Abstract
MFN1 (Mitofusin 1) and MFN2 (Mitofusin 2) are GTPases essential for mitochondrial fusion. Published studies revealed crucial roles of both Mitofusins during embryonic development. Despite the unique mitochondrial organization in sperm flagella, the biological requirement in sperm development and functions remain undefined. Here, using sperm-specific Cre drivers, we show that either Mfn1 or Mfn2 knockout in haploid germ cells does not affect male fertility. The Mfn1 and Mfn2 double knockout mice were further analyzed. We found no differences in testis morphology and weight between Mfn-deficient mice and their wild-type littermate controls. Spermatogenesis was normal in Mfn double knockout mice, in which properly developed TRA98+ germ cells, SYCP3+ spermatocytes, and TNP1+ spermatids/spermatozoa were detected in seminiferous tubules, indicating that sperm formation was not disrupted upon MFN deficiency. Collectively, our findings reveal that both MFN1 and MFN2 are dispensable for sperm development and functions in mice.
1. Introduction
Each cell contains several hundred to thousands of mitochondria, and each mitochondrion has 2–10 copies of independent mitochondrial DNAs (mtDNAs) [1,2,3]. To allow coordinated responses of individual mitochondria to various developmental cues, physiological changes, and pathological stress, mitochondria need to share their membrane and exchange their contents through continuous fusion and fission, which are collectively known as mitochondrial dynamics [4,5]. In mammals, mitochondrial fusion is mediated by MFN1 and MFN2, two GTPases at the outer membrane, as well as OPA1 GTPase, an inner-membrane GTPase that regulates mitochondrial inner membrane fusion [6,7,8]. Similarly, the mitochondrial fission is catalyzed by a GTPase called DRP1, along with four adaptors (i.e., MFF, MID49, MID51, and FIS1), which recruit DRP1 to mitochondria [6]. Previous reports demonstrated an essential role of mitochondrial dynamics in mammalian development. Deletion of either Mfn1 or Mfn2 causes embryonic lethality due to defects in trophoblast development [9]. When conditional knockouts of Mfn2 or Mfn1 and Mfn2 were used to bypass embryonic placental defects, disrupted cerebellum development and cardiac dysfunction were detected [10].
Recent germ cell studies, including ours, show that disturbed mitochondrial fusion by deletion of either Mfn1 or Mfn2 specifically blocks spermatogonial differentiation, but undifferentiated spermatogonia remain largely unaffected [11,12,13,14]. The difference in sensitivity to disrupted mitochondrial dynamics reflects the importance of differential mitochondrial regulation in various developmental stages of spermatogenesis. Although the physiological importance of Mfn1 and Mfn2 currently remains undefined during late spermatogenesis, several reports suggest that mitochondrial dynamics may participate in sperm development. For example, Honda et al. investigated the Mfn1 and Mfn2 expression patterns in rats by Northern blot analysis and found that Mfn1 and Mfn2 were highly expressed in the pubertal and adult rat testes. Immunohistochemistry analysis revealed a pronounced expression of MFN2 in the round and elongating spermatids in the seminiferous tubules [15]. In sperm, mitochondria line the microtubular axoneme and wrap around the midpiece of sperm flagellum to form the mitochondrial sheath [16]. MFN2 was also found to be localized at the flagellum midpiece and post-acrosomal regions of sperm in the caput and cauda area of the epididymis by immunofluorescence analysis [16]. In addition, deficiency of mitochondrial fission caused by Fis1 or Mff null mutations leads to defective mitochondria, aberrant sperm function, and reduced fertility [17,18,19]. These data suggest a potential role of mitochondrial dynamics at the post-meiosis stage in male mice.
To understand whether mitochondrial fusion participates in late spermatogenesis, we generated sperm-specific knockout mice for Mfn1 and Mfn2 with Prm1-Cre drivers. We found that neither Mfn1 nor Mfn2 deficiency in haploid sperm caused defects in sperm development and male fertility in mice. Furthermore, to exclude functional redundancy of MFN1 and MFN2, compound deletion of Mfn1 and Mnf2 was analyzed, and male fertility remained unaffected, indicating that both genes are dispensable during haploid sperm formation and functions. Our study thus suggests that mitochondrial fusion plays a nonessential role in maintaining sperm development in mice.
2. Results
2.1. Mfn1 and Mfn2 mRNA Are Highly Expressed in Testes
To understand whether mitochondrial fusion participates in late spermatogenesis, we first examined if Mfn1 and Mfn2 were expressed in testes by qPCR analyses on various mouse tissues. We found preferential expression of Mfn1 in spleen and testes, while Mfn2 was highly expressed in mouse testes (Figure 1A,C). Because in spermatogenesis, spermatogonia start to differentiate at postnatal day (P) 6–10, with meiotic pachytene spermatocytes emerging at P14, and haploid spermatids around P21, we further analyzed the expression of Mfn1 and Mfn2 in mouse testes at different ages by qPCR. We found that Mfn1 mRNA level started to increase at week 2 and peaked at week 5 (Figure 1B). Similarly, the highest expression of Mfn2 mRNA level was detected at weeks 4–5 (Figure 1D). These results suggest that Mfn1 and Mfn2 were highly expressed in haploid spermatids, corresponding with the time of post-meiotic germ cell development.
