Abstract
Reactive oxygen species impair the blood vessels, leading to the initiation of atherosclerosis, and migration and proliferation of vascular smooth muscle cells and neovascularization by endothelial cells of vasa vasorum are essential for atherosclerosis development. Obg-like ATPase 1 (OLA1), a negative regulator in cellular responses to oxidative stress, binds to breast cancer susceptibility gene 1 (BRCA1), which protects vascular endothelial and smooth muscle cells against reactive oxygen species. However, it is not known whether OLA1 is genetically correlated with atherosclerosis. Here, we conducted two independent population-based case–control studies to explore the effects of variants in OLA1 genes on preclinical atherosclerosis. A total of 564 and 746 subjects who had thicker and normal carotid intima–media thickness (cIMT), respectively, were enrolled. Among 55 screened SNPs, rs35145102, rs201641962, rs12466587, rs4131583, and rs16862482 in OLA1 showed significant associations with cIMT. SNP rs35145102 is a 3′-utr variant and correlates with the differential expression of OLA1 in immune cells. These five genetic markers form a single closely linked block and H1-ATTGT and H2-GCCTC were the top two most prevalent 5-locus haplotypes. The H1 + H1 genotype negatively and H1 + H2 genotype positively correlated with thicker cIMT. The five identified SNPs in the OLA1 gene showed significant correlations with cIMT. Furthermore, we found that OLA1 was required for migration and proliferation of human aortic endothelial and smooth muscle cells and regulated vascular tube formation by human aortic endothelial cells. Therefore, these genetic variants in the OLA1 gene may serve as markers for risk prediction of atherosclerotic diseases.
1. Introduction
Atherosclerosis is a multi-stage and multifactorial vascular disease. Three types of cells—immune cells, vascular endothelial cells (ECs), and vascular smooth muscle cells (SMCs)—play critical roles in the initiation and progression of athersclerosis and the stability of vascular plaque [1,2,3,4,5,6]. Early stages of atherosclerosis are related to blood flow disturbances that damage the endothelium, followed by platelet adhesion, macrophage penetration into the sub-endothelium, and migration and proliferation of vascular SMCs [1,2,3,4,5,6]. Later stages of atherosclerosis are characterized by the formation of unstable plaques, leading to thrombus formation and, potentially, embolism [1,2,3,4,5,6,7]. In addition to traditional cardio-metabolic risk factors, familial and genetic factors also contribute to the development of atherosclerosis [8,9,10].
The intima-medial thickness (IMT), which is the distance between the lumen–intima and adventitia–media borders of blood vessels, is an early preclinical phenotype of atherosclerosis. Carotid IMT (cIMT) can be measured easily and noninvasively by high-resolution B-mode ultrasound with high reliability. It is significantly correlated with the development of atherosclerosis, as well as the risks of cardiovascular diseases (CVDs), such as myocardial infarction and stroke [11,12]. Currently, cIMT is widely recognized as a valid indicator of vascular health.
Breast cancer susceptibility gene 1 (BRCA1) binds to and recruits PALB2 (the partner and localizer of BRCA2) to the DNA damage sites, and the BRCA1–PALB2 interaction is required to activate the G2/M checkpoint upon DNA damage [13,14]. BRCA1 also forms a heterodimer with BRCA1-associated RING-domain protein 1 (BARD1) and functions as an ubiquitin ligase [15]. The BRCA1–BARD1 complex is required for homologous DNA recombination to repair DNA double-strand breaks [15]. Defects in BRCA1 are responsible for the incidences of familial breast and ovarian cancers. It had been estimated that the average cumulative risks by age 80 years in carriers of BRCA1 germline mutation were 72% and 44% for breast cancer and ovarian cancer, respectively [16]. In addition, BRCA1 has been shown to possess cardioprotective and anti-inflammatory roles in mice [17,18]. Further, BRCA1 is constitutively expressed in human vascular ECs and SMCs [19,20]. BRCA1-overexpressing human vascular ECs and SMCs achieved protection against inflammation- and toxin-induced reactive oxygen species (ROS) by impairing TNFα-induced ROS generation in vascular ECs and inhibiting NADPH oxidase 1 (Nox1)-dependent ROS generation in vascular SMCs, respectively [19,20]. As compared to samples from adjacent plaque-free areas of human carotid artery, BRCA1 mRNA and protein levels were significantly lower in the samples from plaque regions of human atherosclerotic carotid artery [20]. These observations suggest that BRCA1 also acts as a regulator of cardio-vascular health and its deficiency is correlated with development of atherosclerosis.
Obg-like ATPase 1 (OLA1) is a translation factor-related class that is part of the Obg family and the YchF subfamily of P-loop GTPases [21]. P-loop GTPases are involved in the regulation of diverse cellular functions, including protein translation, intracellular transport, signal transduction, and cell proliferation [22,23]. Unlike other Obg family proteins, OLA1 binds and hydrolyzes ATP more efficiently than GTP [24]. OLA1 functions as a negative regulator in the cellular responses to oxidative stress in HeLa cells [25]. OLA1 binds to and protects heat shock protein 70 (HSP70) from C-terminus of Hsp70-binding protein (CHIP)-mediated ubiquitin-dependent degradation in response to heat shock stress [26]. Recently, it has been shown in mice that OLA1, via its binding to HSP70, stabilizes mitochondria SOD2 in pulmonary artery SMCs and that loss of OLA1 causes SOD2 deficiency and an increase in the protein level of X-linked inhibitor of apoptosis (XIAP), an anti-apoptotic protein, thereby increasing proliferation of pulmonary artery SMCs in mice [27]. However, OLA1 is required for proliferation of mouse embryonic fibroblasts due to its attenuation of the translation of p21, a CDK inhibitor [28]. Moreover, OLA1 promotes proliferation of hepatocellular carcinoma cells by binding with p21 and upregulating CDK2 expression [29]. Furthermore, OLA1 plays a role in promoting cell migration through regulating expression of E-cadherin, which is required for intercellular adhesion, and focal adhesion kinase (FAK) in nonvascular cells [30,31]. Previous observations have also shown that overexpression of OLA1 promotes tumor progression and causes poor survival in patients bearing hepatocellular carcinoma, colorectal cancer, endometrial cancer and lung cancer and that overexpression of OLA1 enhances drug resistance by inducing the epithelial-to-mesenchymal transition through activation of the TGF-β/Smad signaling pathway in breast cancer [29,30,32,33,34]. Furthermore, a recent study revealed that OLA1 expression is upregulated in cultured human ventricular cardiomyocytes and mouse heart after angiotensin II-induced hypertrophic response and that loss of OLA1 leads to activation of the GSK3β/β-catenin signaling pathway and, thereby, attenuation of angiotensin II-induced hypertrophic response in human ventricular cardiomyocytes [35]. However, there are no reports demonstrating the physiological role of OLA1 in human vascular ECs and SMCs.
In addition to traditional cardio-metabolic risk factors, such as high blood pressure and lipids, up to 50% of cIMT variations could be explained by genetic factors [9]. It has been shown that the methylation changes at specific CpG sites in BRCA1 are associated with cIMT and that BRCA1 protects human vascular ECs and SMCs from oxidative stresses [19,20,36]. Further, OLA1 has been shown to bind to the amino-terminal region of BRCA1 and the carboxy-terminal region of BARD1 for centrosome regulation [37]. OLA1 plays a role in cell migration and cell proliferation and regulates the cellular responses to oxidative and heat shock stresses [25,26,27,28,29,30,31]. These observations raise the possibility that defects in or altered expression of OLA1 is involved in the development of atherosclerosis and prompted us to determine whether polymorphisms of common genetic variants in the BRCA1, BARD1, and OLA1 genes are correlated with cIMT thickening and could be used as indicators of cardiovascular risk.
