Accurate Receptor-Ligand Binding Free Energies from Fast QM Conformational Chemical Space Sampling
Abstract
1. Introduction
2. Results and Discussion
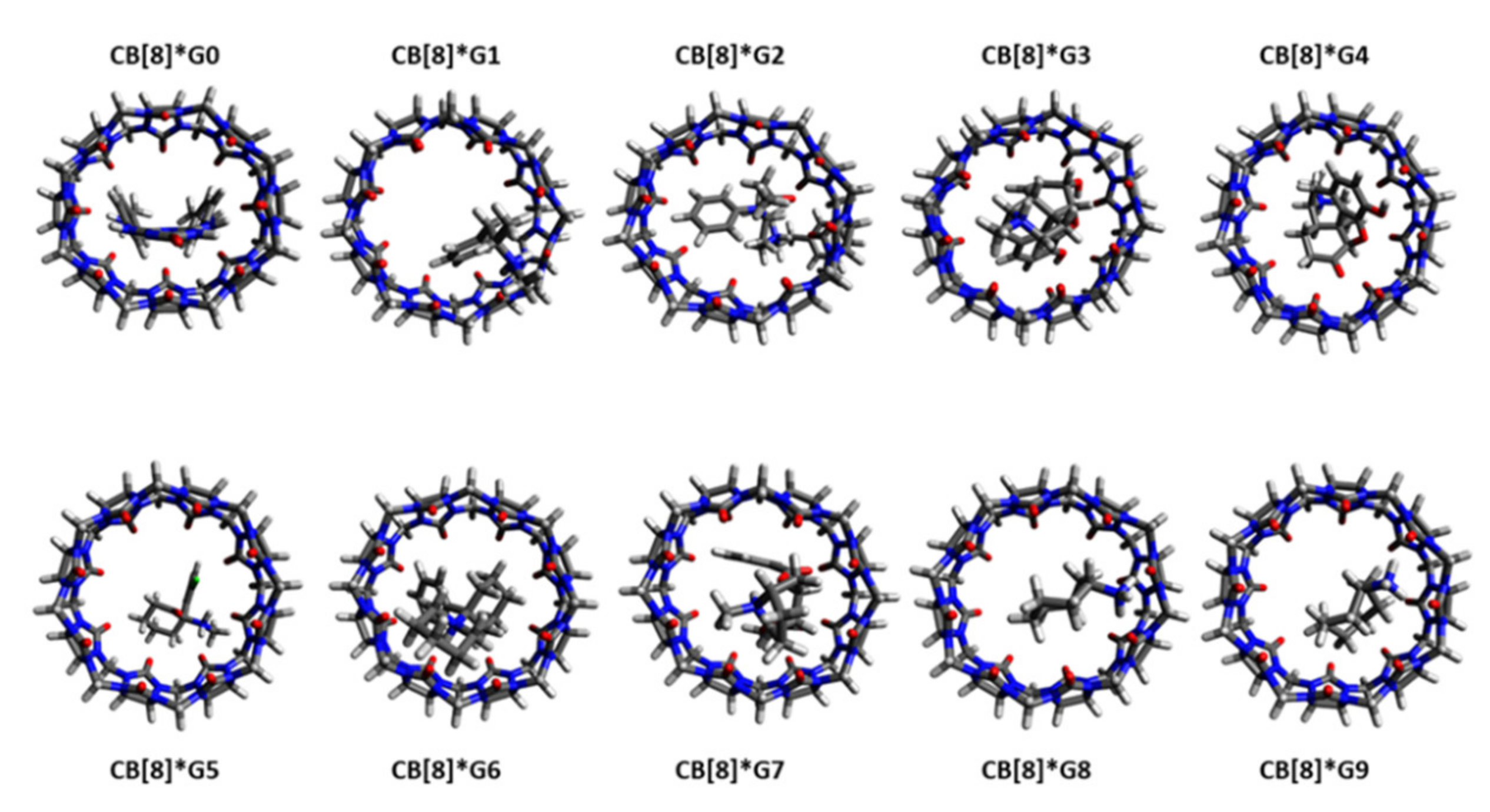
2.1. Structural Parameters of Host and Host-Guest Complexes
2.2. The Protonation State of G0
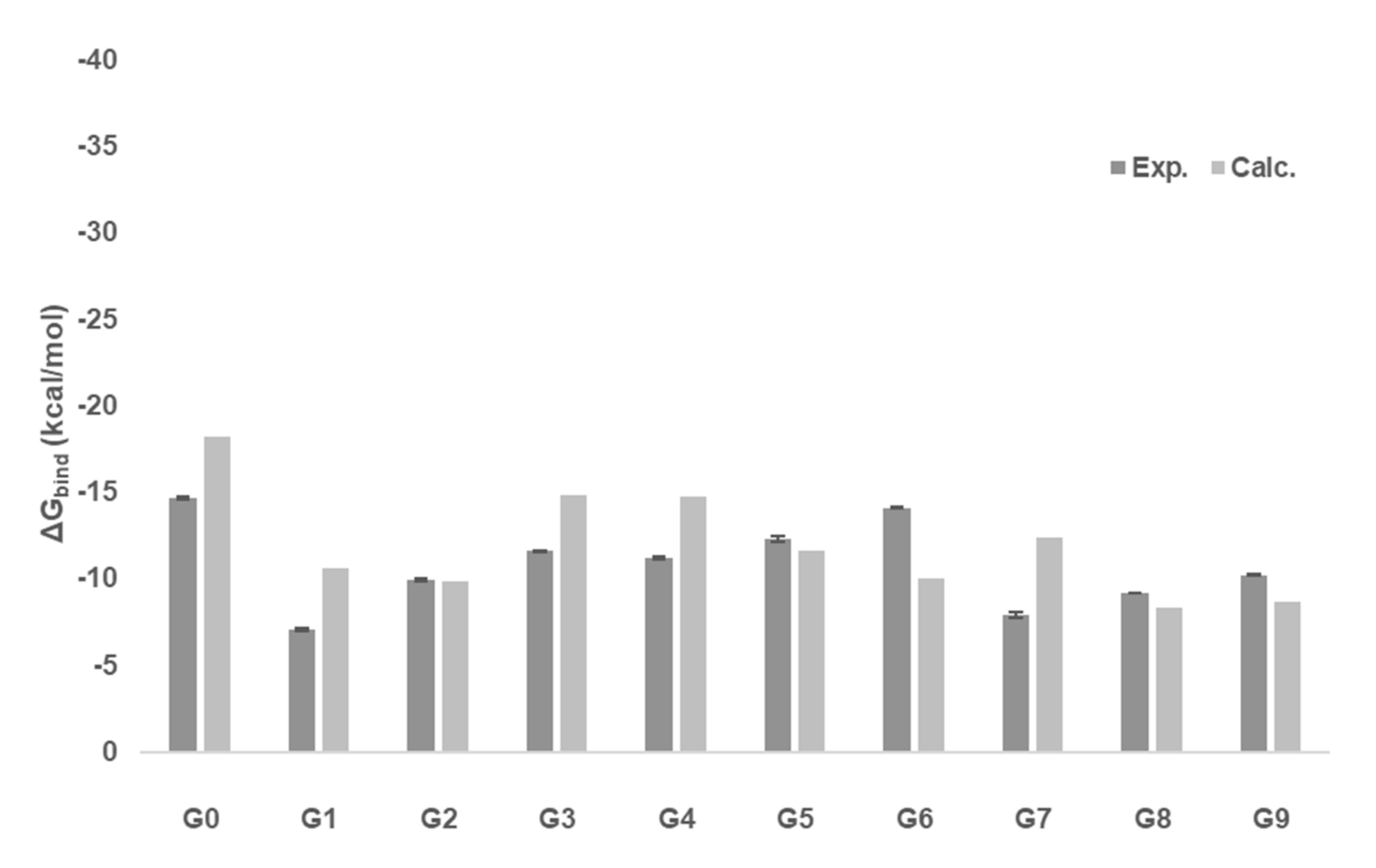
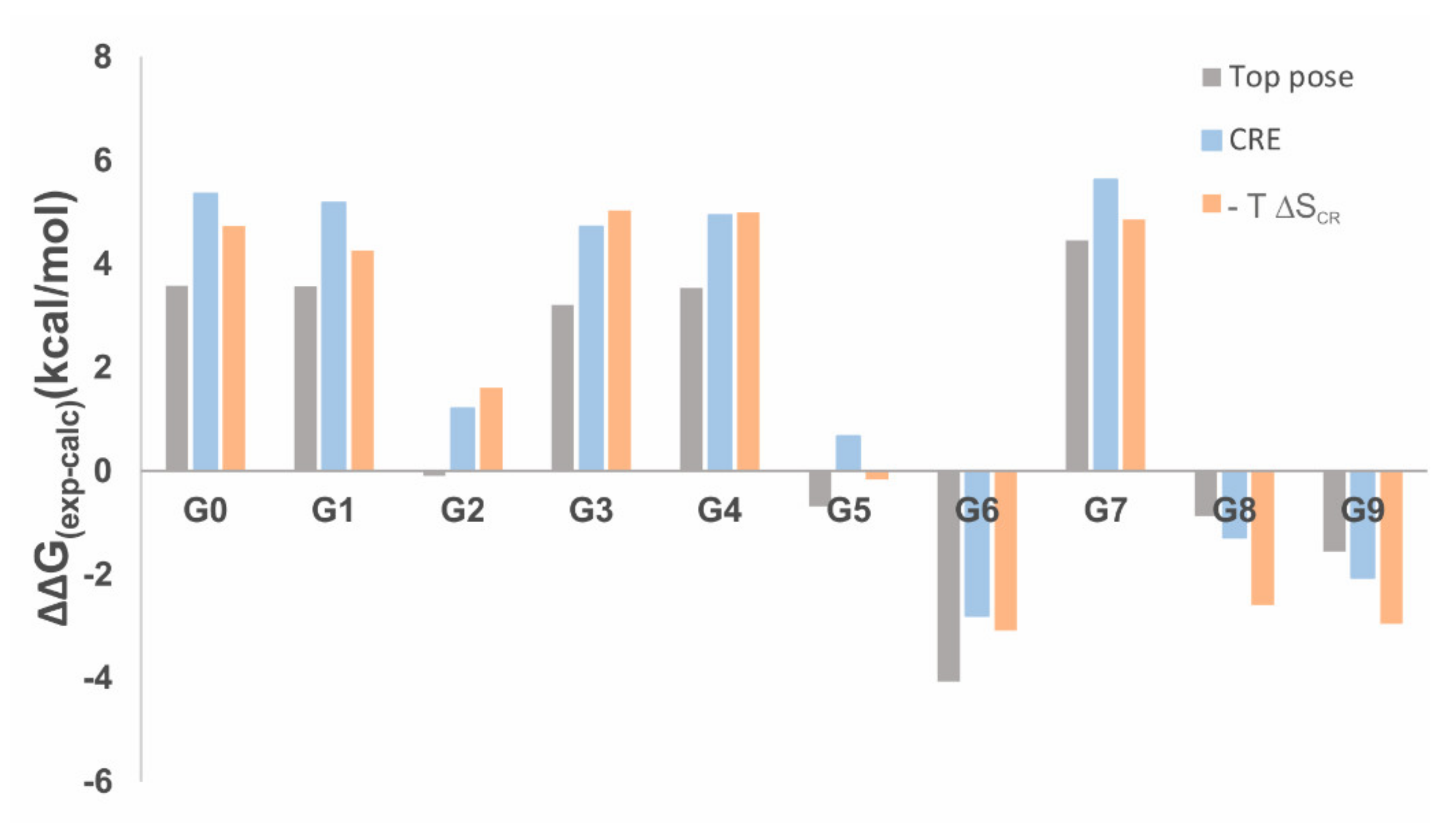
2.3. Gibbs Free Energies of Ligand-Receptor Binding
2.4. Free Energies of Binding of Cyclic Anamines to CB[8]
2.5. Comparison of GFN2-xTB Results with DFT
3. Materials and Methods
3.1. Host-Guest Starting Structures
3.2. Structural Optimization and Free Energies of Binding
3.3. The Conformer/Rotamer Ensemble
3.4. DFT Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mobley, D.L.; Gilson, M.K. Predicting binding free energies: Frontiers and benchmarks. Annu. Rev. Biophys. 2017, 46, 531–558. [Google Scholar] [CrossRef]
- Rosenbaum, M.I.; Clemmensen, L.S.; Bredt, D.S.; Bettler, B.; Strømgaard, K. Targeting receptor complexes: A new dimension in drug discovery. Nat. Rev. Drug Discov. 2020, 19, 884–901. [Google Scholar] [CrossRef]
- Yang, S.Q.; Ye, Q.; Ding, J.J.; Yin, M.Z.; Lu, A.P.; Chen, X.; Hou, T.J.; Cao, D.S. Current advances in ligand-based target prediction. WIREs Comput. Mol. Sci. 2020, e1504. [Google Scholar] [CrossRef]
- Rubio-Ruiz, B.; Serrán-Aguilera, L.; Hurtado-Guerrero, R.; Conejo-García, A. Recent advances in the design of choline kinase α inhibitors and the molecular basis of their inhibition. Med. Res. Rev. 2020, 41, 902–927. [Google Scholar] [CrossRef] [PubMed]
- Homeyer, N.; Stoll, F.; Hillisch, A.; Gohlke, H. Binding free energy calculations for lead optimization: Assessment of their accuracy in an industrial drug design context. J. Chem. Theory Comput. 2014, 10, 3331–3344. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, A.; Hayes, J.M.; Stein, M.; Piras, P.; Roussel, C. Theoretical reassessment of Whelk-O1 as an enantioselective receptor for 1-(4-halogeno-phenyl)-1-ethylamine derivatives. Chirality 2004, 16, S1–S11. [Google Scholar] [CrossRef]