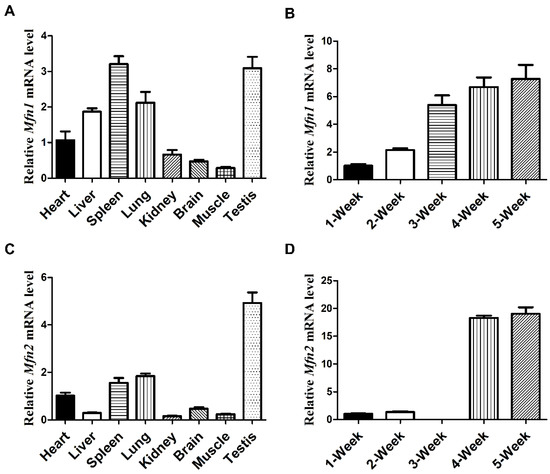
Figure 1.
Mfn1 and Mfn2 are highly expressed in testes. (A,C) qPCR analyses of Mfn1 mRNA (A) and Mfn2 mRNA (C) expression levels in various organs collected from wild-type adult mice. (B,D) qPCR analyses of Mfn1 (B) or Mfn2 (D) mRNA expression levels in developing testes at 1, 2, 3, 4, and 5-week. Mfn1 or Mfn2 mRNA expression levels in heart (A,C) and week 1 (B,D) were used as baseline control. Gapdh served as the cDNA loading control. Data are presented as mean ± SEM.
2.2. Generation of Mfn1 and Mfn2 Conditional Knockout (cKO) Mice
We previously revealed that either Mfn1 or Mfn2 conditional knockout in early germ cells resulted in male infertility [11,13]. To understand whether Mfn1 and Mfn2 regulate post-meiotic germ cell development, we crossed Mfn1 and Mfn2 conditional knockout mice with a Prm1-Cre line (Figure 2A,B), in which CRE recombinase under the sperm-specific promoter of Prm1 deletes Mfn1 or Mfn2 in haploid spermatids [20,21]. Because antibodies against MFN1 or MFN2 for immunohistofluorescence did not work well in our hands, we examined the Mfn1 and Mfn2 deletion efficiency in sperm by the germline transmission of mutant alleles. In this assay, Mfn1f/f, Prm1-Cre and Mfn2 f/f, and Prm1-Cre male mice were crossed with wild-type females. If the CRE deletion is incomplete in sperm, some progenies generated from these mice would contain the Mfn1f/+ (or Mfn2f/+) allele instead of Mfn1−/+(or Mfn2−/+). The results demonstrated that 100% of progeny mice generated from paternal Mfn1f/f; Prm1-Cre (or Mfn2f/f; Prm1-Cre) were carrying the Mfn1−/+(or Mfn2−/+) allele. Thus, these results confirm that Prm1-Cre was effective, deleting floxed Mfn1 or Mfn2 alleles with 100% efficiency (Table 1).
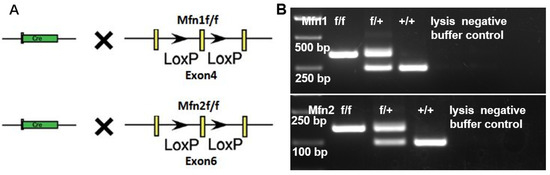
Figure 2.
Deletion of Mfn1 and Mfn2 in haploid sperm by Prm1-Cre. (A) Strategy to delete Mfn1 and Mfn2 in haploid sperm by Prm1-Cre. The exon 4 of Mfn1 and exon 6 of Mfn2 were flanked by two loxP sequences for deletion by Cre/loxP-mediated recombination. Yellow rectangles represent exons; black arrows represent loxP sequences. (B) Examples of genotyping on offspring from Mfn1f/+ and Mfn2f/+ male mice crossed with Mfn1f/+, Prm1-Cre and Mfn2f/+, Prm1-Cre female. PCR on lysed mouse tail snips amplified the Mfn1 genomic locus to generate a 450 bp DNA band from the mutated region, or a 350 bp band from wild-type locus. In Mfn2 mice, PCR amplified a 180 bp DNA band from the mutated region, or a 145 bp band from wild-type locus. +/+: wild-type; f/+: heterozygous; f/f: homozygous.

Table 1.
Transmission efficiency of mutant alleles.