2. Results
2.1. Five SNPs in OLA1 Gene Are Significantly Correlated with cIMT
To present our approaches to determining whether OLA1, BRCA1, and BARD1 are genetically correlated with cIMT, a schematic illustration of this study is shown in Figure 1. The anthropometric and clinical characteristics of the study subjects in the discovery and validation stages have been described previously [38]. In the discovery stage, the mean (SD) ages at enrollment for cases and controls were 58.9 (8.8) and 52.6 (8.7) years, respectively. The percentages of male participants were 55.7% and 45.9% in the case and control groups, respectively. In the validation study, the mean (SD) ages at enrollment for cases and controls were 58.5 (8.5) and 58.0 (8.5) years, respectively. The percentage of male participants in both groups was 50.7%. The detailed anthropometric and clinical profiles of the study subjects are showed in Supplementary Table S1. The results of multivariate logistic regression analyses showed that older age, male sex, higher SBP, higher BMI, higher LDL-C/HDL-C ratio, and cigarette smoking were significantly correlated with higher likelihoods of having thicker cIMT [38].
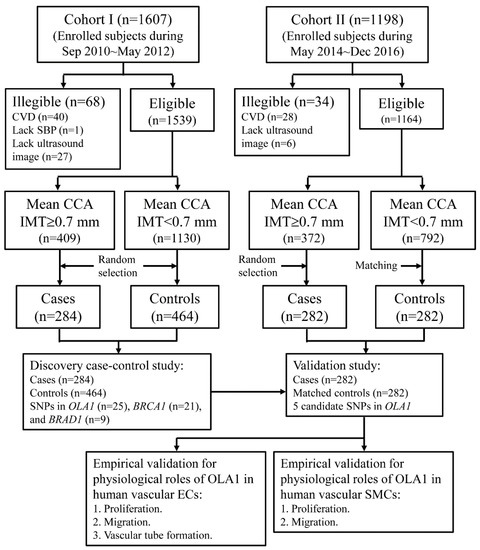
Figure 1.
Schematic illustration of approaches to determine genetic association of OLA1, BRCA1, and BARD1 with cIMT.
Among 59 screened SNPs, one SNP (BARD1 rs10498020) with a p-value in the HWE test <0.001 and another three SNPs (OLA1 rs13019401 and rs16862465 and BARD1 rs34066909) with call rates <95% were excluded (Supplementary Table S2). There were 25, 21, and 9 eligible SNPs in OLA1, BARD1, and BRCA1, respectively, for association analyses. The pre-set critical values of the candidate genetic markers were 0.0040, 0.0048, and 0.011, respectively. A total of five SNPs in OLA1 were eligible for the validation study. None of SNPs in BRCA1 or BRAD1 had a p-value less than the pre-set critical values of candidacy. The primers for PCR reaction and annealing of these five candidate OLA1 SNPs are shown in Supplementary Table S3.
In the discovery case–control study, the minor allele frequencies of these five candidate OLA1 SNPs in the controls ranged from 0.198 to 0.268 and were 0.227 to 0.310 in the cases (Table 1). The minor alleles of all candidate SNPs were significantly more prevalent in the cases. After adjustment for all significant anthropometric and clinical characteristics, heterozygous genotypes were correlated with significantly higher ORs of having thicker cIMT. The multivariate-adjusted ORs ranged from 1.59 to 1.86. Similar results were observed in the validation study.

Table 1.
Association analyses for five candidate SNPs with thicker carotid IMT.
The results of the Breslow–Day test showed that the estimated ORs for all candidate SNPs in the discovery case–control study were not statistically different from those in the validation study (all p-values > 0.2). Accordingly, it was appropriate to obtain the pooled ORs for each candidate SNPs. The pooled ORs of thicker cIMT ranged from 1.55 to 1.72.
Linkage analyses of these five OLA1 SNPs in the controls of the discovery case–control study showed that they formed a single LD block (Figure 2). However, SNP rs12466587 was nearly completely linked with rs16862482 (r2 = 0.97) and SNP rs35145102 showed stronger linear trends with rs201641962 and rs4131583. The most prevalent rs35145102–rs201641962–rs12466587–rs4131583–rs16862482 haplotype was H1-ATTGT, followed by haplotypes H2-GCCTC and H3-GCTTT (Table 2). The estimated ORs for these prevalent haplotypes in the discovery case–control study were not statistically different from those in the validation study (all p-values > 0.2). The pooled OR for H1 haplotype was significantly decreased (OR = 0.72, 95% CI: 0.59–0.86) and for the H2 haplotype it was significantly increased (OR = 1.37, 95% CI: 1.12–1.69).
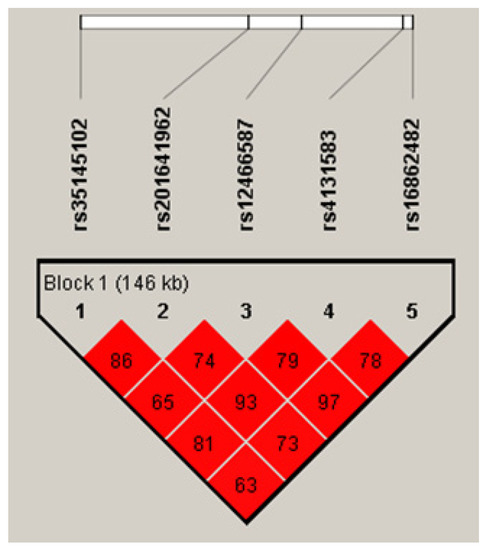
Figure 2.
LD map of the five OLA1 candidate SNPs (values are r2).

Table 2.
Association analyses for rs35145102–rs201641962–rs12466587–rs4131583–rs16862482 haplotypes with thicker carotid IMT.
The results of association analyses of multi-locus genotypes with thicker IMT are shown in Table 3. In the controls of the discovery case–control study, four multi-locus genotypes had frequencies ≥3.0%, including H1 + H1 (53.7%), H1 + H2 (27.2%), H1 + H3 (4.5%), and H2 + H2 (4.7%). The H1 + H1 genotype was negatively and the H1 + H2 genotype positively correlated with thicker cIMT. The estimated ORs for all four prevalent multi-locus genotypes were not statistically different between studies at the discovery and the validation stages (all p-values > 0.2). The pooled ORs of having thicker cIMT were 0.58 (95% CI: 0.46–0.75) and 1.60 (95% CI: 1.23–2.08) for the H1 + H1 and H1 + H2 genotypes, respectively.

Table 3.
Association analyses for multi-locus genotypes with thicker carotid IMT.
2.2. OLA1-Depleted Human Aortic SMCs Are Defective in Proliferation and Migration
Vascular ECs and SMCs play the key roles in the initiation and progression of athersclerosis and the stability of vascular plaque (see reviews in references [1,2,3,4,7]). Endothelium forms a barrier between blood and tissues, and its lesion increases its permeability to and accumulation in the subendothelial matrix of apoprotein-B-containing lipoproteins (apoB LPs), which in turn leads to fibrinolysis and migration and proliferation of vascular SMCs (see reviews in references [1,2,3,4]). Based upon our observations that the common genetic variations in the OLA1 gene we identified were statistically significantly associated with cIMT (Table 1, Table 2 and Table 3), we further investigated the physiological roles of OLA1 in human vascular SMCs and ECs, as described in this section and the subsequent section, respectively.
