Abstract
Plants regularly face the changing climatic conditions that cause biotic and abiotic stress responses. The abiotic stresses are the primary constraints affecting crop yield and nutritional quality in many crop plants. The advances in genome sequencing and high-throughput approaches have enabled the researchers to use genome editing tools for the functional characterization of many genes useful for crop improvement. The present review focuses on the genome editing tools for improving many traits such as disease resistance, abiotic stress tolerance, yield, quality, and nutritional aspects of tomato. Many candidate genes conferring tolerance to abiotic stresses such as heat, cold, drought, and salinity stress have been successfully manipulated by gene modification and editing techniques such as RNA interference, insertional mutagenesis, and clustered regularly interspaced short palindromic repeat (CRISPR/Cas9). In this regard, the genome editing tools such as CRISPR/Cas9, which is a fast and efficient technology that can be exploited to explore the genetic resources for the improvement of tomato and other crop plants in terms of stress tolerance and nutritional quality. The review presents examples of gene editing responsible for conferring both biotic and abiotic stresses in tomato simultaneously. The literature on using this powerful technology to improve fruit quality, yield, and nutritional aspects in tomato is highlighted. Finally, the prospects and challenges of genome editing, public and political acceptance in tomato are discussed.
1. Introduction
Tomato (Solanum lycopersicum) is an economically important crop with almost 160 million tons produced in 2016 (FAO, 2016). Tomato is a diploid plant with 12 chromosomes and a genome size of ~950 Mb. It is one of the most important horticultural crops worldwide because of its nutritional value and derived industrial products. It is also an optimal bridge between the model plant (e.g., Arabidopsis) and other crops due to the availability of enormous genetic and genomic resources. Tomato and its 12 wild relatives are native to western and central South America and are widespread throughout diverse habitats, contributing to high genetic variability. Little is known about the history of tomato domestication [1], but it was at an advanced level before it reached the old world. During the domestication of tomatoes, intense levels of improvement occurred worldwide. Consequently, many morphologically distinct cultivars have been developed from the single species of S. lycopersicum. Unfortunately, some important traits, such as resistance to biotic and abiotic factors, which existed in the wild tomatoes, have been compromised during the domestication. As a result, conventional breeding has resulted in improved traits accompanied by loss of fitness and genetic diversity. However, it is a highly time-consuming and laborious task due to backcrosses [2]. Technological advancement in genomics improved the genetic engineering of crops in the last two decades. The genetic engineering also referred to as recombinant DNA technology involves the transfer of desired gene from one species to another, thereby broadening the chances for crop improvement [3]. The transgenic plants developed using this technology are named as genetically modified organisms (GMOs). However, the regulatory approval of GMOs is a major drawback as the release of GM crops to the public market is costly and often delayed [4].
Recent efforts have ensured the conservation of landraces and wild species that allowed re-introducing resistance traits [5,6]. Besides, artificial mutagenesis offers breeders with genotypes containing novel genetic and phenotypic diversity that helps in enlarging the narrowed genetic base [7,8,9]. Mutant collections also provide a complementary alternative for trait discovery in tomato, providing an allelic series in a uniform genetic background [10]. In the last decade, various tomato cultivars were used for generating mutant collections [8,9,11,12,13,14,15,16,17,18,19,20,21,22,23].
Modern genetics and breeding methods have contributed to the understanding and developing of structural and functional aspects of tomato genomes [10]. In tomato, quantitative trait loci (QTL) mapping has assisted in mapping of genes associated with heat and salt tolerance [24,25,26] and various fruit-related traits [27,28,29]. Further, genome-wide association study (GWAS) has been used to map loci related to traits such as plant architecture, fruit shape, and fruit weight in tomato [30,31,32], and fruit metabolites [33,34,35]. GWAS also helped identify loci related to drought and salt tolerance in tomato [36].
With the advancement in high-throughput sequencing technologies, genomes of tomato [37,38,39,40] have been fully sequenced, including several wild tomato species and landraces [39,41,42,43,44]. The information on genomic sequences of wild tomato cultivars and other tomato accessions are available in Sol Genomics Network (https://solgenomics.net/) and Solanum pennellii genome project (https://www.plabipd.de/project_spenn/start.ep). The availability of a high-quality genome sequence and Rapid improvement in molecular biology and genomics techniques enabled researchers to precisely edit any desired genomics locus in the form of insertions/deletions or base substitution. The availability of pan-genome could facilitate gene-editing tools to test the effect of target gene modifications in tomato breeding and development.
Here we review the structure, mechanism of various genome editing tools, and genome editing approaches for major breeding goals in tomato, such as resistance to various biotic and abiotic stresses and traits improvement. We also would like to shed some light on future applications in the field concerning GMOs public and political acceptance.
2. Structure and Mechanism of Genome Editing Tools
Diversification of organisms is based on variations in the genetic pool [45]. Genetic variation is the basis for improving an organism’s traits and is valuable for the production of new cultivars in plant breeding [46]. During evolution, genetic variations occur spontaneously due to DNA damage or errors in the replication process, which are termed as mutations. This process is called mutagenesis [47,48]. The natural mutations are spontaneous, sporadic, and random; therefore, it is impossible to rely only on natural variations for crop breeding [49,50]. Thus, artificial mutagenesis is needed to increase genetic variation, an essential step for the breeding program [49].
Genome editing (GE) techniques have revolutionized the biological world by facilitating precise, efficient, and targeted modification at genomic loci of the living organisms, including microbes, animals, humans, and plants. The primary mechanism of GE includes double-stranded breaks in the DNA (dsDNA) by specific engineered nucleases. The dsDNA break is repaired, either by non-homologous end joining (NHEJ) or homology-dependent recombination (HDR). HDR allows generating accurate point mutations, deletions, or gene knock-in useful for crop breeding but with extremely low editing frequencies. In contrast, NHEJ is error-prone and allows random small insertions or deletions as well as substitutions, preferably causing a gene knockout [51].
Several genome editing approaches are available for inducing site-specific dsDNA breaks such as induced mutagenesis, Oligonucleotide directed mutagenesis (ODM), epigenome editing, transposable elements, Zinc finger nucleases (ZFNs), transcriptional activator like effector nucleases (TALENs), and more recently clustered regularly interspaced short palindromic repeat (CRISPR/Cas9) systems can be exploited to decipher the role of unannotated and uncharacterized genes [12,52,53,54]. The timeline of the genome editing tools is represented in Figure 1.
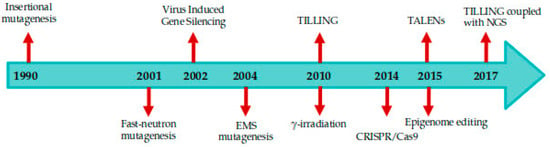
Figure 1.
Timeline of the breakthrough of genome editing in tomato. Insertional mutagenesis [55], Virus Induced Gene Silencing (VIGS) [56], Targeted Induced Local Lesions In Genomes (TILLING) [15], TALENs [57], TILLING coupled with NGS [58], Fast-neutron mutagenesis [59], ethyl methane sulphonate (EMS) mutagenesis [9], γ-irradiation [60], CRISPR/Cas9 [61], Epigenome editing [62].
2.1. Induced Mutagenesis
Induced mutagenesis allows the introduction of novel genetic alleles and facilitates novel genetic resources for crop improvement and gene function discovery. Since the initial reports of mutation breeding by Stadler in 1928, remarkable progress has been accomplished in genetic breeding techniques [63]. It has been widely used in crop plants with low genetic variability and those species that are not amenable to conventional breeding (Figure 2a). The mutagenized population, once generated, becomes an everlasting resource. Due to their long safety record and absence of foreign DNA (Figure 2b), mutagenic plants are exempted from the EU GMO legislation. However, the generation of a mutant population is laborious and time-consuming. Based on the inducing agent, the mutagenesis is further classified into physical and chemical mutagenesis (Figure 2c).
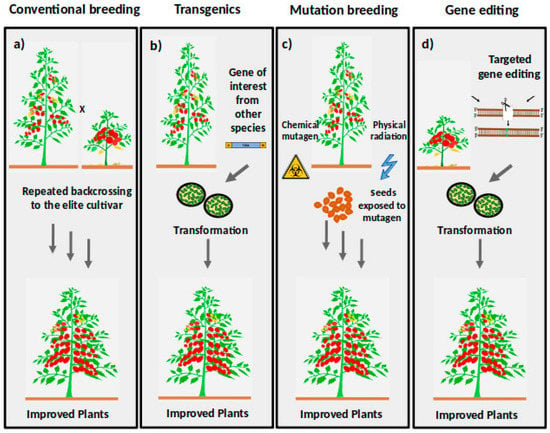
Figure 2.
Represents the breeding techniques employed for crop improvement. (a) Conventional breeding involves selective breeding of two tomato species with desired traits. For example, a tall plant with low yield and stress tolerance is crossed to a small plant with bigger fruits. Repeated backcrossing is performed to the elite cultivar to generate plants with desired traits. Conventional breeding is mostly accompanied with loss of genetic diversity due tote selection process. (b) Transgenic breeding involves the introduction of a desired gene (transgene) from other species into the selected plant by the transformation process. (c) Mutation breeding involves physical radiation or a chemical mutagen to induce mutations. The mutated populations (M1) are generated, and to reduce chimerism M2 or higher populations are produced. The mutant is then screened either by forward or reverse genetics. (d) Targeted genome editing (detailed described in Section 2.7), schematic describes the procedure for generating a wide variety of improved plant traits. After generation of edited plants are then screened phenotypic and genotypic for discovering the plant with desirable traits.
2.1.1. Physical Mutagenesis
Since the 1920s, the physical mutagenic sources found to be effective for inducing mutations are ionizing radiations. The physical mutagens include ionizing (electromagnetic) radiations such as X-rays, cosmic rays, α-rays and β-rays, γ-rays, neutrons, and protons while non-ionizing source includes UV rays [64]. Even though physical mutagenesis has proved to be very effective particularly for producing large DNA fragment deletions, its application in inducing mutagenesis is mostly directed towards the generation of knockout mutants and rearrangement of genes [65]. X-rays and γ-rays are the most commonly used. Exposure of plants to the γ-radiation causes damage such as double-strand breaks and also produces a range of damage to DNA due to the production of free radicals. Fast-neutron mainly causes deletion mutations ranging from few base pairs to few kilo bases while γ-rays irradiation can cause large deletions along with chromosomal rearrangements [12,66,67].
In addition, ion beams such as protons, helium, and heavy charged particles are known to be highly mutagenic. They are accelerated at higher speeds with high linear energy transfer that induce larger DNA lesions [68,69]. These ion beams radiations induce single- and double-stranded breaks, which lead to inversions, deletions, point mutations, and translocations [68,70]. In Arabidopsis, various novel mutations were identified using high-speed carbon ions [67]. Although various sources of physical mutagenesis are available, majority of the mutants induced using physical mutagens were generated by γ-irradiation.
