Abstract
Pathogenic variants in CRB1 lead to diverse recessive retinal disorders from severe Leber congenital amaurosis to isolated macular dystrophy. Until recently, no clear phenotype-genotype correlation and no appropriate mouse models existed. Herein, we reappraise the phenotype-genotype correlation of 50 patients with regards to the recently identified CRB1 isoforms: a canonical long isoform A localized in Müller cells (12 exons) and a short isoform B predominant in photoreceptors (7 exons). Twenty-eight patients with early onset retinal dystrophy (EORD) consistently had a severe Müller impairment, with variable impact on the photoreceptors, regardless of isoform B expression. Among them, two patients expressing wild type isoform B carried one variant in exon 12, which specifically damaged intracellular protein interactions in Müller cells. Thirteen retinitis pigmentosa patients had mainly missense variants in laminin G-like domains and expressed at least 50% of isoform A. Eight patients with the c.498_506del variant had macular dystrophy. In one family homozygous for the c.1562C>T variant, the brother had EORD and the sister macular dystrophy. In contrast with the mouse model, these data highlight the key role of Müller cells in the severity of CRB1-related dystrophies in humans, which should be taken into consideration for future clinical trials.
1. Introduction
Inherited retinal dystrophies (IRDs), despite clinical and genetic heterogeneity, share progressive degeneration of photoreceptors and/or retinal pigment epithelium cells. IRDs affect about 2 million people worldwide among the 300 million patients with inherited diseases. They account for 10% of the causes of visual impairment in Western countries. IRDs are classified according to the predominant cell type affected (rod-cone dystrophy, RCD, versus cone-rod dystrophy, CRD) and/or topography of the lesions (maculopathy versus generalized retinal dystrophy). Thus, full-field electroretinogram (ff-ERG) and multimodal imaging are both essential for IRD classification. RCD (also known as retinitis pigmentosa, RP) is the most frequent phenotype with an estimated prevalence of 1/4000. Maculopathies account for 20% of cases, with Stargardt disease being the most prevalent [1,2]. A distinction is made between IRDs with an early onset (early onset retinal dystrophy, EORD), including Leber congenital amaurosis (LCA), and those with a later onset after the age of 5 years. All modes of inheritance have been reported in these disorders, autosomal recessive (aR), dominant (aD), X-linked or, more rarely, mitochondrial.
Since the advent of next-generation sequencing techniques, LCA-related genes, such as CRX or RDH12, have been unexpectedly linked to isolated maculopathies [3,4,5,6]. Tsang et al. reported the first cases of macular dystrophy with bi-allelic pathogenic variants in CRB1 [7]. Prior to this, CRB1 had only been associated with EORD (where it was responsible for 10 to 13% of LCA cases) or RP (responsible for 8% of cases) with specific clinical features, such as para-arteriolar preservation of the retinal pigment epithelium, peripheral nummular pigmentation or Coats-like disease [8,9,10,11,12,13,14,15,16].
The CRB1 (Crumbs homologue 1) protein belongs to the CRB complex, an apical complex important for cell junctions, and acts as a cell polarity regulator (see [17] for review); it also plays a major role during retinal development [13,18,19,20]. The CRB1 gene contains 12 exons spanning 210 kb with 12 predicted transcripts using alternative exons (https://www.ensembl.org (accessed on 25 October 2021)). In the retina, two major transcripts, which encode cell-surface proteins, are mainly expressed in Müller cells and in photoreceptors (Figure 1 and Figure 2) [21]. Müller cells express exclusively the canonical transcript named CRB1-A [MT470365] encoding a 1406 amino acid protein with 19 EGF-like domains and 3 laminin G domains. Photoreceptors mainly express the CRB1-B [MT470366] transcript encoding a 1003 amino acid protein. Transcript A is the most abundant during development and transcript B is predominant in adult retina [21]. A third transcript named CRB1-C [MT470367] encoding a 754 amino acid protein without transmembrane and intracellular domains was also reported but is less abundant than the CRB1-A and CRB1-B transcripts [21]. To date, 457 CRB1 pathogenic variations have been described in HGMD professional 2021.3 database (https://hgmd/pro/ (accessed on 25 October 2021)): 333 missense/nonsense variants, 28 splice variants, 1 regulatory substitution, 86 small insertions/deletions and 9 large insertions/deletions. One specific pathogenic variation, c.498-506del, which is an in-frame deletion, is specifically encountered in CRB1 related macular phenotypes [7,22].
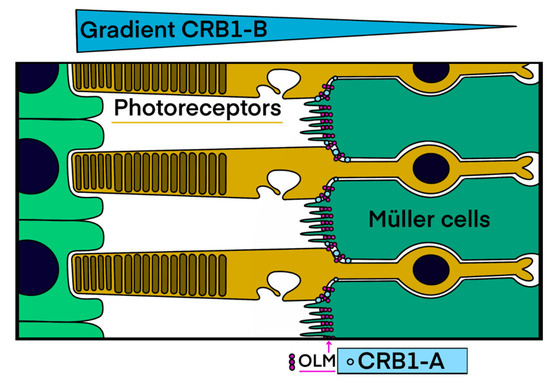
Figure 1.
CRB1 transmembrane isoforms: cell specificity and localization. Isoform A is located at the apical zone of Müller cells and isoform B is localized in photoreceptors with a higher expression reported in the outer segments on the side of the retinal pigment epithelium. Blue circles = CRB1-A transmembrane isoforms of Müller cells. Pink circles = OLM = outer limiting membrane (drawn by M.P.) formed by the adherens junctions between neighboring Müller cells and photoreceptors.
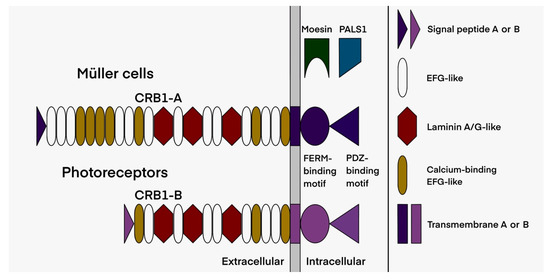
Figure 2.
Schematic representation of the two main retinal isoforms encoded by the CRB1 gene. Isoform B of the photoreceptors differs from the canonical isoform A of the Müller cells by a lower number of EGF-like domains, its specific signal peptide, as well as its transmembrane and intracellular domains (drawn by M.P.).
In this retrospective multicenter observational study, we reappraise for the first time our cohort of 50 patients with bi-allelic pathogenic variants in CRB1 to refine phenotype-genotype correlations in light of recent knowledge on CRB1 isoforms and their retinal cell expression.
2. Results
2.1. Clinical Data
Fifty patients (45 families) with bi-allelic pathogenic variants were included in the study. The group comprised 25 men and 25 women. Thirty-seven patients were reexamined with a median follow-up of 3.9 years (range 3 months to 19 years). Clinical and imaging data are summarized in Table 1. Twenty-eight patients were classified as EORD, thirteen as RP and nine as isolated macular dystrophy based on age of onset, symptomatology, multimodal imaging and ff-ERG responses. In one family (M-1640), the brother had EORD and the sister macular dystrophy (Figure 3).

Table 1.