It is known that vascular SMCs and SMC-derived extracellular matrix accumulate in fibrous plaques (see reviews in references [1,2,4]), indicating that vascular SMCs migrate from media to intima and, thereafter, proliferate at the intima where fibrous plaques form. Furthermore, OLA1 plays a role in promoting cell migration and proliferation through regulating expression of E-cadherin, FAK, and/or p21 in nonvascular cells [28,29,30,31]. Thus, we applied the RNAi technique to determine whether OLA1 is crucial for proliferation and migration of human aortic SMCs. The OLA1 siRNA duplexes we used did efficiently deplete OLA1 in human aortic SMCs (Figure 3A,B). Using the nuclear BrdU incorporation assay (see the Materials and Methods section (Section 4.8)), we found that, compared to non-silencing treatment, depletion of OLA1 caused a reduced proliferation of human aortic SMCs (Figure 3C). Using the trans-well assay (see the Material and Methods section (Section 4.8)), we found that, compared to non-silenced human aortic SMCs, OLA1-depleted human aortic SMCs had reduced migration ability (Figure 3D). These observations indicate that OLA1 is required for migration and proliferation of human aortic SMCs.
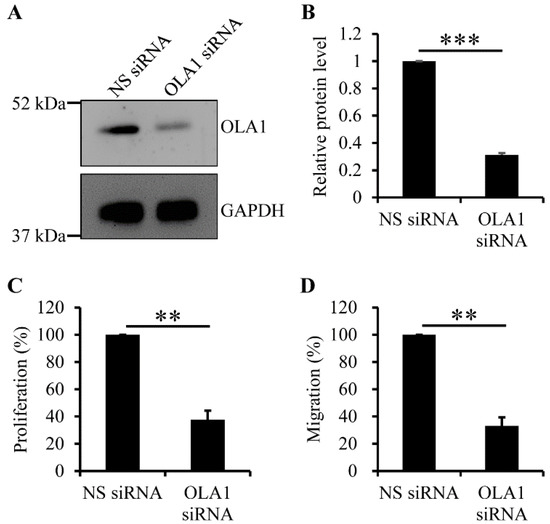
Figure 3.
Effect of OLA1 RNAi (RNA interference) on proliferation and migration of human aortic SMCs. (A,B) Depletion of OLA1 in human aortic SMCs. After human aortic SMCs were transfected with non-silencing (NS) or OLA1 siRNA duplexes for 42 h, cells were collected and lysed for immunoblot analysis with antibodies against OLA1 and GAPDH (loading control). Immunoblot images of both OLA1 and GAPDH are shown in (A). The band intensities of OLA1 and GAPDH from (A) were determined using LS software from VisionWorks (UVP, LLC). The band intensity of OLA1 was normalized to that of GAPDH, and the relative protein levels of OLA1 normalized to the non-silencing treatment in the graph shown in (B) are the means ± s.d. (error bars) from three independent experiments. ***, p < 5 × 10−7. (C) Depletion of OLA1 causes a reduced proliferation of human aortic SMCs. After human aortic SMCs were transfected with the indicated siRNA duplexes for 42 h, 1.5 × 104 cells were incubated with the BrdU-containing medium for 8 h. The BrdU-incorporated cells were stained using 3,3′-diaminobenzidine tetrahydrochloride for analysis (see the Materials and Methods section (Section 4.8)). Each independent experiment was quadruplicated. In the graphs, values are the relative percentages of the amounts of incorporated BrdU in cells normalized to the non-silencing treatment and the means ± s.d. (error bars) from three independent experiments. ***, p ˂ 5 × 10−4 (Student’s t-test). (D) Depletion of OLA1 causes reduced migration ability in human aortic SMCs. After human aortic SMCs were transfected with the indicated siRNA duplexes for 42 h, 3 × 104 cells were resuspended in 100 μL of 0.5% FBS medium for the trans-well assay (see the Materials and Methods section (Section 4.8)). The cells stuck in the pores of the filter were imaged and quantitated. Each independent experiment was triplicated. In the graphs, values are the relative percentages of migrating cells normalized to the non-silencing treatment and the means ± s.d. (error bars) from three independent experiments. **, p ˂ 5 × 10−5 (Student’s t-test).
2.3. OLA1-Depleted Human Aortic ECs Are Defective in Proliferation, Migration, and Tube Formation
ECs of vasa vasorum migrate to and proliferate to form the angiogenic buds in intimal hyperplasia and atherosclerotic lesions and, after bud expansion, the angiogenic buds merge and develop new vessels, which is required for plaque growth, plaque destabilization, and thromboembolic events (see review in reference [7]). Thus, we applied the RNAi technique to determine whether OLA1 is crucial for proliferation and migration of human aortic ECs. The OLA1 siRNA duplexes we used did efficiently deplete OLA1 in human aortic ECs (Figure 4A,B). Using the nuclear BrdU incorporation assay, we found that, compared to non-silenced human aortic ECs, OLA1-depleted human aortic ECs were defective in proliferation (Figure 4C). Using the trans-well assay, we found that, compared to non-silenced human aortic ECs, OLA1-depleted human aortic ECs had reduced migration ability (Figure 4D). These observations indicate that OLA1 is required for migration and proliferation of human aortic ECs.
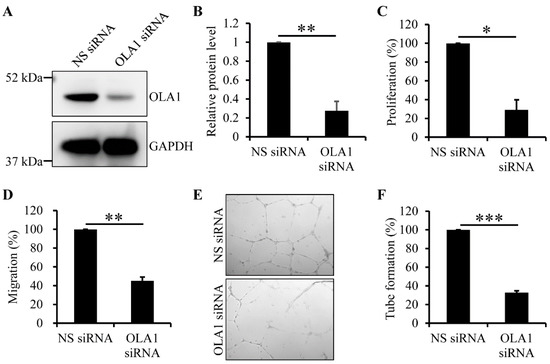
Figure 4.
Effect of OLA1 RNAi on proliferation and migration of and tube formation by human aortic ECs. (A,B) Depletion of OLA1 in human aortic ECs. After human aortic ECs were transfected with non-silencing (NS) or OLA1 siRNA duplexes for 42 h, cells were collected and lysed for immunoblot analysis with antibodies against OLA1 and GAPDH (loading control). Immunoblot images of both OLA1 and GAPDH are shown in (A). The band intensities of OLA1 and GAPDH from (A) were determined using LS software from VisionWorks (UVP, LLC). The band intensity of OLA1 was normalized to that of GAPDH, and the relative protein levels of OLA1 normalized to the non-silencing treatment in the graph show in (B) are the means ± s.d. (error bars) from four independent experiments. **, p < 5 × 10−5. (C) Depletion of OLA1 causes a reduced proliferation of human aortic ECs. After human aortic ECs were transfected with the indicated siRNA duplexes for 42 h, 1.5 × 104 cells were incubated with the BrdU-containing medium for 8 h. The BrdU-incorporated cells were stained using 3,3′-diaminobenzidine tetrahydrochloride for analysis. Each independent experiment was duplicated. In the graph, values are the relative percentages of the amounts of incorporated BrdU in cells normalized to the non-silencing treatment and the means ± s.d. (error bars) from three independent experiments. *, p ˂ 5 × 10−4 (Student’s t-test). (D) Depletion of OLA1 causes reduced migration ability in human aortic ECs. After human aortic ECs were transfected with the indicated siRNA duplexes for 42 h, 3 × 104 cells were resuspended in 100 μL of 0.5% FBS medium for the trans-well assay. The cells stuck in the pores of the filter were imaged and quantitated. Each independent experiment was triplicated. In the graphs, values are the relative percentages of migrating cells normalized to the non-silencing treatment and the means ± s.d. (error bars) from three independent experiments. **, p ˂ 5 × 10−5 (Student’s t-test). (E,F) Depletion of OLA1 causes reduced capillary tube formation by human aortic ECs. After human aortic ECs were transfected with the indicated siRNA duplexes for 42 h, 5 × 104 ECs were used for Matrigel tube formation assay (see the Materials and Methods section (Section 4.8)). The formed capillary tubes were imaged as shown in (E) and quantitated as shown in (F). Each independent experiment was triplicated. In the graph shown in (F), values are the relative percentages of the formed capillary tubes normalized to the non-silencing treatment and the means ± s.d. (error bars) from three independent experiments. ***, p ˂ 1 × 10−6 (Student’s t-test).