2.3. Either Mfn1 or Mfn2 Deletion in Sperm Does Not Affect Sperm Development and Functions
Conditional knockouts of Mfns offer powerful models to understand the functional contribution of MFNs to mouse sperm development and male fertility. We thus analyzed the sperm development upon deletion of Mfns by Prm1-Cre.
We found that the testes in Mfns-cKO (i.e., Mfn1f/f, Prm1-Cre or Mfn2f/f, Prm1-Cre) mice were morphologically normal at 56 dpp compared to wild littermate controls (Figure 3A,C). The weight of Mfns-cKO testes showed no significant difference from controls either (Figure 3B,D). We next examined germ cell development in control and Mfns-cKO mice at 42 dpp by immunohistofluorescence (IHF) with antibodies against TRA98, a pan-germ cell marker [22], and SYCP3, a spermatocyte-specific marker [23]. TRA98 staining revealed normal meiosis and post-meiotic round spermatid development in Mfns-cKO mice (Figure S1A,B). Similarly, SYCP3 signals were comparable between Mfns-cKO and control mice (Figure S1A,B), indicating a normal spermatocyte development. TNP1, a protein that only marks haploid germ cells, was also readily detectable in testis sections from Mfns-cKO mice, similar to those in control littermates (Figure S1C,D), indicating the existence of spermatids and spermatozoa. We further analyzed the epididymides and found no obvious difference in the numbers of spermatozoa in Mfns-cKO mice compared to their littermate controls (Figure 3E). Finally, we evaluated the impact of Mfns deletion on male fertility using a breeding test, in which Mfns-cKO mice and their littermate control male were crossed with wild-type female mice, and the average number of pups per litter over 2 months was calculated. We did not detect any obvious change in numbers of pups per litter produced from breeding pairs with Mfns-cKO male mice compared to those from their littermate controls (Figure 3F). Taken together, these results suggest that Mfns conditional knockout in post-meiotic haploid germ cells does not affect sperm development and functions in mice.
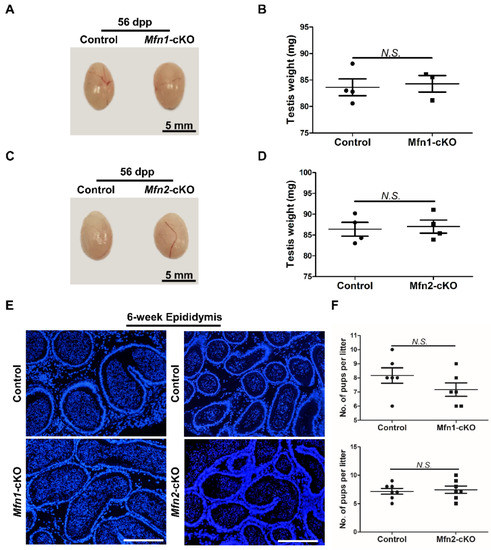
Figure 3.
Either Mfn1 or Mfn2 deletion in haploid germ cells does not affect sperm development and functions. (A,C) Gross morphology of testes from control and Mfn1-cKO mice (A) or Mfn2-cKO mice and their littermate controls (C) at 56 dpp. (B,D) Average testis weight was calculated from Mfn1-cKO (B) or Mfn2-cKO (D) mice and control littermate at 56 dpp. (E) DAPI stained sections of epididymides from control, Mfn1-cKO, and Mfn2-cKO mice at 6 weeks. Scale bars: 200 μm. (F) Average numbers of pups per litter were calculated based on 6 litters from 3 control and 6 litters from 3 Mfn1-cKO male mice (upper panel), or based on 7 litters from 3 control and 7 litters from 3 Mfn2-cKO male mice (lower panel). Each male mouse was bred with 2 wild-type females for over 2 months. Data are presented as mean ± SEM. N.S.: no significance.
2.4. Combined Deletion of Mfn1 and Mfn2 in Sperm Does Not Affect Sperm Development and Functions
MFN1 and MFN2 share more than 81% homology and similar topology [9,24,25,26,27]. In fibroblasts, MFN1 and MFN2 compensate the function deficiency of mitochondrial fusion for each other [9]. We showed that deletion of either Mfn1 or Mfn2 does not affect sperm development and functions in late spermatogenesis. To exclude the possibility that this is due to functional redundancy between MFN1 and MFN2 in spermatozoa, we further generated Mfn1 and Mfn2 double knockout mice (Mfn1f/f, Mfn2f/f, Prm1-Cre, abbreviated as Mfn1&2-cKO). Testes from Mfn1&2-cKO and control littermates at 56 dpp were analyzed. We did not find any significant differences in morphology and weight of testes between the two groups (Figure 4A,B). Moreover, IHF staining with TRA98, SYCP3, and TNP1 antibodies revealed the presence of all types of germ cells from spermatogonia to spermatozoa in Mfn1&2-cKO mice (Figure S2A,B). In addition, mature spermatozoa were readily detected in epididymides of adult Mfn1&2-cKO mice (Figure 4C). We further found that deletion of both Mfn1 and Mfn2 in haploid germ cells did not affect sperm functions and male fertility. In the breeding assay, the average numbers of pups per litter from adult Mfn1&2-cKO mice showed no significant difference from those in the control group (Figure 4D). Taken together, these data demonstrated that the compound knockout of Mfn1 and Mnf2 in haploid germ cells did not affect sperm development and male fertility in mice.