There are over 3200 varieties of induced mutant lines available at the Food and Agriculture Organization of the United Nations and the International Atomic Energy Agency (Joint FAO/IAEA) in Vienna [71]. Several mutants are obtained using physical mutagenesis in various plant species such as Arabidopsis [72,73,74,75], rice [76,77], and tomato [9,14,60,78].
The tomato tangerine mutant (carotenoid isomerase, CRTISO) was identified from fast-neutron mutagenesis [79] by map-based cloning in Micro-Tom [14,59]. In a different study, 6301 mutant lines were generated using γ-ray irradiation in Micro-Tom with variable phenotypes such as fruit size, color, ripening, flower and leaf morphology, brix, etc., which provides a valuable genetic resource for breeding and functional genomics in tomato [78]. Another study generated 865 mutants by fast neutron and 2552 mutants induced by EMS in tomato cultivar M82 and traits like plant height, fruit size, fruit color, ripening, sterility, and plant stress response (for example, Leaf curl disease) were examined [9].
2.1.2. Chemical Mutagenesis
The chemicals used for mutagenesis in plants include alkylating agents, purine analogs, oxidizing agents, sulphonic esters, and epoxides [80]. Chemical mutagenesis is advantageous over physical as it does not require sophisticated pieces of equipment. This mutagenesis induces point mutations or single base substitutions that often lead to gain or loss of functions giving rise to novel allelic mutants instead of large deletions or chromosomal rearrangements.
Different alkylating agents such as ethyl methane sulphonate (EMS), Diethyl sulfate (DES), ethyleneimine (EI), ethyl nitro urethane (ENU), 1-methyl-1-nitrosourea (MNU), ethyl nitrosourea (ENU), and azides are used for mutagenesis [81]. Among these, EMS is the most desired chemical mutagen in plants [9,15,82,83] that particularly alkylates guanine bases and transfers reactive alkyl groups to other molecules [81]. It predominantly induces point mutations randomly in the genome of the species with the majority being G/C to A/T base pair transitions [84]. Like EMS, MNU mainly induces G/C to A/T transitions and also induces translocations and inversions at lower frequencies [85,86]. In contrast to MNU, ENU significantly induces G/C to A/T transitions together with A/T to G/C transitions including transversions [87]. In addition, analogues of nitrogenous bases such as maleic hydrazide, 5-bromouracil, and 2-aminopurine also possess mutagenic activity. However, these bases are rarely used in plants [86].
EMS has been successfully exploited in tomato cultivars such as Moneymaker [88], M82 [9,89], Lycopersicon esculentum Mill. [90], and Red setter [7,15,91], Arka Vikas [58]. In tomato, the first mutant identified from EMS (60 mM) was adh-1, which encodes for alcohol dehyrogenase1 (ADH1) [88]. Adh-1 is a biochemical mutant, which renders the likelihood of screening large populations as only the mutants survive in the presence of allyl alcohol [88]. In 2007, Kostovś group generated an EMS (1.5%) population in L. esculentum Mill. and identified 16 plants resistant to Orabanche ramosa and this mutant population can be used to study Broomrape resistance in tomato breeding [90]. In another study, 0.7% and 1% EMS was used in Red setter cultivar to produce 5508 lines and studied seven fruit quality traits to identify 66 point mutations. This mutant population was developed for various forward and reverse genetic screening [15]. The miniature dwarf tomato cultivar Micro-Tom was used to create EMS mutant population with two different doses (0.5% and 1%) [16,82], out of which 1% EMS was found to be efficient to generate mutant population to perform functional genomic studies in tomato [7].
To screen a mutation population for mutation detection within genes of interest forward genetics (correlate phenotype to gene) or reverse genetic (correlate gene to phenotype) can be used. Targeted induced local lesions in genomes (TILLING) is a high-throughput reverse genetic tool and a well-known approach to identify point mutations in specific genes in the mutagenized population and also to study gene function [15,16]. Three essential steps are necessary for optimal results in TILLING approach; first, the right pooling strategies, second, a good gene model and protein conservation model, which could help to select the gene region with the highest number of possible variations, and third, a suitable PCR primer pairs for a product of approximately 1000–2000 bp. Finally, identified mutations will be confirmed and evaluated by sequencing [92,93]. Ultimately, it can provide an allelic series of silent, mis-sense, non-sense, and splice site mutations to examine the effect of various mutations in a gene. TILLING has been used in Arabidopsis [94], maize [95], barley [96], wheat [97], tomato [15,16,58], lotus [98], etc.
Currently, the conventional TILLING is being replaced by next-generation sequencing (NGS)-based mutation detection, as it allows rapid and accurate screening of a large number of amplicons in a short duration of time through sequencing of smaller amplicons (about 300 bp) [99,100,101,102]. In the first application of NGS in tomato, Rigola and group identified two novel SlelF4E alleles in using 3D pooling, named the approach as “Keypoint” technology [100]. In 2017, TILLING coupled with NGS technology was used to screen a tomato EMS population of 2300 lines and identified 64 mutations with a mutation frequency of 1 in 367 Kb [58].
2.1.3. Limitation of Physical and Chemical Mutagenesis
Physical and chemical mutagenesis are random, and their mutations spectra are not well known. Besides, the optimal dose rate of each mutagen needs to be determined for each genotype. They are time and cost-intensive since they require large populations (5000–10,000 individuals) to select desired phenotype and high-throughput methods for screening mutation rate at the genetic level. Moreover, physical and chemical mutagenesis requires highly sophisticated equipment and infrastructure to ensure safety use, which can be afforded only in labs with specialized laboratory setup. Some physical mutagens such as γ-ray are highly radioactive, and chemical mutagens are hazardous [93,103].
2.2. Oligonucleotide Directed Mutagenesis (ODM)
ODM is known as site-directed mutagenesis or site-specific mutagenesis. It is also called gene targeting or directed gene modification and recently termed precision gene editing [104]. It is a method to introduce specific variations in the target gene of interest. The specific DNA changes include substitutions, insertions, and deletions. The ODM can induce mutations in a particular gene of interest, study the protein function as a result of alterations in the DNA, and introduce or remove sites of restriction enzymes [105].
This technique was first successfully illustrated in mammalian systems in 1996 [106,107], and later it was implemented by researchers in plants [108]. The primary mechanism of ODM came from research on prokaryotes and eukaryotes. ODM’s basic procedure involves the transport of oligonucleotide (that is complementary to the gene of interest) carrying a mutation into the cell via cell membrane and nuclear membrane. It reaches the nucleus where the oligonucleotide binds to the complementary DNA [108]. The host mismatch repair system corrects the DNA damage, and the mutations get incorporated into the genome, inducing a site-specific mutation.
Although ODM has been successfully executed in plants such as maize [109], tobacco [110], rice [111], and wheat [112] that are resistant to herbicides, ODM techniques have shown a relatively low correction rate [104]. However, there are no reports of ODM in tomato yet.
2.3. Epigenome Editing
Epigenetic gene regulation is another important aspect of gene regulation. DNA methylation is a conserved mechanism to regulate gene expression and repress transposon activity [113]. Epigenome editing or engineering refers to employing tools to induce epigenetic changes at a particular location on the genome. This editing is dependent on the methylation status and chromatin organization of the genome. De novo DNA methylation in plants is mediated by RNA-dependent DNA methylation (RdDM) pathway, which involves RNA pol IV and RNA pol V [114,115]. MET1 is the major methyltransferase along with chromomethylases (CMTs), and Domains rearranged methyltransferases (DRM), which maintain the CG/CHG/CHH cytosine methylation in the plant genome [116,117].
De novo DNA methylation in plants involves two sequential steps, biogenesis of short interfering RNAs (siRNAs) and targeting the methylated sequences [118]. The RdDM methylation occurs in two ways-canonical pathway (naturally occurring signaling pathway inside the living system) and non-canonical pathway (induced signaling pathway by chemicals or xenobiotics) [119]. In the canonical pathway, the RNA pol IV transcribes the heterochromatin to single-stranded RNA (ssRNA). The RNA-dependent RNA polymerase (RDR2) converts the ssRNA to transcribe into double standard RNA (dsRNA) [120,121]. The physical interaction of RNA pol IV and RDR2 generates 26–45 nucleotide fragments of dsRNA, which are further cleaved by Dicer-like3 (DCL3) to yield 24 nucleotides siRNAs [121,122]. The non-canonical pathway also exists but is not very common [119]. Several studies have been reported on epigenomic editing in various plant species such as Arabidopsis [123,124,125,126,127], tobacco [128], maize [129], potato [130], and rice [131].
Although the fruit epigenome and full complement of METs, CMTs, and DRMs are known in tomato, there are limited studies on epigenome editing in tomato. In tomato, the first evidence of DNA methylation status affecting fruit ripening came from the studies of colorless non-ripening (CNR), which encodes a SQUAMOSA promoter binding protein3-like (SPB3-like). The normal ripening is hindered in cnr epimutant producing colorless pericarp where SPB3-like gene is hypermethylated, repressing the fruit ripening transcription factors (TFs) and carotenoid biosynthesis [132]. The methylation status of the ripening associated genes is an important factor controlling the transition of the tomato fruit development [133]. Further, another group demonstrated that the DNA methylation in tomato is in turn regulated by DEMETER –like DNA methylases (DMLs) [62]. Knockdown of SlDML2 inhibited the tomato fruit ripening by hypermethylation of the key TFs involved in fruit ripening such as RIN, CNR, and NOR [62]. AlkB homolog 2 (SlAlkBH2) binds to and stabilizes the transcripts of SlDML2 by demethylation of 6-methyl adenosine. The CRISPR/Cas9 knockout of SlAlkBH2 also resulted in reduced transcript levels of SlDML2 and delay in fruit ripening [134]. AlkBH2 is α-ketoglutarate-dependent dioxygenase and repairs the alkylated bases in DNA and mRNA by oxidative demethylation [135]. However, there are limited studies on epigenetic changes governing stress conditions. Rossi and Iusem [136] were the first to show that Asr1 is induced by ABA, water stress, and ripening. Asr1 stands for ABA, stress, and ripening [136]. Later, another gene called Asr2 was identified to be stimulated in leaves and roots of tomato plants upon water stress [137]. Interestingly, Asr2 promoter was demethylated at CNN sites upon exposure to water deficit stress [138]. The emerging high-throughput technologies would enable the discovery of epialleles required for stress tolerance in crop plants in the future.