Given that neovascularization by ECs of vasa vasorum in atherosclerotic lesions is critical for plaque growth, plaque destabilization, and thromboembolic events (see review in reference [7]), we determined whether depletion of OLA1 may interfere with the ability of human aortic ECs to form vascular tubes and found that, compared to non-silenced human aortic ECs, OLA1-depleted human aortic ECs were defective in tube formation (Figure 4E,F). This indicates that OLA1 regulates vascular tube formation by human aortic ECs.
3. Discussion
In this study, we first identified five candidate SNPs—OLA1 rs35145102, rs201641962, rs12466587, rs4131583, and rs16862482—of cIMT in a case–control study and then validated their effects in another independent frequency-matched case–control study. The relationships between these SNPs and cIMT remained statistically significant after adjustment for the effects of traditional cardio-metabolic risk factors. We also found that these five SNPs formed a single LD block and the homozygosity of the most prevalent haplotype was correlated with significantly decreased OR of thicker cIMT. Further, we found that loss of OLA1 caused reduced ability in human aortic ECs and SMCs to migrate and proliferate and reduced ability in human aortic ECs to form vascular tubes in cell culture system (Figure 3 and Figure 4).
OLA1 is ubiquitously expressed in brain, thyroid, esophagus, testis, and 23 other tissues (see information at https://www.ncbi.nlm.nih.gov/gene/29789#gene-expression) (accessed on 15 March 2021). OLA1 belongs to the YchF subfamily of P-loop GTPases, which regulate diverse cellular functions, including protein translation, intracellular transport, signal transduction, and cell proliferation [22,39]. However, to our knowledge, there are no reports on OLA1 association with preclinical traits of atherosclerosis or cardiovascular events. Further, there are no reports showing that OLA1 directly influences the functions of human immune cells and vascular ECs and SMCs. Based upon the previous observations and our observations in this study, several lines of evidence reveal that the genetic variants in OLA1 may correlate with the development of atherosclerosis.
The first line of evidence is that SNP rs35145102 is located at the 3′-untranslated region (3′-UTR) of the OLA1 mRNA, while the others are intron variants. Given that the 3′-UTR is often targeted by microRNA for regulation of mRNA stability [40,41], we searched for the possible microRNA targeting the 3′-UTR of OLA1 mRNA in the TargetScan database (release 7.1) at the website http://www.targetscan.org/ (accessed on 3 March 2022; see reference [42]) and found that rs35145102 resides in a possible but less conserved target sequence of hsa-miR-6770-5p at the 3′-UTR of OLA1 mRNA (Supplementary Figure S1). This raises the possibility that rs35145102 is correlated with the expression level of OLA1. Thus, we retrieved the expression data in human immune cells by using the Ensemble Genome Browser (see the website at http://asia.ensembl.org/Homo_sapiens/Info/Index) (accessed on 15 April 2021) and found that the expression levels of OLA1 in monocytes are significantly correlated with SNPs rs35145102 and rs4131583, while those of OLA1 in macrophages are significantly correlated with SNPs rs12466587 and rs16862482 [43]. Furthermore, knockdown of OLA1 elicited an increased resistance to oxidizing agents in HeLa cells, whereas overexpression of OLA1 increased cellular sensitivity to oxidizing agents [25]. Therefore, the significant associations between these five OLA1 SNPs and cIMT may result from the differential expression of OLA1 in immune cells, which confer the differential resistance to oxidative stresses.
The second line of evidence is that OLA1 interacts with several proteins involved in the development of atherosclerosis. Among those molecules that interact with OLA1, BRCA1 is the most promising candidate. Although none of the nine BRCA1 SNPs in this study were correlated with cIMT thickening (Supplementary Table S2), a previous study has shown that the cytosine methylation changes at specific CpG sites in BRCA1 are associated with cIMT [36]. It is known that DNA cytosine methylation in the CpG dinucleotide usually results in gene silencing [44]. Additionally, BRCA1 mRNA and protein levels in samples of plaque-containing segments of human carotid arteries were significantly lower than those in the control samples of adjunct plaque-free segments [20]. Similarly, BRCA1 can protect human aortic smooth muscle cells against inflammation- and toxin-induced reactive oxygen species [19]. As compared with ad-null-treated ApoE−/− mice, ad-BRCA1-treated ApoE−/− mice developed significantly fewer aortic plaque lesions, exhibited reduced macrophage infiltration, and generated fewer reactive oxygen species [20]. These observations raise the possibility that a reduced expression of BRCA1 contributes to development of atherosclerosis. Further, given that OLA1 and BRCA1 regulate the cellular responses to oxidative stresses [19,20,25], it can be speculated that OLA1 and BRCA1 work together in the development of atherosclerosis.
The third line of evidence is that vascular endothelium forms a barrier between blood and tissues, and its lesion leads to fibrinolysis and migration and proliferation of vascular SMCs (see reviews in references [1,2,3,4]). Further, migration and proliferation of ECs, and then their neovascularization of vasa vasorum, in atherosclerotic lesions are crucial for plaque growth, plaque destabilization, and thromboembolic events (see review in reference [7]). In this study, we found that loss of OLA1 caused reduced ability in human aortic ECs to migrate, proliferate, and form vascular tubes (Figure 4). Therefore, it is possible that a reduced expression of or a defective mutation in OLA1 may lead to a lesion in the vascular endothelium, which occurs at the early stages of the development of atherosclerosis, or that overexpression of OLA1 promotes neovascularization by ECs of vasa vasorum, leading to plaque growth, plaque destabilization, and thromboembolic events in atherosclerotic lesions.
The last line of evidence is that it is known that one of the key steps for the development of atherosclerosis is that vascular smooth muscle cells migrate from media to intima and then proliferate in intima [2,45]. In this study, we found that OLA1 was required for migration and proliferation of human aortic SMCs (Figure 3B,C), suggesting that OLA1 should play a role in the development of atherosclerosis. However, contrary to our observation, loss of OLA1 causes an increased proliferation of mouse pulmonary artery SMCs [27]. Indeed, OLA1 is required for migration and proliferation of mouse embryonic fibroblasts [28]. Therefore, it is possible that OLA1 exerts an effect on the proliferation of cells in either a species- or cell type-dependent manner. It has been shown that OLA1 binds to and inhibits GSK3 to negatively regulate E-cadherin expression and, thereby, promotes cell migration [30]. Further, OLA1 negatively regulates cell adhesion and spreading by attenuating FAK expression and increasing cofilin phosphorylation [31]. FAK controls proliferation and migration of rat aortic SMCs, and that phosphorylation of cofilin controls platelet-derived growth factor-induced migration of human aortic SMCs [46,47,48]. Furthermore, OLA1 negatively regulates translation of p21, a CDK inhibitor, and, thereby, maintains optimal cell proliferation for developmental progression in mice [28]. Additionally, p21 can inhibit proliferation of vascular ECs and migration of vascular SMCs [49,50]. Based upon these observations, it is possible that OLA1 regulates expression of E-cadherin, FAK, and/or p21 and, thereby, controls migration and proliferation of human aortic SMCs.