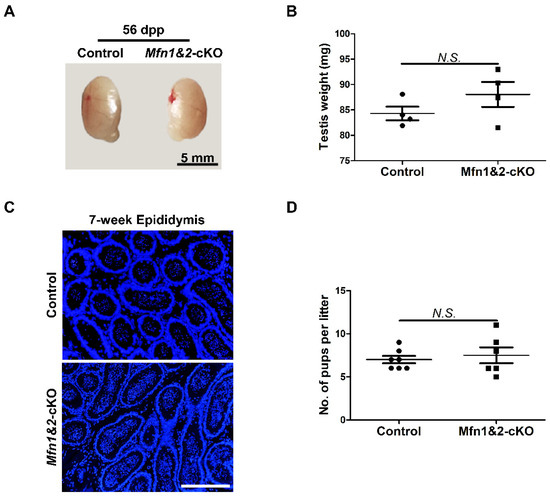
Figure 4.
Compound knockout of both Mfn1 and Mfn2 in haploid sperm does not affect sperm development and mice fertility. (A) Gross morphology of testes from control and Mfn1&2-cKO mice at 56 dpp. (B) Averaged testis weight was calculated from 4 control and 4 Mfn1&2-cKO littermates at 56 dpp. (C) DAPI stained sections of epididymides from control and Mfn1&2-cKO mice at 7 weeks. Scale bars, 200 μm. (D) Averaged pup numbers per litter were calculated based on 7 litters from 3 control and 6 litters from 3 Mfn1&2-cKO male mice. Each male mouse was bred with 2 wild-type females for at least 2 months. (B,D) Data are presented as mean ± SEM. N.S.: no significance.
3. Discussion
Both mitofusins, MFN1 and MFN2, are broadly expressed across various tissues [26]. Recent evidence suggests that MFN1 and MFN2 have both redundant and distinct functions, depending upon cell types and developmental stages [9,28,29,30]. Although roles of mitochondrial fusion in early spermatogenesis before meiosis have been reported, the exact expression pattern and physiological importance of Mfn1 and Mnf2 in mouse haploid spermatids have yet to be characterized. Here we examined the functions of Mfn1 and Mfn2 in sperm development and male fertility.
Published studies demonstrate that Mfn1-null or Mfn2-null mouse embryonic fibroblasts (MEFs) contain highly fragmented mitochondria, and MFN1 and MFN2 compensate the function deficiency of mitochondrial fusion for each other [9]. To understand the functional roles of mitochondrial fusion in sperm formation, we generated conditional knockout of Mfns, using two haploid-specific Prm1-Cre drivers. The first Prm1-Cre driver came from Jackson Laboratory and displayed a partial excision of Mfns based on our analyses on transmission efficiency of mutant alleles (Table S1). To examine the effects of complete MFN deficiency on sperm formation, we further used the second Prm1-Cre (RIKEN BRC) strain, which efficiently removed Mfns from haploid spermatids (Table 1). Notably, although both Prm1-Cre mice were generated through the pronuclear microinjection of transgene into fertilized eggs, the regulatory elements of these two Prm1-Cre transgenes are different [20,21,31]. In addition, the expression of randomly inserted transgenes at different genomic locations is often subject to chromatin positional effects. The genomic location of both the Prm1-Cre (Jackson) and the Prm1-Cre (RIKEN BRC) transgene are unknown. It is thus possible that Prm1-Cre from Jackson Laboratory is not transcriptionally active in a subset of haploid spermatids. Consistent with our findings on those Prm1-Cre mouse models, Batista et al. also observed the inefficient recombinase activity when using Prm1-Cre from Jackson Laboratory [32]. However, another study used the same Prm1-Cre strain from Jackson Laboratory but did not find such incomplete Cre recombinase activity [33]. One potential explanation for this inconsistent observation may be due to different genomic loci of various floxed alleles [32]. The chromatin status of certain floxed genomic loci may prevent CRE from accessing the loxP sites. Alternatively, a genetic drift of the Cre transgene may occur over time. As a mouse line is continuously intra-bred within the colony, mutations may arise and lead to reduced CRE expression. Therefore, properly maintained and characterized mouse models are crucial to permit successful gene manipulation in reproductive research.