- Hayes, J.M.; Stein, M.; Weiser, J. Accurate calculations of ligand binding free energies: Chiral separation with enantioselective receptors. J. Phys. Chem. A 2004, 108, 3572–3580. [Google Scholar] [CrossRef]
- Ragusa, A.; Rossi, S.; Hayes, J.M.; Stein, M.; Kilburn, J.D. Novel enantioselective receptors for N-protected glutamate and aspartate. Chem. A Eur. J. 2005, 11, 5674–5688. [Google Scholar] [CrossRef] [PubMed]
- Gilson, M.K.; Zhou, H.X. Calculation of protein-ligand binding affinities. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 21–42. [Google Scholar] [CrossRef]
- Lamb, M.L.; Jorgensen, W.L. Computational approaches to molecular recognition. Curr. Opin. Chem. Biol. 1997, 1, 449–457. [Google Scholar] [CrossRef]
- Jorgensen, W.L. The many roles of computation in drug discovery. Science 2004, 303, 1813–1818. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, A.; Jensen, T.; Slochower, D.R.; Aldeghi, M.; Gapsys, V.; Ntekoumes, D.; Bosisio, S.; Papadourakis, M.; Henriksen, N.M.; de Groot, B.L.; et al. The SAMPL6 sampling challenge: Assessing the reliability and efficiency of binding free energy calculations. J. Comput. Aided Mol. Des. 2020, 34, 601–633. [Google Scholar] [CrossRef]
- Rizzi, A.; Murkli, S.; McNeill, J.N.; Yao, W.; Sullivan, M.; Gilson, M.K.; Chiu, M.W.; Isaacs, L.; Gibb, B.C.; Mobley, D.L.; et al. Overview of the SAMPL6 host-guest binding affinity prediction challenge. J. Comput. Aided Mol. Des. 2018, 32, 937–963. [Google Scholar] [CrossRef]
- Yin, J.; Henriksen, N.M.; Slochower, D.R.; Shirts, M.R.; Chiu, M.W.; Mobley, D.L.; Gilson, M.K. Overview of the SAMPL5 host-guest challenge: Are we doing better? J. Comput. Aided Mol. Des. 2017, 31, 1–19. [Google Scholar] [CrossRef]
- Muddana, H.S.; Daniel Varnado, C.; Bielawski, C.W.; Urbach, A.R.; Isaacs, L.; Geballe, M.T.; Gilson, M.K. Blind prediction of host–guest binding affinities: A new SAMPL3 challenge. J. Comput. Aided Mol. Des. 2012, 26, 475–487. [Google Scholar] [CrossRef]