RNA Interference (RNAi)
RNA silencing is the natural mechanism exploited by a diverse range of organisms such as protozoa, fungi, animals, and crop plants as a defense response to pathogens like viruses [139]. This phenomenon was first discovered in Caenorhabditis elegans in 1998 [140]. The gene silencing occurs in two ways—transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS) [141,142]. TGS represses mRNA through promoter methylation while in PTGS, dsRNA induces mRNA degradation [64,143,144,145]. In plants, there are several approaches to silence the target gene expression. Virus-induced gene silencing (VIGS) is once such a tool in PTGS for the functional characterization of genes in plants [146]. It can be used both as forward and reverse genetic tools in plants. Plants infected by virus uses the PTGS mechanism to induce dsRNA and destroy the viral RNAs as a defense response [147,148,149].
The term VIGS was coined by Van Kammen in 1997 [150]. VIGS also follows the same mechanism as RNA-induced gene silencing, where dsRNAs are produced from the target gene by host RNA dependent RNA polymerase (RdRP). The DICER-like enzyme cleaves the dsRNA into short interfering RNA (siRNA) of 21–25 nucleotides in length with 2 nucleotides 3′ overhangs [151,152]. Subsequently, these siRNAs are introduced into the RNA-induced silencing complex (RISC) complex. This complex consequently targets the siRNA to the complementary RNA resulting in RNA degradation, abolishing the translation of the mRNA [153,154] (Figure 3).
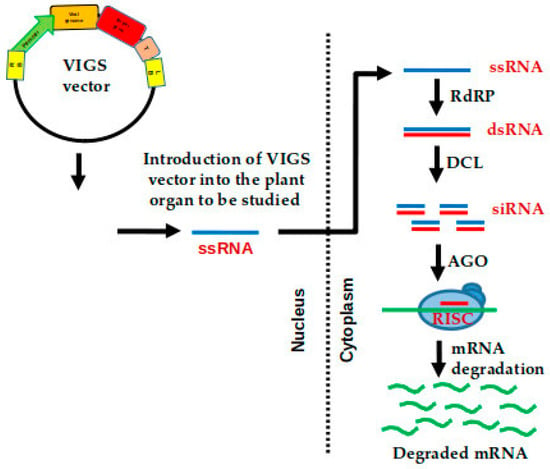
Figure 3.
Represents the virus-induced gene silencing (VIGS) mechanism in tomato. This technique allows transient expression of the target gene introduced through viral vectors into the plant. Once the vector is released in the plant, the single-stranded RNA (ssRNA) is converted to double-stranded RNA (dsRNA), which is further cleaved by dicer-like (DCL) enzymes and loaded to protein Argonaute (AGO) to generate short interfering RNAs (siRNA). The siRNA enters the RNA-induced silencing complex (RISC) to initiate the degradation of targeted mRNA, thereby silencing the gene function. RNA dependent RNA polymerase (RdRP). VIGS vector contains: Left Border (LB) and Right Border (RB) highlighted in yellow; Promoter in green; Viral genome in orange; Target gene in red; Terminator (T).
Tobacco Mosaic Virus (TMV) was the first modified virus vector used for VIGS to suppress the expression of phytoene desaturase (PDS) in Nicotiana benthamiana [155]. Later, other viruses such as Tobacco Rattle Virus (TRV), Turnip yellow Mosaic Virus (TYMV), and Potato virus X (PVX) were modified for VIGS studies in plants. Modified TRV-based was used for efficient gene silencing in tobacco [156] and tomato [56]. TRV is advantageous over other VIGS vectors as it is easy to introduce TRV-based VIGS vector in plants, especially in Solanaceous plants [56]. Another advantage of using TRV is that TRV infection spreads more rapidly all over the plant. However, TRV symptoms are low [56,157]. Potato virus X (PVX) has a limited host range of three plant families, whereas only nine plant families are susceptible to TMV virus [158].
VIGS is advantageous over other genome editing tools as it is a rapid and efficient method to study the gene function. As it is a transient method, there is no need to generate transgenic plants, screen large populations, and no plant transformations are required. As it is performed at the early stage of the plant, the target gene’s role in plant development is rapidly known. If a conserved sequence of the multigene family is chosen for the VIGS, it will silence all the genes in the family, and their role in the plant growth and development would be obvious. Otherwise, a gene member also can be targeted by choosing the sequence for VIGS. If the gene homologies are nearly the same, the same VIGS vector construct can be used to study the gene function in different plant species [56,156,158,159]. Most commonly studied genes using VIGS technology belong to defense responses and other developmental traits in different crop plants [160]. Several examples of RNAi in biotic and abiotic stress responses in tomato are presented in Tables 1–3.
2.4. Transposons
Transposable elements (TEs) or transposons are also known as “jumping genes,” are found in large proportions in most of the species’ genome. TEs were first reported as controlling elements in maize by Barbara McClintock in the 1950s [161]. TEs are one of the sources of the spontaneous mutations which can induce the genetic rearrangements in the genome, such as transposition, translocations, inversions, and duplication by excising from one place to another and integration into another chromosome [162,163] The TEs abundances vary from species to species. For example, TEs contribute to 80% of the maize genome, 66.8% of wheat, 38.8% of cabbage, 59% of soybean, 80% of barley, and 20% of Arabidopsis thaliana genome [164,165].
The transposons are classified into retrotransposons and DNA transposons based on the DNA and RNA intermediate. The retrotransposons are the Class I transposons that are amplified throughout the genome where RNA is reverse transcribed to cDNA and transpose themselves to different locations in the genome. The DNA transposons are the class II transposons where DNA is mobilized and integrated into other sites following a “cut and paste” mechanism in the genome via DNA intermediate [166,167]. The Ac/Ds (Activator/Dissociate) elements are class II transposons and were first identified in maize by Barbara McClintock in 1948, for which she was awarded a Nobel prize in 1983 [161,168,169]. Both the class I and II transposons have autonomous and non-autonomous elements. The autonomous elements use their own encoded proteins to mobilize transposons, while the non-autonomous uses the host machinery [167].
The plants generated through insertional mutagenesis using transposon DNA (T-DNA) are generally knockout mutants. It mostly creates a loss of function mutants, which will enable us to identify the gene function. Once the insertional mutants are generated, the transposed DNA will remain in the same location, even in the next generations. The T-DNA insertion may act as a marker for the identification of the mutant. T-DNA insertions have several disadvantages. If the T-DNA is placed in the intron, the insertion would be curated in the RNA splicing event. Furthermore, when the insertion of T-DNA is in the exon/intron splice site, it may lead to a truncated version of the protein. It may also lead to chromosomal dislocations [166].
The regulation of TEs in plant development is not well understood. However, few reports suggest the role of TEs in biotic and abiotic stresses. For example, ONSEN Ty1-Copia retrotransposon regulates the temperature stress while Ty3-gypsy retrotransposon mediates the escaping of epigenetic silencing [170,171]. In the genome of many plant species, TEs constitute a significant proportion. TEs would be better tools for improved crop breeding [172]. Insertional mutagenesis of T-DNA through Agrobacterium-mediated gene generates an inactivation gene, which is confirmed by a PCR-based approach called as site-selected insertion [173]. T-DNA insertions of maize transposable elements, activator/dissociation (Ac/Ds) have been exploited for insertional mutagenesis in tomato [55,174,175,176,177,178]. A study used site-selected insertion of Polygalacturonase (PG) and Dihydroflavonol 4-reductase (DHFR) in tomato [179]. This study showed that the progeny from Ds plants exhibited a high rate of insertion in PG than other genes [179]. Meissner and group [8] also created a similar mutant resource by using Ac/Ds elements in Micro-Tom and reported several lines carrying at least 2 or 3 Ds inserts. Similarly, several mutants sensitive to drought and salt stress have been identified in Solanum pennellii using T-DNA insertions [180]. Transposon tagging in tomato led to FEEBLY (fb) mutant isolation that displays high sensitivity to a herbicide called phosphinothricin [181].
2.5. Zinc Finger Nucleases (ZFNs)
Among the modern genome-editing tools, ZFNs are the pioneer that enable site-specific modifications in different organisms like the fruit fly, zebrafish, and plants such as A. thaliana, N. benthamiana, and Zea mays [182,183,184]. ZFNs have approximately 30 amino acids in length, which is stabilized by coordinating zinc ions to conserve the Cys2His2 motif [185]. The ZFN arrays can bind to the target DNA by inserting its α-helix into the DNA helix’s major groove and can recognize triple tandem nucleotides. Once ZFNs bind to the target DNAs, it introduces a double-stranded break in the DNA by its cleavage domain, FokI restriction endonuclease [186]. The Fok I cleavage domain was isolated from the bacterium Flavobacterium okeanokoites [187] and must dimerize to catalyze DNA cleavage and become active. Therefore it is necessary to design two ZFNs, one for the complementary and one for the non-complementary DNA strand [188,189]. The DSBs activate the DNA repair system, which results in small insertions, deletions, or base substitutions [190] (Figure 4). After its discovery in 1996 [191], it has been successfully used in several plant species such as maize [184], tobacco [192], Arabidopsis [183,193] and soybean [194]. In Z. mays, exon 2 of Inositol-1,3,4,5,6-pentakisphosphate 2-kinase (IPK1) gene is disrupted, leading to herbicide resistance [184]. However, there is no literature available on tomato.
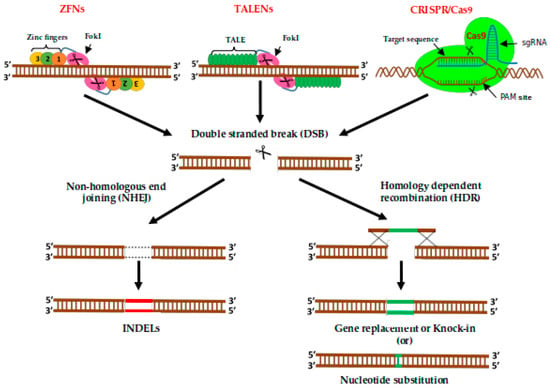
Figure 4.
Represents the genome editing process in Zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and CRISPR/Cas9. A pair of ZFNs bound to DNA. ZFNs are synthetic proteins with separate DNA-cleavage and DNA-binding domains, connected by a short linker sequence. By designing the recognition domain, it is possible to control the site of cleavage. The DNA-binding domain consists commonly of three ZFs. Each connects with 3 bp of target DNA. TALENs consist of a programmable DNA-binding domain and an unspecific FokI cleavage domain. The TALENs recognition domain consists of two TALE DNA-binding sites, which contain arrays of multiple 34-amino acid repeat sequences. A single DNA nucleotide can be recognized by the amino acids at the 12th and 13th position. Or the CRISPR/Cas9 system, the first 20 bp guide sequence can quickly design and fused to the scaffold (gRNA backbone) sequence immediately upstream of a protospacer adjacent motif (PAM). The designed gRNA makes it possible to precisely guide the Cas9 RNA-guided endonucleases (RGENs) to induce a double-stranded DNA break (DSB) in the gene of interest. DBS in the target DNA is repaired either by NHEJ method resulting in small insertions or deletions (INDELs), or by HDR leading to knock-in or nucleotide substitutions.