To further confirm that these five OLA1 SNPs are the major genetic determinants of cIMT, we searched for functional OLA1 SNPs with a minor allele frequency >3.0% in the Taiwan Biobank (see the website at https://taiwanview.twbiobank.org.tw/index) (accessed on 20 April 2021). A total of six functional SNPs were identified: rs35145102, rs3739153, rs3739152, rs6737629, rs1063613, and rs10209474 (see the website at https://taiwanview.twbiobank.org.tw/index) (accessed on 20 April 2021). Except for SNP rs10209474, a 5′-UTR variant, the others are 3′-UTR variants. The LD data in the 1000 Human Genome Project Phase 3-Southern Han Chinese (see reference [51]) showed that SNPs rs3739153 and rs3739152 were in complete LD (r2 = 1.0), and these two SNPs were also in complete LD with two screened SNPs (rs12693034 and rs6741764). SNP rs6737629 was in complete LD with two screened SNPs (rs17239055 and rs2358443). SNP rs10209474 was closely linked with one screened SNP (rs12479030; r2 = 0.802). Our discovery case–control study showed that SNPs rs12693034, rs6741764, rs17239055, rs2358443, and rs12479030 were not correlated with cIMT. Accordingly, rs3739153, rs3739152, rs6737629, and rs10209474 are unlikely to play critical roles in the development of atherosclerosis. Of note, SNP rs1063613 was not included in our screened SNPs and was not linked with any other SNPs located 50 Kb down- or up-stream of its location. Its physiological significance needs further exploration.
In the study, we conducted two independent case–control studies and used two different genotyping methods to explore the relationships between common genetic variants in OLA1, BRCA1, and BRAD1. Additionally, we found that the minor allele frequencies of the five OLA1 candidate SNPs in the controls were similar to the corresponding frequencies in the Taiwan Biobank, which enrolls subjects from the general population. However, there were potential limitations in the present study. The negative finding for the relationship between SNPs in BRCA1 and BARD1 genes and cIMT does not necessarily preclude the possibility that these two genes may play important roles in atherosclerotic diseases. In the study, only nine SNPs in BRCA1, in contract to 25 SNPs in OLA1 and 21 SNPs in BARD1, were screened for their association with cIMT. Accordingly, BRCA1 seems more likely to suffer from false negative bias. Additionally, to control the overall type I error and to reduce the likelihood of false negative findings at the same time, only SNPs with a p-value less than twofold the corrected significance level were included in the validation study. However, false-negative and false-positive findings may still exist.
There were two further potential limitations in this study. The first was that, before the start of cohort I, we conducted a prior study to evaluate the feasibility of obtaining data associated with pharmacological treatments. Most interviewees could provide the information related to the classes of medicines they took. However, many could not provide the names of medicines and the duration of medical treatment. Therefore, we did not collect detailed data associated with treatments and were unable to assess the effects of these treatments. However, based on the following reasons, our findings were more likely to be conservative. The reasons include: (a) genotypes of subjects were not influenced by treatment. (b) None of the subjects had ever received carotid ultrasonographic scans before. Therefore, it is very unlikely that individuals would take medicines to control carotid IMTs. Finally, (c) all technicians who operated the ultrasonographic systems and measured the carotid IMTs were blinded to examinees’ clinical profiles. Variations in measurements of carotid IMTs tend to be non-differential. The second limitation is that, although our study revealed that OLA1 was required to promote proliferation and migration of human aortic ECs and SMCs and vascular tube formation by human aortic ECs in a cell culture system, we cannot clarify whether loss of OLA1 in vascular ECs or SMCs would cause development of atherosclerosis or whether overexpression of OLA1 in vascular ECs or SMCs would cause development of atherosclerosis. Therefore, vascular EC- or SMC-specific inducible OLA1 knockout mice are needed to verify whether loss of OLA1 in vascular ECs or SMCs may cause development of atherosclerosis in mice. Similarly, vascular EC- or SMC-specific inducible OLA1-overexpressing mice are also needed to verify whether overexpression of OLA1 in vascular ECs or SMCs may cause development of atherosclerosis in mice.
In conclusion, we identified five OLA1 SNPs showing significant associations with cIMT. These genetic markers formed a closely linked LD block. The homozygosity of the most prevalent multi-locus haplotype was correlated with significantly lower ORs of having thicker cIMT. Furthermore, we found that OLA1 was required for migration and proliferation of human aortic ECs and SMCs and regulated vascular tube formation by human aortic ECs (Figure 3 and Figure 4). Given that vascular ECs and SMCs play the key roles in development of atherosclerosis [1,2,3,4,7], these genetic markers may be used for individual risk prediction of atherosclerotic diseases. These genetic markers may possess the potential for use in the development of therapeutic agents for the secondary prevention of atherosclerotic diseases.
4. Materials and Methods
4.1. Study Subjects
We used two case–control studies to screen and validate the relationships of common genetic variants in BRCA1, BARD1 and OLA1 genes with cIMT thickening. The cases and controls of the case–control studies were selected from among the participants of our community-based studies and have been described previously [38]. In brief, the subjects in the discovery stage were selected from among the participants of our previous community study (cohort I), which recruited middle-aged adults and elderly subjects from September 2010 to May 2012. The case group was a random sample of 284 subjects whose mean common carotid artery (CCA) IMT was ≥ 0.70 mm, the 75th percentile of the distribution of the mean far-wall CCA IMT. The control group was a random sample of 464 subjects who had a mean CCA IMT < 0.70 mm (Figure 1).
The study subjects of the validation study were selected from an ongoing community-based study (cohort II) that enrolled residents aged 40–74 years [38]. From May 2014 to December 2016, 1164 residents who had no CVD history and had good quality recorded carotid ultrasound images were enrolled. We selected a random sample of 282 subjects from those who had thicker cIMTs (CCA IMT ≥ 0.70 mm) as the case group. Based on the age (40–49, 50–59, and 60–74 years) and sex distributions of the case group, we performed stratified random sampling and selected 282 subjects who had normal cIMTs (CCA IMT < 0.70 mm) as the control group (Figure 1).
All participants of the two community-based studies voluntarily provided informed consent. The studies complied with the 1975 Helsinki Declaration on ethics in medical research and were reviewed and approved by the Institution Review Board of Mackay Memorial Hospital (14MMHIS075).
4.2. Measurements of Anthropometric and Cardio-Metabolic Profiles
The measurements of anthropometric characteristics and cardio-metabolic profiles have been described previously [52]. The anthropometric characteristics included age at enrollment, sex, body weight and height, and circumferences of the waist and hip. Blood pressure was measured three times after 10 min of rest. The cardio-metabolic profiles included systolic blood pressure (SBP), diastolic blood pressure (DBP), total cholesterol, high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), fasting triglycerides (FTG), and fasting plasma glucose (FPG). A structured questionnaire was used to collect personal health behaviors, such as cigarette smoking, alcohol drinking, and dietary pattern. Personal histories of common diseases, including hypertension, hyperlipidemia, hyperglycemia, diabetes mellitus (DM), and CVDs, were also collected by the structured questionnaire.
4.3. The cIMT Measurement
The measurement of cIMT was performed as described previously [52]. In short, two experienced technicians who were blinded to patients’ anthropometric and clinical characteristics obtained CCA images by using high-resolution B-mode ultrasonography systems (GE Healthcare Vivid 7 and Vivid E9; General Electric Company, Milwaukee, WI, USA). A well-trained technician retrieved the digital images and measured the far-wall IMTs blindly by using automatic contouring software (GE Healthcare EchoPAC version 112.0.2; General Electric-Vingmed, Horten, Norway). The IMT was defined as the distance between the lumen–intima and media–adventitia interfaces and included plaques. The average, minimum, and maximum IMTs of the distal 1 to 2 cm of the left and right CCA were recorded. Mean cIMT was calculated as the mean of the left and right average IMTs and was used for correlation and regression analyses.