- Kim, J.; Jung, I.-S.; Kim, S.-Y.; Lee, E.; Kang, J.-K.; Sakamoto, S.; Yamaguchi, K.; Kim, K. New cucurbituril homologues: Syntheses, isolation, characterization, and X-ray crystal structures of cucurbit[n]uril (n = 5, 7, and 8). J. Am. Chem. Soc. 2000, 122, 540–541. [Google Scholar] [CrossRef]
- Barrow, S.J.; Kasera, S.; Rowland, M.J.; del Barrio, J.; Scherman, O.A. Cucurbituril-based molecular recognition. Chem. Rev. 2015, 115, 12320–12406. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, F.; Scherman, O.A. Cucurbit[8]uril mediated donor-acceptor ternary complexes: A model system for studying charge-transfer interactions. J. Phys. Chem. B 2012, 116, 2842–2849. [Google Scholar] [CrossRef]
- Liu, S.; Ruspic, C.; Mukhopadhyay, P.; Chakrabarti, S.; Zavalij, P.Y.; Isaacs, L. The cucurbit[n]uril family: Prime components for self-sorting systems. J. Am. Chem. Soc. 2005, 127, 15959–15967. [Google Scholar] [CrossRef]
- Murkli, S.; McNeill, J.N.; Isaacs, L. Cucurbit 8 uril center dot guest complexes: Blinded dataset for the SAMPL6 challenge. Supramol. Chem. 2019, 31, 150–158. [Google Scholar] [CrossRef]
- Guan, D.; Lui, R.; Matthews, S. LogP prediction performance with the SMD solvation model and the M06 density functional family for SAMPL6 blind prediction challenge molecules. J. Comput. Aided Mol. Des. 2020, 34, 511–522. [Google Scholar] [CrossRef]
- Sun, Z.X.; He, Q.L.; Li, X.; Zhu, Z.D. SAMPL6 host-guest binding affinities and binding poses from spherical-coordinates-biased simulations. J. Comput. Aided Mol. Des. 2020, 34, 589–600. [Google Scholar] [CrossRef]
- Papadourakis, M.; Bosisio, S.; Michel, J. Blinded predictions of standard binding free energies: Lessons learned from the SAMPL6 challenge. J. Comput. Aided Mol. Des. 2018, 32, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Eken, Y.; Patel, P.; Diaz, T.; Jones, M.R.; Wilson, A.K. SAMPL6 host-guest challenge: Binding free energies via a multistep approach. J. Comput. Aided Mol. Des. 2018, 32, 1097–1115. [Google Scholar] [CrossRef] [PubMed]