Using these haploid-specific Prm1-Cre drivers, our study showed that mice with single deletion of either Mfn1 or Mfn2 in spermatids had normal sperm development and male fertility, either when Mfns were partially deleted (Figure S3) or when efficient Prm1-Cre driver from RIKEN BRC completely removed Mfns (Table 1, Figure 2 and Figure 3) in sperm. To exclude the possibility of functional compensation between MFN1 and MFN2 in sperm, we further analyzed combined knockouts of Mfn1 and Mfn2, using the efficient Prm1-Cre from RIKEN BRC. Unexpectedly, adult mice with complete deficiency of both MFNs displayed well-developed and functional sperm. The male fertility remained unaffected. This is unlikely due to any existing truncated functional MFN proteins in sperm. Deletion of exon 4 of Mfn1 and of exon 6 in Mfn2 by CRE recombinase leads to a frameshift of Mfn genes. In addition, CRE deletes the exon four or exon six encoding the canonical GTPase motif of MFNs, thus preventing the formation of any functional protein. Notably, the same mouse Mfn cKO lines (Mfn1: MMRRC # 029901-UCD and Mfn2: MMRRC # 029902-UCD) have been widely validated to be absent of functional proteins upon CRE deletion in cells from these mice, including neurons [10], skeletal muscle [34], and early-stage germ cells in our hands [11,13]. Therefore, our data suggest that mitochondrial fusion is dispensable for sperm development during late spermatogenesis.
Mitochondria are dynamic organelles that continually undergo fusion and fission. These two opposing processes work in concert to maintain the shape, size, and number of mitochondria and their physiological functions [6]. Previous evidence suggests that two mitofusins form three distinct molecular complexes to promote mitochondrial fusion: MFN1 homotypic oligomers, MFN2 homotypic oligomers, and MFN1-MFN2 heterotypic oligomers [9]. The type of complex that plays a dominant action depends on the cell type. For example, MEFs appear to use all three oligomeric complexes [9]. By contrast, trophoblast giant cells rely on MFN2 homotypic oligomers. Placental defects were detected in Mfn2 but not Mfn1 mutants [9]. Our results suggest none of these complexes are required for proper sperm functions. The morphology, number, and mitochondrial organization of germ cell mitochondria change throughout spermatogenesis. Spermatogonia and early spermatocytes harbor “orthodox” mitochondria, while late spermatocytes and early spermatids contain a condensed form of mitochondria with enlarged matrix and vesiculate cristae, randomly distributing throughout the cytoplasm [35]. In elongated spermatids, mitochondria with “intermediate” configuration are aligned along the outer dense fibers in the forming tail [36,37,38]. During spermiogenesis, a large fraction of mitochondria is degraded, leaving far fewer mitochondria (~50–75 per cell) and mtDNAs (~1.4 per mitochondrion) in the sperm [37,38,39,40]. Although sperm need mitochondrial metabolism for energy production to support their mobility and fertilization, the activities of mitochondrial dynamics in sperm are likely kept minimal to ensure proper mitochondrial organization. Indeed, low levels of mitochondrial fusion and fission in sperm have been reported [41]. Therefore, the haploid spermatids likely have high tolerance for perturbations in mitochondrial dynamics. This is different from differentiating spermatogonial and spermatocytes; loss of Mfn1 or Mfn2 alone will result in germ cell development defects, leading to male sterility in mice [11,12,13,14].
4. Materials and Methods
4.1. Experimental Design
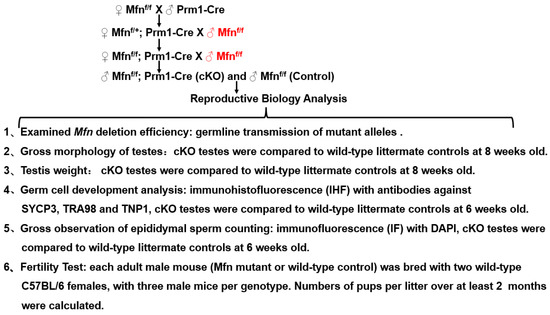
Figure 5.
Summary of experimental design and methods used in this study.
4.2. Mouse Lines, Animal Care, and Fertility Test
Sperm harboring the floxed Mfn1 and Mfn2 alleles were obtained from Mutant Mouse Regional Resource Center (MMRRC at UC Davis, USA, resource identifier no. MMRRC # 029901-UCD and MMRRC # 029902-UCD) [10]. Mfn1f/+ and Mfn2f/+ mice were derived by intracytoplasmic injection of sperm into oocytes and maintained by crossing with C57BL/6 mice. Two Prm1-Cre mouse lines were used. One was obtained from Shanghai Model Organisms Center Inc (originally developed from Jackson Laboratory, Bar Harbor, ME, USA, Stock ID: 003328), and another was purchased from RIKEN BioResource Research Center (ID: RBRC02182) [20,21]. To generate the post-meiotic haploid spermatids specific deletion of Mfn1, male conditional Mfn1 knockout mice (Mfn1f/+) were bred with female Mfn1f/+ and Prm1-Cre, and the progeny mice were genotyped with PCR analyses. Conditional Mfn2 knockout mice were generated in the same way as described above. For the fertility test, adult males were housed with wild-type C57BL/6 females for at least 2 months. The size of the litters generated by these males was recorded. All animal experimental procedures (Protocol ID: m20190403) were conducted in accordance with the local Animal Welfare Act and Public Health Service Policy (consistent with the WMA Statement on Animal Use in Biomedical Research) and approved on 01-04-2019 by the Committee of Animal Experimental Ethics at East China Normal University.