Clinical and genetic data of the 50 patients with bi-allelic pathogenic variants in CRB1 classified according to phenotype. Early onset retinal dystrophy (EORD) in 28 patients, retinitis pigmentosa (RP; or rod-cone dystrophy) in 13 patients and macular dystrophy (MD) in 9 patients. HM: hand motion, LP: light perception; ND: not done; PPARPE: preserved para-arteriolar retinal pigment epithelium.
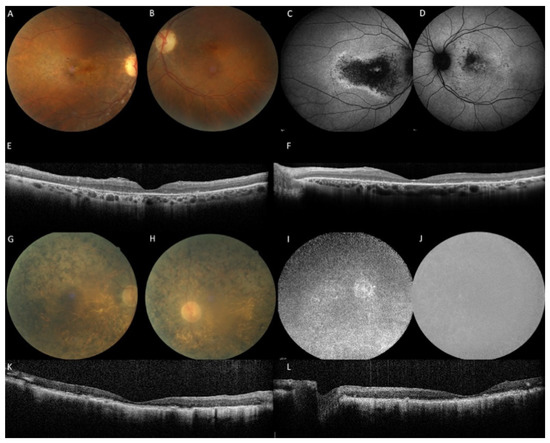
Figure 3.
Family M-1640: the sister with an aR macular phenotype and the brother with an aR EORD. (A–F): The sister at the age of 45 years with a disease onset at 42 years complained of slow bilateral visual loss (RE 20/60, LE 20/30). No photophobia, no night blindness. Note the multiple yellow granular spots on the color frames (A,B). Peripheral retina and vessels are preserved. On fundus autofluorescence frames (FAF), the macular hypoautofluorescence is asymmetric and more extensive in the right eye. On SD-OCT scans (E,F), the macular is thickened with cystic degeneration and hyperreflective dots, in line with CRB1-related dystrophy. EZ and IZ lines are not visible except within the foveola in the left eye. (G–L): The brother: the visual acuity is hand motion in both eyes at the age of 46 years, night blindness appeared at 3 years, reading loss at 20 years, major difficulties to move alone at 30 years. On color frames, note the optic disc pallor, vascular attenuation and pigmentary changes of the posterior pole and the entire peripheral retina with atrophic zone. FAF (I,J) revealed severe diffuse hypoautofluorescence patches. SD-OCT (K,L) shows major alterations of the retinal segmentation with a retinal thickening and a complete loss of EZ and IZ lines.
Considering the classical anomalies encountered in CRB1 cases, twelve patients had papillary drusen and fourteen had para-arteriolar preservation of the retinal pigment epithelium. In generalized retinal dystrophy with exclusion of LCA cases (no relevant OCT measurements), we observed a macular thickening with loss of the retinal reflectivity segmentation pattern. This thickening was noted during the first three decades compared to a control group (Table 2). Such a retinal thickening is not the rule in retinal dystrophies, which are characterized by a constant and progressive cell loss. This is related to the specific role of CRB1 during retinal development, thus, the retina is initially thickened and disorganized. Macular cystoid edema occurred mainly in the first two decades in the EORD or RP patients, and in the third decade in maculopathy patients (Figure 4). In the macular group, visual anomalies were noted during the third decade (mean age of onset: 23.5; range 16 to 42 years). In the family of proband L-93112991 with initial maculopathy, the proband progressed with time to a generalized retinal dystrophy whereas his brother had a less severe progression.

Table 2.
SD-OCT macular measurements. Retinal thickness in microns. EORD: early onset retinal dystrophy. Controls were matched by age and sex. SD = standard deviation. T1500 and T3000 = retinal thickness at 1500 and 3000 μ temporal of the fovea, N1500 = retinal thickness at 1500 μ nasal of the fovea. * SD not calculated because measurement was made only on one eye.
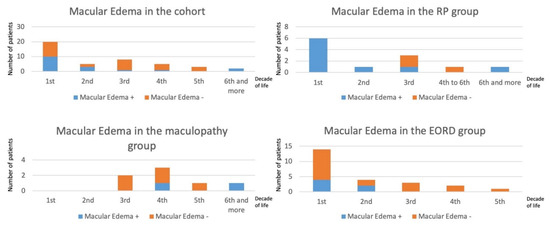
Figure 4.
Prevalence of macular edema in EORD, RP and maculopathy by decades. The number of patients per decade is indicated in the vertical axes. The age of patients (in years) is indicated in the horizontal axes. The more severe the phenotype, the earlier the cystic changes occurred.
2.2. Genetic Results
All genetic data are summarized in Table 3. In our cohort, 41 different pathogenic variants are reported: 26 missense (63% of the total variants), nine indel, four nonsense variants, one splice variation and one leading to the loss of the final stop codon, resulting in a protein extended by 110 amino acids. Most of these variants (27/41) are localized in the exons 6, 7 and 9, thus affecting both CRB1-A and CRB1-B isoforms (Figure 2 and Figure 5).

Table 3.
Pathogenic variants in CRB1 in each affected patient and probable impact on specific Müller cell and photoreceptor isoforms. From left to the right: Patient ID; allele 1 (protein annotation whenever possible), impact on isoform A and isoform B (protein domain level) from allele 1; allele 2, impact on isoform A and isoform B from allele 2 (protein domain level); final impact on protein CRB1 in Müller cells and final impact on CRB1 in photoreceptors (both in terms of amount of WT and mutated proteins). WT = wild type. NMD = presumed nonsense-mediated decay when stop codon or frameshift is present before the penultimate exon.
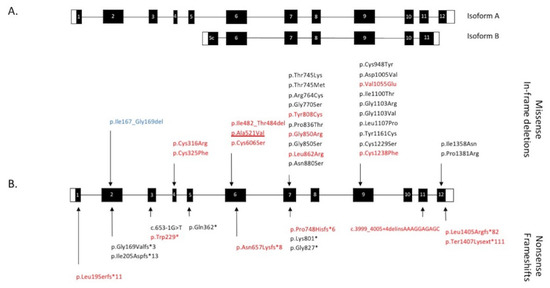
Figure 5.
Schematic representation of the CRB1 gene and pathogenic variants. (A) The 12 exons of the canonical tran-script encoding isoform A and the equivalent exons comprising the smaller transcript encoding isoform B. (B) Distribu-tion of CRB1 variants (protein or cDNA level when the consequences on the protein are unknown) found in our 50-patient cohort. Missense variations and in-frame deletions are indicated above the gene structure, nonsense varia-tions and frameshifts are noted below. In red: novel variants. Underlined: specific variant responsible for both EORD and maculopathy. In blue: in-frame deletion encountered in macular dystrophy. White boxes: 5’ and 3’ UTR. Black box-es: coding parts of exons. White numbers in black boxes indicate exon number related to the canonical transcript (CRB1-A).
Bi-allelic indel or nonsense pathogenic variants leading to loss of function, due to nonsense mediated decay (NMD) activation, were noted in seven EORD cases versus none in RP cases (Table 3). Bi-allelic missense pathogenic variants were observed in 14 EORD and in nine RP cases. The recurrent pathogenic variant c.2290C>T, p.(Arg764Cys) [23] was frequent and carried by ten patients. Four of them were homozygous for this variation, three presenting EORD and one RP. All patients with a macular phenotype, except one, carried one allele with the c.498_506del, p.(Ile167_Gly169del) variant. The remaining patient with a macular phenotype was homozygous for the c.1562C>T, p.(Ala521Val) variant, whereas his brother had an EORD.
