Abstract
Although it is known that rice 14-3-3 family genes are involved in various defense responses, the functions of OsGF14f in response to diseases have not been reported. Here, we showed that the transcription of OsGF14f was significantly induced by leaf blast infection, and the overexpression of OsGF14f quantitatively enhanced resistance to leaf blast and bacterial blight in rice. Further analysis showed that the expression levels of salicylic acid (SA) pathway-associated genes (PAL1, NH1, PR1a and PR10) in the OsGF14f-overexpressing plants, were higher than those in wild-type plants after inoculation with the blast isolate (Magnaporthe oryzae Barr). In addition, the expression level of OsGF14f was significantly induced after SA treatment, and higher endogenous SA levels were observed in the OsGF14f-overexpressing plants compared with that in wild-type plants, especially after blast challenge. Taken together, these results suggest that OsGF14f positively regulates leaf blast and bacterial blight resistance in rice via the SA-dependent signaling pathway.
1. Introduction
As sessile organisms, plants are continuously subjected to various types of stresses such as pathogen attacks, insect herbivory, and environmental stresses [1]. These stresses have adverse effects on plant growth and seed production. To survive, plants have developed intricate mechanisms to efficiently perceive external signals and to tailor their responses to the precise environmental conditions encountered [1,2]. Many genes were reported as important regulators in various responses to stresses [2]. Among these genes, the genes encoding 14-3-3 proteins are modulated by several biotic and abiotic stresses [3,4].
14-3-3 proteins belong to a family of regulatory proteins that are uniquely eukaryotic, and evolutionarily conserved across all eukaryotes [5]. In plants, 14-3-3s have long been thought to play important roles in defense responses [6,7,8,9,10,11]. For example, early research indicated that 14-3-3 genes are differentially modulated during various disease defense responses [6,7,12]. Additionally, 14-3-3s are also involved in R-gene mediated plant disease resistance, as well as in reactive oxygen species (ROS)-mediated plant defense responses [9,13,14,15]. In addition, 14-3-3s have been implicated in programmed cell death (PCD) in tomatoes through directly interacting with mitogen-activated protein (MAP) kinase kinase kinase (MAPKKKa) and SIMKK2 [8,16]. Recently, 14-3-3 proteins have been reported to be the targets of bacterial effectors, and the interactions between 14-3-3 proteins and effectors affect bacterial virulence [10,17,18].
Rice (Oryza sativa L.) is the staple food for over half of the world population, but its production and quality are severely threatened by various diseases. Bacterial blight and blast, caused by the bacteria Xathomonas oryzae pv. oryzae (Xoo) and the fungus Magnaporthe oryzae Barr (M. oryzae), respectively, are the most destructive rice diseases, and can lead to a 30%-50% yield loss when these diseases become epidemic [19]. In our previous studies, we found that the overexpression of OsGF14e enhances resistance to panicle blast, whereas silencing OsGF14e results in increased susceptibility to panicle blast in rice [20]. OsGF14b positively regulates panicle blast resistance but negatively regulates leaf blast. Further results showed that OsGF14b is a target of WRKY71, which positively regulates rice resistance to panicle blast [21]. Moreover, a multi-omics study revealed that OsGF14b strongly activated the gibberellin, auxin and jasmonic acid signaling pathways during pathogen infection [22]. An earlier study demonstrated that OsGF14f was significantly up-regulated during the rice–M. oryzae and rice–Xoo interactions [4], suggesting OsGF14f may play important roles in biotic stresses in rice. However, all these suggested roles are based on its changes in expression level, and the actual functions of OsGF14f in biotic stresses in rice remain to be confirmed.
In the present study, the biological functions of OsGF14f in leaf blast and bacterial blight resistance and the underlying molecular mechanisms were investigated using OsGF14f-overexpressing rice plants and their wild-type plants. Our results suggest that OsGF14f functions as a positive regulator to modulate disease resistance in a quantitative manner, and OsGF14f-mediated disease resistance is associated with the activation of the SA signaling pathway.
2. Results
2.1. Expression Patterns of OsGF14f and Subcellular Localization of OsGF14f
To provide insight into the function of OsGF14f during different physiological processes, tissue-specific expression pattern was examined in various tissues. As shown in Figure 1A, OsGF14f was ubiquitously expressed in all tested tissues, with the highest expression in 14-day-old leaf, and the lowest expression in 10-15 cm panicle. To understand the functions of OsGF14f in blast resistance, quantitative RT–PCR analysis was performed to monitor OsGF14f gene expression at 6, 12, 24 and 48 h after leaf blast inoculation. The qRT–PCR data showed that the transcription of OsGF14f was rapidly and significantly induced by the infection of the blast pathogen (Figure 1B). To examine the subcellular localization of OsGF14f, we transiently co-expressed a GF14f-GFP fusion construct with a nuclear marker (NLS-mCherry) in Arabidopsis protoplasts. As shown in Figure 1C, GF14f-GFP was mainly localized in the cytoplasm, with some occurrence in the nucleus (Figure 1C).