Although, ZFNs are more specific resulting in fewer off-targets compared to CRISPR/Cas9 due to dimerization of FokI domains [195], this technique also possesses several disadvantages. Unlike TALENs and CRISPR-Cas9, constructing zinc-finger arrays is difficult, hindering their widespread use in unspecialized laboratories [196]. Designing ZFNs is quite a cumbersome strategy and usually takes several months. As it is highly specific, a new cloning strategy is employed every-time, and is highly expensive [195,197,198].
2.6. Transcription Activator-Like Effector Nucleases (TALENs)
TALENs have emerged as an alternative tool for genome editing, similar to ZNFs [199]. Like ZFNs, TALENs also use FokI domain as the DNA cleavage domain, cutting within a 12- to 19-bp spacer sequence that separates each TALE binding site; whereas the DNA binding domain consists of a tandem repeat of 33–35 amino acids, with highly variable amino acids at 12th and 13th positions [200,201] (Figure 4). These variable amino acids are referred to as repeat variable diresidue (RVD), which are specific to recognize particular nucleotides. The tandem repeat proteins from TALEs are effectors of the Xanthomonas bacteria, used to recognize DNA [200,201]. Unlike ZFNs, TALENs are easy to target any DNA sequence because of its simple interaction of the TALENS and DNA interaction [197,202]. However, there is a chance of off-targets in TALENS, which may lead to double-stranded break elsewhere in the genome [203,204]. In addition, TALENs are markedly larger than ZFNs, making their efficient delivery into cells challenging.
Nevertheless, TALENS is an efficient strategy to generate plants with efficient and economically improved traits [205]. TALENS has been successfully used in Arabidopsis [206,207], tobacco [208,209], rice [210,211], barley [212] and Brachypodium [213]. For example, in Arabidopsis, five genes (ADH1, MAPKKK1, DSK2B, TT4, and NATA2) were targeted using seven TALENS. They observed somatic mutations in the transgenic plants with a mutation frequency of 2–15% and the mutations were transferred to the transgenic progeny with a frequency of 1.5–12% [207]. The somatic mutagenesis was also reported in rice and barley [210,212]. High mutation frequency was reported in rice and generated transgenic plants that are resistant to plant pathogens [210]. In tomato, TALENS was used to edit Anthocyanin gene (ANT1), which encodes a MYB transcription factor [57]. Using Geminivirus for genetic transformation, they could obtain precise insertions with no off-targets at all. They also used CRISPR-Cas along with TALENs, which showed similar efficiencies in editing of ANT1 gene. They overexpressed the ANT1 transgene resulting in purple plant tissue [57].
2.7. CRISPR/Cas9
CRISPR/Cas9 is a rapidly emerging genome editing tool, immensely used in various organisms, including plants. Unlike ZFNs and TALENs, CRISPR/Cas9 made the genome editing much convenient and effective to generate knockout mutants [190]. ZFNs and TALENS use FokI endonuclease, which forms a dimer for better specificity to bind to the target DNA. The designing of active FokI nucleases is tedious and very expensive. CRISPR/Cas9 requires a guide RNA (gRNA) to target the gene and a Cas9 endonuclease, which is an RNA dependent DNA endonuclease to edit. Cas9 forms a complex with gRNA and recognizes a specific protospacer adjacent motif (PAM) with a consensus sequence of 5′ NGG 3′ at the 3′ end of the target sequence. The Cas9 induces a double-stranded break, 2–5 bp upstream of the PAM [211,214,215].
The basic principle of CRISPR/Cas9 is taken from a bacterial immune system where DNA segments of the invaded virus are arranged in an array called CRISPR array [216,217]. Upon exposure to the virus again, the bacteria generate RNA segments of the CRISPR array to bind and destroy the genome of the virus [214,218,219,220]. The Cas9 protein is composed of two connected lobes, a nuclease (NUC) lobe, and a recognition (REC) lobe. The NUC lobe has a PAM-interacting (PI) domain and two nickase domains, HNH is responsible for cleavage of the complementary strand and RuvC responsible for cleavage the non-complementary strand. The REC lobe is responsible for Cas9-sgRNA complex formation. During gRNA loading the confirmation of Cas9 changes from an inactive to an active form by building a central channel where the RNA-DNA heteroduplex will be positioned. The Cas9-gRNA complex scans the double-stranded DNA. Once the PI domain recognizes a three-bp-long PAM sequence, the DNA is melted, and the complementary DNA strand connects with the gRNA (RNA-DNA heteroduplex formation). After heteroduplex formation, the HNH and RuvC nickase domains cleave the double-stranded DNA three bases upstream of the PAM sequence [221,222,223] (Figure 4).
According to [224], the CRISPR/Cas systems are separated into two classes with several types and subtypes. The class 1 CRISPR systems (type I, III, and IV) use numerous Cas proteins to form a complex with crRNAs (tracrRNA), whereas the class 2 CRISPR systems (type II, V, and VI) use a single Cas protein. The Cas9 enzyme commonly used in most CRISPR studies, belongs to type II, is acquired from Streptococcus pyogenes [214]. The CRISPR/Cas induces a DSB at the target gene, which is repaired by the DNA repair system of the host [214,225,226]. The CRISPR knockouts generated are looked for the presence of the mutation and the removal of the transgene. Although CRISPR/Cas9 is very precise, it is not an unfailing system. Unexpected mutations, so-called off-targets effects, could be detected in various studies, which lead to concerns for biomedical and clinical applications [227]. However, the off-target effects in plants are very rare [228]. Nevertheless, different strategies were developed to increase the specificity of Cas9 cleavage (e.g., using double nicking, or truncate gRNA).
Compared to ZFNs and TALENs, the CRISPR/Cas9 editing system is easy to perform as it requires gRNA of 18–20 bp while ZFNs and TALENs need designing of specific nucleases, which is cumbersome and laborious. Besides gene knockout, the CRISPR/Cas9 tool can be used for several applications. The deletion of whole gene clusters is possible by the simultaneous expression of two or more gRNAs. Due to the cell’s homology-directed repair (HDR) of DSBs, genes can be knocked-in by providing template DNA with overlapping flanking regions [221,229]. For CRISPR/Cas9 system, the major limitation is the PAM motif requirement. The sporadic presence of PAM sequence in the genome or sometimes its complete absence in the desired coding parts of the genomes to be edited restricts the application of CRISPR/Cas9 [214,230]. To be more versatile by searching target sites in the gene of interest it is possible to use Cas9 proteins from other bacteria like S. aureus, S. thermophilus, Neisseria meningitides, or Brevibacillus laterosporus, which have different PAM specificity [231,232,233,234]. Another CRISPR tool specific to the RNA is CRISPR/C2c2, a type VI CRISPR/Cas system [235,236].
Being precise, efficient, and highly cost-effective technology, CRISPR/Cas9 is extensively used to edit several crop plants since its first report in 2013. Since then, this technology has been revolutionizing crop breeding. Several studies have reported genome editing using CRISPR/Cas9 in Arabidopsis, tobacco, rice, cucumber, maize, tomato, wheat, cassava, and potato to develop improved crop varieties in terms of biotic and abiotic stress responses as well as nutritional and other aspects [237]. Some of the most prominent examples of CRISPR/Cas9 editing are listed in Tables 1, 2 and 4.
3. Genome Editing for Resistance Breeding in Tomato
3.1. Biotic Stress Resistance Breeding in Tomato
The biotic factors include diseases caused by the attack of pathogens that reduces the crop yield by 20–40% worldwide (http://www.fao.org/news/story/en/item/280489/icode/). Several strategies have been developed for controlling diseases. The pesticides are used to contain the pathogens, but the pesticide spray is harmful to both humans and the environment [238]. The spray of chemicals also destroys some useful organisms, hence disturbing the ecological balance. In order to overcome diseases, the development and use of more pesticides are often exploited as a preventive measure. However, the application of pesticides is successful in resisting pathogens to some extent. Therefore, the dependence of crop productivity on chemicals/pesticides is not the best solution due to their harmful side-effects that may lead to ecological imbalance over long-term usage [239].
Genetic breeding of disease resistance crops provides an efficient and eco-friendly strategy to resist diseases. Initially, conventional breeding methods were successful in creating genetic resources with disease resistance. However, there are various disadvantages to traditional breeding. Firstly, it can be used for plants that can mate with each other. Secondly, the plant population must have sufficient genetic variation, especially in disease resistance. Thirdly, the time required and linkage drag often introduces several unwanted traits such as decreased yield. It is challenging to keep pace with fast-evolving pathogens with conventional breeding techniques [240,241].
The plants produced by targeted gene editing can be exploited to overcome the above challenges. The genes that negatively regulate disease resistance represent promising targets for genome editing. The examples of biotic stress responses in tomato are listed in Table 1.

Table 1.
Genome editing of tomato cultivars in response to biotic stress. APETALA2-domain transcription factor (SlSHN3), basic helix–loop–helix transcription factor (SlMYC2), Chitinase 1 gene from Helicoverpa armigera (HaCH1), coat protein (CP) or replicase (Rep) of TYLCV, Dicer-like (DCL1), DNA binding protein from starved cells of Ralstonia solanacearum (RsDps), Downy mildew resistance 6-1 (SlDMR6-1), early TYLCV replication associated protein gene (C1), Fusarium MAP kinase (Fmk1), High osmolarity glycerol pathway genes (Hog1 and Pbs2), hairpin RNA (hpRNA) construct derived from Potato spindle tuber viroid (PSTVd), JASMONATE ZIM DOMAIN2 (SlJAZ2), MAP kinase3 (SlMAPK3), MILDEW RESISTANT LOCUS O1 (SlMLO1), nucleotide-binding site (NBS) and leucine-rich repeat (LRR) gene (SlNL33), one of the disrupted genes in reduced mycorrhizal colonization (rmc) tomato mutant (Solyc08g075770), Pectate lyase (SlPL), Powdery Mildew Resistance 4 (PMR4), RESPIRATORY BURST OXIDASE HOMOLOG1 (SlRBOH1), HD-Zip I family transcription factor (SlHZ24), Sucrose non-fermenting 1-related protein kinase 2 (SnRK2.1 and SnRK2.2), ToLCV Replicase (Rep), ToLCV replication initiator protein (AC4).
3.1.1. Fungal Infections
Some of the most common and prevalent fungal infections in tomato are gray mold (Botrytis cinerea), early blight (Alternaria solani), late blight (Phytophthora infestans), and Fusarium wilt (Fusarium oxysporum f. sp. lycopersici). Here are a few examples of disease resistance for fungal infections using genome editing (Table 1).