2.3. Assessment of the Functional Effect of CRB1 Variants
We identified a number of known pathogenic CRB1 variants in our cohort: p.Gly169Valfs*3 [24]; p.Ile205Aspfs*13 [25]; c.653-1G > T [26]; p.Gln362* [27]; p.Thr745Met [23]; p.Thr745Lys [28]; p.Arg764Cys [23]; p.Gly770Ser [29]; p.Lys801* [30]; p.Gly827* [31]; p.Pro836Thr and p.Gly850Ser [9]; p.Asn880Ser [32]; p.Cys948Tyr [23]; p.Asp1005Val [16]; p.Ile1100Thr [33]; p.Gly1103Arg [34]; p.Gly1103Val [26]; p.Leu1107Pro [31]; p.Tyr1161Cys [35]; p.cys1229Ser [36]; p.Ile1358Asn [37]; p.Pro1381Arg [38].
In addition, we identified 17 novel variants (Table 4). Among these, three indels and the nonsense variant in exon 3 probably lead to NMD, as they are situated before the penultimate exon. This is not the case for the two novel variants in exon 12 that presumably lead to an extension of the intracellular part of the protein, a region interacting with other partners, such as PALS1 (protein associated with lin seven 1) [17,39,40,41], which stabilizes the CRB complex, and Moesin [17,42], which combines with the FERM-binding domain and is involved in epithelial to mesenchymal transition. We also describe nine novel missense variations. Two of these variants, p.(Ala521Val) and p.(Cys606Ser), are predicted to destabilize the laminin domain by bringing a larger hydrophobic residue valine into a buried position or by breaking a conserved disulfide bridge (Figure 6A). A similar situation is encountered for two variants, p.(Gly850Arg) and p.(Leu862Arg), in the second laminin domain (Figure 6B). The large and polar arginine residues can hardly be accommodated in the buried and compact environment of Gly850 and Leu862. By contrast, the variant p.(Tyr808Cys) is predicted to occur at the surface of this domain. However, the added cysteine would favor improper disulfide bridge formation in the extracellular environment, resulting in incorrect macromolecular assembly. The variant p.(Val1055Glu) again brings a larger and polar residue into a position buried in the third laminin domain, and hence, it is also predicted to strongly destabilize the corresponding domain (Figure 6C). Finally, the variant p.(Cys1238Phe) will abolish a predicted disulfide bridge in another EGF-like domain and again prevent its proper folding or impact its overall stability (Figure 6F).

Table 4.
Novel CRB1 pathogenic variants found in our 50-patient cohort. For each variant, we indicate the cDNA and protein position relative to the transcript NM_201253.3, the exon number, the type of variant (indel, insertion and deletion of one or several nucleotides, Del, deletion), the frequency of the variant in the database gnomAD, the result of in silico pathogenicity predictions (pathogenic versus benign predictions) and number of programs, the ACMG classification (P, pathogenic; LP, likely pathogenic) and the domain impacted in the canonical protein (TM, transmembrane domain).
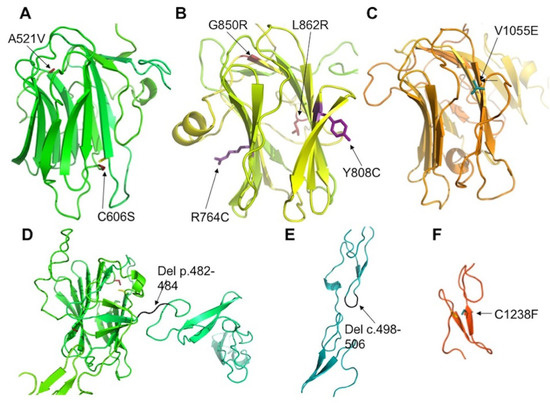
Figure 6.
Positions of mutated residues in CRB1. Models of the 1st (A), 2nd (B) and 3rd (C) laminin domains of CRB1 shown as colored ribbons with mutated residues indicated as sticks colored firebrick (A), violet (B) or cyan (C). (D,E): Deletions in EGF-like modules are shown in black. Variant p.(Cys1238Phe) (C1238F) is shown in cyan. In addition, cysteines involved in disulfide bridge with mutated cysteines are shown as yellow sticks in (A,F).
2.4. Impact of CRB1-A and CRB1-B Variants on the Phenotype
We then reappraised the clinical data regarding the localization and the consequences of the genomic variants on both isoforms: the specific Müller isoform CRB1-A (encoded by 12 exons) and the photoreceptor isoform CRB1-B (7 exons from 5c to 11). The two isoforms differ in their N- and C-termini (Figure 2). The predicated impact of the different pathogenic variants on the isoforms, in terms of the estimated levels of mutant and wild type isoforms in Müller cells and in photoreceptors, is described in Table 3. The CRB1 isoform B has no impact on the severity of the phenotype as EORD, RP and macular dystrophy can occur regardless of the level of isoform B (Figure 7). For example, patients M-116 and M-4621 presenting an EORD, carried two pathogenic variants (c.4219T>A and c.4142C>G, respectively) that did not affect isoform B. These variants are localized in exon 12 and affect the cytoplasmic C-terminal domains. The patient M-4621 was homozygous for the pathogenic variant c.4142C>G, p.(Pro1381Arg) which modifies the FERM-binding domain interacting with the protein membrane Moesin (Figure 8). In patient M-116, no isoform A was produced, as both alleles carried loss-of-function variants affecting specifically the Müller cell isoform. The p.(Trp229*) variant in exon 3 is predicted to lead to a shorter CRB1-A protein truncated after the 5th EGF-like domain, thus, if the protein were expressed, it would be likely degraded. On the other allele, the p.(Ter1407Lysext*111) variant leads to an abnormal C-terminal elongated isoform, affecting the PDZ domain. The second example of a dystrophy associated with the normal isoform B is the maculopathy presented by the two patients (M-2183 and M-3343) carrying the specific c.498_506del macular variant. Both patients carried on the second allele variants located in exons 4 and 12, respectively, that did not affect isoform B. Patient M-3343 also carried the pathogenic variant p.(Pro1381Arg) modifying the FERM-binding domain. Furthermore, all patients with the specific c.498_506del macular variant developed a macular dystrophy regardless of the impact on isoform B.
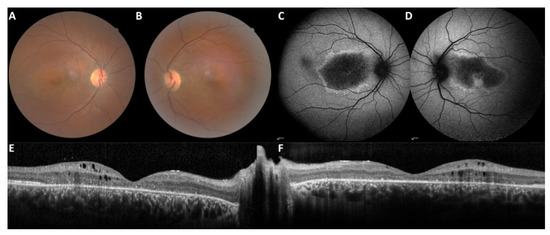
Figure 7.
Autosomal recessive macular dystrophy in patient M-3989 carrying the c.498_506del on one allele. This patient, seen at the age of 33 years (disease onset at 17 years), had a bilateral visual loss still in progress. No photophobia, no night blindness. VA 20/400 right eye, 20/25 left eye. Note on the color frames (A,B), the multiple yellow granular macular pattern and, on the SD-OCT scan (E,F), macular thickening, cystic degeneration and hyperreflective dots in line with CRB1-related diseases. On FAF (C,D), the macular lesion is deeply hypoautofluorescent and surrounded by a hyperautofluorescent border. Note the foveal preservation in the left eye.
