Abstract
Plant growth and development are challenged by biotic and abiotic stresses including salinity and heat stresses. For Populus simonii × P. nigra as an important greening and economic tree species in China, increasing soil salinization and global warming have become major environmental challenges. We aim to unravel the molecular mechanisms underlying tree tolerance to salt stress and high temprerature (HT) stress conditions. Transcriptomics revealed that a PsnNAC036 transcription factor (TF) was significantly induced by salt stress in P. simonii × P. nigra. This study focuses on addressing the biological functions of PsnNAC036. The gene was cloned, and its temporal and spatial expression was analyzed under different stresses. PsnNAC036 was significantly upregulated under 150 mM NaCl and 37 °C for 12 h. The result is consistent with the presence of stress responsive cis-elements in the PsnNAC036 promoter. Subcellular localization analysis showed that PsnNAC036 was targeted to the nucleus. Additionally, PsnNAC036 was highly expressed in the leaves and roots. To investigate the core activation region of PsnNAC036 protein and its potential regulatory factors and targets, we conducted trans-activation analysis and the result indicates that the C-terminal region of 191–343 amino acids of the PsnNAC036 was a potent activation domain. Furthermore, overexpression of PsnNAC036 stimulated plant growth and enhanced salinity and HT tolerance. Moreover, 14 stress-related genes upregulated in the transgenic plants under high salt and HT conditions may be potential targets of the PsnNAC036. All the results demonstrate that PsnNAC036 plays an important role in salt and HT stress tolerance.
1. Introduction
Various abiotic stresses such assoil salinity, drought, extreme temperature, and heavy metals affect plant growth, development, and productivity [1]. Many important economic trees are particularly sensitive to these environmental stresses, such as Populus. simonii × P. nigra. P. simonii × P. nigra is an endemic plant found in the Yellow River basin and the northern part of mainland in China [2]. It has been used as an urban afforestation tree species in northeast and northwest China. Importantly, the wood is widely used for paper, fiber, matchsticks, and building materials [3]. In recent years, the influence of adverse environmental conditions on the growth of P. simonii × P. nigra has become more and more severe. Therefore, it is urgent to improve the stress tolerance of this important wood plant through molecular genetics and breeding.
The NAC (NAM, ATAF, and CUC) family is one of the most important transcription factor (TF) gene families in plants. Most NAC TFs are reported to participate in regulation of plant growth and developmental processes [4], including shoot apical meristem formation [5], seed and embryo development [6], lateral root development [7], cell division [8], and leaf senescence [9]. Besides development, NAC TFs also play vital roles in plant response to abiotic stresses, such as salinity, heat, cold, and drought [10]. There are 138 NAC TFs in Arabidopsis thaliana, 289 in Populus trichocarpa, and 42 in Nicotiana tabacum according to PlantTFDB (http://planttfdb.gao-lab.org/family.php?fam=NAC accessed on 4 March 2021). Regarding Arabidopsis, there are a large number of studies of NAC TFs in stress responses. For example, ANAC019, ANAC055, and ANAC072 were induced by drought, salinity, and abscisic acid (ABA), and they play a vital role in ABA signaling and osmotic stress [11]. ANAC078 was confirmed to positively regulate anthocyanin biosynthesis under high light [12]. ANAC002 (ATAF1) can be induced by drought, salinity, ABA, methyl jasmonate, and wounding. Overexpression of the ATAF1 led to increased Arabidopsis sensitivity to ABA, salt and oxidative stresses [13]. In recent years, NAC TFs have been characterized in poplar to be associated with a stress response. For example, NAC13 was significantly induced in the roots of 84K poplar by salt stress, and NAC13 overexpression enhanced poplar salt tolerance [14]. In addition, PeNAC036 in P. euphratica was strongly induced by drought, ABA, and salt, and played a positive role in abiotic stress responses [15]. Transgenic Arabidopsis overexpressing a poplar NAC57 gene displayed higher seed germination, superoxide dismutase, and peroxidase activities under salt stress than wild type (WT) plants [16]. Therefore, studies of poplar NAC TFs are important to understand the molecular mechanisms of stress response and how to enhance stress tolerance in woody plants.
NAC proteins contain a highly conserved NAM domain at the N-terminus, which can be divided into five subdomains known as A-E, and a highly divergent activation domain at the C-terminus [17]. Based on the conserved N-terminal NAM domain, the NAC family was divided into 18 subfamilies from NAC-a to r in populous [10]. The PsnNAC036 in this study belongs to the NAC-d subfamily. It was cloned and its relative expression levels were analyzed under various treatments. A total of eight transgenic tobacco lines overexpressing the TF were obtained. Morphological and biochemical measurements indicate that PsnNAC036 can enhance salinity and HT tolerance of transgenic tobacco plants. RT-qPCR results showed that overexpression of PsnNAC036 upregulated the expression of 14 stress-related genes in tobacco. These results demonstrate that PsnNAC036 plays an important role in improving plant salinity and HT tolerance.
2. Results
2.1. Transcript Analysis of 289 NAC TF Genes in Populus
To analyze changes of expression levels of the NAC TFs, the fragment per kilobases per million reads (FPKM) of 289 NAC transcription factor genes from Populus simonii × P. nigra leaves were retrieved from an RNA-seq dataset [18], which has been deposited in NCBI SRA (accession SRP267437). The salt-stress responsive NAC TFs were visualized using a heatmap (Figure 1A). Based on a fold change of more than 1.2 and a false discovery rate (FDR) smaller than 0.05, 37 differentially expressed NACs were identified (Figure 1B). Among these genes, PsnNAC036 was significantly upregulated after salt stress treatment of the P. simonii × P. nigra seedlings compared to control.
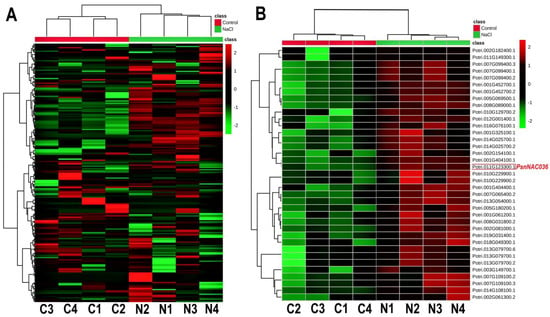
Figure 1.
Heatmaps of the relative expression of NAC genes in control and NaCl treated P. simonii × P. nigra seedlings. (A) Relative expression patterns of 289 NACs in salt stress versus control. (B) Heatmap of the 37 differentially expressed NACs. Red and green colors indicate high and low expression, respectively. The colored scale bar represents fold changes of transcription levels betweenNaCl stress and control. C1, C2, C3, and C4 represent the four biological replicates under control conditions; N1, N2, N3, and N4 represent the four biological replicates under NaCl stress.