4.4. SNP Selection and Genotyping
In the discovery study, we used a plate (Axiom® CHB 1 Array Plate; Affymetrix Ltd., Santa Clara, CA, USA) to screen the associations of common genetic variants in OLA1, BRCA1, and BARD1 with cIMT. In the plate, there were 59 SNPs within 25 Kb up- or downstream of the OLA1 (n = 27), BARD1 (n = 23), and BRCA1 (n = 9) genes. The eligibility of SNPs for association analyses were a call rate >95%, a p-value in the Hardy–Weinberg equilibrium (HWE) test in the controls that was >0.001, and a minor allele frequency >3%.
The significance level of the discovery study, designated αcorrected, was calculated as 0.05 divided by the number of eligible SNP in the locus. To reduce the influence of over-adjustment, we used 2 × αcorrected as the pre-set critical value for candidate genetic markers. Eligible SNPs with a p-value less than the pre-set critical value for candidacy were subject to a validation study using the Sequenom iPLEX MassARRAY system (Sequenom, San Diego, CA, USA). All genotyping was performed at the National Center for Genome Medicine, Academic Sinica, Taiwan.
4.5. Cell Culture and Transfection
Human aortic endothelial cells (HAoEC, Promocell Inc., Heidelberg, German) were cultured in endothelial growth medium MV2 (C-22022, Promocell Inc., Heidelberg, Germany) with the addition of growth medium MV2 supplement mix (C-39226, Promocell Inc., Heidelberg, Germany), and human aortic smooth muscle cells (HAoSMC, Promocell Inc., Heidelberg, Germany) were cultured in vascular smooth muscle cell basal medium (M231500, Thermo Fisher Scientific Inc., Waltham, MA, USA) with the addition of smooth muscle growth supplement (S00725, Thermo Fisher Scientific Inc., Waltham, MA, USA). Cell transfection with siRNA duplexes was performed using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific Inc., Waltham, MA, USA) following the manufacturer’s instruction.
4.6. siRNA Duplexes
The OLA1 siRNA duplexes used in this study were the same as previously described [29]. The sense strand sequences of non-silencing and OLA1 siRNA duplexes were 5′-r(UUCUCCGAACGUGUCACGU)dTdT-3′ and 5′-r(GCUGCUGGAAAGUACAGAC)dTdT, respectively. The siRNA duplexes were synthesized by and purchased from GE Healthcare Dharmacon Inc., Lafayette, CO, USA.
4.7. Immunoblot Assay
After cells were transfected with the indicated siRNA duplexes for 42 h, cells were lysed with RIPA buffer and 200 μg cell lysates were used for the immunoblot assay. To perform the immunoblot assay, OLA1 was detected using rabbit anti-OLA1 antibodies (Abcam Inc., Cambridge, UK) and GAPDH (internal loading control) using rabbit anti-GAPDH (Genetex Inc., Irvine, CA, USA), and they were visualized by chemiluminescence (Genetex Inc., Irvine, CA, USA) and imaged with a ChemiDoc-It® 815 Imager (Analytik Jena, An Endress + Hauser Company, Jena, Germany) after incubation with HRP-conjugated secondary antibodies (Genetex Inc., Irvine, CA, USA).
4.8. Assays for Cell Proliferation, Cell Migration, and Tube Formation
Proliferation of human aortic endothelial and smooth muscle cells was determined with the nuclear bromodeoxyuridine (BrdU) incorporation assay using a BrdU immunochemistry kit (Merk Millipore Inc., Burlington, MA, USA). The assay was performed following the manufacturer’s instruction (Millipore Inc.). In brief, cells (1.5 × 104/well) were seeded on coverslips in a 24-well plate and incubated with 10 μM BrdU for 8 hours. After being washed with 1 × PBS buffer, cells were fixed with ice-cold 70% ethanol at 4 °C for 30 min. The BrdU-incorporated cells were then stained using 3,3′-diaminobenzidine tetrahydrochloride, and the stained intensity was determined by a microplate reader (Molecular Devices Inc., San Jose, CA, USA).
The migration assay for human aortic ECs and SMCs was performed using a 24-well trans-well plate with a pore size of 8 μm (Corning Inc., Corning, NY, USA). A total of 3 × 104 cells were resuspended in 100 μL of 0.5% FBS medium and plated in the upper chamber, and 600 μL of 2.5% FBS medium was added into the lower chamber. After incubation at 37 °C for 4 h, cells were fixed with −20 °C methanol for 5 min and stained with 18.7 μM bisbenzimide (Sigma-Aldrich Inc., Saint Louis, MO, USA) in the dark for 15 min. After the filter was washed with 1 × PBS three times, the cells on the upper surface of filter were completely wiped away with the cotton swab. The cells stuck in the pore of the filter were imaged under a fluorescence microscope (Leica Microsystems Inc., Wetzlar, Germany) using a 40 × objective lens and the image analyzed using the QWIN software (Leica Microsystems Inc., Wetzlar, German).
A Matrigel tube formation assay was performed following the manufacturer’s instruction (BD Biosciences Inc., Mississauga, ON, Canada). The Matrigel (BD Biosciences Inc., Mississauga, ON, Canada) was dissolved at 4 °C overnight before use. Each well in the 48-well plate was coated with 150 µL Matrigel and then incubated at 37 °C for 30 min. A total of 5 × 104 ECs were seeded per well on the layer of polymerized Matrigel for 24 h. The formed capillary tubes were imaged under an inverted phase contrast microscope (Zeiss Inc., Oberkochen, Germany) using a 20 × objective lens and quantified by measuring the long axis of each tube in three randomly selected fields per well using MacBiophotonics Image J software.
4.9. Statistical Analyses
In the study, the chi-square test was used to compare whether there were significant differences in the frequency distributions of genotypes between cases and controls. The relationships with thicker cIMT for each eligible SNP were assessed by minor allele dominant, recessive, and additive and (minor and major allele) co-dominant genotypic models and adjusted for age, sex, and other significant cardio-metabolic factors. Assessment of the pairwise LD of candidate SNPs in the control group of the discovery study was performed in Haploview 4.2 software [53].
Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated with multivariate logistic regression models. The homogeneity of the estimated ORs between the discovery and validation studies was evaluated with the Breslow–Day test. The pooled ORs were obtained by summation of the study-specific ORs, weighting by the inverse of their variances when the estimated ORs were not statistically different between studies. Except for the assessment of pairwise LD, all statistical analyses were performed using SAS 9.4 (SAS Institute Inc., Cary, NC, USA).
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231911511/s1.
Author Contributions
Conceptualization, Y.-C.C., T.-W.W., Y.-L.J. and L.-Y.W.; Data curation, L.-Y.W.; Formal analysis, L.-Y.W.; Funding acquisition, L.-Y.W.; Investigation, T.-F.L., C.-L.C., C.-J.H. and S.-X.L.; Project administration, Y.-L.J. and L.-Y.W.; Resources, Y.-J.W.; Supervision, Y.-L.J. and L.-Y.W.; Validation, T.-F.L. and L.-Y.W.; Visualization, Y.-L.J. and L.-Y.W.; Writing—original draft, T.-F.L., Y.-L.J. and L.-Y.W.; Writing—review and editing, T.-F.L., C.-L.C., Y.-J.W., Y.-C.C., T.-W.W., Y.-L.J. and L.-Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the research grants from the Ministry of Science and Technology of Taiwan (MOST 107-2314-B-715-003 and MOST 108-2314-B-715-005-MY3 to Li-Yu Wang), MacKay Medical College (1081B15 and 1091B03 to Li-Yu Wang), and the Wang Jhan-Yang Public Charitable Trust Fund (WJY 2016-HR-01 and WJY 2017-HR-01 to Li-Yu Wang). The funding agencies played no role in the research.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Boards of MacKay Medical College (No. P990001, granted date: 5 July 2010) and MacKay Memorial Hospital (No. 14MMHIS075, granted date: 23 May 2014).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used and/or analyzed in the current study are available from the corresponding author (Li-Yu Wang) on reasonable request.