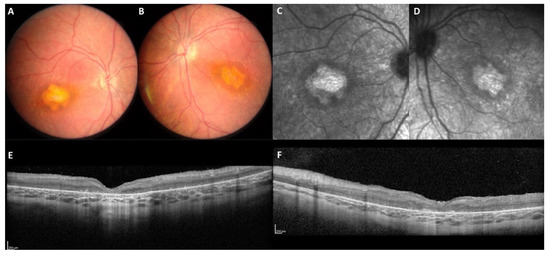
Figure 8.
Autosomal recessive EORD in patient M-4621 homozygous for the c.4142C>G p.(Pro1381Arg) in exon 12. This patient was seen at the age of 4.5 years for low vision in both eyes. Isoform B was not impacted by the homozygous variant (WT isoform B in photoreceptors). On color frames (A,B), the macula was altered with an abnormal yellowish appearance. On infrared reflectance imaging (C,D), note the atrophic pattern of the abnormal whitish macular lesion. On the SD-OCT scans (E,F), the retina is completely thickened and disorganized, no correct segmentation is visible.
Among the 13 patients with RP, nine had missense variants on both alleles and four had one missense and one frameshift variant. None of the RP patients presented a pathogenic variant in the intracytoplasmic domain of the isoform CRB1-A nor in the EGF-like 8 domain encoded by exon 4.
Interestingly, the p.(Lys801*) variant, which leads to either a shorter protein or to the absence of protein synthesis via NMD activation, was associated with two different phenotypes when present in trans with a variant specific to the Müller cell isoform. Patients M-3073-1 and 3073-3 carrying p.(Lys801*) in trans with the c.498_506del variant affecting the EGF-like 4 domain (exon 2) manifested a maculopathy, whereas patient M-1732 carrying p.(Lys801*) in trans with c.974G>T, p.(Cys325Phe) had EORD. This latter variant disrupts a disulfide bond in the EGF-like 8 domain. Another cysteine change in the EGF-like 8 domain of exon 4 in position 316 was observed in patient M-2183, also a carrier in trans of c.498_506del. The patient manifested a maculopathy with an abnormal ERG showing reduced (50%) rod and cone responses.
3. Discussion
We demonstrate that CRB1-related maculopathy is not a rare phenotype. In this large cohort, it occurs in nine patients, of which all except one carried the specific c.498_506del pathogenic variant in one allele. Bi-allelic nonsense and frameshift variants can be noted in EORD, including LCA, as well as in RP, but NMD is more frequently predicted in EORD (22/28) than in RP (2/13). In this cohort, macular dystrophy and EORD occurs even if the CRB1-B isoform is not impacted by pathogenic CRB1 variants.
3.1. CRB1-Related Retinal Dystrophies
All the patients studied necessarily had bi-allelic variants modifying CRB1-A, the canonical long isoform, as the next generation sequencing (NGS) gene panel used for molecular diagnosis has not been designed to screen the specific region of the shorter isoform B. These two isoforms have a specific cell distribution and a differential expression pattern in the retina during the embryonic and adult periods [21]. CRB1-A is localized to the apical tips of Müller cells and CRB1-B in the inner and outer segments with a gradient (Figure 1). These cell-surface proteins are both present at the level of the outer limiting membrane (OLM), which is formed by the adherens junctions between the adjacent Müller cells and photoreceptors, creating a distinct barrier between the neural retina and the inner/outer segments. CRB1 expression is variable: CRB1-A is the predominant isoform during developmental stages. By contrast, CRB1-B is by far the most abundant isoform in the adult human, as well as murine retina. Thus, there is a relative CRB1-A depletion in mature human retina.
3.1.1. The Macular Phenotype Is Determined by the c.498_506del Variant Specific to CRB1-A
All patients with macular dystrophy, except one, had the variant c.498_506del specific to isoform A. It appears that a maculopathy can also be observed even if isoform B is normal, such as in patients M-2183 and M-3343. The remaining patients with a macular dystrophy carried c.498_506del with in trans missense and nonsense variants also found in EORD. The maculopathy is not the consequence of the impairment of CRB1 at developmental stages, as the lamination of the retina is preserved outside the macular zone on SD-OCT and its onset occurs most frequently in the third decade of life.
The specific macular c.498_506del variant leads to the formation of a protein lacking 3 amino acids belonging to the Ca2+ EGF-like 4 domain. With the lack of a corresponding mouse model, the mechanism of this variant is not clearly known. Recently, human induced pluripotent stem cells [43] were generated from a patient with a macular dystrophy carrying the c.498_506del in trans with the c.613_619del variant in the aim of studying the dysfunction and changes in Müller cells and photoreceptors in human retinal organoids. These patient-specific organoids will provide valuable insights into the specific macular pathophysiology specifically associated with the c.498_506del variant. We can hypothesize that this in-frame deletion abolishes a calcium binding site in two consecutive EGF-like modules (EGF-4 and EGF-5) of CRB1. The deletion is also expected to strongly destabilize the local folding, as well as the relative orientation of those two EGF-like modules, and hence the proper structural organization of the whole protein.
Despite the lack of a precise mechanism, our data underline that the variant c.498_506del drives the macular phenotype independently of CRB1-B photoreceptor impairment.
3.1.2. CRB1 Variants and Structure Function Correlation in EORD and in RP
Twenty-six patients with EORD had a combined impairment of one or two alleles of the CRB1-B isoform. A haploinsufficiency in CRB1-B, the photoreceptor isoform, is not the mechanism that leads to a generalized retinal phenotype, as patients M-3073-1 and -3 who had 50% of the WT CRB1-B isoform only presented with a macular dystrophy. Moreover, it should be noted that two patients with bi-allelic variants only belonging to isoform A, and thus producing a normal photoreceptor isoform B, still presented with EORD.
Taken together, these data suggest that the phenotype in CRB1 patients is also dependent on the severity of the Müller cell impairment related to specific CRB1-A variant combinations. The two patients with EORD and WT CRB1-B both have variants in exon 12. One patient is homozygous for the variant c.4142C>G p.(Pro1381Arg) in exon 12. The second patient is pseudo-homozygous for the variant c.4219T>A p.(Ter1407Lysext*111) in exon 12 with an abnormal elongation of the protein in its intracellular part, as the variant in trans, c.687G>A (p.Trp229*) in exon 3, leads to NMD. It appears that there is a high severity associated with variants modifying the transmembrane or intracellular domains (exon 12).
Mutations in exon 12 likely affect the functioning of CRB1 in different manners. First, p.(Ile1358Asn) is predicted to destabilize the transmembrane segment due to the replacement of a hydrophobic isoleucine with a polar asparagine. This could lead to improper membrane insertion or alternatively to improper multimerization to hide the polar sidechain. The variant p.(Pro1381Arg) occurs in the vicinity of the phosphorylation site Thr1378 (see https://www.phosphosite.org/ (accessed on 25 October 2021)). The positively charged arginine could either (i) prevent recognition by the protein-kinase or (ii) stabilize the phosphorylated form of CRB1 and hence prevent association with other partners, such as PALS1 and other PDZ-containing proteins. The two other mutants have lost their PDZ binding motif in the C-terminus of CRB1 due to sequence extension and/or substitution/addition of a charged residue (arginine or lysine). Such variants would directly prevent interactions with various PDZ-containing partners, such as PALS1. This likely impacts the function of CRB1 to the same extent as the two other mutations discussed above.