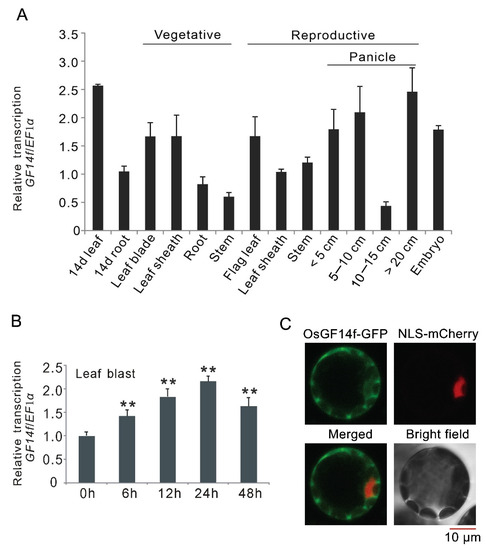
Figure 1.
Expression patterns of OsGF14f and subcellular localization of OsGF14f. (A) Expression patterns of OsGF14f in different tissues of Nipponbare plants. (B) Quantitative RT-PCR analysis of the response of OsGF14f to leaf blast infection. The values 3 h, 6 h, 9 h, 12 h, 24 h and 48 h indicate the time after blast inoculation. ** p < 0.01. (C) Subcellular localization of OsGF14f. GF14f-GFP and the nuclear marker (NLS-mCherry) were co-transformed into Arabidopsis protoplasts. Fluorescence signal was detected by laser confocal microscopy after 16 h of transfection.
2.2. Overexpression of OsGF14f Enhances Resistance to Leaf Blast in Rice
To confirm the function of OsGF14f in blast resistance in rice, transgenic rice plants constitutively overexpressing OsGF14f (OXGF14f), under the control of the CaMV 35S promoter, were produced in Nipponbare. Three independent homozygous lines (OX-2, OX-3 and OX-5), with increased transcription of GF14f were selected for disease evaluation (Figure 2A). We inoculated 2-week-old OXGF14f plants with the blast isolate GD08-T13, which showed strong virulence to Nipponbare plants, by the spray-inoculation method. The diseased leaf area ranged from 29.61% to 56.02% (average, 39.23%) in transgenic plants, significantly lower than that in wild-type plants (77.35%) (p < 0.01) (Figure 2B,C). To precisely quantify the OsGF14f-mediated quantitative blast resistance, we inoculated the OXGF14f plants with the same isolate by the punch method. Our results showed that the lesions on the leaves of OXGF14f plants were significantly smaller than that on the leaves of Nipponbare plants (p < 0.01) (Figure 2D,E). Moreover, the spore number on the leaves of OXGF14f plants was also significantly less than that on Nipponbare leaves (p < 0.01) (Figure 2F). These results suggest that the overexpression of OsGF14f can improve quantitative resistance against leaf blast in rice.
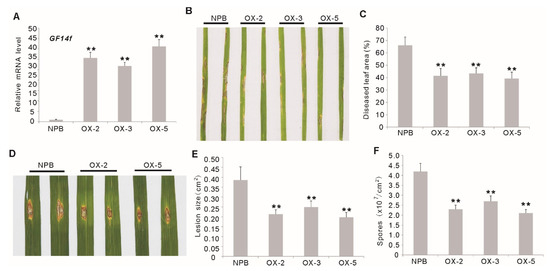
Figure 2.
Disease phenotypes of the OsGF14f-overexpressing (OXGF14f) plants (OX-2, OX-3, OX-5) after leaf blast infection. (A) Transcription analysis of OsGF14f in the OXGF14f plants by quantitative RT-PCR. ** p < 0.01. (B) Disease phenotypes of the OXGF14f plants and Nipponbare plants after spraying inoculation with M. oryzae isolate GD08-T13 for six days. (C) Diseased leaf area (DLA) in the OXGF14f plants and Nipponbare plants after leaf blast inoculation for 6 days. Error bars indicate the SD from at least ten biological replicates. ** p < 0.01. (D) Disease phenotypes of the OXGF14f plants and Nipponbare plants after inoculation with M. oryzae isolate GD08-T13 using punch method at 6 weeks after sowing. (E) Relative lesion size in the OXGF14f plants and Nipponbare plants after leaf blast inoculation. Error bars indicate the SD from at least ten biological replicates. ** p < 0.01. (F) Numbers of spores produced on the OXGF14f plants and Nipponbare plants after punch inoculation. Error bars indicate the SD from three biological replicates. ** p < 0.01.