4.3. Genomic DNA Extraction and Genotyping
Genomic DNA was isolated as described previously [42,43]. Briefly, tail tips were digested in alkaline lysis reagent (25 mM NaOH, 0.2 mM EDTA at pH = 12) at 95 °C for 30 min, and the reactions were then stopped by neutralizing reagent (40 mM Tris-HCl, pH = 5). The supernatant was collected for PCR genotyping with Taq Master Mix (Vazyme Biotech, Nanjing, China, P112-AA) according to the manufacturer′s instructions. Sequences of primers and size of products are provided in Supplementary Table S2.
4.4. Total RNA Extraction, Reverse Transcription, and Real-Time PCR
As previously described [42], total RNA from various mouse tissues was extracted using RNAiso Plus solution (TAKARA, Dalian, China, 9109), and cDNAs were synthesized using a cDNA Synthesis Kit (TAKARA, RR037A). Quantitative Real-Time PCR was performed on Thermo Scientific QuantStudio 3 Real-Time PCR System. Three technical replicates were conducted for each independent experiment. Sequences of primers used in this study are listed in Supplementary Table S3.
4.5. Histology and Immunohistofluorensce (IHF)
IHF was performed as described previously [42]. Testes were fixed in 4% paraforaldehyde (PFA) in PBS at 4 °C overnight and embedded in paraffin. The 5 μm thick testicular sections were incubated with primary antibodies and washed three times with PBS containing 0.1% Tween 20. Nuclei were stained in 0.5 mg/mL DAPI (Beyotime Biotechnology, Shanghai, China, C1002) after blotting with Alexa Fluor® conjugated secondary antibodies. Slides were imaged under a fluorescent microscope (Olympus BX53, Olympus Life Science, Japan) and processed with Image J software. Antibodies and fluorescent probes used in this study: TNP1 (ab73135), TRA98 (ab82527; 1:500), and SYCP3 (ab15093; 1:400) were purchased from Abcam (Cambridge, MA, USA), Alexa Fluor® conjugated secondary antibodies came from Jackson ImmunoResearch Laboratories (West Grove, PA, USA).
4.6. Statistics
The data were evaluated for significant differences using Student’s t-test, calculated with GraphPadPrism5 software (GraphPad Software, La Jolla, CA, USA). A p-value < 0.05 was considered statistically significant.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222413507/s1.
Author Contributions
S.W. and Y.W. designed research, S.W. performed experiments and wrote manuscript, S.W. analyzed the data, J.M., W.C., X.Z. and L.W. contributed with the methodology, P.W. contributed to the mouse keeping and Y.W. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants the National Natural Science Foundation of China (31771655), and the Science and Technology Commission of Shanghai Municipality (19JC1412300). Yuan Wang is partially supported by USDA National Institute of Food and Agriculture Hatch Project (1015058).
Institutional Review Board Statement
The study was conducted in accordance with the local Animal Welfare Act and Public Health Service Policy (consistent with the WMA Statement on Animal Use in Biomedical Research) and approved by the Ethics Committee of East China Normal University (Protocol ID: m20190403) on 1 April 2019.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the article and Supplementary Materials.
