–1795. [Google Scholar] [CrossRef]
- Seok, S.; Fu, T.; Choi, S.E.; Li, Y.; Zhu, R.; Kumar, S.; Sun, X.; Yoon, G.; Kang, Y.; Zhong, W.; et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 2014, 516, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, J.; Tong, Q.; Lin, L. SIRT3 Acts as a Positive Autophagy Regulator to Promote Lipid Mobilization in Adipocytes via Activating AMPK. Int. J. Mol. Sci. 2020, 21, 372. [Google Scholar] [CrossRef]
- Jordan, T.X.; Randall, G. Dengue Virus Activates the AMP Kinase-mTOR Axis To Stimulate a Proviral Lipophagy. J. Virol. 2017, 91, e02020-16. [Google Scholar] [CrossRef]
- Chao, X.; Wang, S.; Zhao, K.; Li, Y.; Williams, J.A.; Li, T.; Chavan, H.; Krishnamurthy, P.; He, X.C.; Li, L.; et al. Impaired TFEB-Mediated Lysosome Biogenesis and Autophagy Promote Chronic Ethanol-Induced Liver Injury and Steatosis in Mice. Gastroenterology 2018, 155, 865–879. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.K.; Marcinko, K.; Desjardins, E.M.; Lally, J.S.; Ford, R.J.; Steinberg, G.R. Treatment of nonalcoholic fatty liver disease: Role of AMPK. Am. J. Physiol. Endocrinol. Metab. 2016, 311, E730–E740. [Google Scholar] [CrossRef]
- Lu, W.; Mei, J.; Yang, J.; Wu, Z.; Liu, J.; Miao, P.; Chen, Y.; Wen, Z.; Zhao, Z.; Kong, H.; et al. ApoE deficiency promotes non-alcoholic fatty liver disease in mice via impeding AMPK/mTOR mediated autophagy. Life Sci. 2020, 252, 117601. [Google Scholar] [CrossRef]
- Heaton, N.S.; Randall, G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host. Microbe. 2010, 8, 422–432. [Google Scholar] [CrossRef]
- Martin-Acebes, M.A.; Vazquez-Calvo, A.; Saiz, J.C. Lipids and flaviviruses, present and future perspectives for the control of dengue, Zika, and West Nile viruses. Prog. Lipid Res. 2016, 64, 123–137. [Google Scholar] [CrossRef]
- Monson, E.A.; Helbig, K.J. Host upregulation of lipid droplets drives antiviral responses. Cell Stress. 2021, 5, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Monson, E.A.; Crosse, K.M.; Duan, M.; Chen, W.; O’Shea, R.D.; Wakim, L.M.; Carr, J.M.; Whelan, D.R.; Helbig, K.J. Intracellular lipid droplet accumulation occurs early following viral infection and is required for an efficient interferon response. Nat. Commun. 2021, 12, 4303. [Google Scholar] [CrossRef] [PubMed]
- Monson, E.A.; Whelan, D.R.; Helbig, K.J. Lipid Droplet Motility Increases Following Viral Immune Stimulation. Int. J. Mol. Sci. 2021, 22, 4418. [Google Scholar] [CrossRef]
- Mohamed, B.; Mazeaud, C.; Baril, M.; Poirier, D.; Sow, A.A.; Chatel-Chaix, L.; Titorenko, V.; Lamarre, D. Very-long-chain fatty acid metabolic capacity of 17-beta-hydroxysteroid dehydrogenase type 12 (HSD17B12) promotes replication of hepatitis C virus and related flaviviruses. Sci. Rep. 2020, 10, 4040. [Google Scholar] [CrossRef] [PubMed]
- Cloherty, A.P.M.; Olmstead, A.D.; Ribeiro, C.M.S.; Jean, F. Hijacking of Lipid Droplets by Hepatitis C, Dengue and Zika Viruses-From Viral Protein Moonlighting to Extracellular Release. Int. J. Mol. Sci. 2020, 21, 7901. [Google Scholar] [CrossRef]
- Shavinskaya, A.; Boulant, S.; Penin, F.; McLauchlan, J.; Bartenschlager, R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 2007, 282, 37158–37169. [Google Scholar] [CrossRef]
- Oliveira, E.R.A.; Mohana-Borges, R.; de Alencastro, R.B.; Horta, B.A.C. The flavivirus capsid protein: Structure, function and perspectives towards drug design. Virus Res. 2017, 227, 115–123. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Jia, R.; Wang, M.; Yin, Z.; Cheng, A. Structure and function of capsid protein in flavivirus infection and its applications in the development of vaccines and therapeutics. Vet. Res. 2021, 52, 98. [Google Scholar] [CrossRef]
- Martins, I.C.; Gomes-Neto, F.; Faustino, A.F.; Carvalho, F.A.; Carneiro, F.A.; Bozza, P.T.; Mohana-Borges, R.; Castanho, M.A.; Almeida, F.C.; Santos, N.C.; et al. The disordered N-terminal region of dengue virus capsid protein contains a lipid-droplet-binding motif. Biochem. J. 2012, 444, 405–415. [Google Scholar] [CrossRef]
- Saumya, K.U.; Kumar, D.; Kumar, P.; Giri, R. Unlike dengue virus, the conserved 14–23 residues in N-terminal region of Zika virus capsid is not involved in lipid interactions. Bioc.Chim. Biophys. Acta Biomembr. 2020, 1862, 183440. [Google Scholar] [CrossRef]
- Zhang, J.; Lan, Y.; Sanyal, S. Modulation of Lipid Droplet Metabolism-A Potential Target for Therapeutic Intervention in Flaviviridae Infections. Front. Microbiol. 2017, 8, 2286. [Google Scholar] [CrossRef] [PubMed]
- Hyrina, A.; Meng, F.; McArthur, S.J.; Eivemark, S.; Nabi, I.R.; Jean, F. Human Subtilisin Kexin Isozyme-1 (SKI-1)/Site-1 Protease (S1P) regulates cytoplasmic lipid droplet abundance: A potential target for indirect-acting anti-dengue virus agents. PLoS ONE 2017, 12, e0174483. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).