2.3. Overexpression of OsGF14f Enhances Resistance to Bacterial Blight in Rice
Bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo), is another devastating rice disease worldwide. Previous research has suggested that blast resistance and bacterial blight resistance might share common pathways to some extent [23]. To see if the OsGF14f gene also functions in bacterial blight resistance, we evaluated the bacterial blight resistance of the OXGF14f plants by inoculation with an isolate from Chinese Xoo race 4. The results showed that the OXGF14f plants showed significantly enhanced resistance to Chinese Xoo race 4 (p < 0.01) (Figure 3A), with the lesion area percentage ranging from 14.9 to 18.3% versus 30% for Nipponbare (Figure 3B). The growth rate of bacteria on the leaves of OXGF14f plants was much slower than that of the wild-type plants at 16 days after inoculation (Figure 3C). These results suggest that the overexpression of OsGF14f enhances quantitative resistance to bacterial blight in rice.
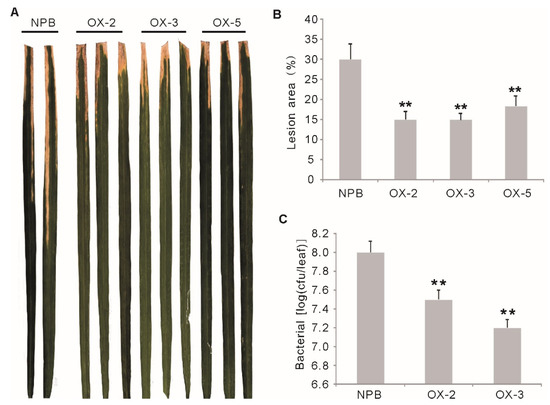
Figure 3.
Disease phenotypes of the OXGF14f plants after bacterial blight infection. (A) Disease phenotypes of the OXGF14f plants and Nipponbare plants after Xoo inoculation. (B) Relative lesion area in the OXGF14f plants and control plants after Xoo inoculation. Error bars indicate the SD from at least ten biological replicates. ** p < 0.01. (C) The growth of the bacterial from Xoo race 4 in leaves of the OXGF14f plants and Nipponbare plants. Bacterial populations were determined from three leaves at 14 days after inoculation by counting colony-forming units (cfu). Similar results were obtained in two independent biological experiments. Error bars indicate the SD from six biological replicates. ** p < 0.01.
2.4. OsGF14f Positively Regulates the SA-Dependent Pathway Instead of JA Pathway
Jasmonic acid (JA) and salicylic acid (SA) are the major defense signaling compounds mediating disease resistance in plants [24]. To determine whether these signaling compounds are involved in OsGF14f-mediated defense responses, we analyzed the expression patterns of some well-characterized defense-related genes, which are associated with JA- or SA-dependent pathways, in both the Nipponbare and the OsGF14f transgenic plants before and after blast infection. Two LOX (lipoxygenase, LOX1 and LOX11) genes and AOS2 (allene oxide synthase 2), which are involved in the JA-dependent pathway, and two pathogenesis-related PR (PR1a and PR10) genes, NHI (Arabidopsis NPR1 homolog 1) and PAL1 (phenylalanine ammonia-lyase), which are associated with the SA-dependent pathway [25] were selected for the study. The blast pathogen infection strongly induced the expression of PAL1, NH1, PR1a and PR10, both in Nipponbare and in OsGF14f-overexpressing plants (Figure 4). The expression levels of PAL1, PR10 and PR1a were significantly higher in OXGF14f plants than in Nipponbare plants, either before or after infection (p < 0.01) (Figure 4). Although the expression level of NH1 was not significantly changed in the transgenic plants compared to that in control plants before inoculation (Figure 4), its transcript was strongly induced in OXGF14f plants compared to that in control plants after inoculation (Figure 4). There was no obvious difference in the expression levels of LOX1, LOX11 and AOS2 between transgenic plants and wild-type plants, either before or after pathogen inoculation (Supplemental Figure S1). These results suggest that OsGF14f-mediated blast resistance may be involved in SA-dependent instead of JA-dependent signaling pathways.