- Song, L.F.; Bansal, N.; Zheng, Z.; Merz, K.M. Detailed potential of mean force studies on host-guest systems from the SAMPL6 challenge. J. Comput. Aided Mol. Des. 2018, 32, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Mikulskis, P.; Genheden, S.; Rydberg, P.; Sandberg, L.; Olsen, L.; Ryde, U. Binding affinities in the SAMPL3 trypsin and host-guest blind tests estimated with the MM/PBSA and LIE methods. J. Comput. Aided Mol. Des. 2012, 26, 527–541. [Google Scholar] [CrossRef]
- Grimme, S. Supramolecular binding thermodynamics by dispersion-corrected density functional theory. Chem. A Eur. J. 2012, 18, 9955–9964. [Google Scholar] [CrossRef]
- Sure, R.; Antony, J.; Grimme, S. Blind prediction of binding affinities for charged supramolecular host–guest systems: Achievements and shortcomings of DFT-D3. J. Phys. Chem. B 2014, 118, 3431–3440. [Google Scholar] [CrossRef]
- Pracht, P.; Bohle, F.; Grimme, S. Automated exploration of the low-energy chemical space with fast quantum chemical methods. Phys. Chem. Chem. Phys. 2020, 22, 7169–7192. [Google Scholar] [CrossRef]
- Schwabe, T.; Grimme, S. Double-hybrid density functionals with long-range dispersion corrections: Higher accuracy and extended applicability. Phys. Chem. Chem. Phys. 2007, 9, 3397–3406. [Google Scholar] [CrossRef] [PubMed]
- Murkli, S.; Klemm, J.; Brocket, A.T.; Shuster, M.; Briken, V.; Roesch, R.M.; Isaacs, L. In vitro and in vivo sequestration of phencyclidine by Me4Cucurbit [8] uril. Chem. A Eur. J. 2021, 27, 3098–3105. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, L. CB8-DOA-SAMPL-Answer-Sheet-20201014.Pdf. Available online: https://github.com/samplchallenges/SAMPL8/blob/master/host_guest/Analysis/ExperimentalMeasurements/CB8-DOA-SAMPL-Answer-Sheet-20201014.pdf (accessed on 13 March 2021).