B. cinerea is the most cosmopolitan and destructive fungal infection that causes gray mold in several plants, including tomato. Buxdorf et al. [242] have reported that RNAi silencing of SlSHN3 TF resulted in reduced susceptibility to the necrotrophic foliar pathogen B. cinerea in Micro-Tom tomato. In contrast, over-expression of SlSHN3 caused resistance to B. cinerea infection. Another similar study reported reduced tolerance to B. cinerea upon RNAi silencing of tomato pectate lyase (SlPL) [243]. SlNL33 is a gene that encode an NL type of NBS-LRR resistance proteins. Silencing of SlNL33 not only increased the tolerance to gray mold disease by B. cinerea but also enhanced the tolerance to oxidative stress [244].
The fungus F. oxysporium is a soil-borne parasite that penetrates the vascular tissues of the roots through hyphal branching and invades the xylem causing a characteristic wilting phenotype. This fungus destroys many economically important crops. Fusarium mitogen-activated protein kinase (MAPK) signaling genes (FMK1, HOG1), and PBS2) are involved in plant surface hydrophobicity (sensing) and pathogenesis [245]. The silencing of these three genes in F. oxysporium resulted in reduced mycelial growth on tomato fruits leading to reduced pathogenicity compared to the unsilenced fungus [246]. Zhang et al. [247] reported that knockout mutants of SlMAPK3 are susceptible to grey mold disease.
Prihatna et al. [248] reported the involvement of a novel gene expressed in the roots of reduced mycorrhizal colonization (rmc) mutant. This mutant is incapable of forming mycorrhiza colonization but is vulnerable to Fusarium wilt. The rmc mutant has a chromosomal deletion of five genes, where one of them is CYCLOPS, which is non-mycorrhizal similar to rmc mutant but is not susceptible to Tfw. Solyc08g075770 is one of the other four genes in the deleted fragment. This gene function is not yet annotated but is expressed in roots. CRISPR/Cas9 knockouts of this gene resulted in some mutants with rmc phenotype (susceptible to Tfw) and rmc mutants with 76R (wild type) phenotype.
Nekrasov et al. [249] generated a CRISPR/Cas9 mutant of Mildew resistance locus (Mlo) in Moneymaker. The wild type allele of Mlo is susceptible to powdery mildew, whereas the knockout mutants of mlo are resistant to the fungus causing powdery mildew infection. In a similar study, CRISPR/Cas9 mutants of Powdery Mildew Resistance 4 (PMR4) were resistant to the powdery mildew infection [250]. RNA silencing of SlPMR4 also showed enhanced resistance to powdery mildew [251]. A recent study used four guide RNAs to generate a full knockout CRISPR/Cas9 mutant of SlPMR4. However, the knockout mutants exhibited reduced susceptibility to the fungus Oidium neolycopersici but not complete resistance.
3.1.2. Viral Diseases
Several viruses like Gemini viruses are a threat to most of the crop plants, which affect crop productivity. Here are a few examples. Tashkandi et al. [252] targeted the coat protein (CP) and replicase (Rep) of Tomato Yellow leaf Curl Virus (TYLCV) using the CRISPR/Cas9 system in tomato cultivar Moneymaker. The CRISPR edited plants were resistant to the TYLCV infection, which was apparent by low levels of the TYLCV genome in the edited plants. The CP region of the genome was efficiently targeted than Rep. It could be due to the competition between the viral replication machinery and the Cas9 endonuclease to the Rep.
Another viral pathogen causing significant damage to tomato and other crop plants is Tomato leaf curl virus (ToLCV), which is transmitted by whitefly. In a study by [253], intron-hairpin RNA was used for silencing the ToLCV infection. Seven hundred and twenty-seven nucleotides at the C1 gene were used in sense and antisense orientation separated by an intron to form a hairpin. C1 gene is the ToLCV replication-associated protein. RNAi silencing of the C1 gene resulted in resistance against ToLCV. Similarly, siRNA mediated silencing of conserved sequences of ToLCV antisense replicase (AC4) gene efficiently inhibited the geminivirus ToLCV in the most ToLCV sensitive tomato cultivar, Campbell [254]. A related study of the silencing of Rep gene of ToLCV showed that a threshold level of dsRNA is needed for gene silencing resulting in inhibition of the virus in the ToLCV-infected tomato plants [255].
3.1.3. Bacterial Diseases
Pseudomonas syringae is a cosmopolitan bacterial pathogen affecting the most important crop plants. Upon infection, P. syringae pv. Tomato (Pto) DC3000 releases coronatine (COR) that induces a stomatal opening for the bacteria invasion. COR needs a coreceptor JAZ2 for a stomatal response. CRISPR/Cas9 edited SlJAZ2 lacked Jas domain at the C-terminal end of JAZ2, which is now SlJaz2 Δjas, which are resistant to the bacterial speck disease [256]. The study also conferred resistance to necrotrophs such as B. cinerea by uncoupling SA-JA pathways [255].
3.1.4. Oxidative Stress
Ralstonia solanacearum is a soil-borne pathogen that infects the host roots to cause a bacterial wilt disease. The pathogen infests through mechanical injury or wounds. Once infected, the pathogen enters the xylem of host roots to acquire the essentials nutrients leading the plant to death. The host roots release reactive oxygen species (ROS) in response to stress. As a result, the pathogen R. solanacearum experiences oxidative stress. Then pathogen upregulates the DPS gene (DNA binding protein from starved cells) to acclimatize to the host’s conditions. The DPS helps to maintain DNA integrity under stressful situations, including lack of nutrients, in E. coli and also protects the bacteria from oxidative stress [257,258,259,260,261,262,263]. The dps mutant (insertional) exhibited increased hydrogen peroxide levels and increased mutation rate upon starvation. The oxidative stress response regulator (OxyR) upregulated the expression of dps. R. solanacearum requires DPS to overcome the oxidative stress in the host roots, and the mutant shows decreased virulence in the host [264].
A tomato HD-ZIP1 TF (SlHZ24), regulates the transcription of GDP-D-mannose pyrophosphorylase 3 (SlGMP3) by binding to the promoter element of SlGMP3. The over-expression of SlGMP3 increased the ascorbic acid A levels by 1.6-fold. The ascorbic acid is produced under stress conditions to protect the plants from ROS. The over-expression of SlHZ24 elevated the ascorbic acid in the transgenic plants improving stress tolerance. While RNAi silenced, plants are sensitive to oxidative stress [265].
3.2. Abiotic Stress Resistance Breeding in Tomato
The abiotic stress conditions include drought, salinity, light intensity, temperature (hot/cold), nutrient availability, and biotic stress that involve an attack by pathogens. The examples of abiotic stress response are listed in Table 2.

Table 2.
Genome editing of tomato cultivars in response to abiotic stress tolerance. A putative Na+/H+ antiporter gene (SlSOS1), AGAMOUS-LIKE6 (SlAGL6), Altered response to salt stress 1 (SlARS1), ASCORBATE OXIDASE (SlAO), Basic region/Leucine zipper transcription factor (SlbZIP1), BRASSINAZOLE RESISTANT1 (SlBZR1), Chilling-tolerance divergence1 (LeCOLD1), C-repeat binding factor (SlCBF1), Cys2/His2-type zinc-finger protein (SlZF3), DNA-binding with one finger 22 (SlDof22), Fructose 1,6 biphospahte aldolase (SlFBA1), Glutamate decarboxylases (SlGADs), GABA transaminases (SlGABA-Ts), Glutamate receptor-like (SlGLR3.3 and SlGLR3.5), Guanine nucleotide-binding protein alpha-1 subunit (LeGPA1), invertase inhibitor1 (SlINVINH1), JUNGBRUNNEN1 (SlJUB1), MADS-BOX transcription factor (SlMBP11), MADS-BOX transcription factor (SlMBP8), MAP kinase1 (SlMPK1), MAP kinase3 (SlMAPK3), Mitochondrial Alternate oxidase 1a (SlAOX1a), MIXTA-like MYB transcription factor (SlMX1), non-expressor of pathogenesis-related gene 1 (SlNPR1), Phytol kinase (SlVTE5), Plant specific NAC Transcription factor (SlNAC11), Plant specific Transcription factor (SlGRAS4), Proline-,lysine-,glutamic-rich protein gene (SpPKE1), RING-H2 finger gene (ShATL78- Like), SUMO E3 ligase1 (SlSIZ1), Tomato 2-oxoglutarate-dependent dioxygenase gene (SlF3HL), Tomato Ethylene response factor genes (TERF2/LeERF2), Tomato osmotic sensitive-1 (tos1), Tomato salt sensitive-2 (tss2), uridine diphosphate glucosyltransferase (SlUGT75C1), Whirly1 (SlWHY1).
3.2.1. Freezing Stress
Ethylene response factors (ERF) family members play a role in plant stress responses [276], Briefings in functional biology). In tomato, TERF2/LeERF2 regulates the ethylene biosynthesis transcriptionally and is induced by cold temperature [277,278]. In tobacco, the over-expression of TERF2/LeERF2 enhanced the tolerance to freezing with increased expression of cold response genes. While RNAi silenced lines showed decreased expression of cold-responsive genes and reduced tolerance to freezing stress [279].
3.2.2. Chilling Stress
Cold temperatures affect plant growth and development, limiting crop productivity. The invertases convert sucrose to glucose and fructose. These hexoses act as signals molecules in plant stress responses [280,281,282,283]. Tomato cell wall invertase inhibitor (SlINVINH1) inhibits the invertase activity, thus limiting hexose production. Cold stress suppressed the expression of SlINVINH1 and induced the expression of invertases (Lin6 and Lin8). The RNAi mediated silencing of SlINVINH1 increased the invertase activity and improved the tolerance to chilling, whereas over-expression of SlINVINH1 suppressed the cell wall invertase activity, making the plants sensitive to cold stress. Glucose is known to induce the expression of ABA biosynthesis and signaling genes. Therefore, they checked for the expression of tomato 9-cis-epoxy carotenoid dioxygenases1 (SlNCED1) in silenced SlINVINH1 mutants and observed that cold stress-induced the SlNCED1 expression in the silenced lines and wild type [284].
Fructose 1,6 biphosphate aldolases (FBA) play an important role in the Calvin-Benson cycle and FBA expression vary with response to heat or cold stress in tomato seedlings [285,286,287]. Eight FBA genes are present in tomato. All FBA members are susceptible to cold temperatures in tomato seedlings [285]. Cai et al. [288] suppressed the SlFBA7 and observed that the RNAi silenced lines were found to show severe injury symptoms in response to chilling stress, which could be due to increased levels of hydrogen peroxide (H2O2) and superoxide anions (O2-). Similar studies of silenced plants of several other genes show susceptibility to chilling stress (Table 2), which could be possible candidates to overcome the chilling stress.
3.2.3. Heat Stress
Like cold, heat stress is also major environmental stress to plants affecting plant growth and development. In the coming years, heat stress is a serious threat to the environment due to global warming, making climate warmer [289]. Plants evolve mechanisms to tolerate these adverse conditions. In response to these situations, several genes responsive to heat stress have been identified and are summarized in Table 2.