Among the patients with the c.2401A>T, p.(Lys801*) variant and 50% WT isoform B (M-1732, M-3073-1 and M-3073-3), patient M-1732 who carries in trans the variant c.974G>T, p.(Cys325Phe) had EORD. In the same manner, patient M-2183 with the c.946T>C, p.(Cys316Arg) had a moderate decrease in rod and cone responses. These cases could suggest that variants affecting the EGF-like 8 domain in exon 4 could drive a more severe Müller cell impairment. Regarding the two mutations in exon 4, p.(Cys325Phe) and p.(Cys316Arg), modeling of the corresponding EGF-like domain suggests that both would be deleterious for the proper folding and/or stability of the local structure: p.(Cys325Phe) would prevent the formation of a predicted disulfide bridge important for the EGF-like structure; p.(Cys316Arg) would replace a small and buried residue with a large and polar arginine, likely preventing proper folding of the corresponding domain. It is also predicted to break up a conserved disulfide bridge, similar to p.(Cys325Phe).
Patients with RP have mainly missense variants and none of them carry a pathogenic variant in CRB1 exons 4 or 12. This again highlights that the EGF-like 8 and the intracellular domains are essential for the proper function of Müller cells.
3.2. Intrafamilial Variation Is an Exception
Among the 45 families studied, a heterogeneity in phenotype was only noted in one consanguineous family. In family M-1640, homozygous for the novel c.1562C>T, p.(Ala521Val) variant in exon 6, the proband had EORD with indiscernible responses of ff-ERG, whereas his sister had a maculopathy with discernible responses. There was no additional pathogenic variant in any of the 230 retinal dystrophy genes analyzed on this NGS panel, which included the CRB2 gene. As the c.1562C>T variant is located in an exon shared by both isoforms A and B, we cannot rule out that the phenotypic difference could be explained by an additional variant present in the CRB1 isoform B-specific regions not covered by the NGS panel (N-terminus and C-terminal parts including 58 amino acids). Lastly, a splice defect of this variant was ruled out according to MobiDetails prediction.
3.3. Limitations of Mouse Models, Concordance and Discordance
Diverse Crb1 mouse models have been reported to date [17]. Recently, Ray et al. designed specific novel mutant mouse models to evaluate each isoform separately (Crb1ex1, Crb1delB/delB), as well as their combined functions (Crb1null) [21]. All the CRB1 transcripts are effectively disrupted in the novel Crb1null mouse model (CRISPR-mediated deletion of exons common to all Crb1 isoforms) [21]. Similarly, Crb1-A and Crb1-B are both disrupted in previously described rd8 mouse models (1-bp frameshift deletion in exon 9 leading to the loss of the transmembrane and intracellular domains), whereas Crb1-A isoform is specifically lost in the Crb1ex1 knockout model (deletion of the promotor region of the first exon of canonical isoform A) [17]. By contrast, isoform B is specifically lost in the novel designed Crb1delB/null and in Crb1delB/delB models (CRISPR-mediated deletion in the promotor and the first exon of isoform B) [21]. Photoreceptor displacement and rosette formation were described in rd8 and Crb1ex1 knockout model, due to the loss of adhesion between photoreceptors and Müller cells [17]. Ray et al. [21] reported similar OLM gaps with loss of adherens junctions between photoreceptors and Müller cells, and with an abnormal shift of the photoreceptor nucleus to its inner segment in Crb1null (all retinal isoforms A, B and C are disrupted), as well as in Crb1delB/null and in Crb1delB/delB models (CRISPR-mediated deletion in the promotor and the first exon of isoform B) but with a lower frequency [21]. Structural alterations in number and size of the apical microvilli of Müller cells were also described in rd8 and knockout models [17]. All these models indicate that the lack of one or both isoforms induces OLM changes with loss of photoreceptor-Müller cell contacts combined with structural changes in both cell types.
Beyond Müller cell-photoreceptor adhesion and interaction, photoreceptor loss was not significant in mice lacking only one of the two isoforms, except in aged mice. A retinal degeneration with photoreceptor loss occurs only in Crb1null mice lacking all CRB1 isoforms.
Based on the characterization of the previously known and novel models, Ray et al. stated that, in mice, Crb1-B is essential and predominant, as retinal anomalies can develop even when Crb1-A is strictly normal. However, our human data differs from that of mouse models. In humans, the retinal degeneration appears early, and the phenotype is mainly driven by the Müller cell isoform A impairment. For instance, patients with 100% WT isoform B can develop a generalized retinal degeneration and those with the c.498_506del variant always develop a macular dystrophy. A human phenotype equivalent to the Crb1delB/null or Crb1delB/delB mouse models has never been reported, as available NGS panels do not cover the specific promotor and 5′and 3′exons of isoform B.
3.4. Future Directions
In the future, human iPSC-derived retinal organoids will be required to understand the different impacts of bi-allelic CRB1 variants. This will allow a better comprehension on the specific and combined role of CRB1 isoforms in developmental and post adult retinae, as well as their interactions within the CRB complex. We should also screen for cases heterozygous for mutated isoform A and heterozygous or compound heterozygous for mutated isoform B. To this end, genetic analyses should be completed in patients with only one variant in CRB1 via a novel design of the retinal NGS gene panels to accommodate screening of the specific CRB1-B transcript.
4. Materials and Methods
This study conforms to the Declaration of Helsinki and to approved protocols of the Montpellier, Lille and Rennes University hospitals. Signed informed consent for clinical examination and genetic analysis was obtained from all participants and from the parents of minors. In this retrospective study, the clinical databases of the three different national departments dedicated to inherited retinal dystrophies (Lille, Rennes, Montpellier, France) were screened to identify all patients with bi-allelic pathogenic variants in CRB1. The Ministry of Public Health accorded approval for biomedical research under the authorization number 11018S.
4.1. Clinical Investigation
Age of onset and initial symptoms were recorded for all patients. Best-corrected visual acuity was in LogMar. Color photographs were performed with a Nidek non-mydriatic automated fundus camera (AFC 330, Nidek Inc, Tokyo, Japan). Near infrared reflectance (NIR), autofluorescence and spectral domain optical coherence tomography (SD-OCT) imaging were completed with a Combined Heidelberg Retina Angiograph + Spectralis OCT device (Heidelberg Engineering, Dossenheim, Germany). Full-field electroretinography (ff-ERG) was performed with a contact lens electrode using Ganzfeld apparatus (Ophthalmologic Monitor Metrovision, Pérenchies, France) according to the guidelines of the International Society for Electrophysiology of Vision. Goldmann visual fields were classified based on the criteria of the World Health Organization (WHO: mild visual impairment for a central visual field between 20° to 70°, low vision if limited to the central 20° and blindness if inferior to 10°).
Based on the clinical, imaging and ff-ERG data, patients were divided into one of the three following groups. The first group comprised all patients with EORD or LCA, both characterized by early onset (before the age of 5 years). In LCA, a nystagmus and oculo-digital reflexes were frequently noted within the first twelve months of life. The second group comprised patients with RCD and onset after the age of five years. The third group comprised patients with a macular dystrophy and anomalies restricted to the posterior pole and eventually the peripapillary retina (no night blindness, no constriction of peripheral visual field).