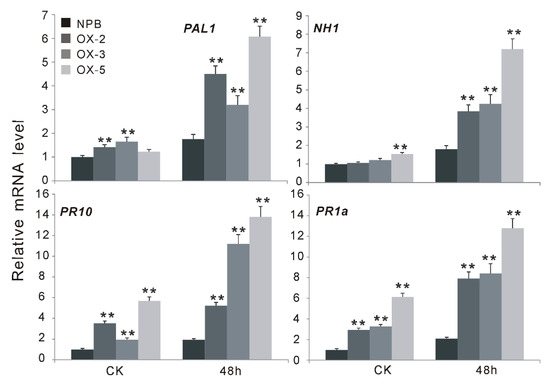
Figure 4.
The expression changes of the genes associated with the salicylic acid (SA) signaling pathway before and after blast infection. Overexpression of OsGF14f significantly induced the expression of SA responsive genes and the SA synthesis-related genes. ** p < 0.01.
2.5. OsGF14f Is Induced by SA Treatment and Overexpression of OsGF14f Can Increase Endogenous SA Levels
To further support the involvement of OsGF14f in SA-dependent signaling pathways, exogenous SA treatment was applied, and the expression changes of OsGF14f in wild-type plants at different time points (3, 6, 12 and 24 h) were monitored using qRT-PCR. Our results showed that the transcription levels of OsGF14f increased at all four time points after SA treatment in Nipponbare, with the peak at 24 h (Figure 5). In addition, we also observed that the endogenous SA level in OXGF14f plants was significantly increased after blast infection, while it remained at the same level in wild-type plants; and the SA level was significantly higher (p < 0.05) in OXGF14f plants than that in wild-type plants, both before and after blast infection (Figure 6). Together, these results suggest that OsGF14f positively regulates the SA-dependent pathway.
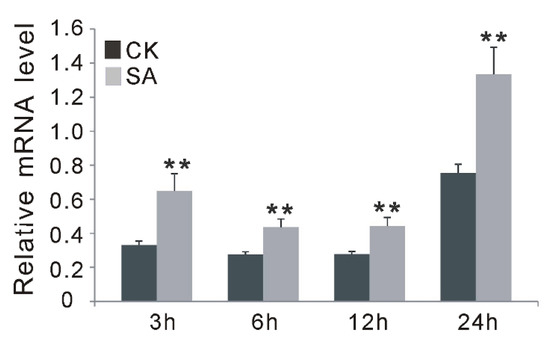
Figure 5.
Time-course expression analysis of OsGF14f after SA and water (CK) treatments by quantitative RT-PCR analysis in Nipponbare plants. ** p < 0.01.
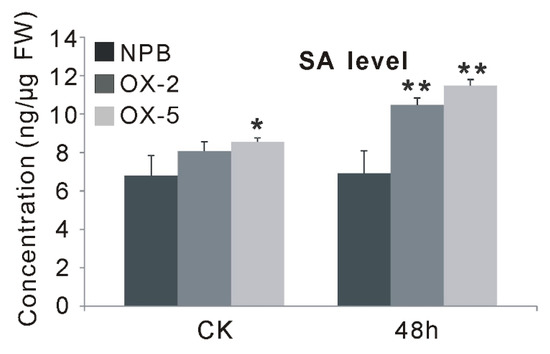
Figure 6.
The SA levels of the OXGF14f and wild-type plants before and after blast infection. * p < 0.05, ** p < 0.01.
3. Discussion
3.1. OsGF14f Enhances Blast and Bacterial Blight Resistance in Rice in a Quantitative Manner
It is well-known that plants fight against blast invasion via two different resistance strategies: qualitative (complete) resistance, mediated by major disease resistance (R) genes, and quantitative (partial) resistance, contributed by multiple genes or quantitative trait loci (QTL) [26,27]. In most cases, qualitative resistance mediated by R genes is highly efficient, but it is race-specific and easily overcome, owing to the rapid evolution of pathogens [27,28]. In contrast, quantitative resistance conferred by quantitative trait loci (QTL) is presumably non-race-specific and is generally considered to be more broad-spectrum and durable under natural conditions [26]. Thus, quantitative resistance has been considered as a preferred strategy in disease control in rice [29]. Over the past two decades, numerous QTLs for quantitative blast and bacterial blight resistance have been identified. However, these sources have not been effectively used for the improvement of blast and bacterial blight resistance in rice because the genes of underlying resistance QTLs are unknown. Therefore, the identification and isolation of the genes of underlying resistance QTLs is the key for effective molecular breeding for quantitative disease resistance. In the present study, our results showed that enhanced disease resistance correlated with increasing transcription levels of OsGF14f (Figure 1, Figure 2 and Figure 3). Moreover, the OsGF14f-overexpressing rice plants displayed less diseased leaf area, smaller lesion size, fewer spores during blast infection, lower percent lesion length and fewer spores during bacterial blight infection compared with the wild-type plants (Figure 2 and Figure 3), exhibiting the nature of quantitative disease resistance. These results suggest that OsGF14f confers blast and bacterial blight resistance in a quantitative manner, and is a good target in molecular breeding for durable disease resistance in rice.