Acknowledgments
We thank the staffs in the district health station of Sanzhi District, Tamsui District, and Shimen District, New Taipei City, for their administrative supports. We also thank the National Center for Genome Medicine, Academia Sinica, Taiwan, for their skillful techniques and highly efficient services. This center was supported by grants from the National Core Facility Program for Biotechnology.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lusis, A.J. Atherosclerosis. Nature 2000, 407, 233–241. [Google Scholar] [PubMed]
- Bennett, M.R.; Sinha, S.; Owens, G.K. Vascular Smooth Muscle Cells in Atherosclerosis. Circ. Res. 2016, 118, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Gimbrone, M.A., Jr.; García-Cardeña, G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ. Res. 2016, 118, 620–636. [Google Scholar]
- Lu, H.; Daugherty, A. Atherosclerosis. Arter. Thromb. Vasc. Biol. 2015, 35, 485–491. [Google Scholar]
- Wolf, D.; Ley, K. Immunity and Inflammation in Atherosclerosis. Circ. Res. 2019, 124, 315–327. [Google Scholar] [PubMed]
- Tabas, I.; Lichtman, A.H. Monocyte-Macrophages and T Cells in Atherosclerosis. Immunity 2017, 47, 621–634. [Google Scholar]
- Camaré, C.; Pucelle, M.; Nègre-Salvayre, A.; Salvayre, R. Angiogenesis in the atherosclerotic plaque. Redox. Biol. 2017, 12, 18–34. [Google Scholar]
- Lee, K.; Sung, J.; Lee, S.C.; Park, S.W.; Kim, Y.S.; Lee, J.Y.; Ebrahim, S.; Song, Y.M. Segment-specific carotid intima-media thickness and cardiovascular risk factors in Koreans: The Healthy Twin Study. Eur. J. Prev. Cardiol. 2012, 19, 1161–1172. [Google Scholar] [CrossRef]
- Zhao, J.; Cheema, F.A.; Bremner, J.D.; Goldberg, J.; Su, S.; Snieder, H.; Maisano, C.; Jones, L.; Javed, F.; Murrah, N.; et al. Heritability of carotid intima-media thickness: A twin study. Atherosclerosis 2008, 197, 814–820. [Google Scholar] [CrossRef]
- Medda, E.; Fagnani, C.; Schillaci, G.; Tarnoki, A.D.; Tarnoki, D.L.; Baracchini, C.; Meneghetti, G.; Fanelli, F.; Alaeddin, A.; Pucci, G.; et al. Heritability of arterial stiffness and carotid intima-media thickness: An Italian twin study. Nutr. Metab. Cardiovasc. Dis. 2014, 24, 511–517. [Google Scholar] [CrossRef]
- Tschiderer, L.; Klingenschmid, G.; Seekircher, L.; Willeit, P. Carotid intima-media thickness predicts carotid plaque development: Meta-analysis of seven studies involving 9341 participants. Eur. J. Clin. Investig. 2020, 50, e13217. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, T.Z.; Lee, M.S. Carotid intima-media thickness and plaque in cardiovascular risk assessment. JACC Cardiovasc. Imaging 2014, 7, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Simhadri, S.; Vincelli, G.; Huo, Y.; Misenko, S.; Foo, T.K.; Ahlskog, J.; Sørensen, C.S.; Oakley, G.G.; Ganesan, S.; Bunting, S.F.; et al. PALB2 connects BRCA1 and BRCA2 in the G2/M checkpoint response. Oncogene 2019, 38, 1585–1596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ma, J.; Wu, J.; Ye, L.; Cai, H.; Xia, B.; Yu, X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 2009, 19, 524–529. [Google Scholar] [CrossRef]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299. [Google Scholar]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.A.; Mooij, T.M.; Roos-Blom, M.J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar]
- Shukla, P.C.; Singh, K.K.; Quan, A.; Al-Omran, M.; Teoh, H.; Lovren, F.; Cao, L.; Rovira, I.I.; Pan, Y.; Brezden-Masley, C.; et al. BRCA1 is an essential regulator of heart function and survival following myocardial infarction. Nat. Commun. 2011, 2, 593. [Google Scholar] [CrossRef]
- Teoh, H.; Quan, A.; Creighton, A.K.; Bang, K.W.A.; Singh, K.K.; Shukla, P.C.; Gupta, N.; Pan, Y.; Lovren, F.; Leong-Poi, H.; et al. BRCA1 gene therapy reduces systemic inflammatory response and multiple organ failure and improves survival in experimental sepsis. Gene 2013, 20, 51–61. [Google Scholar]
- Lovren, F.; Pan, Y.; Quan, A.; Singh, K.K.; Khan, R.; Gupta, N.; Brezden-Masley, C.; Teoh, H.; Wheatcroft, M.D.; Al-Omran, M.; et al. BRCA1 shields vascular smooth muscle cells from oxidative stress. J. Thorac. Cardiovasc. Surg. 2014, 147, 1946–1955.e1. [Google Scholar] [CrossRef]
- Singh, K.K.; Shukla, P.C.; Quan, A.; Al-Omran, M.; Lovren, F.; Pan, Y.; Brezden-Masley, C.; Ingram, A.J.; Stanford, W.L.; Teoh, H.; et al. BRCA1 is a novel target to improve endothelial dysfunction and retard atherosclerosis. J. Thorac. Cardiovasc. Surg. 2013, 146, 949–960.e4. [Google Scholar] [CrossRef]
- Sun, H.; Luo, X.; Montalbano, J.; Jin, W.; Shi, J.; Sheikh, M.S.; Huang, Y. DOC45, a novel DNA damage-regulated nucleocytoplasmic ATPase that is overexpressed in multiple human malignancies. Mol. Cancer Res. 2010, 8, 57–66. [Google Scholar]
- Paduch, M.; Jeleń, F.; Otlewski, J. Structure of small G proteins and their regulators. Acta Biochim. Pol. 2001, 48, 829–850. [Google Scholar] [CrossRef]
- Sprang, S.R. G protein mechanisms: Insights from structural analysis. Annu. Rev. Biochem. 1997, 66, 639–678. [Google Scholar]
- Koller-Eichhorn, R.; Marquardt, T.; Gail, R.; Wittinghofer, A.; Kostrewa, D.; Kutay, U.; Kambach, C. Human OLA1 defines an ATPase subfamily in the Obg family of GTP-binding proteins. J. Biol. Chem. 2007, 282, 19928–19937. [Google Scholar]
- Zhang, J.; Rubio, V.; Lieberman, M.W.; Shi, Z.Z. OLA1, an Obg-like ATPase, suppresses antioxidant response via nontranscriptional mechanisms. Proc. Natl. Acad. Sci. USA 2009, 106, 15356–15361. [Google Scholar]
- Mao, R.F.; Rubio, V.; Chen, H.; Bai, L.; Mansour, O.C.; Shi, Z.Z. OLA1 protects cells in heat shock by stabilizing HSP70. Cell Death Dis. 2013, 4, e491. [Google Scholar] [CrossRef]