4.2. SD-OCT Measurements
On SD-OCT, central foveal thickness was measured independently in each eye with automated segmentation using Heidelberg HRA software. Retinal thickness, defined as the distance between the basal retinal pigment epithelium/Bruch’s membrane and the internal limiting membrane, were measured at the fovea, at 1500 μm and 3000 μm nasal of the fovea and at 1500 μm and 3000 μm temporal of the fovea on the horizontal 9 mm long section, using the automated software and, if necessary, manually by two independent operators (KM, VS). The horizontal ellipsoid line width was measured on the horizontal 9 mm long section by two independent operators (KM, VS). Disease progression was assessed in 27 patients having undergone at least two examinations with SD-OCT. This device has only been available since 2010 in Montpellier, Lille and Nantes departments.
4.3. Molecular Investigation
Genomic DNA was extracted from leukocytes using a FlexiGene kit (Qiagen). We used dedicated large IRD gene panels including 230 genes associated with generalized retinal and macular dystrophies to screen for pathogenic variants [44]. The design of this panel does not allow the analysis of the specific 5′ and 3′ exons of CRB1 isoform B.
4.4. Variant Analysis and Protein Modelling
Familial variant segregation analysis was carried out for each case (at least one relative). Variant classification was assessed according to the American College of Medical Genetics and Genomics guidelines [45] and using Varsome (https://varsome.com/ (accessed on 25 October 2021 )) and MobiDetails (https://mobidetails.iurc.montp.inserm.fr/MD/ (accessed on 25 October 2021)). The protein sequence of the various mutants was submitted to the webserver @TOME for sequence-structure alignment and model building using SCWRL 3.0 [46]. The Ab initio model of CRB1 available at https://alphafold.ebi.ac.uk/entry/P82279 (accessed on 25 October 2021) was also scrutinized [47]. Resulting models were visualized and analyzed using Pymol (www.pymol.org (accessed on 25 October 2021)).
Author Contributions
Conceptualization, K.M., V.S., B.B., G.L., C.A., S.D.-D., X.Z., D.H., D.D., T.D., M.P., H.H., A.-F.R., V.K., C.-M.D. and I.M.; data curation, K.M., V.S., B.B., D.H., D.D., M.-C.P., T.D., O.G., A.-F.R., V.K., C.-M.D. and I.M.; investigation, K.M., V.S., B.B., S.D.-D., X.Z., O.G., M.P., H.H., V.K., C.-M.D. and I.M.; methodology, K.M., V.S., B.B., G.L., C.A., S.D.-D., X.Z., D.H., D.D., M.-C.P., T.D., M.P., H.H., A.-F.R., V.K., C.-M.D. and I.M.; supervision, I.M.; validation, B.B., G.L., C.A., M.-C.P., O.G. and I.M.; writing—original draft, K.M., V.S., B.B., G.L., C.A., S.D.-D., X.Z., D.H., D.D., M.-C.P., T.D., O.G., M.P., H.H., A.-F.R., V.K., C.-M.D. and I.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. This study was funded by the University Hospital of Montpellier and by the Institute for Neurosciences of Montpellier, INSERM.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinski and approved by the Ethics committee of the University Hospital of Montpellier. The Ministry of Public Health accorded approval for biomedical research under the authorization number 11018S.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent was obtained from the patients to publish this paper.
Data Availability Statement
All data are contained within the article.
Acknowledgments
We are grateful to the patients and their family members participating in this study, to different patient associations (IRRP: Information for Research on Retinitis Pigmentosa, SBKB: Sébastien Joachim Kick Blindness), and to the Sensgene and ERN-Eye networks.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| CRB1 | Crumbs homologue 1 |
| EORD | Early onset retinal dystrophy |
| IRD | Inherited retinal dystrophy |
| LCA | Leber congenital amaurosis |
| MD RP, RCD | Macular dystrophy Retinitis pigmentosa, rod cone dystrophy |
| SD-OCT | Spectral domain optical coherence tomography |
| EORD | Early onset retinal dystrophy |
References
- Hamel, C.P. Cone Rod Dystrophies. Orphanet J. Rare Dis. 2007, 2, 7. [Google Scholar] [CrossRef] [Green Version]
- Bocquet, B.; Lacroux, A.; Surget, M.-O.; Baudoin, C.; Marquette, V.; Manes, G.; Hebrard, M.; Sénéchal, A.; Delettre, C.; Roux, A.-F.; et al. Relative Frequencies of Inherited Retinal Dystrophies and Optic Neuropathies in Southern France: Assessment of 21-Year Data Management. Ophthalmic Epidemiol. 2013, 20, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahim, A.T.; Bouzia, Z.; Branham, K.H.; Kumaran, N.; Vargas, M.E.; Feathers, K.L.; Perera, N.D.; Young, K.; Khan, N.W.; Heckenlively, J.R.; et al. Detailed Clinical Characterisation, Unique Features and Natural History of Autosomal Recessive RDH12-Associated Retinal Degeneration. Br. J. Ophthalmol. 2019, 103, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
- Scott, H.A.; Place, E.M.; Ferenchak, K.; Zampaglione, E.; Wagner, N.E.; Chao, K.R.; DiTroia, S.P.; Navarro-Gomez, D.; Mukai, S.; Huckfeldt, R.M.; et al. Expanding the Phenotypic Spectrum in RDH12-Associated Retinal Disease. Cold Spring Harb. Mol. Case Stud. 2020, 6, a004754. [Google Scholar] [CrossRef] [Green Version]
- Hull, S.; Arno, G.; Plagnol, V.; Chamney, S.; Russell-Eggitt, I.; Thompson, D.; Ramsden, S.C.; Black, G.C.M.; Robson, A.; Holder, G.E.; et al. The Phenotypic Variability of Retinal Dystrophies Associated with Mutations in CRX, with Report of a Novel Macular Dystrophy Phenotype. Invest Ophthalmol. Vis. Sci. 2014, 55, 6934–6944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birtel, J.; Eisenberger, T.; Gliem, M.; Müller, P.L.; Herrmann, P.; Betz, C.; Zahnleiter, D.; Neuhaus, C.; Lenzner, S.; Holz, F.G.; et al. Clinical and Genetic Characteristics of 251 Consecutive Patients with Macular and Cone/Cone-Rod Dystrophy. Sci. Rep. 2018, 8, 4824. [Google Scholar] [CrossRef] [PubMed]