3.2. OsGF14f-Mediated Disease Resistance Is Involved in the SA-Dependent Pathway
SA is an archetypal defense hormone and its importance in the hard wiring of the plant innate immune system is well documented, particularly in the model plant Arabidopsis [30]. In this study, we showed that the expression levels of the defense-related genes, including PAL1, NH1, PR1a and PR10 which are involved in the SA-dependent pathway, were significantly higher in OsGF14f transgenic plants than in Nipponbare plants, both before and after pathogen infection (Figure 4). Moreover, the transcript of OsGF14f was significantly induced by exogenous SA, and the endogenous SA levels were significantly higher (p < 0.05) in OXGF14f plants than those in wild-type Nipponbare plants, both before and after fungal infection (Figure 5 and Figure 6). These results suggest that OsGF14f mediated disease resistance is involved in the SA-dependent pathway. This is consistent with the previous findings, that the resistance to biotrophic and hemi-biotrophic pathogens is frequently controlled by the SA-dependent pathway [31]. We also found that the expression of LOX1, LOX11 and AOS2 is not changed between transgenic plants and wild-type plants, either before or after pathogen inoculation (Supplemental Figure S1), indicating that the JA-dependent pathway might function scarcely in OsGF14f-mediated disease resistance. Interestingly, in our previous study, we identified that OsGF14b, another member of the 14-3-3 family, was induced by blast infection and positively regulates panicle blast resistance in rice [20]. However, OsGF14b-mediated disease resistance is associated with the activation of the JA-dependent pathway and suppression of the SA-dependent pathway [21], different to the OsGF14f-mediated disease resistance pathway. These results suggest that there might be divergent functions and regulatory mechanisms in disease resistance among the members of the 14-3-3 family.
In conclusion, we have confirmed the functions of OsGF14f in response to blast and bacterial blight infection through gene expression analysis, a transgenic method and physiology analysis, and its possible regulatory mechanisms underlying these processes were also investigated. Our results indicate that OsGF14f positively regulates leaf blast and bacterial blight resistance in rice. OsGF14f-mediated disease resistance is dependent on the SA signaling pathway. Since blast and bacterial blight are two of the most devastating diseases in rice, and quantitative disease resistance is more durable, OsGF14f is a good target in rice improvement for disease resistance.
4. Materials and Methods
4.1. Plant Materials and Pathogens
The OsGF14f-overexpressing rice plants and their wild-type plants, Nipponbare, were used in this study. Rice seeds were surface-sterilized and transferred to Murashige and Skoog medium [32] and incubated in a growth chamber under light of 200 μmol/m2/s with a 12 h photoperiod at 26 °C. After germination, rice seedlings were transplanted into soil and kept in a greenhouse. Blast (M. oryzae) isolate GD08-T13 collected from Guangdong, China, was used for rice blast inoculation, and Chinese Xoo race 4 isolate collected from Guangdong, China, was used for bacterial blight inoculation.
4.2. Pathogen Inoculation and Evaluation of Disease Resistance
The inoculum of M. oryzae isolate GD08-T13 was prepared as described by Liu et al. (2018) [33]. The revived isolate GD08-T13 was cultured on prune agar plates (three pieces prune, 5 g lactose, 1 g yeast extract, and 20 g agar bar in 1 L, pH of 6–6.5, autoclaved) for 7 d at room temperature, and mycelial growth was scraped with a sterilized spatula and exposed to fluorescent light for 4–5 days to induce sporulation. Spores were collected and suspended in water to reach a concentration of 1 × 106 mL−1. To examine the effect of rice blast inoculation on the expression of OsGF14f, conidia of GD08-T13 were washed and suspended at 1 × 106 cells mL−1 in sterile water, and sprayed on the 14-day-old Nipponbare plants, which were then incubated at 26 °C in a growth chamber (16,000 Lux and 100% relative humidity) in the dark for 24 h, followed by 12 h light and 12 h dark cycles.