- Bannwarth, C.; Ehlert, S.; Grimme, S. GFN2-xTB—an accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J. Chem. Theory Comput. 2019, 15, 1652–1671. [Google Scholar] [CrossRef]
- Mobley, D.L.; Amezcua, M. The SAMPL8 CB8 “Drugs of Abuse” Challenge. Available online: https://github.com/samplchallenges/SAMPL8/blob/master/host_guest/CB8/README.md (accessed on 21 February 2021).
- Grimme, S.; Pracht, P. CREST Conformer-Rotamer Ensemble Sampling Tool Based on the GFN-; Mulliken Center for Theoretical Chemistry, University of Bonn: Bonn, Germany, 2020. [Google Scholar]
- Bannwarth, C.; Caldeweyher, E.; Ehlert, S.; Hansen, A.; Pracht, P.; Seibert, J.; Spicher, S.; Grimme, S. Extended tight-binding quantum chemistry methods. WIREs Comput. Mol. Sci. 2020, 11, e1493. [Google Scholar] [CrossRef]
- Grimme, S. Semi-empirical Extended Tight-Binding Program Package xtb.v- 6.3.2; Mulliken Center for Theoretical Chemistry, University of Bonn: Bonn, Germany, 2020. [Google Scholar]
- Schmitz, S.; Seibert, J.; Ostermeir, K.; Hansen, A.; Göller, A.H.; Grimme, S. Quantum chemical calculation of molecular and periodic peptide and protein structures. J. Phys. Chem. B 2020, 124, 3636–3646. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S. Exploration of chemical compound, conformer, and reaction space with meta-dynamics simulations based on tight-binding quantum chemical calculations. J. Chem. Theory Comput. 2019, 15, 2847–2862. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S. Accurate description of van der Waals complexes by density functional theory including empirical corrections. J. Comput. Chem. 2004, 25, 1463–1473. [Google Scholar] [CrossRef]
- Ernzerhof, M.; Scuseria, G.E. Assessment of the Perdew–Burke–Ernzerhof exchange-correlation functional. J. Chem. Phys. 1999, 110, 5029–5036. [Google Scholar] [CrossRef]
- Grimme, S. Semiempirical hybrid density functional with perturbative second-order correlation. J. Chem. Phys. 2006, 124, 034108. [Google Scholar] [CrossRef] [PubMed]
- Goerigk, L.; Grimme, S. Efficient and accurate double-hybrid-meta-gga density functionals—evaluation with the extended GMTKN30 database for general main group thermochemistry, kinetics, and noncovalent interactions. J. Chem. Theory Comput. 2011, 7, 291–309. [Google Scholar] [CrossRef]
- Neese, F. The Orca program system. WIREs Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Neese, F. Software update: The Orca program system, version 4.0. WIREs Comput. Mol. Sci. 2018, 8, e1327. [Google Scholar] [CrossRef]
- Neese, F. An improvement of the resolution of the identity approximation for the formation of the Coulomb matrix. J. Comput. Chem. 2003, 24, 1740–1747. [Google Scholar] [CrossRef] [PubMed]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Eichkorn, K.; Treutler, O.; Öhm, H.; Häser, M.; Ahlrichs, R. Auxiliary basis sets to approximate coulomb potentials. Chem. Phys. Lett. 1995, 240, 283–290. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal solvation model based on solute electron density and on a continuum model of the solvent defined by the bulk dielectric constant and atomic surface tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef] [PubMed]
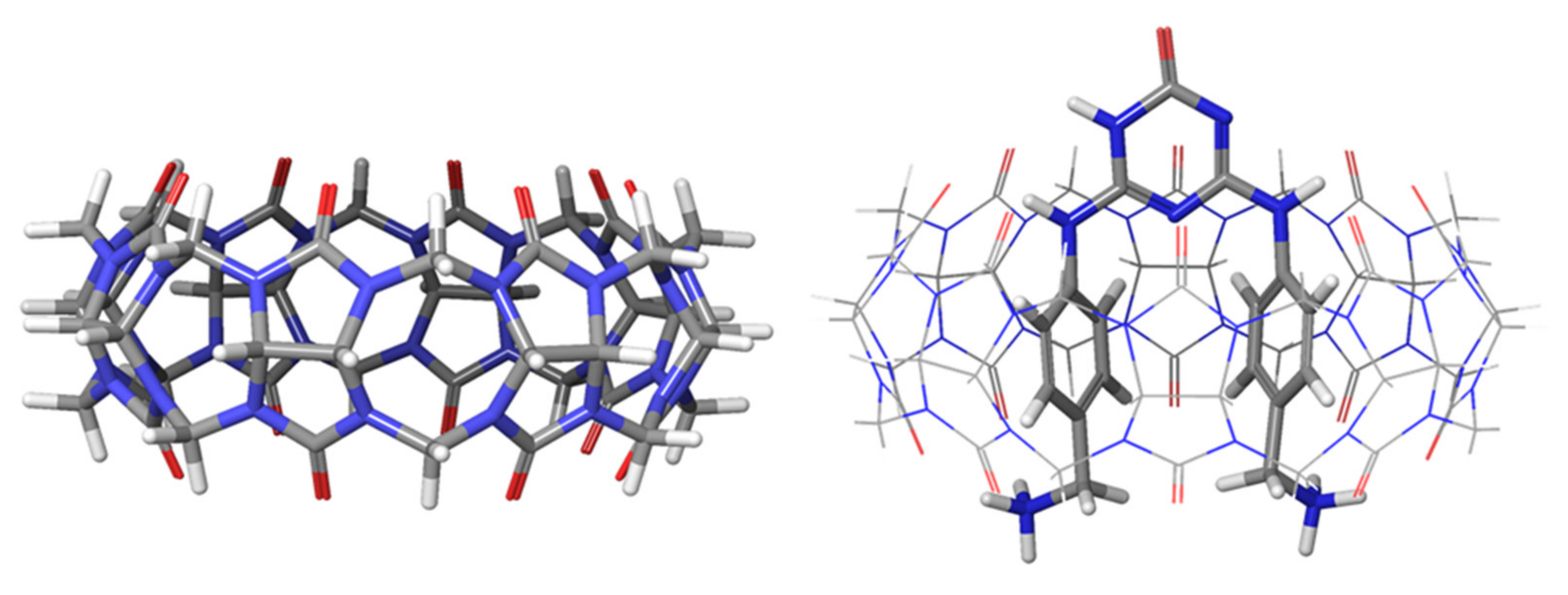
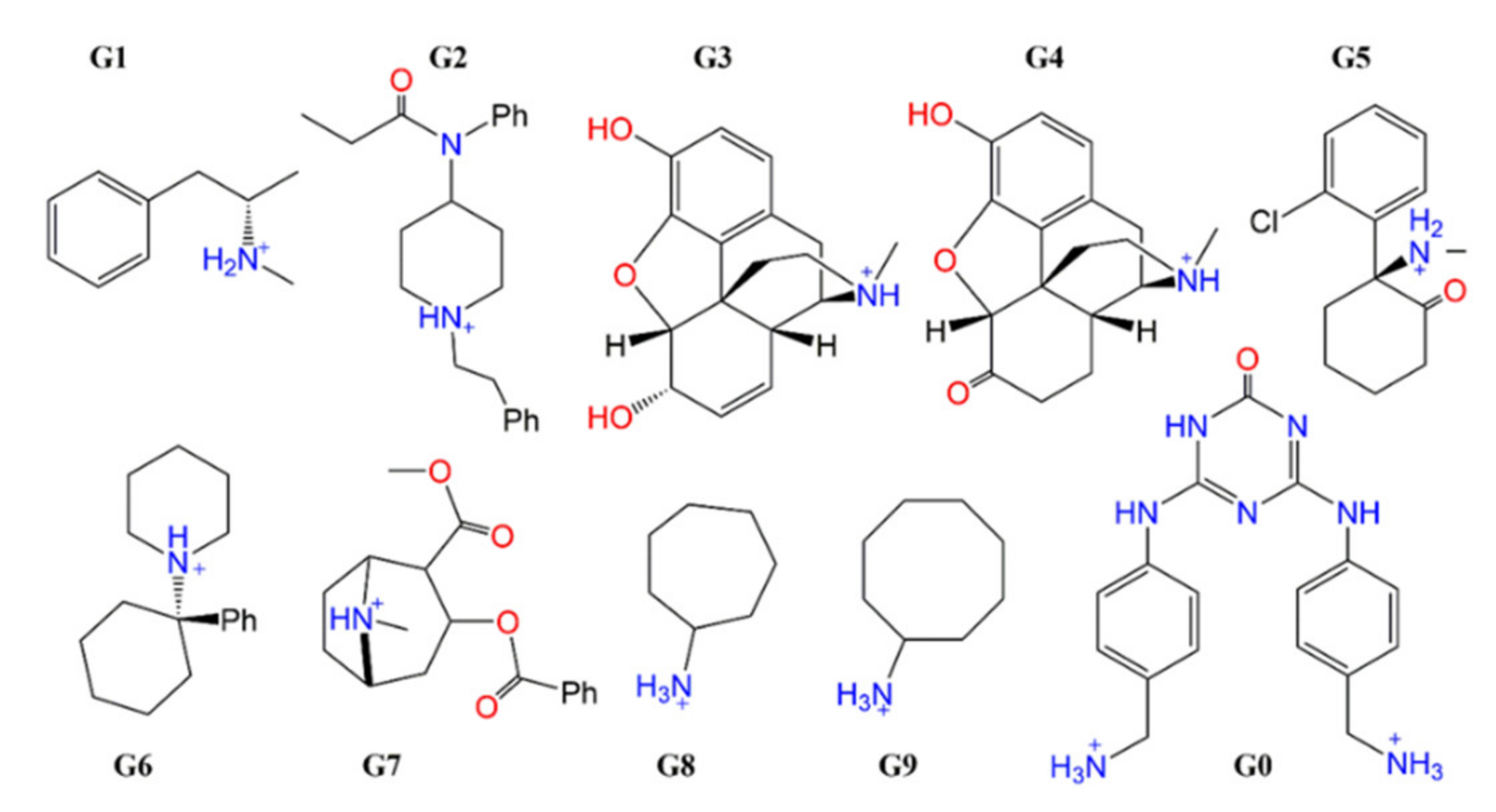
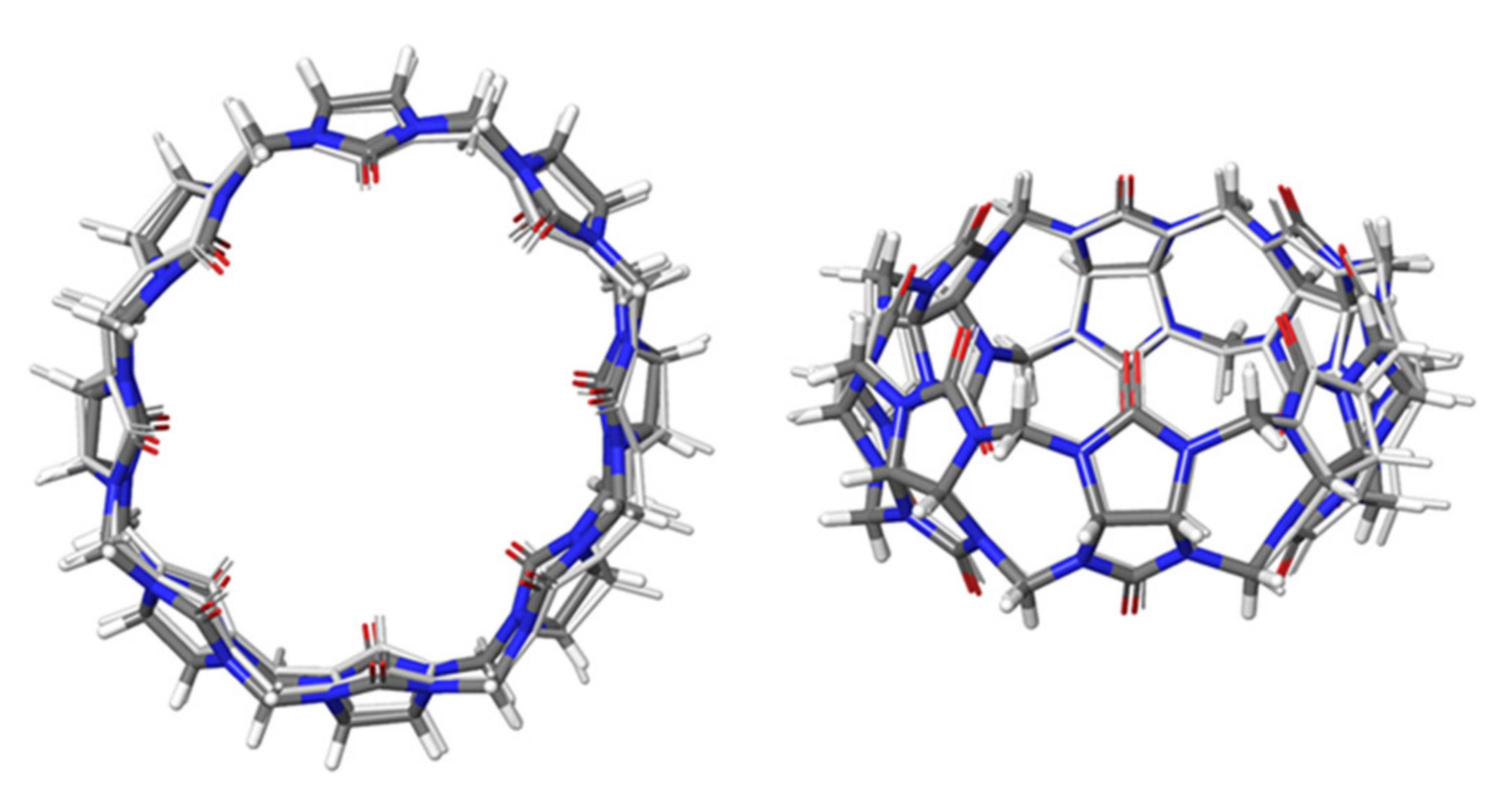
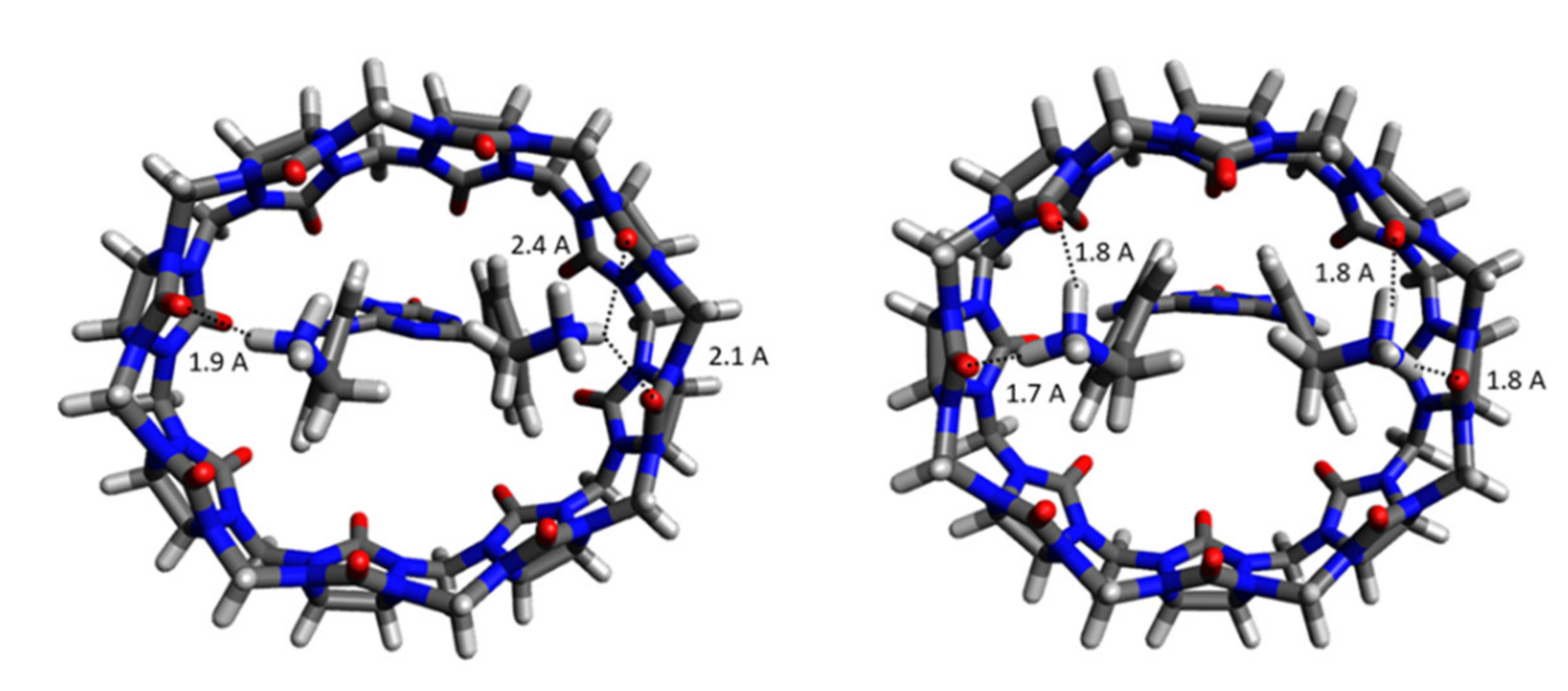

| Number of Unique Entries in CRE of Free Ligand | Number of Unique Entries in the CRE of the CB[8]*Ligand Complex | |
|---|---|---|
| G0 | 9 | 12 |
| G1 | 11 | 21 |
| G2 | 158 | 137 |
| G3 | 5 | 4 |
| G4 | 4 | 8 |
| G5 | 9 | 10 |
| G6 | 14 | 19 |
| G7 | 16 | 39 |
| G8 | 17 | 90 |
| G9 | 22 | 175 |
| Ligand | Experiment | ΔGbind | ΔGbind,ens |
|---|---|---|---|
| G0 | −14.67 ± 0.1a | −18.25 | −18.25 |
| G1 | −7.05 ± 0.1 | −10.61 | −12.22 |
| G2 | −9.94 ± 0.06 | −9.84 | −11.14 |
| G3 | −11.6 ± 0.04 | −14.80 | −16.31 |
| G4 | −11.2 ± 0.1 | −14.74 | −16.13 |
| G5 | −12.3 ± 0.16 | −11.62 | −12.95 |
| G6 | −14.1 ± 0.04 | −10.03 | −11.30 |
| G7 | −7.93 ± 0.15 | −12.38 | −13.54 |
| G8 | −9.18 ± 0.03 | −8.30 | −7.90 |
| G9 | −10.2 ± 0.04 | −8.64 | −8.13 |
| ΔGtop-pose | ΔGensemble | ΔGensemble−T ΔSCR | |
|---|---|---|---|
| RMSE | 2.97 | 3.86 | 3.77 |
| MAE | 2.56 | 3.38 | 3.43 |
| SD | 1.4 | 2.0 | 1.6 |
| CB[8]* Ligand | GFN2-xTB | PBE0 | B2PLYP | PWPB95 |