- Tsang, S.H.; Burke, T.; Oll, M.; Yzer, S.; Lee, W.; Xie, Y.; Allikmets, R. Whole Exome Sequencing Identifies CRB1 Defect in an Unusual Maculopathy Phenotype. Ophthalmology 2014, 121, 1773–1782. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Sun, W.; Xiao, X.; Li, S.; Jia, X.; Wang, P.; Zhang, Q. Clinical and Genetic Analysis of 63 Families Demonstrating Early and Advanced Characteristic Fundus as the Signature of CRB1 Mutations. Am. J. Ophthalmol. 2021, 223, 160–168. [Google Scholar] [CrossRef]
- den Hollander, A.I.; Davis, J.; van der Velde-Visser, S.D.; Zonneveld, M.N.; Pierrottet, C.O.; Koenekoop, R.K.; Kellner, U.; van den Born, L.I.; Heckenlively, J.R.; Hoyng, C.B.; et al. CRB1 Mutation Spectrum in Inherited Retinal Dystrophies. Hum. Mutat 2004, 24, 355–369. [Google Scholar] [CrossRef]
- Bujakowska, K.; Audo, I.; Mohand-Saïd, S.; Lancelot, M.-E.; Antonio, A.; Germain, A.; Léveillard, T.; Letexier, M.; Saraiva, J.-P.; Lonjou, C.; et al. CRB1 Mutations in Inherited Retinal Dystrophies. Hum. Mutat. 2012, 33, 306–315. [Google Scholar] [CrossRef] [Green Version]
- Lotery, A.J.; Malik, A.; Shami, S.A.; Sindhi, M.; Chohan, B.; Maqbool, C.; Moore, P.A.; Denton, M.J.; Stone, E.M. CRB1 Mutations May Result in Retinitis Pigmentosa without Para-Arteriolar RPE Preservation. Ophthalmic Genet. 2001, 22, 163–169. [Google Scholar] [CrossRef]
- Nguyen, X.-T.-A.; Talib, M.; van Schooneveld, M.J.; Wijnholds, J.; van Genderen, M.M.; Schalij-Delfos, N.E.; Klaver, C.C.W.; Talsma, H.E.; Fiocco, M.; Florijn, R.J.; et al. CRB1-Associated Retinal Dystrophies: A Prospective Natural History Study in Anticipation of Future Clinical Trials. Am. J. Ophthalmol. 2021, 234, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, S.G.; Cideciyan, A.V.; Aleman, T.S.; Pianta, M.J.; Sumaroka, A.; Schwartz, S.B.; Smilko, E.E.; Milam, A.H.; Sheffield, V.C.; Stone, E.M. Crumbs Homolog 1 (CRB1) Mutations Result in a Thick Human Retina with Abnormal Lamination. Hum. Mol. Genet. 2003, 12, 1073–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulou Laiou, C.; Preising, M.N.; Bolz, H.J.; Lorenz, B. [Genotype-Phenotype Correlations in Patients with CRB1 Mutations]. Klin. Monbl. Augenheilkd 2017, 234, 289–302. [Google Scholar] [PubMed]
- Talib, M.; van Schooneveld, M.J.; van Genderen, M.M.; Wijnholds, J.; Florijn, R.J.; Ten Brink, J.B.; Schalij-Delfos, N.E.; Dagnelie, G.; Cremers, F.P.M.; Wolterbeek, R.; et al. Genotypic and Phenotypic Characteristics of CRB1-Associated Retinal Dystrophies: A Long-Term Follow-up Study. Ophthalmology 2017, 124, 884–895. [Google Scholar] [CrossRef] [Green Version]
- Corton, M.; Tatu, S.D.; Avila-Fernandez, A.; Vallespín, E.; Tapias, I.; Cantalapiedra, D.; Blanco-Kelly, F.; Riveiro-Alvarez, R.; Bernal, S.; García-Sandoval, B.; et al. High Frequency of CRB1 Mutations as Cause of Early-Onset Retinal Dystrophies in the Spanish Population. Orphanet J. Rare Dis. 2013, 8, 20. [Google Scholar] [CrossRef] [Green Version]
- Alves, C.H.; Pellissier, L.P.; Wijnholds, J. The CRB1 and Adherens Junction Complex Proteins in Retinal Development and Maintenance. Prog. Retin Eye Res. 2014, 40, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Mehalow, A.K.; Kameya, S.; Smith, R.S.; Hawes, N.L.; Denegre, J.M.; Young, J.A.; Bechtold, L.; Haider, N.B.; Tepass, U.; Heckenlively, J.R.; et al. CRB1 Is Essential for External Limiting Membrane Integrity and Photoreceptor Morphogenesis in the Mammalian Retina. Hum. Mol. Genet. 2003, 12, 2179–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Pavert, S.A.; Kantardzhieva, A.; Malysheva, A.; Meuleman, J.; Versteeg, I.; Levelt, C.; Klooster, J.; Geiger, S.; Seeliger, M.W.; Rashbass, P.; et al. Crumbs Homologue 1 Is Required for Maintenance of Photoreceptor Cell Polarization and Adhesion during Light Exposure. J. Cell Sci. 2004, 117, 4169–4177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, S.; Cho, M.; Lee, J. Crumbs Proteins Regulate Layered Retinal Vascular Development Required for Vision. Biochem Biophys Res. Commun. 2020, 521, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Ray, T.A.; Cochran, K.; Kozlowski, C.; Wang, J.; Alexander, G.; Cady, M.A.; Spencer, W.J.; Ruzycki, P.A.; Clark, B.S.; Laeremans, A.; et al. Comprehensive Identification of MRNA Isoforms Reveals the Diversity of Neural Cell-Surface Molecules with Roles in Retinal Development and Disease. Nat. Commun. 2020, 11, 3328. [Google Scholar] [CrossRef]
- Khan, K.N.; Robson, A.; Mahroo, O.A.R.; Arno, G.; Inglehearn, C.F.; Armengol, M.; Waseem, N.; Holder, G.E.; Carss, K.J.; Raymond, L.F.; et al. A Clinical and Molecular Characterisation of CRB1-Associated Maculopathy. Eur. J. Hum. Genet. 2018, 26, 687–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- den Hollander, A.I.; ten Brink, J.B.; de Kok, Y.J.M.; van Soest, S.; van den Born, L.I.; van Driel, M.A.; van de Pol, D.J.R.; Payne, A.M.; Bhattacharya, S.S.; Kellner, U.; et al. Mutations in a Human Homologue of Drosophila Crumbs Cause Retinitis Pigmentosa (RP12). Nat. Genet. 1999, 23, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Glöckle, N.; Kohl, S.; Mohr, J.; Scheurenbrand, T.; Sprecher, A.; Weisschuh, N.; Bernd, A.; Rudolph, G.; Schubach, M.; Poloschek, C.; et al. Panel-Based next Generation Sequencing as a Reliable and Efficient Technique to Detect Mutations in Unselected Patients with Retinal Dystrophies. Eur. J. Hum. Genet. 2014, 22, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Lotery, A.J.; Jacobson, S.G.; Fishman, G.A.; Weleber, R.G.; Fulton, A.B.; Namperumalsamy, P.; Héon, E.; Levin, A.V.; Grover, S.; Rosenow, J.R.; et al. Mutations in the CRB1 Gene Cause Leber Congenital Amaurosis. Arch. Ophthalmol. 2001, 119, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Martin-Merida, I.; Avila-Fernandez, A.; Del Pozo-Valero, M.; Blanco-Kelly, F.; Zurita, O.; Perez-Carro, R.; Aguilera-Garcia, D.; Riveiro-Alvarez, R.; Arteche, A.; Trujillo-Tiebas, M.J.; et al. Genomic Landscape of Sporadic Retinitis Pigmentosa: Findings from 877 Spanish Cases. Ophthalmology 2019, 126, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Yzer, S.; Leroy, B.P.; De Baere, E.; de Ravel, T.J.; Zonneveld, M.N.; Voesenek, K.; Kellner, U.; Ciriano, J.P.M.; de Faber, J.-T.H.N.; Rohrschneider, K.; et al. Microarray-Based Mutation Detection and Phenotypic Characterization of Patients with Leber Congenital Amaurosis. Invest Ophthalmol. Vis. Sci. 2006, 47, 1167–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanks, M.E.; Downes, S.M.; Copley, R.R.; Lise, S.; Broxholme, J.; Hudspith, K.A.; Kwasniewska, A.; Davies, W.I.; Hankins, M.W.; Packham, E.R.; et al. Next-Generation Sequencing (NGS) as a Diagnostic Tool for Retinal Degeneration Reveals a Much Higher Detection Rate in Early-Onset Disease. Eur. J. Hum. Genet. 2013, 21, 274–280. [Google Scholar] [CrossRef] [Green Version]