Acknowledgments
We would like to thank Shuiping Li (Shanghai Jiao Tong University School of Medicine) for image processing.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Mfn1 | Mitofusin 1 |
| Mfn2 | Mitofusin 2 |
| SYCP3 | Synaptonemal complex protein 3 |
| TRA98 | Germ cell specific antigen 98 |
| mtDNAs | Mitochondrial DNAs |
| OPA1 | Optic atrophy 1 |
| DRP1 | Dynamin-related protein 1 |
| Prm1 | Protamine 1 |
| cKO | Conditional knockout |
| IHF | Immunohistofluorescence |
| dpp | Days postpartum |
| MEFs | Mouse embryonic fibroblasts |
| PCR | Polymerase chain reaction |
| PFA | Paraformaldehyde |
References
- Chen, H.; Chan, D.C. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metab. 2017, 26, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Mishra, P.; Chan, D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 634–646. [Google Scholar] [CrossRef] [Green Version]
- Samiec, M.; Skrzyszowska, M. Extranuclear Inheritance of Mitochondrial Genome and Epigenetic Reprogrammability of Chromosomal Telomeres in Somatic Cell Cloning of Mammals. Int. J. Mol. Sci. 2021, 22, 3099. [Google Scholar] [CrossRef]
- Shaw, J.M.; Nunnari, J. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 2002, 12, 178–184. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Chan, D.C. Mitochondrial Dynamics in Mammals. Curr. Top. Dev. Biol. 2004, 59, 119–144. [Google Scholar] [CrossRef]
- Chan, D.C. Fusion and Fission: Interlinked Processes Critical for Mitochondrial Health. Annu. Rev. Genet. 2012, 46, 265–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labbé, K.; Murley, A.; Nunnari, J. Determinants and Functions of Mitochondrial Behavior. Annu. Rev. Cell Dev. Biol. 2014, 30, 357–391. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C. Mitochondrial Fusion and Fission in Mammals. Annu. Rev. Cell Dev. Biol. 2006, 22, 79–99. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003, 160, 189–200. [Google Scholar] [CrossRef]
- Hsiuchen, C.J. Michael McCaffery, David C Chan. Mitochondrial fusion protects against neurodegeneration in the cere-bellum. Cell 2007, 130, 548–562. [Google Scholar]
- Zhang, J.; Wang, Q.; Wang, M.; Jiang, M.; Wang, Y.; Sun, Y.; Wang, J.; Xie, T.; Tang, C.; Tang, N.; et al. GASZ and mi-tofusin-mediated mitochondrial functions are crucial for spermatogenesis. EMBO Rep. 2016, 17, 220–234. [Google Scholar] [CrossRef] [Green Version]
- Varuzhanyan, G.; Rojansky, R.; Sweredoski, M.J.; Graham, R.L.; Hess, S.; Ladinsky, M.S.; Chan, D.C. Mitochondrial fusion is required for spermatogonial differentiation and meiosis. eLife 2019, 8, 51601. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Sun, Y.; Sun, Q.; Zhang, J.; Jiang, M.; Chang, C.; Huang, X.; Wang, C.; Wang, P.; Zhang, Z.; et al. MFN2 Plays a Distinct Role from MFN1 in Regulating Spermatogonial Differentiation. Stem Cell Rep. 2020, 14, 803–817. [Google Scholar] [CrossRef]
- Wang, X.; Wen, Y.; Zhang, J. Grace Swanson, Shuangshuang Guo, Congcong Cao, Stephen A Krawetz, Zhibing Zhang, Shuiqiao Yuan. Mitochondrial fusion and fission in mammals. Development 2021, 148, dev196295. [Google Scholar] [CrossRef]
- Honda, S. Shigehisa Hirose. Stage-specific enhanced expression of mitochondrial fusion and fission factors during sper-matogenesis in rat testis. Biochem. Biophys. Res. Commun. 2003, 11, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Vadnais, M.L.; Lin, A.M.; Gerton, G.L. Mitochondrial fusion protein MFN2 interacts with the mitostatin-related protein MNS1 required for mouse sperm flagellar structure and function. Cilia 2014, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Chan, D.C. The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol. Biol. Cell 2015, 26, 4466–4477. [Google Scholar] [CrossRef]
- Varuzhanyan, G.; Chen, H.; Rojansky, R.; Ladinsky, M.S.; Michael, J. McCaffery, David C Chan. Mitochon-drial fission factor (Mff) is required for organization of the mitochondrial sheath in spermatids. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129845. [Google Scholar] [CrossRef]
- Varuzhanyan, G.; Ladinsky, M.S.; Yamashita, S.-I.; Abe, M.; Sakimura, K.; Kanki, T.; Chan, D.C. Fis1 ablation in the male germline disrupts mitochondrial morphology and mitophagy, and arrests spermatid maturation. Development 2021, 148, 199686. [Google Scholar] [CrossRef]
- Hecht, N.B.; Bower, P.A.; Waters, S.H.; Yelick, P.C.; Distel, R.J. Evidence for haploid expression of mouse testicular genes. Exp. Cell Res. 1986, 164, 183–190. [Google Scholar] [CrossRef]
- Matsumura, H.; Hasuwa, H.; Inoue, N.; Ikawa, M.; Okabe, M. Lineage-specific cell disruption in living mice by Cre-mediated expression of diphtheria toxin A chain. Biochem. Biophys. Res. Commun. 2004, 321, 275–279. [Google Scholar] [CrossRef]