As discussed in the above section (biotic stress), MAP kinases are involved in plant growth and developmental processes, disease resistance, and various stress responses such as drought, salt, and cold. The first report describing the role of MAPKs in heat stress came from the study of Link et al. [290]. Heat stress by activating MAP kinase was found to regulate tomato plant response by phosphorylating heat stress TF HsfA3. Similarly, in Arabidopsis, MAPK activates the HsfA2 in response to heat stress [291]. In a recent study, RNAi silencing of SlMPK1 (MAP kinase1) in tomato resulted in improved heat tolerance [292]. In contrast, its over-expression resulted in a compromised tolerance. Likewise, Mishra et al. [293] used the post-transcriptional gene silencing of SlHsfA1 by short interfering RNAs. Tandem inverted repeats of SlHsfA1 was used for co-suppression. The co-suppressed lines of SlHsfA1 exhibited sensitivity to heat stress, while over-expressor lines showed enhanced tolerance to heat.
The plant hormone brassinosteroids (BRs) play an important role in various plant developmental and physiological processes such as seed germination, photomorphogenesis, cell elongation, cell division, xylem differentiation, and plant reproduction [294]. BRs also play a role in biotic and abiotic stress responses [295,296,297]. BR signaling is majorly regulated through two TFs, Brassinazole resistant1 (BZR1) and BZR1-EMS-suppressor1 (BES1 or BZR2). CRISPR/Cas9 knockout plants of SlBZR1 was susceptible to heat stress with a decreased quantum efficiency of PSII. In response to heat stress, ROS and heat shock proteins are induced but downregulated in the knockout mutant. It also led to the curtailed production of apoplastic H2O2 (which activates the ROS-scavenging enzymes) in the knockout mutant in response to heat stress [298].
3.2.4. Drought Stress
ABA is conjugated to glucose by uridine diphosphate glycosyltransferases (UGTs) and is required for ABA homeostasis [299]. Among three UGTs in tomato, [300] silenced SlUGT75C1 using RNAi and observed that SlNCED1 (key enzyme in ABA biosynthesis) levels were unaltered while SlCYP707A2 (a key enzyme in ABA degradation) was up-regulated in the silenced line. Consistent with the down-regulation of SlUGT75C1 in the silenced line, ABA levels significantly increased, whereas Glc-conjugated ABA (ABA-GE) levels were decreased. Increased ABA and ethylene in the silenced fruits hastened fruit ripening. The knockdown mutants also exhibited tolerance to drought stress.
bZIP has an important role in various physiological and signaling processes and also in stress response. bZIP proteins such as ABA-responsive element-binding proteins (AREBs) and ABRE binding factors (ABFs) regulate the ABA-dependent transcription and abiotic stress responses [301,302]. ABA and salt stress induce the expression of bZIP1. RNAi silencing of bZIP1 downregulated the genes involved in ABA biosynthesis such as SlNCED1, SlNCED2, and signal transduction genes such as SlABF2, SlABF4 were down-regulated. Further, the silenced plants exhibited reduced tolerance to both drought and salt stresses [303]. Therefore, as the bZIP act as essential regulator of stress tolerance, these could be promising candidates for crop improvement in response to stress.
3.2.5. Salt Stress
A tomato MADS-box TF, SlMBP11, is implicated in tolerance to salinity [304]. Similarly, SlMBP8 is involved in abiotic stress responses such as salinity, drought, wounding, cold, and heat stresses. SlMBP8 is also induced by methyl, jasmonates, 1-amino cyclopropane-1-carboxylic acid, indole acetic acid, and ABA [305]. RNAi knockdown lines of SlMBP8 showed improved tolerance to drought and salinity [305].
SlZF3 is a C2H2 zinc-finger protein TF. Zinc finger TFs are involved in plant stress response/tolerance. Salt stress induces the expression of SlZF3. RNAi silenced lines of SlZF3 are susceptible to salt stress, whereas its over-expression lines are tolerant to salt stress [306].
To elucidate the role of GABA in salt stress, Bao et al. [307] silenced the GABA pathway genes, Glutamate decarboxylase (GAD), GABA transaminase (GABA-T), and succinic semialdehyde dehydrogenase (SSADH) using the VIGS approach. Silencing of SlGAD and SlGABA-T showed increased sensitivity to salinity, while that of SlSSADH showed reduced sensitivity to salt stress.
3.2.6. Osmotic Stress
Osmotic stress is mainly caused by drought, salinity, and cold stresses where the plant cells lose its water potential, reducing crop productivity [308]. The osmotic stress involves oxidative damage and ROS production. Excessive production of ROS induces cell damage and tissue death [309]. The osmotic stress causes the SNF1-related protein kinase2 (SnRK2) family, which are essential for stress signaling. SnRK2 family has seven genes out of which SnRK2.1, 2.2, and 2.3 were suppressed while the expression of the remaining four members of the family was initially decreased then increased [310]. The over-expression of SnRK2.1 and SnRK2.2 resulted in plants with reduced tolerance to salt stress, while RNAi silenced lines showed enhanced tolerance to salt stress [310].
Another study by Borsani et al. [311] screened EMS population based on sensitivity to osmotic stress. They isolated two osmotic hypersensitive mutants, tomato osmotic sensitive-1 (tos1) and tomato salt sensitive-2 (tss2) [311]. When checked for seed germination, tos1 showed reduced sen-sitivity to ABA while tss2 was hypersensitive to ABA. The study further suggests that ABA perception and signaling are essential for osmotic stress response [311].
3.3. Genes Regulating Combined Biotic and Abiotic Stress Response
There are very few studies on tomato where editing of genes regulated both biotic and abiotic stresses. Here we discussed the selected examples below and further summarized the entire list in Table 3.

Table 3.
Genome editing of tomato cultivars in response to combined biotic and abiotic stresses. Abscisic acid-induced MYB1 (SlAIM1), Hybrid proline-rich protein1 (HyPRP1), Stress-related NAC1 (SlSRN1), Tomato ethylene response factor (SlPti4), Tomato lipoxygenase gene (SlTomLoxD), Tomato SR/CAMTA transcription factors (SlSR1 and SlSR3L).
NAC TFs are involved in various plant growth and developmental processes and participate in response to various biotic and abiotic stresses. Liu et al. [337] identified SlSRN1 (Stress Related NAC1). Infection of tomato plants with B. cinerea and Pto DC3000 triggered SlSRN1 expression. Silencing of SlSRN1 through VIGS resulted in more susceptibility of plant infections caused by B. cinerea and Pto DC3000. However, the VIGS silenced plants were more tolerant to drought and oxidative stress [337].
Another gene, SlTomLoxD, is a lipoxygenase that plays a key role in jasmonic acid (JA) biosynthesis [338]. The expression of SlTomLoxD is induced in response to wounds and pathogen infections by stimulating JA and systemins [339]. RNAi silencing of SlTomLoxD resulted in reduced susceptibility to heat and highly susceptible to Cladosporium fulvum [340].
AbuQamar et al. [341] reported that ABA-induced R2R3MYB1 (AIM1) TF is induced in response to oxidative stress, pathogens, salt stress, and plant hormones. RNAi silenced plants of SlAIM1 were highly sensitive to salinity and oxidative stress and also susceptible to B. cinerea.
A family of transcription activators, called SR/CAMTA (signal responsive/Calmodulin transcription activators), are involved in the response to biotic and abiotic factors. In Arabidopsis and rice, these proteins play an important role in response to several biotic factors such as Golovinomyces cichoracearum, P. syringae pv. tomato (Pto) DC3000 and B. cinerea [342,343,344,345,346] and abiotic factors such as temperature and drought [347,348,349,350]. In tomato, seven members constitute this gene family [351]. VIGS silencing of SlSR1L led to increased susceptibility to drought stress along with the reduced expression of drought-responsive genes [352]. Similarly, VIGS mediated silencing of SlSR1, and SlSR3L genes exhibited enhanced tolerance to necrotrophic fungus B. cinerea and Pst DC3000 [352].
Hybrid proline-rich proteins (HyPRP) are well-known proteins in plant response to various biotic and abiotic factors [353,354,355,356,357]. In tomato, HyPRPs are downregulated under stress conditions such as heat, cold, salt, drought, oxidative stress, etc. Li et al. [358] reported that RNAi knockdown plants showed increased resistance to salt, dehydration, and oxidative stress.
SlPti4 is a tomato TF of the ERF gene family, and it plays an essential role in plant disease resistance [359,360,361]. The RNAi knockdown of SlPti4 resulted in the transgenic plants with decreased tolerance to drought and enhanced susceptibility to B. cinerea [362].
3.4. Genome Editing for Yield and Fruit Quality Improvement in Tomato
Fruit quality and quantity are certain key traits that are necessary for crop improvement. While early flowering, multiple flowers, determinate growth habit, fruit size, and locule number are the major traits deciding the yield, fruit quality and nutritional value are rated based on fruit ripening, fruit pigmentation, and fruit metabolites. The examples of fruit quality and yield improvement in tomato using genome editing are listed in Table 4.

Table 4.
List of gene editing for yield and fruit quality improvement in tomato. AGAMOUS-LIKE6 (SlAGL6), Alcobaca (ALC), Anthocycanin1 (ANT1), ARGONAUTE7 (AGO7), Aux/IAA9 transcription factor (SlIAA9), BLADE-ON-PETIOLE family (BOP1, BOP2, BOP3), mitochondrial transcription factor A (TFAM1, TFAM2), Carotenoid isomerase (CRTISO), CLAVATA3 (CLV3), Defective chloroplasts and leaves (DCL), DELLA (aspartic acid–glutamic acid–leucine–leucine–alanine), fasciated and branched (fab) and fasciated inflorescence (fin), FEEBLY (FB), GDP-L-GALACTOSE PHOSPHORYLASE (GGP1), LEAFY-COTYLEDON1-LIKE4 (L1L4), LYCOPENE BETA CYCLASE (CycB), MULTIFLORA (MULT), organelle RNA recognition motif-containing protein4 (ORRM4), OVATE (O), Phytoene desaturase (PDS), pyruvate-dependent GABA-T family proteins (GABA-TP1, GABA-TP2, GABA-TP3), cationic amino acid transporter 9 (CAT9), succinate semialdehyde dehydrogenase (SSADH), RIPENING INHIBITOR (RIN), SELF PRUNING (SP), SELF PRUNING 5G (SP5G), SHORT ROOT (SHR), stay-green 1 (SGR1), lycopene ε-cyclase (LCY-E), beta-lycopene cyclase (Blc), lycopene b-cyclase 1 (LCY-B2), WUSCHEL (WUS).