2.2. Bioinformatics and Gene Expression Analysis of the PsnNAC036 Gene
To analyze the cDNA and encoded amino acids sequences of PsnNAC036, we isolated PsnNAC036 gene from P. simonii × P. nigra. The results showed that the coding sequence of the PsnNAC036 is 1029 bp in length (Supplementary Figure S1A), and encodes a 343 aa protein with 16.2% alpha helix, 15.8% extended strand, 4.0% beta turn, and 64.0% random coil. Five highly homologous genes were found in the NCBI database according to amino acid sequence blast, including PtrNAC036 from Populus trichocarpa, XP_011029436 from Populus euphratica which has 155 NAC TFs, PtrNAC044 from Populus trichocarpa, ANAC072 (AT4G27410.2) from A. thaliana and Nta009260 from Nicotiana tabacum (Supplementary Figure S1B). They shared 98.5, 97.1, 79.4, 70.5, and 62.0% amino acid sequence similarity with the PsnNAC036, respectively (Supplementary Figure S1C). According to the sequence alignment, these NACs all contain five highly conserved motifs MA, MB, MC, MD, and ME, which constitute a NAM domain at the N-terminus. Among them, MC and MD were known to bind to DNA [19]. In addition, they all have a predicted nuclear localization signal (NLS) region between 119–150 amino acids (Supplementary Figure S1C).
To investigate temporal expression patterns of PsnNAC036 under different stress conditions, we treated the P. simonii × P. nigra seedlings with NaCl, ABA, HT, cold, or drought, and harvested the leaves and roots at 0, 3, 6, 12, 24, and 48 h, respectively. The RT-qPCR results showed that the relative expression level of PsnNAC036 was upregulated in the leaves after different treatments, and it peaked at 12 h. In particular, the gene expression was significantly upregulated after salinity and HT treatments. The PsnNAC036 expression showed similar expression patterns in the root tissues and the relative expression level of the gene in the root reached 15.2 and 8.8 times higher under NaCl and HT at 12 h than control, respectively (Figure 2).
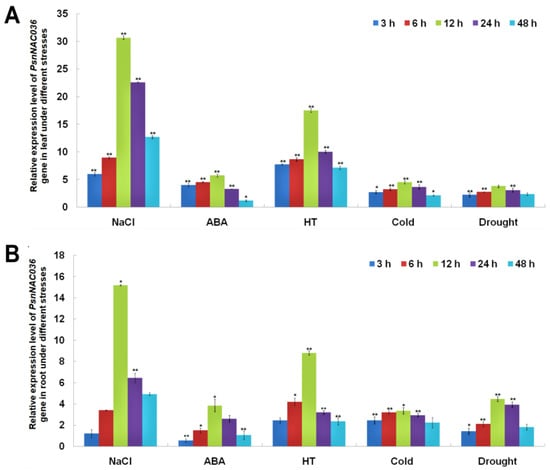
Figure 2.
Expression analysis of the PsnNAC036 under various treatments. Relative expression levels of PsnNAC036 in leaves (A) and roots (B) after NaCl (150 mM), ABA (50 µM), high temperature (HT, 37 °C), cold (4 °C), or drought treatments. Expression levels in the control samples were normalized to 1. The data are from three independent experiments. Student’s t-test: t: * p < 0.05; ** p < 0.01. Error bars indicate mean ± standard deviation.
2.3. Characterization of the PsnNAC036 Promoter Sequence
Promoters contain different transcriptional regulatory elements that can be recognized and bound by RNA polymerases and transcription factors. Some of the regulatory elements play important roles in response to external stimuli [20]. To explore the structure of PsnNAC036 promoter, the promoter sequence of PsnNAC036 (from −1 to −1726 bp) in length was isolated from the P. simonii × P. nigra. Different cis-elements in the promoter sequence were predicted (Supplementary Figure S2), including ABRE, Box 4, CGTCA-motif, G-box, I-box, W-box, MYC, O2-site, etc. by PlantCARE (Supplementary Table S1). Then the promoter was cloned into the pBI121 vector to drive the GUS gene expression. To further investigate the promoter activity, we obtained stable transgenic tobacco lines expressing GUS under the control of the PsnNAC036 promoter. GUS histochemical staining showed that only young leaves of transgenic tobacco showed a light blue color (Figure 3). The same pattern was observed under cold and drought stresses. After ABA treatment, the GUS activity was mainly expressed in the roots, while after 150 mM NaCl treatment for 12 h the GUS activity was found throughout the seedlings, especially in the leaves. A similar phenomenon was observed under HT stress, in spite of lower staining intensity than with NaCl treatment. The results confirmed that the PsnNAC036 gene was responsive to NaCl and HT stresses.
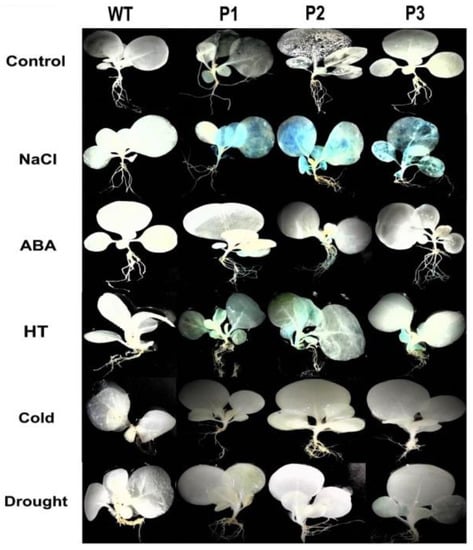
Figure 3.
GUS activity analysis of the PsnNAC036 promoter under different treatments.
2.4. PsnNAC036 Protein Is Localized to the Nucleus and Potent Activation Domain in C-terminal Domain
To determine the predicted nuclear localization of the PsnNAC036 protein (http://cello.life.nctu.edu.tw/cello2go/ accessed on 4 March 2021), the PsnNAC036 ORF without the termination codon was fused with GFP (Figure 4A). 35S::PsnNAC036-GFP and positive control 35S::GFP were transformed into onion epidermal cells by particle bombardment, respectively. As shown in Figure 4B, the fluorescent signal of PsnNAC036-GFP was only found in the nucleus; however, the fluorescent signal of the GFP control was distributed throughout the cell. The results clearly showed that the PsnNAC036 protein was indeed targeted to the nucleus.
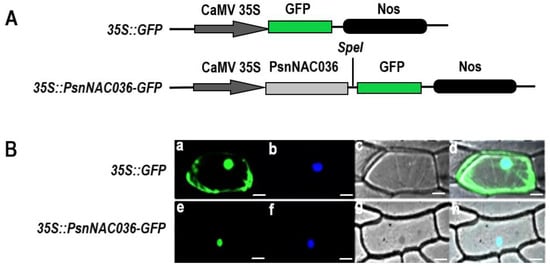
Figure 4.
Subcellular localization of the PsnNAC036 protein. (A) Schematic map of the T-DNA inserted in the 35S::GFP binary vector. (B) The 35S::PsnNAC036-GFP fusion construct and the positive control 35S::GFP plasmid were introduced into onion epidermal cells by particle bombardment. GFP fluorescence was observed by confocal laser scanning microscopy. (a,e) are fluorescence images observed in a dark field (green); (b,f) are 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) staining, which is specific for the nucleus (blue); (c,g) are light images observed in bright field; (d,h) are merged images of dark field and bright field. Scale bar = 20 μm.