Two inoculation methods were used in evaluation of blast resistance in this study. For spray inoculation, two-week-old rice seedlings were sprayed with a spore suspension of GD08-T13 (1 × 106 spores/mL) containing 0.05% Tween-20. Inoculated plants were maintained in the same growth chamber used for transcription analysis at 26 °C, 16,000 Lux and 100% relative humidity in the dark for 24 h. Subsequently, the growth chamber was set to a photoperiod of 2 h light and 12 h dark at 26 °C and 100% relative humidity. Disease was assessed 6 d after inoculation by measuring the diseased leaf area. For punch inoculation, six-week-old plants were punched as described previously [34,35]. Briefly, 5 μL of spore suspension of GD08-T13 (5 × 105 spores/mL) containing 0.05% Tween-20 was added to the press-injured spots on fully expanded rice leaves; the inoculated spots were wrapped with transparent Scotch tape, and inoculated leaves were photographed at 14 days after inoculation, and lesion size was measured using ImageJ (http://rsbweb.nih.gov/ij/, accessed on 8 July 2021). For determination of in planta sporulation after punch inoculation, leaf strips containing a lesion spot were excised and submerged in 100 μL of distilled water, in a 1.5 mL microcentrifuge tube. After the suspension was vigorously mixed, spores were counted with a microscope. To evaluate bacterial blight disease, plants were inoculated with Chinese Xoo race 4 (OD600 = 0.3) isolate at the booting stage by the leaf-clipping method [36]. Disease was scored by measuring the percent lesion length (lesion length/leaf length) at 12 d after inoculation.
4.3. Plasmid Construction and Rice Transformation
To generate the overexpression vector of OsGF14f, we obtained the full-length coding region of the gene from Nipponbare leaf cDNA by PCR using primers GF14f-OE-F/R (Supplemental Table S1), and cloned the fragment into the PHQSN (modified from pCAMBIA1390), harboring a CaMV 35S promoter. The plasmid was transferred into the calli induced from the immature seeds of Nipponbare, using the Agrobacterium tumefaciens-mediated rice transformation method as previously described [37].
4.4. Subcellular Localization
The full-length cDNA of OsGF14f was constructed into the pCAMBIA1300-GFP to generate pUBQ10: OsGF14f-GFP construct. The OsGF14f-GFP and NLS-mCherry plasmid were co-transformed into Arabidopsis protoplasts for transient expression following previously described method [38]. After incubation in darkness for 16 h, images were captured using a confocal fluorescence microscope (LSM 710).
4.5. Gene Expression Analysis
Total RNA was extracted from rice tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and was treated with DNaseI (Takara, Dalian, China) to remove genomic DNA contamination. The first strand of cDNA was synthesized from 1 μg of total RNA using the primescriptTM RT reagent kit (Takara, Dalian, China) according to the manufacturer’s instructions. Quantitative PCR was performed using SYBR Premix ExTaqTM (Takara, Dalian, China) on a CFX Connect real-time PCR detection system (Bio-Rad, Hercules, CA, USA), and data were analyzed using Bio-Rad CFX Manager 3.1 (Bio-Rad, Hercules, CA, USA). EF1a was used as the internal control. Gene-specific primers that were used are listed in Supplemental Table S1.
4.6. SA Treatment and Measurement
To examine the effect of rice blast inoculation on the expression of OsGF14f, two-week-old rice seedlings were sprinkled with 100 μM hormone solution, and sampled at 3 h, 6 h, 12 h and 24 h after treatment.
The hormones SA were extracted from the rice leaf [39], and then 10 μL of SA sample solution was injected onto a C18 column (AQUITY UPLC BEH 130, 1.7 μm, 2.1 by 100 mm, Waters) at a flow rate of 0.1 mL/min, and the column was maintained at 30 °C. The sample solution was separated by reversed-phase ultra-fast LC (Shimadzu, Kyoto, Japan) with a multi-step linear gradient elution over 30 min. The eluate was then introduced into the electrospray ion source of a tandem triple quadrupole MS analyzer (API4000, AB SCIEX, Foster City, CA, USA), and the SA compound was quantified in the multiple reaction monitoring (MRM) mode using optimized MS/MS conditions, which are listed in Supplemental Table S2. The Analyst 1.5.2 software (AB SCIEX, Foster City, CA, USA) was used to control the instrument, and to acquire and process all of the MS data.
4.7. Statistical Analysis
All the graphic data are presented as mean ± SD. Student’s t-test was used to test the significance of difference: * and ** indicate a significant difference at p < 0.05 and p < 0.01, respectively.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23137440/s1.