- Schultz, A.; Olorundami, O.A.; Teng, R.J.; Jarzembowski, J.; Shi, Z.Z.; Kumar, S.N.; Pritchard, K., Jr.; Konduri, G.G.; Afolayan, A.J. Decreased OLA1 (Obg-Like ATPase-1) Expression Drives Ubiquitin-Proteasome Pathways to Downregulate Mitochondrial SOD2 (Superoxide Dismutase) in Persistent Pulmonary Hypertension of the Newborn. Hypertension 2019, 74, 957–966. [Google Scholar] [CrossRef]
- Ding, Z.; Liu, Y.; Rubio, V.; He, J.; Minze, L.J.; Shi, Z.Z. OLA1, a Translational Regulator of p21, Maintains Optimal Cell Proliferation Necessary for Developmental Progression. Mol. Cell Biol. 2016, 36, 2568–2582. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, C.; Sun, C.; Hou, Y.; Zhang, Y.; Tam, N.L.; Wang, Z.; Yu, J.; Huang, B.; Zhuang, H.; et al. Obg-like ATPase 1 (OLA1) overexpression predicts poor prognosis and promotes tumor progression by regulating P21/CDK2 in hepatocellular carcinoma. Aging 2020, 12, 3025–3041. [Google Scholar] [CrossRef]
- Bai, L.; Yu, Z.; Zhang, J.; Yuan, S.; Liao, C.; Jeyabal, P.V.; Rubio, V.; Chen, H.; Li, Y.; Shi, Z.Z. OLA1 contributes to epithelial-mesenchymal transition in lung cancer by modulating the GSK3β/snail/E-cadherin signaling. Oncotarget 2016, 7, 10402–10413. [Google Scholar] [CrossRef]
- Jeyabal, P.V.; Rubio, V.; Chen, H.; Zhang, J.; Shi, Z.Z. Regulation of cell-matrix adhesion by OLA1, the Obg-like ATPase 1. Biochem. Biophys. Res. Commun. 2014, 444, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kong, X.X.; He, J.J.; Xu, Y.B.; Zhang, J.K.; Zou, L.Y.; Ding, K.F.; Xu, D. OLA1 promotes colorectal cancer tumorigenesis by activation of HIF1α/CA9 axis. BMC Cancer 2022, 22, 424. [Google Scholar]
- Dong, Y.; Yin, A.; Xu, C.; Jiang, H.; Wang, Q.; Wu, W.; Guo, S. OLA1 is a potential prognostic molecular biomarker for endometrial cancer and promotes tumor progression. Oncol. Lett. 2021, 22, 576. [Google Scholar] [CrossRef]
- Liu, J.; Miao, X.; Xiao, B.; Huang, J.; Tao, X.; Zhang, J.; Zhao, H.; Pan, Y.; Wang, H.; Gao, G.; et al. Obg-Like ATPase 1 Enhances Chemoresistance of Breast Cancer via Activation of TGF-β/Smad Axis Cascades. Front. Pharmacol. 2020, 11, 666. [Google Scholar] [CrossRef]
- Narasimhan, G.; Henderson, J.; Luong, H.T.; Rajasekaran, N.S.; Qin, G.; Zhang, J.; Krishnamurthy, P. OBG-like ATPase 1 inhibition attenuates angiotensin II-induced hypertrophic response in human ventricular myocytes via GSK-3beta/beta-catenin signalling. Clin. Exp. Pharmacol. Physiol. 2019, 46, 743–751. [Google Scholar] [CrossRef]
- Istas, G.; Declerck, K.; Pudenz, M.; Szic, K.S.V.; Lendinez-Tortajada, V.; Leon-Latre, M.; Heyninck, K.; Haegeman, G.; Casasnovas, J.A.; Tellez-Plaza, M.; et al. Identification of differentially methylated BRCA1 and CRISP2 DNA regions as blood surrogate markers for cardiovascular disease. Sci. Rep. 2017, 7, 5120. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa, A.; Kanno, S.-I.; Nakayama, M.; Mochiduki, H.; Wei, L.; Shimaoka, T.; Furukawa, Y.; Kato, K.; Shibata, S.; Yasui, A.; et al. The BRCA1/BARD1-Interacting Protein OLA1 Functions in Centrosome Regulation. Mol. Cell 2014, 53, 101–114. [Google Scholar] [CrossRef]
- Wu, T.W.; Chou, C.L.; Chen, Y.C.; Juang, Y.L.; Wang, L.Y. Associations of Common Genetic Variants on IL-17 Genes and Carotid Intima-Media Thickness. J. Atheroscler. Thromb. 2018, 25, 1156–1167. [Google Scholar] [CrossRef]
- Leipe, D.D.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002, 317, 41–72. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Schmiedel, B.J.; Singh, D.; Madrigal, A.; Valdovino-Gonzalez, A.G.; White, B.M.; Zapardiel-Gonzalo, J.; Ha, B.; Altay, G.; Greenbaum, J.A.; McVicker, G.; et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018, 175, 1701–1715.e16. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Newby, A.C. Dual role of matrix metalloproteinases (matrixins) in intimal thickening and atherosclerotic plaque rupture. Physiol. Rev. 2005, 85, 1–31. [Google Scholar]
- Martín, A.S.; Lee, M.Y.; Williams, H.C.; Mizuno, K.; Lassègue, B.; Griendling, K.K. Dual regulation of cofilin activity by LIM kinase and Slingshot-1L phosphatase controls platelet-derived growth factor-induced migration of human aortic smooth muscle cells. Circ. Res. 2008, 102, 432–438. [Google Scholar] [CrossRef]
- Morla, A.O.; Mogford, J.E. Control of smooth muscle cell proliferation and phenotype by integrin signaling through focal adhesion kinase. Biochem. Biophys. Res. Commun. 2000, 272, 298–302. [Google Scholar] [CrossRef]
- Taylor, J.M.; Mack, C.P.; Nolan, K.; Regan, C.P.; Owens, G.K.; Parsons, J.T. Selective expression of an endogenous inhibitor of FAK regulates proliferation and migration of vascular smooth muscle cells. Mol. Cell. Biol. 2001, 21, 1565–1572. [Google Scholar] [CrossRef]
- Akimoto, S.; Mitsumata, M.; Sasaguri, T.; Yoshida, Y. Laminar shear stress inhibits vascular endothelial cell proliferation by inducing cyclin-dependent kinase inhibitor p21(Sdi1/Cip1/Waf1). Circ. Res. 2000, 86, 185–190. [Google Scholar] [CrossRef]
- Fukui, R.; Shibata, N.; Kohbayashi, E.; Amakawa, M.; Furutama, D.; Hoshiga, M.; Negoro, N.; Nakakouji, T.; Ii, M.; Ishihara, T.; et al. Inhibition of smooth muscle cell migration by the p21 cyclin-dependent kinase inhibitor (Cip1). Atherosclerosis 1997, 132, 53–59. [Google Scholar] [CrossRef]
- Yates, A.; Akanni, W.; Amode, M.R.; Barrell, D.; Billis, K.; Carvalho-Silva, D.; Cummins, C.; Clapham, P.; Fitzgerald, S.; Gil, L.; et al. Ensembl 2016. Nucleic Acids Res. 2016, 44, D710–D716. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.W.; Hung, C.L.; Liu, C.C.; Wu, Y.J.; Wang, L.Y.; Yeh, H.I. Associations of Cardiovascular Risk Factors with Carotid Intima-Media Thickness in Middle-Age Adults and Elders. J. Atheroscler. Thromb. 2017, 24, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).