- Tanaka, H.; Pereira, L.A.V.D.; Nozaki, M.; Tsuchida, J.; Sawada, K.; Mori, H.; Nishimune, Y. A germ cell-specific nuclear antigen recognized by a monoclonal antibody raised against mouse testicular germ cells. Int. J. Androl. 1998, 20, 361–366. [Google Scholar] [CrossRef]
- Liu, J.-G.; Yuan, L.; Brundell, E.; Bjorkroth, B.; Daneholt, B.; Höög, C. Localization of the N-terminus of SCP1 to the Central Element of the Synaptonemal Complex and Evidence for Direct Interactions between the N-termini of SCP1 Molecules Organized Head-to-Head. Exp. Cell Res. 1996, 226, 11–19. [Google Scholar] [CrossRef]
- Santel, A.; Fuller, M.T. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 2001, 114, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Legros, F.; Lombès, A.; Frachon, P.; Rojo, M. Mitochondrial Fusion in Human Cells Is Efficient, Requires the Inner Membrane Potential, and Is Mediated by Mitofusins. Mol. Biol. Cell 2002, 13, 4343–4354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojo, M.; Legros, F. Danielle Chateau, Anne Lombès. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 2002, 115, 1663–1674. [Google Scholar] [CrossRef] [PubMed]
- Santel, A.; Frank, S.; Gaume, B.; Herrler, M.; Youle, R.J.; Fuller, M.T. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J. Cell Sci. 2003, 116, 2763–2774. [Google Scholar] [CrossRef] [Green Version]
- Cipolat, S.; De Brito, O.M.; Zilio, B.D.; Scorrano, L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA 2004, 101, 15927–15932. [Google Scholar] [CrossRef] [Green Version]
- Ishihara, N.; Eura, Y.; Mihara, K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 2004, 117, 6535–6546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Chomyn, A.; Chan, D.C. Disruption of Fusion Results in Mitochondrial Heterogeneity and Dysfunction. J. Biol. Chem. 2005, 280, 26185–26192. [Google Scholar] [CrossRef] [Green Version]
- O’Gorman, S.; Dagenais, N.A.; Qian, M.; Marchuk, Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 1997, 94, 14602–14607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batista, F.; Lu, L.; Williams, S.A.; Stanley, P. Complex N-Glycans Are Essential, but Core 1 and 2 Mucin O-Glycans, O-Fucose Glycans, and NOTCH1 Are Dispensable, for Mammalian Spermatogenesis1. Biol. Reprod. 2012, 86, 179. [Google Scholar] [CrossRef] [PubMed]
- Diao, F.; Jiang, C.; Wang, X.-X.; Zhu, R.-L.; Wang, Q.; Yao, B.; Li, C.-J. Alteration of protein prenylation promotes spermatogonial differentiation and exhausts spermatogonial stem cells in newborn mice. Sci. Rep. 2016, 6, 28917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Vermulst, M.; Wang, Y.E.; Chomyn, A.; Prolla, T.A.; McCaffery, M.J. David C Chan. Mito-chondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 2010, 141, 280–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Martino, C.; Floridi, A.; Marcante, M.L.; Malorni, W.; Barcellona, P.S.; Bellocci, M.; Silvestrini, B. Morphological, histochemical and biochemical studies on germ cell mitochondria of normal rats. Z. Zellforsch. Mikrosk. Anat. 1979, 196, 1–22. [Google Scholar] [CrossRef]
- Ramalho-Santos, J.; Varum, S.; Amaral, S.; Mota, P.C.; Sousa, A.P.; Amaral, A. Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Updat. 2009, 15, 553–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaral, A.; Lourenço, B.; Marques, M.; Ramalho-Santos, J. Mitochondria functionality and sperm quality. Reproduction 2013, 146, R163–R174. [Google Scholar] [CrossRef] [Green Version]
- Varuzhanyan, G.; Chan, D.C. Mitochondrial dynamics during spermatogenesis. J. Cell Sci. 2020, 133, 23593. [Google Scholar] [CrossRef] [PubMed]
- Otani, H.; Tanaka, O.; Kasai, K.-I.; Yoshioka, T. Development of mitochondrial helical sheath in the middle piece of the mouse spermatid tail: Regular dispositions and synchronized changes. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 1988, 222, 26–33. [Google Scholar] [CrossRef]
- Ho, H.-C.; Wey, S. Three-dimensional rendering of the mitochondrial sheath morphogenesis during mouse spermi-ogenesis. Microsc. Res. Tech. 2007, 70, 719–723. [Google Scholar] [CrossRef]
- Pham, A.H.; McCaffery, J.M.; Chan, D.C. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genes 2012, 50, 833–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Wang, P.; Liang, D.; Wang, Y. BRG1 Is Dispensable for Sertoli Cell Development and Functions in Mice. Int. J. Mol. Sci. 2020, 21, 4358. [Google Scholar] [CrossRef] [PubMed]
- Truett, G.; Heeger, P.; Mynatt, R.; Truett, A.; Walker, J.; Warman, M. Preparation of PCR-Quality Mouse Genomic DNA with Hot Sodium Hydroxide and Tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).