To test the transcriptional activity of PsnNAC036, the full length ORF of the PsnNAC036 sequence was inserted into the pGBKT7 vector.As shown in the transactivation result (Figure 5A), PsnNAC036 clearly acted as a TF. To determine the functional domains required for activating transcription, we used a yeast assay system to test the activation domains of PsnNAC036. Deletion analysis indicates that the constructs including fragments with amino acids 191–241, 242–292, or 293–343 resulted in transactivation, strongly suggesting that the entire region of 191–343 possesses transactivation activity (Figure 5B).
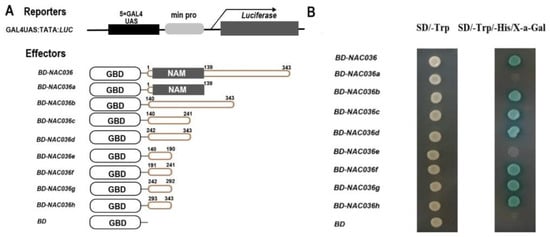
Figure 5.
Transactivation analysis of PsnNAC036 protein. Transactivation analysis of PsnNAC036 protein. (A) Schematic diagrams of the effector and reporter constructs. The reporter construct includes GAL4 binding sites UAS and minimal CaMV35S promoter (min pro) upstream from the luciferase reporter gene. The effector constructs including BD-NAC036 and GAL4-BD were fused with full-length PsnNAC036; BD-NAC036a, N-terminal domain (1–139 aa); BD-NAC036b, C-terminal domain (140–343 aa); BD-NAC036c, half of BD-NAC036b (140–241 aa); BD-NAC036d, 242–343 aa; BD-NAC036e, half of BD-NAC036c (140–190 aa); BD-NAC036f, 191–241 aa; BD-NAC036g, half of BD-NAC036d (242–292 aa); and BD-NAC036h, 51 aa of C-terminal domain (293–343 aa). BD was the negative control for pGBKT7. (B) Yeast assay.
2.5. PsnNAC036 Enhanced Tolerance to Salt and HT Stresses in Transgenic Tobacco
Transgenic tobacco lines overexpressing the PsnNAC036 (Figure 6A) and WT were screened for the presence of transgenes by gDNA PCR with the gene specific primers F1 (5′-ATGGGACTGCAAGAAACAGACC-3′) and R1 (5′-TCACTGCCTAAACCCATACCCA-3′) and RT-PCR with primers F2 (5′- CTTGAATCCTCTCGCAAAAGTG-3′) and R2 (5′-GAGCCGGTCATCAATCTCTGTC-3′). The housekeeping gene actin (aF: 5′-GCTTGCTTACATTGCTCTCGAC-3′, aR: 5′- TGCTTCCGGCTCTGATGTTGTG-3′) for RT-PCR was designed from internal actin gene (GenBank accession number: X69885). As shown in Supplementary Figure S3, PsnNAC036 fragments were amplified in the transgenic tobacco lines and had the same length as the positive control, but were not detected in the non-transgenic WT.
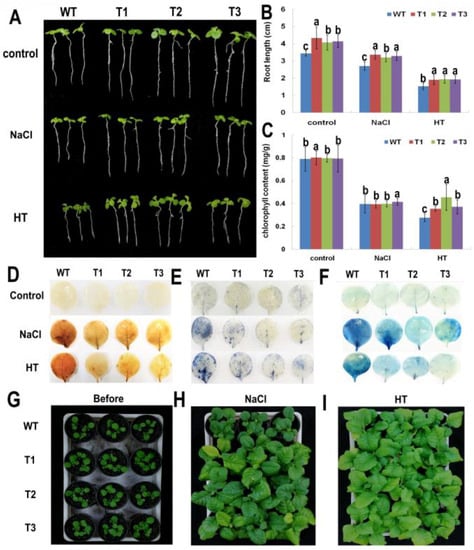
Figure 6.
Morphological analysis and histochemical staining of transgenic tobacco lines under NaCl and HT stresses. (A) Growth of PsnNAC036 transgenic lines T1, T2, and T3 seedlings in comparison with WT under control, 150 mM NaCl and 37 °C high temperature (HT) conditions. (B) Root length assay of the transgenic lines, compared to WT under control, 150 mM NaCl and HT conditions. (C) Chlorophyll content of transgenic tobacco lines and WT plants. The statistical analysis was done using a one-way analysis of variance (ANOVA) with a post-hoc with Tukey lines with an alpha value of 0.05. Different letters indicate significant difference between sites. (D) 3,3′-Diaminobenzidine (DAB) staining; (E) Nitroblue tertazolium (NBT) staining; (F) Evans blue staining; (G) Four-week-old tobacco plants in soil right before treatments; (H) 200 mM NaCl treatment for two weeks; (I) HT treatment for two weeks.
To investigate the functions of the PsnNAC036, transgenic lines T1, T2, T3, and WT were grown in MS, MS with 150 mM NaCl, and MS under 37 °C for two weeks. Under the control condition the plants all grew well. However, the root length of the transgenic lines was about 1.12 ± 0.07 times longer than WT. Under high NaCl or HT treatments, the growth of WT was obviously affected, and the size of WT was smaller than the transgenic lines (Figure 6A). Additionally, the root length of transgenic lines was approximately 1.22 ± 0.03 and 1.26 ± 0.01 times longer than WT under NaCl and HT treatments, respectively (Figure 6B). Besides, after salt treatment, the root length ratios of WT, T1, T2, and T3 (to the transgenic lines) were 72.29%, 77.58%, 78.61%, and 79.40%, respectively, compared to the control plants. Among them, T1 and T3 were more significant than WT. After HT treatment, the root length ratios of WT, T1, T2, and T3 (to the transgenic lines) were 40.95%, 43.95%, 47.53%, and 46.50%, respectively, compared to the control plants. These results demonstrate that the transgenic tobacco plants overexpressing PsnNAC036 exhibited significant NaCl and HT tolerance.
2.6. Changes in Leaf Chlorophyll Content and Physiological Indexes
Leaf chlorophyll is central for light capture and energy exchange between the biosphere and the atmosphere [21], and it serves as a physiological index closely related to plant metabolism and stress resistance [22]. To detect the changes of leaf chlorophyll content and physiological indexes in different tobacco lines, chlorophyll a and chlorophyll b were extracted and quantified from leaves of the transgenic lines and WT plants after salt and HT treatments.The results of chlorophyll content from T1, T2, and T3 lines compared with WT showed that there is no significant difference under normal and high salt conditions, while after HT treatment, the total chlorophyll content of transgenic tobacco lines was 1.29–1.65 fold higher than that of the WT plants (Figure 6C). The results of physiological parameters showed that under normal conditions, the activities of superoxide dismutase (SOD) and peroxidase (POD), as well as proline content were nearly the same in the transgenic lines and WT. However, under salt treatment, SOD and POD activities and proline content were all significantly higher in transgenic lines than in WT. MDA contents of WT were also approximately 1.2- and 1.6-times larger than those of the transgenic plants under normal and salt stress conditions, respectively (Supplementary Figure S4).