Many genome editing experiments resulted in a change in floral morphology and flowering time. Xu et al. [363] identified one EMS mutant for fab (fasciated and branched) and two EMS mutants, and three fast neutrons (FN) mutants for fin (fasciated inflorescence). These mutants were characterized with enlarged meristems resulting in branched inflorescence and fasciated flowers, leading to fruits with a higher number of locules. Map-based cloning identified the mutations in CLAVATA1 (CLV1) for FAB and an arabinosyltransferase gene for FIN. To generate quantitative variants by editing the Cis-regulatory elements (CRE), Rodríguez-Leal et al. [364] generated CRISPR/Cas9 mutants in S. pimpinellifolium. Mutations in the promoter of CLV3 resulted in a series of variants for floral organs and locule number. They also generated promoter mutants for WUSCHEL-RELATED HOMEOBOX 9, WOX9 (S), and SELF PRUNING (SP), resulting in many quantitative variations in plant architecture and inflorescence modifications. These variants allow studying specific CREs in the regulation of gene expression. They are also promising sources to explore the dominant and semi-dominant effects of genes with possibilities for orphan crops’ domestications. CRISPR/Cas9 mediated gene editing for the flowering repressor SELF-PRUNING 5G (SP5G) leads to compact determinate growth habit, with day-length-independent early flowering facilitating early harvest [365]. They introduced the mutation into the sp background, which has a determinate growth habit, resulting in ‘double-determinate’ plants with early yield. This study was later supported by another similar gene-editing effort by [366]. Since S. pimpinellilfolium is tolerant of many bacterial diseases and salt stress, they generated single and double mutants for SP and SP5G through CRISPR/Cas9-engineering, confirming the effect of sp in determinate growth habit, sp5G in day neutrality, and the double mutant with synchronized fruit ripening. They also generated mutants targeting the promoter region of WUSCHEL (WUS) and CLV3. While mutation in the WUS promoter enlarged the fruit size, the CLV3 promoter mutation did not increase the locule numbers as expected, possibly because the targeted site was not necessary for gene expression regulation. This study’s highlight was that the retention of the disease and salt tolerance of S. pimpinellilfolium in the CRISPR/Cas9 mutants. Efforts to domesticate S. pimpinellilfolium was also carried out by Zsögön et al. [367] using multiplex CRISPR/Cas9 editing of coding sequences of SP, OVATE (O), CLV3, MULTIFLORA (MULT), and LYCOPENE BETA CYCLASE (CycB). This multiplex editing resulted in diverse combinations of mutant alleles. The possibility of off-target editing was excluded by sequencing of two closely related targets for each gene. This study successfully introduced several domestication traits, including determining growth habits, more fruits (10-fold increase) with a bigger size (3-fold) of the fruits, and a 500% increase in fruit lycopene content [362].
Fruit ripening is another important fruit trait that has been targeted using CRISPR/Cas9 mediated gene editing. Ito et al. [368] targeted three regions in the coding sequence of the MADS-box TF RIN (RIPENING INHIBITOR), resulting in the generation of multiple mutated alleles with altered ripening phenotypes ranging from low pigmentation and incomplete ripening in varying degrees. The actual rin mutant was thought to be a null mutant, but the CRISPR knockout of RIN is rather a gain of function mutant repressing the fruit ripening process [369]. The knockout mutant of RIN shows induction of fruit ripening associated with physiological changes such as lycopene accumulation albeit much lower than wild type and expression of fruit softening enzymes such as pectate lyase and polygalacturonase. This study suggests that RIN is essential for fruit ripening but not for induction of fruit ripening [370]. Virus-induced gene silencing of 11 putative tomato fruit ripening-related factors indicated that organelle RNA recognition motif-containing protein4 (ORRM4) positively regulates tomato fruit ripening. The coding sequence of SlORRM4 was edited using the CRISPR/Cas9 method, leading to a delay in fruit ripening associated with low ethylene production and respiratory rate. This study concluded that ORRM4 is a Mitochondria-localized RNA editing factor that has a significant role in regulating the fruit ripening process [371].
The long shelf life of the fruits is another desirable trait in tomato. Yu et al. [372] reported knock-out of ALC (Alcobaca) gene using CRISPR/Cas9 mediated editing, resulting in delayed pigment accumulation. However, it did not affect the fruit ripening, harvest time, or the ripened fruit’s color. Most importantly, it was observed that alc mutation significantly improved the fruit shelf life. They also generated Cas9-overexpressing (Cas9-OE) S. lycopersicum cv. M82 transgenic lines for “virus-mediated genome editing system”, where a TRV RNA2 genome-derived vector facilitates the gRNA delivery. Another study reported that SlGH3.2 expression is induced during fruit ripening. The RNAi silencing of SlGH3.2 elevated the shelf life of fruits possibly due to increased IAA and IBA levels in the transgenic fruits [373].
Klap et al. [329] identified a parthenocarpic mutant from an EMS mutagenized population of M82 cultivar. They identified a candidate gene AGAMOUS-LIKE 6 (SlAGL6) and created gene knockout lines targeting the coding sequence using CRISPR/Cas9. The mutant exhibited facultative parthenocarpy, which resulted in fruit production under heat stress conditions. Another gene-editing study on SlIAA9 resulted in parthenocarpy and altered leaf morphology [374].
There are few gene-editing studies carried out for carotenoids and anthocyanins in tomato. Hayut et al. reported the PHYTOENE SYNTHASE (PSY1) gene-editing resulting in yellow fruits with red sectors. They also demonstrated somatically induced double-strand breaks by crossing the psy1 mutant with S. pimpinellifolium. Multiplex CRISPR/Cas9 genome editing was carried out for stay-green 1 (SGR1), lycopene ε-cyclase (LCY-E), beta-lycopene cyclase (Blc), lycopene b-cyclase 1 (LCY-B1), and LCY-B2 in Solanum lycopersicum cv. AC [375]. This resulted in various combinations of mutations. The knock-out lines with only SGR1 mutation exhibited the highest lycopene (5.1-fold) and increased other carotenoids like phytoene, prolycopene, α-carotene, and lutein compared to the remaining combinations. All the mutants with the SGR1 mutation showed the characteristic rust color after the breaker stage. In another study, fast neutron-induced mutagenesis, followed by map-based cloning, identified a tangerine mutant with a 282 bp deletion in the coding sequence [14]. Čermák et al. [57] reported the use of both TALENs and CRISPR/Cas9 to edit Anthocycanin1 (ANT1) gene, which codes for an MYB TF using geminivirus replicons. Instead of silencing, they overexpressed the endogenous ANT1 coding sequence by inserting a cauliflower mosaic virus 35S promoter upstream. They could achieve precise insertion without sequence modifications in more than two-thirds of the insertions. Almost the same efficiency was observed for both the methods without any off-target modifications. Moreover, these chromosomal changes showed a Mendelian pattern of transmission to the next generation. Transposon mediated mutagenesis of FEEBLY (FB) gene with an insertion of the Ds element in the intron was characterized by high anthocyanin levels, small, fragile plants, and insensitivity to phosphinothricin [181].
ZFNs-mediated genome editing for LEAFY-COTYLEDON1-LIKE4 (L1L4) resulted in a number of phenotypic variations, including seed storage proteins and fatty acids, fruit shape, moisture content, and fruit metabolite levels including fructose, total polyphenols, antioxidants, β-carotene, oxalic, and citric acid [376,377]. Some other CRISPR/Cas9 gene-editing studies resulted in enhanced GABA content and ascorbic acid content in leaves [378], changes in leaf complexity, shape, and serration [61,379,380]. Two independent reports explained the knockdown of PDS, resulting in the plant’s albino phenotype [366,381]. Transposon-mediated mutagenesis was achieved for defective chloroplasts and leaves (DCL) gene resulting in albino phenotype with green patches [382]. Ron et al. [383] reported the short root phenotype by editing the SHR gene. TALENs mediated knockout of the PROCERA gene resulted in enhanced GA response in tomato [384].
4. Challenges and Future Perspectives
Tomato has been domesticated by selective breeding that often leads to loss of genetic diversity and fitness. Conventional breeding of desired traits into the elite tomato cultivars is quiet a labor-intensive and time-consuming process. The availability of tomato genome sequence has increased the use of reverse genetic tools such as induced mutagenesis, transposons, RNA interference, etc. However, these tools have several limitations such as screening of large populations, cloning and transformation strategies, which are quite labor-intensive and cumbersome. These challenges were overwhelmed by genome editing tools such as ZFNs, TALENs, CRISPR/Cas9 in crop plants such as tomato. These tools offer precise editing of desired genes and multiple genes can be edited simultaneously, which can accelerate the breeding process and greatly reduce costs. However, the genome editing tools also possess disadvantages. For example, TALENs cannot edit methylated regions and the engineering of endonuclease every time is the major limitation in ZFNs and TALENs. CRISPR/Cas9 does not require engineering of Cas9 to target different genes. However, the PAM sequence which is usually 2–5 bp, is found very often in the genome, therefore it is difficult to edit the desired genes with the sequence constraint [386]. This can be overcome by variants of Cas9 with different sequence specificities for PAM. For instance, Cas12a (Cpf1), a class II Type V nuclease requires a PAM sequence rich in thymine (5′-TTTN-3′) [387,388]. The genomes such as tomato are AT-rich, and certain genomic regions are reluctant to gene editing. Such AT-rich regions can be edited by Cas variants like Cas12a. The generation of off-targets has also been a major concern in genome editing and the use of Cas12a is said to decrease the possibility of off-target mutations [53]. The induction of double cas9 nickase mutants or the truncation of the gRNA increase the specificity of Cas9 cleavage and reduce the off-target effects. Besides, to perform precise genome editing or to introduce new elements, it is essential to overcome the limitation of the low frequency of HDR in plant [227].
As a result of selective breeding, much of the tomato genetic diversity is lost in the modern domestic cultivars. This can be restored by introgression of specific traits into the domesticated cultivars, but since this process is quite laborious, genome editing can be useful to help restore the lost genetic traits in the present tomato cultivars. There are various examples of gene editing in tomato that are presented in this review, which show stress resistance and improvement in yield and nutrition. However, significant progress is required to achieve success in combating the changing environmental conditions. New variants of Cas9 with diverse specificities of PAM would greatly enhance the genetic restoration in crop plants such as tomato.
A combination of genome editing technology and conventional breeding can speed up the introduction of the trait of interest. Genetic crosses help remove undesired elements, which is a prerequisite to gain regulatory approval of transgene-free gene-edited plants [389]. Apart from these, there are several regulatory issues associated with genome-edited plants. There exists a gap in public towards the understanding of gene edited plants with and without foreign genetic material. There are several hurdles in releasing the non-GMO plants to the public, therefore the regulatory policies need to be harmonized. In 2016, the United States Department of Agriculture gave an exception to genome-edited mushrooms and corn from the traditional genetic modification policies, whereas the Court of justice of the European Union announced that the gene-edited crops come under genetically modified organisms. Recently, the United States Department of Agriculture released six virus-resistant tomato plants generated by gene-editing (https://www.isaaa.org/kc/cropbiotechupdate/article/default.asp?ID=17661).