|---|---|---|---|---|
| G0 | −38.61 | −31.12 | −37.68 | −33.77 |
| G1 | −28.39 | −19.57 | −21.68 | −19.66 |
| G2 | −30.34 | −18.63 | −23.20 | −20.30 |
| G3 | −35.28 | −22.31 | −27.55 | −24.42 |
| G4 | −35.79 | −19.12 | −24.51 | −21.44 |
| G5 | −31.26 | −22.57 | −28.41 | −25.15 |
| G6 | −28.31 | −25.57 | −29.70 | −28.59 |
| G7 | −28.69 | −11.53 | −15.94 | −12.96 |
| G8 | −22.46 | −20.86 | −21.69 | −19.87 |
| G9 | −24.51 | −22.37 | −23.54 | −21.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boz, E.; Stein, M. Accurate Receptor-Ligand Binding Free Energies from Fast QM Conformational Chemical Space Sampling. Int. J. Mol. Sci. 2021, 22, 3078. https://doi.org/10.3390/ijms22063078
Boz E, Stein M. Accurate Receptor-Ligand Binding Free Energies from Fast QM Conformational Chemical Space Sampling. International Journal of Molecular Sciences. 2021; 22(6):3078. https://doi.org/10.3390/ijms22063078
Chicago/Turabian StyleBoz, Esra, and Matthias Stein. 2021. "Accurate Receptor-Ligand Binding Free Energies from Fast QM Conformational Chemical Space Sampling" International Journal of Molecular Sciences 22, no. 6: 3078. https://doi.org/10.3390/ijms22063078
APA StyleBoz, E., & Stein, M. (2021). Accurate Receptor-Ligand Binding Free Energies from Fast QM Conformational Chemical Space Sampling. International Journal of Molecular Sciences, 22(6), 3078. https://doi.org/10.3390/ijms22063078