2.7. Histochemical Staining and Growth Assay of the PsnNAC036 Transgenic Plants
Histochemical staining including diaminobenzidine (DAB), nitroblue tetrazolium (NBT) and Evans blue was carried out to investigate the degree of oxidative damage and cell death. The transgenic and WT plants were treated with NaCl and HT for 12 h. Under control conditions, there was no significant difference between the transgenic and WT leaves. However, under NaCl and HT conditions, intense coloring was observed in the WT leaves after NBT, DAB, and Evans blue staining, in contrast to light staining in the transgenic lines (Figure 6D–F). The results showed that accumulation of reactive oxygen species (ROS) and cell death in the transgenic plants were greatly reduced compared to the WT NaCl and HT treatments.
Under normal growth conditions, the WT and PsnNAC036 transgenic lines grew similarly. However, after the one-month-old seedlings were treated with 200 mM NaCl or kept at 37 °C for two weeks, the growth of WT was severely retarded, compared to transgenic lines, especially after high salt treatment (Figure 6G–I).
2.8. PsnNAC036 Alters the Expression of Stress-Related Genes
To elucidate molecular functions of the PsnNAC036, relative expression levels of 14 genes related to stress response in the transgenic plants were analyzed after NaCl and HT treatments by RT-qPCR. As shown in Figure 7, these genes were significantly upregulated in the transgenic plants compared to WT under control and stress treatments. Expression of NtSOD that conferred osmotic stress tolerance was found to increase by about 26.8 folds in transgenic lines under control conditions, 85.1-fold under salt stress, and 38.7-fold under HT stress, compared to the WT. The relative expression level of NtPOD was upregulated by 4.2-fold, 31.6-fold, and 15.4-fold compared to the WT under control, salinity, and HT, respectively. Furthermore, NtP5CS and NtLEA5 involved in osmotic adjustment and membrane protection [23] were also upregulated by 1.4- and 4.3-fold in the transgenic tobacco, compared to WT, respectively (Figure 7F,H). Under high salt stress, their expression levels were upregulated by 5.4- and 10.8-fold, respectively. Under HT, their expression was significantly upregulated by 10.1- and 51.7-fold, respectively. NtPPO encodes a polyphenol oxidase involved in plant stress tolerance. Under normal conditions, the transcript of NtPPO in transgenic lines was upregulated by 5.1-fold, while under NaCl and HT, it was upregulated by 2.6- and 1.2-fold, respectively, compared to WT. Moreover, the transcript levels of NtERD10A, NtERD10B, NtERD10C, and NtERD10D [24] also showed higher expression (4.4-, 2.6-, 1.3-, and 6.5-fold, respectively) in transgenic plants compared to the WT (Figure 7H,I). The fold-change values were 3.3, 14.9, 4.0, and 9.3 under salt stress, and 12.2, 10.2, 3.9, and 10.0 under HT treatment, respectively. Also, NtHKT555, NtHKT586, and NtSOS encode Na+/H+ antiporters [25], and their expression levels were all higher in the transgenic lines than in the WT under control or treatments. Interestingly, NtNCED1, involved in the biosynthesis of SA, JA, and ABA [26], was significantly upregulated (41.9, 35.0, and 17.0 fold in the transgenic lines under control, NaCl and HT conditions, respectively). The upregulation of these stress responsive genes may account for the NaCl and HT tolerance of the transgenic plants.
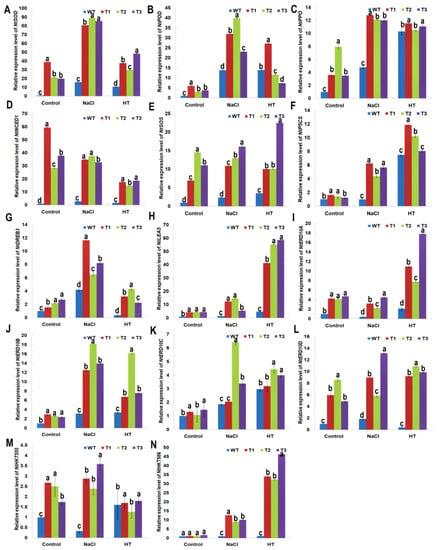
Figure 7.
Genes expression profiling of stress-responsive genes in the PsnNAC036 transgenic plants under NaCl and HT treatments. The relative fold change in expression of (A) NtSOD; (B) NtPOD; (C) NtPPO; (D) NtNCED1; (E) NtSOS; (F) NtP5CS; (G) NtDREB3; (H) NtLEA5; (I) NtERD10A; (J) NtERD10B; (K) NtERD10C; (L) NtERD10D; (M) NtHKT555; (N) NtHKT586 genes in the PsnNAC036 transgenic lines under control, high salt and heat conditions. The statistical analysis was done using a one-way ANOVA with a post-hoc with Tukey lines with an alpha value of 0.05. Different letters indicate significant difference between sites.
3. Discussions
With the intensive farming and climate change, salinity and heat have become serious environmental challenges that reduce agricultural productivity of many crops world-wide. Salinity causes osmotic stress and cellular toxicity to plants [27], and heatstress often leads to protein misfolding and degradation that will affect critical cellular reactions and processes [28]. In this study, we found that salt and heat stresses can negatively affect the growth of P. simonii × P. nigra (Supplementary Figure S5). It is therefore intriguing to investigate molecular mechanisms underlying the stress responses of this woody plant.
The NAC family first identified in Petunia is one of the largest plant-specific TF families [29]. Many NAC family members have been shown to play important roles in gene regulation under environmental stresses [30,31,32,33]. In the reference plant Arabidopsis, many studies have elucidated the functions of NAC TFs in plant response to various abiotic stresses. For example, ANAC019 functions as an upstream regulator of several drought-induced genes such as DREB2A, DREB2B, ARF2, MYB21, MYB24, and thereby plays an important role in stress response and floral development [34]. NAC TFJUNGBRUNNEN1 (ANAC042) was found to respond to H2O2 treatment and enhance plant thermo-tolerance [35]. In addition, overexpression of ANAC069 can decrease plant ROS scavenging capability and proline biosynthesis, while knock down of ANAC069 improves plant salt and osmotic stress tolerance [36], indicating ANAC069 as a negative regulator of plant stress tolerance. In Arabidopsis, the homologous gene of PsnNAC036 is ANAC072 (AT4G27410.2, RD26), which has been reported to respond to drought, salinity, ABA, SA, and MeJA [37,38]. In this study, we provide several lines of evidence, showing that the PsnNAC036 can improve plant salinity and heattolerance. Besides, the results of RT-qPCR of seedlings under ABA, cold and drought stress treatments also showed that PsnNAC036 expression can be induced by these abiotic stresses. Further studies are needed to explore the PsnNAC036 gene response to other abiotic stresses and determine whether it may have similar functions to the Arabidopsis ANAC072 homolog.