Author Contributions
Y.M., Q.L. and B.L. conceived and designed this research; Y.M., Q.L. performed the experiment and analyzed the data; J.Y., J.D., S.Z., W.Y., J.Z., T.Y., L.C., L.Z., J.W., J.C., W.L., W.W. assisted in experiments and discussed the results. Y.M., Q.L. and B.L. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Guangdong Province (grant number 2019A1515010003, 2022A1515012361) to Y.M., the National Natural Science Foundation of China (grant number 31901441) to Y.M., the Innovation Team Project of Guangdong Modern Agricultural Industrial System (grant number 2021KJ106, 2022KJ106) to B.L., Science and Technology Program of Guangzhou (grant number 202002030375) to Y.M., Special Fund for Scientific Innovation Strategy—Construction of High Level Academy of Agriculture Science (grant number R2021PY-QF001 to Y.M., R2020PY-JX001 to Q.L.), Guangdong Key Laboratory of New Technology in Rice Breeding (grant number 2020B1212060047).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sharma, R.; De Vleesschauwer, D.; Sharma, M.K.; Ronald, P.C. Recent Advances in Dissecting Stress-Regulatory Crosstalk in Rice. Mol. Plant 2013, 6, 250–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Capel, J.; Leyva, A.; Salinas, J. Two related low-temperature-inducible genes of Arabidopsis encode proteins showing high homology to 14-3-3 proteins, a family of putative kinase regulators. Plant Mol. Biol. 1994, 25, 693–704. [Google Scholar] [CrossRef]
- Chen, F.; Li, Q.; Sun, L.; He, Z. The Rice 14-3-3 Gene Family and its Involvement in Responses to Biotic and Abiotic Stress. DNA Res. 2006, 13, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Rosenquist, M.; Sehnke, P.; Ferl, R.J.; Sommarin, M.; Larsson, C. Evolution of the 14-3-3 Protein Family: Does the Large Number of Isoforms in Multicellular Organisms Reflect Functional Specificity? J. Mol. Evol. 2000, 51, 446–458. [Google Scholar] [CrossRef]
- Roberts, M.; Bowles, D.J. Fusicoccin, 14-3-3 Proteins, and Defense Responses in Tomato Plants1. Plant Physiol. 1999, 119, 1243–1250. [Google Scholar] [CrossRef] [Green Version]
- Cooper, B.; Clarke, J.D.; Budworth, P.; Kreps, J.; Hutchison, D.; Park, S.; Guimil, S.; Dunn, M.; Luginbühl, P.; Ellero, C.; et al. A network of rice genes associated with stress response and seed development. Proc. Natl. Acad. Sci. USA 2003, 100, 4945–4950. [Google Scholar] [CrossRef] [Green Version]
- Oh, C.-S.; Pedley, K.F.; Martin, G.B. Tomato 14-3-3 Protein 7 Positively Regulates Immunity-Associated Programmed Cell Death by Enhancing Protein Abundance and Signaling Ability of MAPKKK α. Plant Cell 2010, 22, 260–272. [Google Scholar] [CrossRef] [Green Version]
- Manosalva, P.M.; Bruce, M.; Leach, J.E. Rice 14-3-3 protein (GF14e) negatively affects cell death and disease resistance. Plant J. 2011, 68, 777–787. [Google Scholar] [CrossRef]
- Teper, D.; Salomon, D.; Sunitha, S.; Kim, J.-G.; Mudgett, M.B.; Sessa, G. Xanthomonas euvesicatoria type III effector XopQ interacts with tomato and pepper 14-3-3 isoforms to suppress effector-triggered immunity. Plant J. 2013, 77, 297–309. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, S.; Liu, B. 14-3-3 proteins: Macro-regulators with great potential for improving abiotic stress tolerance in plants. Biochem. Biophys. Res. Commun. 2016, 477, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Seehaus, K.; Tenhaken, R. Cloning of genes by mRNA differential display induced during the hypersensitive reaction of soybean after inoculation with Pseudomonas syringae pv. glycinea. Plant Mol. Biol. 1998, 38, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, W.-M.; Coleman, M.; Orgil, U.; Feng, J.; Ma, X.; Ferl, R.; Turner, J.G.; Xiao, S. Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 2009, 60, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Denison, F.C.; Paul, A.-L.; Zupanska, A.K.; Ferl, R.J. 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 2011, 22, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Elmayan, T.; Fromentin, J.; Riondet, C.; Alcaraz, G.; Blein, J.-P.; Simon-Plas, F. Regulation of reactive oxygen species production by a 14-3-3 protein in elicited tobacco cells. Plant Cell Environ. 2007, 30, 722–732. [Google Scholar] [CrossRef]
- Oh, C.-S.; Martin, G.B. Tomato 14-3-3 Protein TFT7 Interacts with a MAP Kinase Kinase to Regulate Immunity-associated Programmed Cell Death Mediated by Diverse Disease Resistance Proteins. J. Biol. Chem. 2011, 286, 14129–14136. [Google Scholar] [CrossRef] [Green Version]