- Kousal, B.; Dudakova, L.; Gaillyova, R.; Hejtmankova, M.; Diblik, P.; Michaelides, M.; Liskova, P. Phenotypic Features of CRB1-Associated Early-Onset Severe Retinal Dystrophy and the Different Molecular Approaches to Identifying the Disease-Causing Variants. Graefes Arch. Clin. Exp. Ophthalmol. 2016, 254, 1833–1839. [Google Scholar] [CrossRef]
- den Hollander, A.I.; Heckenlively, J.R.; van den Born, L.I.; de Kok, Y.J.; van der Velde-Visser, S.D.; Kellner, U.; Jurklies, B.; van Schooneveld, M.J.; Blankenagel, A.; Rohrschneider, K.; et al. Leber Congenital Amaurosis and Retinitis Pigmentosa with Coats-like Exudative Vasculopathy Are Associated with Mutations in the Crumbs Homologue 1 (CRB1) Gene. Am. J. Hum. Genet. 2001, 69, 198–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanein, S.; Perrault, I.; Gerber, S.; Tanguy, G.; Barbet, F.; Ducroq, D.; Calvas, P.; Dollfus, H.; Hamel, C.; Lopponen, T.; et al. Leber Congenital Amaurosis: Comprehensive Survey of the Genetic Heterogeneity, Refinement of the Clinical Definition, and Genotype-Phenotype Correlations as a Strategy for Molecular Diagnosis. Hum. Mutat 2004, 23, 306–317. [Google Scholar] [CrossRef] [Green Version]
- Carss, K.J.; Arno, G.; Erwood, M.; Stephens, J.; Sanchis-Juan, A.; Hull, S.; Megy, K.; Grozeva, D.; Dewhurst, E.; Malka, S.; et al. Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am. J. Hum. Genet. 2017, 100, 75–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernal, S.; Calaf, M.; Garcia-Hoyos, M.; Garcia-Sandoval, B.; Rosell, J.; Adan, A.; Ayuso, C.; Baiget, M. Study of the Involvement of the RGR, CRPB1, and CRB1 Genes in the Pathogenesis of Autosomal Recessive Retinitis Pigmentosa. J. Med. Genet. 2003, 40, e89. [Google Scholar] [CrossRef] [Green Version]
- Zernant, J.; Külm, M.; Dharmaraj, S.; den Hollander, A.I.; Perrault, I.; Preising, M.N.; Lorenz, B.; Kaplan, J.; Cremers, F.P.M.; Maumenee, I.; et al. Genotyping Microarray (Disease Chip) for Leber Congenital Amaurosis: Detection of Modifier Alleles. Invest Ophthalmol. Vis. Sci. 2005, 46, 3052–3059. [Google Scholar] [CrossRef]
- Vallespin, E.; Avila-Fernandez, A.; Velez-Monsalve, C.; Almoguera, B.; Martinez-Garcia, M.; Gomez-Dominguez, B.; Gonzalez-Roubaud, C.; Cantalapiedra, D.; Trujillo-Tiebas, M.J.; Ayuso, C. Novel Human Pathological Mutations. Gene Symbol: CRB1. Disease: Leber Congenital Amaurosis. Hum. Genet. 2010, 127, 119. [Google Scholar] [PubMed]
- Zaneveld, J.; Siddiqui, S.; Li, H.; Wang, X.; Wang, H.; Wang, K.; Li, H.; Ren, H.; Lopez, I.; Dorfman, A.; et al. Comprehensive Analysis of Patients with Stargardt Macular Dystrophy Reveals New Genotype-Phenotype Correlations and Unexpected Diagnostic Revisions. Genet. Med. 2015, 17, 262–270. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Xiao, X.; Li, S.; Jia, X.; Xin, W.; Wang, P.; Sun, W.; Huang, L.; Guo, X.; Zhang, Q. Molecular Genetics of Leber Congenital Amaurosis in Chinese: New Data from 66 Probands and Mutation Overview of 159 Probands. Exp. Eye Res. 2016, 149, 93–99. [Google Scholar] [CrossRef]
- Henderson, R.H.; Mackay, D.S.; Li, Z.; Moradi, P.; Sergouniotis, P.; Russell-Eggitt, I.; Thompson, D.A.; Robson, A.G.; Holder, G.E.; Webster, A.R.; et al. Phenotypic Variability in Patients with Retinal Dystrophies Due to Mutations in CRB1. Br. J. Ophthalmol. 2011, 95, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Kantardzhieva, A.; Gosens, I.; Alexeeva, S.; Punte, I.M.; Versteeg, I.; Krieger, E.; Neefjes-Mol, C.A.; den Hollander, A.I.; Letteboer, S.J.F.; Klooster, J.; et al. MPP5 Recruits MPP4 to the CRB1 Complex in Photoreceptors. Invest Ophthalmol. Vis. Sci. 2005, 46, 2192–2201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, B.; Alves, C.H.; Lundvig, D.M.; Tanimoto, N.; Beck, S.C.; Huber, G.; Richard, F.; Klooster, J.; Andlauer, T.F.M.; Swindell, E.C.; et al. PALS1 Is Essential for Retinal Pigment Epithelium Structure and Neural Retina Stratification. J. Neurosci. 2011, 31, 17230–17241. [Google Scholar] [CrossRef] [PubMed]
- van Rossum, A.G.S.H.; Aartsen, W.M.; Meuleman, J.; Klooster, J.; Malysheva, A.; Versteeg, I.; Arsanto, J.-P.; Le Bivic, A.; Wijnholds, J. Pals1/Mpp5 Is Required for Correct Localization of Crb1 at the Subapical Region in Polarized Muller Glia Cells. Hum. Mol. Genet. 2006, 15, 2659–2672. [Google Scholar] [CrossRef]
- Wei, Z.; Li, Y.; Ye, F.; Zhang, M. Structural Basis for the Phosphorylation-Regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-Binding Protein Moesin. J. Biol. Chem. 2015, 290, 11384–11392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cañibano-Hernández, A.; Valdes-Sanchez, L.; Garcia-Delgado, A.B.; Ponte-Zúñiga, B.; Diaz-Corrales, F.J.; de la Cerda, B. Generation of the Human IPSC Line ESi082-A from a Patient with Macular Dystrophy Associated to Mutations in the CRB1 Gene. Stem Cell Res. 2021, 53, 102301. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, V.; Grunewald, O.; Muller, J.; Zeitz, C.; Obermaier, C.D.; Devos, A.; Pelletier, V.; Bocquet, B.; Andrieu, C.; Bacquet, J.-L.; et al. Novel TTLL5 Variants Associated with Cone-Rod Dystrophy and Early-Onset Severe Retinal Dystrophy. Int. J. Mol. Sci. 2021, 22, 6410. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Canutescu, A.A.; Dunbrack, R.L. SCWRL and MolIDE: Computer Programs for Side-Chain Conformation Prediction and Homology Modeling. Nat. Protoc. 2008, 3, 1832–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).