Plant promoter includes a conservative basic core promoter region and upstream cis-acting elements that are crucial to the specificity and activity of gene transcription [39]. In this study, promoter region of PsnNAC036 gene which is 1726 bp isolated. According to the analysis of PlantCARE software, the promoter contains a variety of cis-elements. Most of these elements are related to stress response, such as as-1 belongs to oxidative stress-responsive element, DRE core is involved in dehydration response, and MYC is related to chilling response. Treating PsnNAC036 promoter transgenic tobacco with salt, cold, heat, ABA, and drought, we found that the GUS activity is mainly expressed under salt and HT treatment. This suggested that there may exist other elements in the PsnNAC036 promoter that contribute to respond high salt and HT stress.
Production of ROS including the superoxide anion (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH−) are associated with plant salt stress responses [40]. MDA is a cellular indicator of lipid peroxidation when plants experience oxidative stress caused by environmental challenges, such as salinity and heat [41]. To maintain the redox homeostasis, plants regulate antioxidant enzymes and molecules [42]. SOD and POD are key antioxidant enzymes in ROS-scavenging [43]. In addition, salt stress also causes osmotic stress.Proline is known to function as an important osmolyte in adjusting cellular osmotic balance [44]. In this study, we measured several physiological parameters including SOD, POD activity, proline, and MDA contents in PsnNAC036 transgenic plants and WT under salt treatment, and found that SOD, POD, and proline content in transgenic lines were increased after stress treatment and MDA content was lower when compared to WT. These results demonstrate that PsnNAC036 can improve the salt tolerance by regulating the redox and osmotic processes. Also, since DAB, NBT, and Evans blue are dyes commonly used to detect ROS levels and cell death in plants [45,46], the staining results clearly show that overexpression of PsnNAC036 reduced ROS accumulation and cell death in plants (Figure 7). Our findings are consistent with previous reports that NAC proteins play a crucial role in mediating gene regulation of the anti-oxidative system under stress conditions, thereby conferring higher stress tolerance [47].
Salinity depresses plant growth through lowering water content and excessive accumulation of predominant ions including sodium (Na+), chloride (Cl−), calcium (Ca2+), potassium (K+), and hydrogen carbonate (HCO3−) [48]. It has been reported that salt stress can cause the accumulation of Na+ and Cl− while inhibiting the uptake of K+ and Ca2+, which would perturb osmotic homeostasis. Here we measured the H+-antiporter-related gene NtSOS and an isoform of high-affinity K+ transporter NtHKT555/586 are critical in improving the salinity tolerance of plants [49,50]. Under stress treatment, the enhanced expression levels of these genes in transgenic tobacco lines showed that PsnNAC036 might affect the expression of the genes participating in ion exchange to maintain the osmotic homeostasis in plants.
To further understand the regulatory functions of the PsnNAC036 TF, relative expression levels of 14 stress-related genes (NtSOD, NtPOD, NtPPO, NtNCED1, NtP5CS, NtDREB3, NtSOS, NtLEA5, NtERD10A/B/C/D, and NtHKT555/586) were quantified in the PsnNAC036 transgenic lines and WT under control, salinity, and heat conditions (Figure 7). NtSOD and NtPOD activities are known to be regulated at their transcription levels [51]. NtPPO is a polyphenol oxidase gene involved in stress tolerance in many plants [52]. NtNECD1 plays an important role in ABA biosynthesis [53]. NtP5CS encodes a key enzyme in proline biosynthesis [54]. NtLEA5 is known to protect osmotic adjustment membrane under stress conditions [55]. NtDREB3 regulates stress-responsive genes by interacting with specific cis-acting elements [56]. NtERD10A/B/C/D are associated with dehydration response [54]. The results clearly showed that PsnNAC036 upregulates these stress responsive genes, leading to enhanced salinity and heat stress tolerance. However, how PsnNAC036 is activated and regulates the stress-related genes is not known. Investigating how PsnNAC036 interacts with DNA and proteins would be an exciting future research direction.
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
P. simonii × P. nigra seedlings were grown on 1/2 MS (Murashige and Skoog medium) plant medium (pH5.8), in a growth chamber with a temperature of 25 ± 1 °C, light of 160 μmol/m2s and 16/8-h light/dark cycle, and relative humidity of approximately 65%. The one-month-old plants were grown hydroponically and those with new roots and leaves of similar sizes were selected for 150 mM NaCl treatment for 24 h. The control (water) and treated leaves from eight plants (four biological replicates each) were frozen in liquid nitrogen and provided to Beijing GENAWIZ Company (www.genewiz.com accessed on 4 March 2021) for RNA sequencing using Illumina HiSeq2500. RNA-Seq data analysiswas carried out as previously described [18] and the heatmap was generated using MetaboAnalyst (https://www.metaboanalyst.ca accessed on 4 March 2021, Version 5.0). The RNA-Seq data have been deposited in NCBI SRA with the accession number SRP267437.
4.2. Cloning and Sequence Analysis of PsnNAC036
Total RNA was extracted from the leaves, and then reversely transcribed into cDNA as a template for PCR amplification. Based on the RNA-Seq analysis, PsnNAC036 was significantly upregulated in response to high salt treatment. From the PlantTFDB (http://planttfdb.gao-lab.org/family.php?sp=Ptr&fam=NAC accessed on 4 March 2021, Version 5.0), we identified a highly homologous gene of PsnNAC036 with gene ID Potri.011G123300.1 and designed a pair of primers F1: 5′-ATGGGACTGCAAGAAACAGACC-3′ and R1: 5′-TCACTGCCTAAACCCATACCCA-3′ to clone the PsnNAC036 gene from P. simonii × P. nigra. The amino acid sequence of PsnNAC036 was used for multi-sequence alignment analysis by BioEdit (Version 7.2). Conserved domain and motifs analyses were conducted using MEME (http://meme-suite.org/tools/meme, Version 5.3.3). Nuclear localization signals (NLS) were predicted with cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi#opennewwindow accessed on 4 March 2021, Version 2012). A phylogenetic tree was constructed using MEGA 7.
4.3. PsnNAC036 Gene Expression Analysis
To analyze the relative expression levels of PsnNAC036 under different stresses, the seedlings were treated with NaCl, ABA, HT, cold, ordrought for 0, 3, 6, 12, 24, and 48 h, respectively. For the salt and ABA treatment, seedlings were incubated in 150 mM NaCl and 50 µM ABA solution, respectively. For drought stress, the seedlings were exposed to air on filter paper for dehydration. For high or low temperature treatment, seedlings were kept in 37 °C or 4 °C illumination incubators. There was only one variable per treatment compared to the control. Leaf and root tissues were harvested with four biological replicatesat each time point for RNA extraction and RT-qPCR. Actin (AF: 5′-ACCCTCCAATCCAGACACTG-3′; AR: 5′- TTGCTGACCGTATGAGCAAG-3′) was used as the reference gene in RT-qPCR. The relative expression level in different samples was calculated using 2−ΔΔCt method.