- Taylor, K.W.; Kim, J.-G.; Su, X.B.; Aakre, C.D.; Roden, J.A.; Adams, C.M.; Mudgett, M.B. Tomato TFT1 Is Required for PAMP-Triggered Immunity and Mutations that Prevent T3S Effector XopN from Binding to TFT1 Attenuate Xanthomonas Virulence. PLOS Pathog. 2012, 8, e1002768. [Google Scholar] [CrossRef] [Green Version]
- Lozano-Durán, R.; Robatzek, S. 14-3-3 Proteins in Plant-Pathogen Interactions. Mol. Plant-Microbe Interact. 2015, 28, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Ashkani, S.; Rafii, M.Y.; Shabanimofrad, M.; Miah, G.; Sahebi, M.; Azizi, P.; Tanweer, F.A.; Akhtar, M.S.; Nasehi, A. Molecular Breeding Strategy and Challenges Towards Improvement of Blast Disease Resistance in Rice Crop. Front. Plant Sci. 2015, 6, 886. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Yang, J.; Zhang, S.; Zhao, J.; Feng, A.; Yang, T.; Wang, X.; Mao, X.; Dong, J.; Zhu, X.; et al. OsGF14e positively regulates panicle blast resistance in rice. Biochem. Biophys. Res. Commun. 2016, 471, 247–252. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, J.; Zhang, S.; Zhao, J.; Feng, A.; Yang, T.; Wang, X.; Mao, X.; Dong, J.; Zhu, X.; et al. OsGF14b Positively Regulates Panicle Blast Resistance but Negatively Regulates Leaf Blast Resistance in Rice. Mol. Plant-Microbe Interact. 2016, 29, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Liu, Q.; Naake, T.; Huang, W.; Chen, M.; Kong, Q.; Zhang, S.; Li, W.; Li, X.; Liu, Q.; et al. OsGF14b modulates defense signaling pathways in rice panicle blast response. Crop J. 2020, 9, 725–738. [Google Scholar] [CrossRef]
- Hu, K.-M.; Qiu, D.-Y.; Shen, X.-L.; Li, X.-H.; Wang, S.-P. Isolation and Manipulation of Quantitative Trait Loci for Disease Resistance in Rice Using a Candidate Gene Approach. Mol. Plant 2008, 1, 786–793. [Google Scholar] [CrossRef] [Green Version]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef]
- Deng, H.; Liu, H.; Li, X.; Xiao, J.; Wang, S. A CCCH-Type Zinc Finger Nucleic Acid-Binding Protein Quantitatively Confers Resistance against Rice Bacterial Blight Disease. Plant Physiol. 2011, 158, 876–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kou, Y.; Wang, S. Broad-spectrum and durability: Understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 2010, 13, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liu, H.; Li, Y.; Yu, H.; Li, X.; Xiao, J.; Wang, S. Manipulating Broad-Spectrum Disease Resistance by Suppressing Pathogen-Induced Auxin Accumulation in Rice. Plant Physiol. 2011, 155, 589–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, N.; Inoue, H.; Kato, T.; Funao, T.; Shirota, M.; Shimizu, T.; Kanamori, H.; Yamane, H.; Hayano-Saito, Y.; Matsumoto, T.; et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010, 64, 498–510. [Google Scholar] [CrossRef]
- De Vleesschauwer, D.; Gheysen, G.; Höfte, M. Hormone defense networking in rice: Tales from a different world. Trends Plant Sci. 2013, 18, 555–565. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2008, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Liu, Q.; Yan, S.; Huang, W.; Yang, J.; Dong, J.; Zhang, S.; Zhao, J.; Yang, T.; Mao, X.; Zhu, X.; et al. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol. Biol. 2018, 98, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Ono, E.; Wong, H.L.; Kawasaki, T.; Hasegawa, M.; Kodama, O.; Shimamoto, K. Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 2001, 98, 759–764. [Google Scholar] [CrossRef]
- Ding, B.; Bellizzi, M.D.R.; Ning, Y.; Meyers, B.; Wang, G.-L. HDT701, a Histone H4 Deacetylase, Negatively Regulates Plant Innate Immunity by Modulating Histone H4 Acetylation of Defense-Related Genes in Rice. Plant Cell 2012, 24, 3783–3794. [Google Scholar] [CrossRef] [Green Version]
- Kauffman, H.E.; Reddy, A.P.K.; Hsieh, S.P.Y.; Merca, S.D.; U.S. Agricultural Research Service Plant Science Research Division. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 1973, 57, 537–541. [Google Scholar]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef]
- Yang, C.; Ma, Y.; He, Y.; Tian, Z.; Li, J. OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling. Plant J. 2017, 93, 489–501. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Welti, R.; Wang, X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 986–992. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).