4.4. GUS Activity of PsnNAC036 Promoter
DNA was extracted from theleaves using a NuCleanPlantGen DNA Kit (CWBIO, Beijing, China). To clone promoter of PsnNAC036 from P. simonii × P. nigra, the promoter sequence of the PtrNAC036 gene (https://phytozome.jgi.doe.gov/jbrowse/index.html accessed on 4 March 2021, Populus trichocarpa v3.0)was taken as a reference for designing primers pF: 5′-GCGAAGCTTGTTAGGTGCCGAATCTCCGGTGTCC-3′ and pR: 5′-CGCGGATCCCGGGTGAAACCGAAATCCCGGTGGC-3′, which contain HindШ and BamHI restriction enzyme sites, respectively (underlined). The promoter sequence was input in PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ accessed on 4 March 2021) for cis-acting element prediction. The promoter sequence was cloned into a pBI121 vector to replace its CaMV35S promoter. Then the recombinant vector was transferred into Agrobacterium tumefaciens EHA105. Using a leaf disc method [57,58], it was then transformed into tobacco plants. Three homozygous transgenic lines P1, P2, and P3 and WT (control) were treated with 150 mM NaCl, 50 µM ABA, HT (37 °C), cold (4 °C), or drought for 12 h, respectively. Histochemical assays of GUS enzyme activity driven by the PsnNAC036 promoter were conducted as previously described [59].
4.5. Subcellular Localization of the PsnNAC036 Protein
The PsnNAC036 open reading frame (ORF) lacking the stop codon was cloned using primers F3:5′-GGGTCGACTGACTAGTATGGGACTGCAAGAAACAGACC-3′, and R3: TGCTCACCATACTAGTCTGCCTAAACCCATACCCACTT-3′, which contain a SpeI restriction site (underlined).Then the sequence was ligated into the linearized pBI121 vector that contains a green fluorescent protein (GFP)using an In-Fusion HD Cloning Kit (TaKaRa, China). The fusion plasmid 35S::PsnNAC036-GFP and the control plasmid 35S::GFP were introduced into onion epidermis according to a published method [14]. The GFP fluorescence was observed using a confocal laser scanning microscope (LSM 700, Zeiss, Germany). The results were reproduced three times, and each one had 10 biological replicates.
4.6. Transcriptional Activation Assay of the PsnNAC036 Protein
The ORF of PsnNAC036 (1–343amino acids (aa)) and the different truncations of the PsnNAC036 encoding the N-terminal NAM domain 1–139 aa, non-conserved domain 140–343 aa, and the short ORF fragments 140–241 aa, 242–343 aa, 140–190 aa, 191–241 aa, 242–292 aa, and 293–343 aa were inserted into the pGBKT7 vector, respectively. These constructs were named as BD-NAC036, BD-NAC036a, BD-NAC036b, BD-NAC036c, BD-NAC036d, BD-NAC036e, BD-NAC036f, BD-NAC036g, and BD-NAC036h, respectively (Figure 3). The negative control pGBKT7 plasmid (BD) and all the constructs were transformed into Y2H yeast strain. Transformants were grown for 3–5 days on a selective medium without Trp and His. β-Galactosidase assay was performed on filter lifts of the colonies to detect activation of the lacZ reporter gene.
4.7. PsnNAC036 Overexpression Vector Construction for Plant Transformation
On the basis of the PsnNAC036 sequence, we designed a pair of primers F4: 5′-GCGTCTAGAATGGGACTGCAAGAAACAGACC-3′ and R4: 5′-GCGGAGCTCTGGGTATGGGTTTAGGCAGTGA-3′, which contain XbaI and SacI restriction sites, respectively (underlined). Both the amplified fragment and the plant binary expression vector pBI121 were double digested by XbaI and SacI and then were ligated together by T4 ligase. The recombinant plasmid PsnNAC036-pBI121 was confirmed by PCR using the vector primers pBI121F1:5′-CCATCGTTGAAGATGCCTCTGC-3′ and pBI121R1:5′-CTCTTCGCTATTACGCCAGCTGG-3′. Positive clones were transferred into Agrobacterium tumefaciens EHA105 for plant genetic transformation.
4.8. Generation of PsnNAC036 Transgenic Tobacco Plants
One-month-old tobacco seedlings were used for PsnNAC036 transformation by a leaf disc method [2,19]. A.tumefaciens EHA105 containing the recombinant vector PsnNAC036-pBI121 were cultured in LB liquid medium with rifampicin (50 mg/mL) and kanamycin (50 mg/mL) until OD600 = 0.6. Tobacco leaves were cut into 1 × 1 cm squares and incubated in the bacterial solution for 15 min.The leaves were then cultured in MS media with kanamycin (100 mg/mL) for selection. Total RNA of each line wasextracted for PCR with the specific primers F1 and R1. WT was regarded as a negative control and the PsnNAC036-pBI121 plasmid as a positive control (P). Three independent homozygous transgenic lines T1, T2, and T3 were selected for subsequent experiments.
4.9. Root Length of the PsnNAC036 Transgenic Tobacco
To determine salinity and HT tolerance of thetransgenic tobacco overexpressing PsnNAC036, seeds of the transgenic lines T1, T2, T3, and WT were grown on the MS medium for seven days. Then they were transplanted into MS containing 150 mM NaCl (salt treatment) or grown in a 37 °C (HT treatment) illumination incubator. Root length was measured two weeks after the transplant.
4.10. Chlorophyll Content and Physiological measurement
Chlorophyll a and chlorophyll b were extracted and quantified from leaves of the transgenic lines and WT plants after salt and HT treatments following a previous method [60]. Total chlorophyll content was calculated according to the following equations: Chlorophyll a = [(12.72 × A663 − 2.69 × A645)V/W], Chlorophyll b = [(22.88 × A645 − 4.68 × A663)V/W], total chlorophyll content = Chlorophyll a +Chlorophyll b, where A663 and A645 are absorbance at 663 nm and 645 nm, respectively. V is the final volume of chlorophyll extract in 80% acetone, and W is fresh weight of leaves. Biochemical analyses of superoxide dismutase (SOD), peroxidase (POD), proline content, and malondialdehyde (MDA) were conducted according to previous methods [14].
4.11. Histochemical Analyses of the PsnNAC036 Transgenic Tobacco
Cell death, accumulation of hydrogen peroxide (H2O2) and superoxide (O2−) in the transgenic and WT plants under high saltor HT were visualized through histochemical staining with Evans blue, 3,3′-diaminobenzidine (DAB) and nitro-blue tetrazolium (NBT), respectively. Two-week-old seedlings were irrigated with water (control), 150 mM NaCl solution or grown in 37 °C HT for 12 h. Leaves were excised for histochemical staining as previously described [10].
4.12. Salt and HT Treatments of the PsnNAC036 Transgenic Tobacco
The seedlings of WT and transgenic tobacco lines on MS medium were transplanted in the soil oncethe fourth leaf appeared. The seedlings were grown under normal growth conditions for a total of two weeks, and then were subjected to 200 mM NaCl irrigation or 37 °C HT treatment for another two weeks.
4.13. Expression Analysis of Stress-Related Genes in PsnNAC036 Transgenic Tobacco
To further understand the functions the PsnNAC036 TF, leaves of the transgenic lines and WT under 200 mMNaCl or 37 °C HT for 12 h were used for RT-qPCR. Relative expression levels ofstress-related genes including superoxide dismutase (NtSOD), peroxidase (NtPOD), polyphenol oxidase (NtPPO), 9-cis-epoxycarotenoid dioxygenase1 (NtNCED1), 1-pyrroline-5-carhoxylate synthetase (NtP5CS), regulatory proteins (NtDREB3), plasmalemma Na+/H+ antiporter (NtSOS), late-embryogenesis-abundant protein5 (NtLEA5), early responsive to dehydration (NtERD10A/B/C/D) and Na+ antiporter genes (NtHKT555/586) were quantified. RT-qPCR was conducted according to a published method [14]. The primer sequences used for the above genes can be found in Supplementary Table S2.
5. Conclusions
In this study, we analyzed the expression changes of NAC TFs in P. simonii × P. nigra under 150 mM salt treatment and isolated a significantly upregulated NAC gene, PsnNAC036. RT-qPCR and promoter GUS analyses showed that the gene was highly responsive to stresses, especially salinity and heat. Overexpression of PsnNAC036 improved salinity and HT tolerance of transgenic plants. RT-qPCR results revealed that overexpression of the PsnNAC036 TF regulated the expression of several stress-related genes in the transgenic plants. Furthermore, we validated that the C-terminal domain (191–343 amino acids) was the potent activator of PsnNAC036. These results have demonstrated that PsnNAC036 functions as a transcriptional activatorand plays an important role in plant salinity and HT tolerance. As such, it is a potential molecular target for crop biotechnology and marker-based breeding for enhanced resilience and yield.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/22/5/2656/s1.
Author Contributions
T.J. designed research. X.Z. conducted experiments and wrote the manuscript. Z.C., K.Z. and X.W. performed in data analysis. W.Y. revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (2572019AA06) and Natural Science Foundation of Jiangsu Province (BK20190748).
Acknowledgments
The authors thank Lisa David from University of Florida for critical reading and editing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ku, Y.; Sintaha, M.; Cheung, M.; Lam, H. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Yao, W.; Wang, S.; Zhou, B.; Jiang, T. Transgenic poplar overexpressing the endogenous transcription factor ERF76 gene improves salinity tolerance. Tree Physiol. 2016, 36, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, J.; Xia, D. Research progress on elasticity modulus nondestructive examination of wood and glulam structures. J. Cent. South Univ. For. Technol. 2013, 33, 149–153. [Google Scholar]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Kim, S.Y.; Park, C.M. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 2007, 226, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Duval, M.; Hsieh, T.-F.; Kim, S.Y.; Thomas, T.L. Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant. Mol. Biol. 2002, 50, 237–248. [Google Scholar] [CrossRef]
- Xie, Q.; Frugis, G.; Colgan, D.; Chua, N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant. Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant. Cell 2011, 23, 873–894. [Google Scholar] [CrossRef]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive Analysis of NAC Domain Transcription Factor Gene Family in Populustrichocarpa. BMC Plant Biol. 2010, 10, 1–23. [Google Scholar] [CrossRef]
- Li, X.; Li, L.; Liu, X.; Zhang, B.; Zheng, W.; Ma, W. Analysis of physiological chaaracteristics of abscisic acid sensitivity and salt resistance in Arabidopsis ANAC mutants (ANAC019, ANAC072 and ANAC055). Biotechnol. Biotechnol. Equip. 2012, 26, 2966–2970. [Google Scholar] [CrossRef]
- Morishita, T.; Kojima, Y.; Maruta, T.; Nishizawa-Yokoi, A.; Yabuta, Y.; Shigeoka, S. Arabidopsis NAC Transcription Factor, ANAC078, Regulates Flavonoid Biosynthesis under High-light. Plant Cell Physiol. 2009, 50, 2210–2222. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cheng, Z.; Zhao, K.; Yao, W.; Sun, X.; Jiang, T. Functional characterization of poplar NAC13 gene in salt tolerance. Plant Sci. 2019, 281, 1–8. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, X.; Duan, H.; Lian, C.; Liu, C.; Yin, W. Three stress-responsive NAC transcription factors from Populuseuphratica differentially regulate salt and drought tolerance in transgenic plants. Physiol. Plant. 2018, 162, 73–97. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Zhao, K.; Cheng, Z.; Li, X.; Zhou, B.; Jiang, T. Transcriptome Analysis of Poplar Under Salt Stress and Over-Expression of Transcription Factor NAC57 Gene Confers Salt Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 1121. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, G.L.; Marques, C.S.; Costa, M.D.B.L.; Reis, P.A.B.; Alves, M.S.; Carvalho, C.M. Complete inventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 2009, 444, 10–23. [Google Scholar] [CrossRef]
- Yao, W.; Li, C.; Lin, S.; Wang, J.; Zhou, B.; Jiang, T. Transcriptome analysis of salt-responsive and wood-associated NACs in Populus simonii × Populus nigra. BMC Plant Biol. 2020, 20, 317. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Croft, H.; Chen, J.M.; Wang, R. The global distribution of leaf chlorophyll content. Remote Sens. Environ. 2020, 236, 111479. [Google Scholar] [CrossRef]
- Wu, X.; Jia, Q.; Ji, S. Gamma-aminobutyric acid (GABA) alleviates salt damage in tomato by modulating Na+ uptake, the GAD gene, amino acid synthesis and reactive oxygen species metabolism. BMC Plant Biol. 2020, 20, 1–21. [Google Scholar] [CrossRef]
- Han, D.; Zhang, Z.; Ding, H.; Chai, L.; Liu, W.; Li, H. Isolation and characterization of MbWRKY2 gene involved in enhanced drought tolerance in transgenic tobacco. J. Plant Interact. 2018, 13, 163–172. [Google Scholar] [CrossRef]
- Jin, C.; Li, K.; Xu, X.; Zhang, H.P.; Chen, H.X. A Novel NAC Transcription Factor, PbeNAC1, of Pyrusbetulifolia Confers Cold and Drought Tolerance via Interacting with PbeDREBs and Activating the Expression of Stress-Responsive Genes. Front. Plant Sci. 2017, 8, 1049. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, G.; Zhou, S.; Ren, Y.; Wang, W. The improvement of salt tolerance in transgenic tobacco by overexpression of wheat F-box gene TaFBA1. Plant Sci. 2017, 259, 71–85. [Google Scholar] [CrossRef]
- Yang, T.; Xu, Z.; Lv, R.; Zhu, L.; Peng, Q.; Qiu, L. N gene enhances resistance to Chili veinal mottle virus and hypersensitivity to salt stress in tobacco. J. Plant Physiol. 2018, 230, 92–100. [Google Scholar] [CrossRef]
- Liang, W.J.; Ma, X.L.; Wan, P.; Liu, L.Y. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Sedaghatmehr, M.; Thirumalaikumar, V.P.; Kamranfar, I.; Marmagne, A.; Masclaux-Daubresse, C.; Balazadeh, S. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019, 42, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.A.; Dennis, E.S.; Llewellyn, D.J.; Wilson, I.W. ATAF NAC transcription factors: Regulators of plant stress signaling. Plant Signal.Behav. 2010, 5, 428–432. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of a Stress-Responsive NAC Transcription Factor Gene ONACO22 Improves Drought and Salt Tolerance in Rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Shen, S.; Zhang, Q.; Shi, Y.; Sun, Z.; Zhang, Q.; Hou, S. Genome-Wide Analysis of the NAC Domain Transcription Factor Gene Family in Theobroma cacao. Genes 2020, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Z.; Lu, M.; Wang, Y. ThNAC13, a NAC Transcription Factor from Tamarix hispida, Confers Salt and Osmotic Stress Tolerance to Transgenic Tamarix and Arabidopsis. Front. Plant Sci. 2017, 8, 635. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; He, K.; Chi, X.; Chai, G.; Wang, Y.; Jia, C. Miscanthus NAC transcription factor MlNAC12 positively mediates abiotic stress tolerance in transgenic Arabidopsis. Plant Sci. 2018, 277, 229–241. [Google Scholar] [CrossRef]
- Sukiran, N.L.; Ma, J.C.; Ma, H.; Su, Z. ANAC019 is required for recovery of reproductive development under drought stress in Arabidopsis. Plant Mol. Biol. 2019, 99, 161–174. [Google Scholar] [CrossRef]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal. Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef]
- He, L.; Shi, X.; Wang, Y.; Guo, Y.; Yang, K.; Wang, Y. Arabidopsis ANAC069 binds to C A/G CG T/G sequences to negatively regulate salt and osmotic stress tolerance. Plant Mol. Biol. 2017, 93, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Zhao, H.; Xiao, Y.; Huang, Y.; Yao, P.; Lei, Y. Cloning, Characterization, and Expression Analysis of Eight Stress-Related NAC Genes in Tartary Buckwheat. Crop Sci. 2019, 59, 266–279. [Google Scholar] [CrossRef]
- Pang, X.; Xue, M.; Ren, M.; Nan, D.; Wu, Y.; Guo, H. Ammopiptanthus mongolicus stress-responsive NAC gene enhances the tolerance of transgenic Arabidopsis thaliana to drought and cold stresses. Genet. Mol. Biol. 2019, 42, 624–634. [Google Scholar] [CrossRef]
- Liu, S.; Liu, C.; Wang, X. Seed-specific activity of the Arabidopsis β-glucosidase 19 promoter in transgenic Arabidopsis and tobacco. Plant Cell Rep. 2020, 40, 213–221. [Google Scholar] [CrossRef]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Kong, W.; Liu, F.; Zhang, C.; Zhang, J.; Feng, H. Non-destructive determination of Malondialdehyde (MDA) distribution in oilseed rape leaves by laboratory scale NIR hyperspectral imaging. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H. Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in Arabidopsis. PLoS Genet. 2014, 10, e1004664. [Google Scholar] [CrossRef]
- Ábrahám, E.; Hourton-Cabassa, C.; Erdei, L. Methods for determination of proline in plants. Plant Stress Toler. 2010, 317–331. [Google Scholar] [CrossRef]
- Szymanska, K.P.; Polkowska-Kowalczyk, L.; Lichocka, M.; Maszkowska, J.; Dobrowolska, G. SNF1-Related Protein Kinases SnRK2.4 and SnRK2.10 Modulate ROS Homeostasis in Plant Response to Salt Stress. Int. J. Mol. Sci. 2019, 20, 143. [Google Scholar] [CrossRef]
- Zafar, S.A.; Hameed, A.; Ashraf, M.; Khan, A.S.; Ziaul, Q.; Li, X. Agronomic, physiological and molecular characterisation of rice mutants revealed the key role of reactive oxygen species and catalase in high-temperature stress tolerance. Funct. Plant Biol. 2020, 47, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Ambastha, V.; Chauhan, G.; Tiwari, B.S.; Tripathy, B.C. Execution of programmed cell death by singlet oxygen generated inside the chloroplasts of Arabidopsisthaliana. Protoplasma 2020, 257, 841–851. [Google Scholar] [CrossRef]
- Dashti, A.; Khan, A.A.; Collins, J.C. Effects of Salinity on Growth, Ionic Relations and Solute Content of SorghumBicolor (L.) Monench. J. Plant Nutr. 2009, 32, 1219–1236. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Barkla, B.J.; Vera-Estrella, R.; Zhu, J.K.; Schumaker, K.S. Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 2003, 132, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Dang, C.; Ye, Y.; Wang, Z.; Hu, L.; Zhang, F. Overexpression of Grapevine VvIAA18 Gene Enhanced Salt Tolerance in Tobacco. Int. J. Mol. Sci. 2020, 21, 1323. [Google Scholar] [CrossRef]
- Aziz, E.; Batool, R.; Akhtar, W.; Rehman, S.; Gregersen, P.L.; Mahmood, T. Expression analysis of the polyphenol oxidase gene in response to signaling molecules, herbivory and wounding in antisense transgenic tobacco plants. 3 Biotech 2019, 9. [Google Scholar] [CrossRef]
- Singh, N.K.; Shukla, P.; Kirti, P.B. A CBL-interacting protein kinase AdCIPK5 confers salt and osmotic stress tolerance in transgenic tobacco. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Li, X.; Zhuang, K.; Liu, Z.; Yang, D.; Ma, N.; Meng, Q. Overexpression of a novel NAC-type tomato transcription factor, SlNAM1, enhances the chilling stress tolerance of transgenic tobacco. J. Plant Physiol. 2016, 204, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhong, M.; Li, S.; Pan, Y.; Jiang, B.; Jia, Y. Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, in tobacco enhances tolerance to salt stress. Plant Physiol. Biochem. 2013, 69, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, B.; Liu, C.; Tong, B.; Guan, T.; Xia, D. Expression of a populus histone deacetylase gene 84KHDA903 in tobacco enhances drought tolerance. Plant Sci. 2017, 265, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, X.; Zhao, K.; Zhou, B.; Jiang, T. Ectopic expression of a poplar gene NAC13 confers enhanced tolerance to salinity stress in transgenic Nicotiana tabacum. J. Plant Res. 2020, 133, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Wang, L.; Zhou, B.; Wang, S.; Li, R.; Jiang, T. Over-expression of poplar transcription factor ERF76 gene confers salt tolerance in transgenic tobacco. J. Plant Physiol. 2016, 198, 23–31. [Google Scholar] [CrossRef]
- Yao, W.; Wang, S.; Zhou, B.; Wang, J.; Jiang, T. Characterization of ERF76 promoter cloned from Populus simonii × P. nigra. Acta Physiol. Plant 2017, 39. [Google Scholar] [CrossRef]
- Shivakrishna, P.; Reddy, K.A.; Rao, D.M. Effect of PEG-6000 imposed drought stress on RNA content, relative water content (RWC), and chlorophyll content in peanut leaves and roots. Saudi J. Biol. Sci. 2018, 25, 285–289. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).