5. Conclusions
According to the Food and Agricultural Organization, the world population would reach approximately 9 billion by 2050. It is estimated that crop productivity must be increased by 70% to feed the ever-increasing population (United Nations World Population Prospects: The 2017 Revision; FAO, 2019). New varieties of vegetables with high yield and stress-tolerance must be developed to address food scarcity for the overexploited population under an ever-changing climate [390]. Over time, humans have successfully developed crops with new or improved traits by transferring desirable genetic variations through conventional breeding techniques. Although conventional breeding methods have improved gradually, there is still an urgent need to enhance crop yield and productivity further quickly in the recent future [391,392].
With the advent of sequencing technologies, functional genomics has revolutionized with new editing tools to create novel allelic series of mutants for crop improvement. Among them, CRISPR-Cas technology has gained much attention over the other genome editing techniques because designing specific nuclease domains each time is a tedious task. However, in the last decade, CRISPR has emerged with different Cas endonucleases rendering the editing process much more precise and easier. In tomato, the genome editing tools is applied to enhance the nutritional value, yield, and tolerance to biotic and abiotic stresses. Further studies are required to enhance the essential traits such as improvement in the resistance to pathogens and abiotic stresses, yield, and nutritional aspects (such as enrichment of lycopene) in tomato and other agronomic crops. With the newly emerging CRISPR/Cas systems, the plant genetic engineering would increase the scope of generating plants with improved flavor, nutrition, and stress tolerance.
Author Contributions
H.S., S.T., V.M., and F.M. wrote the manuscript. H.S. and S.T. prepared the figures. F.M. designed, advised, and improved the Manuscript. All authors (H.S., S.T., V.M, R.K. and F.M.) read and approved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors wish to thank Tarak Ramji Moturu and Gorji Marzban.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AC4 | ToLCV replication initiator protein |
| AGO7 | ARGONAUTE7 |
| ALC | Alcobaca |
| ANT1 | Anthocycanin1 |
| Blc | beta-lycopene cyclase |
| BOP1, BOP2, BOP3 | BLADE-ON-PETIOLE family |
| C1 | early TYLCV replication associated protein gene |
| CAT9 | cationic amino acid transporter 9 |
| CLV3 | CLAVATA3 |
| CMTs | chromomethylases |
| CNR | colourless non-ripening |
| CP and Rep | coat protein and Replicase of TYLCV |
| Cpf1 | CRISPR-associated endonuclease in Prevotella and Francisella 1 |
| CRISPR/Cas9 | Clustered Regularly Interspaced Short Palindromic Repeat/CRISPR- associated protein 9) |
| CRTISO | Carotenoid isomerase |
| CycB | LYCOPENE BETA CYCLASE |
| DCL | Defective chloroplasts and leaves |
| DCL1 | Dicer-like |
| DELLA | aspartic acid–glutamic acid–leucine–leucine–alanine |
| DES | Diethyl sulfate |
| DHFR | Dihydroflavonol 4-reductase |
| DRM | Domains rearranged methyltransferases |
| DSB | Double-stranded DNA break |
| dsDNA | Double stranded DNA |
| EI | ethyleneimine |
| EMS | ethyl methanesulphonate |
| ENU | ethyl nitro urethane |
| ENU | ethyl nitrosourea |
| fab | fasciated and branched |
| FAO | Food and Agricultural Organization |
| FB | FEEBLY |
| fin | fasciated inflorescence |
| Fmk1 | Fusarium MAP kinase |
| GABA-TP1, GABA-TP2, GABA-TP3 | pyruvate-dependent GABA-T family proteins |
| GE | Genome editing |
| GGP1 | GDP-L-GALACTOSE PHOSPHORYLASE |
| GMOs | Genetically modified organisms |
| gRNA | Guide RNA |
| GWAS | Genome Wide Association study |
| HaCH1 | Chitinase 1 gene from Helicoverpa armigera |
| HDR | Homology-dependent recombination |
| Hog1 and Pbs2 | High osmolarity glycerol pathway genes |
| hpRNA | hairpin RNA |
| HyPRP1 | Hybrid proline-rich protein1 |
| INDELs | insertions or deletions |
| L1L4 | LEAFY-COTYLEDON1-LIKE4 |
| LCY-B2 | lycopene b-cyclase 1 |
| LCY-E | lycopene ε-cyclase |
| LeCOLD1 | Chilling-tolerance divergence1 |
| LeGPA1 | Guanine nucleotide-binding protein alpha-1 subunit |
| MET1 | Methyltransferase1 |
| MNU | 1-methyl-1-nitrosourea |
| MULT | MULTIFLORA |
| NGS | Next-generation sequencing |
| NHEJ | Non-homologous end joining |
| O | OVATE |
| ODM | Oligonucleotide directed mutagenesis |
| ORRM4 | organelle RNA recognition motif-containing protein4 |
| PAM | protospacer adjacent motif |
| PDS | Phytoene desaturase |
| PMR4 | Powdery Mildew Resistance 4 |
| PSTVd | Potato spindle tuber viroid |
| PTGS | post-transcriptional gene silencing |
| PVX | Potato virus X |
| QTL | Quantitative trait loci |
| RdDM | RNA-dependent DNA methylation |
| RdRP | RNA dependent RNA polymerase |
| Rep | Replicase of TOLCV |
| RIN | RIPENING INHIBITOR |
| RISC | RNA-induced silencing complex |
| rmc | reduced mycorrhizal colonization |
| RNA pol IV | RNA polymerase IV |
| RNA pol V | RNA polymerase V |
| RNAi | RNA interference |
| RsDps | DNA binding protein from starved cells of Ralstonia solanacearum |
| RVD | Repeat Variable Diresidue |
| SGR1 | stay-green 1 |
| ShATL78- Like | RING-H2 finger gene |
| SHR | SHORT ROOT |
| siRNAs | short interfering RNAs |
| SlAGL6 | AGAMOUS-LIKE6 |
| SlAIM1 | Abscisic acid-induced MYB1 |
| SlAlkBH2 | AlkB homolog 2 |
| SlAO | ASCORBATE OXIDASE |
| SlAOX1a | Mitochondrial Alternate oxidase 1a |
| SlARS1 | Altered response to salt stress 1 |
| SlbZIP1 | Basic region/Leucine zipper transcription factor |
| SlBZR1 | BRASSINAZOLE RESISTANT1 |
| SlCBF1 | C-repeat binding factor |
| SlDML2 | Domains rearranged methyltransferases |
| SlDMR6-1 | Downy mildew resistance 6-1 |
| SlDof22 | DNA-binding with one finger 22 |
| SlF3HL | Tomato 2-oxoglutarate-dependent dioxygenase gene |
| SlFBA1 | Fructose 1,6 biphospahte aldolase |
| SlGABA-Ts | GABA transaminases |
| SlGADs | Glutamate decarboxylases |
| SlGLR3.3 | Glutamate receptor-like |
| SlGLR3.5 | Glutamate receptor-like |
| SlGRAS4 | Plant specific Transcription factor |
| SlHZ24 | HD-Zip I family transcription factor |
| SlIAA9 | Aux/IAA9 transcription factor |
| SlINVINH1 | invertase inhibitor1 |
| SlJAZ2 | JASMONATE ZIM DOMAIN2 |
| SlJUB1 | JUNGBRUNNEN1 |
| SlMAPK3 | MAP kinase3 |
| SlMAPK3 | MAP kinase3 |
| SlMBP11 | MADS-BOX transcription factor |
| SlMBP8 | MADS-BOX transcription factor |
| SlMLO1 | MILDEW RESISTANT LOCUS O1 |
| SlMPK1 | MAP kinase1 |
| SlMX1 | MIXTA-like MYB transcription factor |
| SlMYC2 | basic helix–loop–helix transcription factor |
| SlNAC11 | Plant specific NAC Transcription factor |
| SlNL33 | nucleotide-binding site (NBS) and leucine-rich repeat (LRR) gene |
| SlNPR1 | non-expressor of pathogenesis-related gene 1 |
| SlPL | Pectate lyase |
| SlPti4 | Tomato ethylene response factor |
| SlRBOH1 | RESPIRATORY BURST OXIDASE HOMOLOG1 |
| SlSHN3 | APETALA2-domain transcription factor |
| SlSIZ1 | SUMO E3 ligase1 |
| SlSOS1 | A putative Na+/H+ antiporter gene |
| SlSR1 | Tomato SR/CAMTA transcription factors |
| SlSR3L | Tomato SR/CAMTA transcription factors |
| SlSRN1 | Stress-related NAC1 |
| SlTomLoxD | Tomato lipoxygenase gene |
| SlUGT75C1 | uridine diphosphate glucosyltransferase |
| SlVTE5 | Phytol kinase |
| SlWHY1 | Whirly1 |
| SlZF3 | Cys2/His2-type zinc-finger protein |
| SnRK2.1 | Sucrose non-fermenting 1-related protein kinase 2 |
| SnRK2.2 | Sucrose non-fermenting 1-related protein kinase 2 |
| SP | SELF PRUNING |
| SP5G | SELF PRUNING 5G |
| SPB3-like | SQUAMOSA promoter binding protein3-like |
| SpPKE1 | Proline-,lysine-,glutamic-rich protein gene |
| SSADH | succinate semialdehyde dehydrogenase |
| TALENs | Transcriptional Activator Like Effector Nucleases |
| T-DNA | Transposon DNA |
| TE | Transposable elements |
| TERF2/LeERF2 | Tomato Ethylene response factor genes |
| TFAM1, TFAM2 | mitochondrial transcription factor A |
| TGS | transcriptional gene silencing |
| TILLING | Targeted Induced Local Lesions IN Genomes |
| TMV | Tobacco Mosaic Virus |
| tos1 | Tomato osmotic sensitive-1 |
| TRV | Tobacco Rattle Virus |
| tss2 | Tomato salt sensitive-2 |
| TYMV | Turnip yellow Mosaic Virus |
| VIGS | Virus-induced gene silencing |
| WUS | WUSCHEL |
| ZFNs | Zinc finger nucleases |
References
- Razifard, H.; Ramos, A.; Della Valle, A.L.; Bodary, C.; Goetz, E.; Manser, E.J.; Li, X.; Zhang, L.; Visa, S.; Tieman, D. Genomic evidence for complex domestication history of the cultivated tomato in Latin America. Mol. Biol. Evol. 2020, 37, 1118–1132. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Bawa, A.; Anilakumar, K. Genetically modified foods: Safety, risks and public concerns—A review. J. Food Sci. Technol. 2013, 50, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Gascuel, Q.; Diretto, G.; Monforte, A.J.; Fortes, A.M.; Granell, A. Use of natural diversity and biotechnology to increase the quality and nutritional content of tomato and grape. Front. Plant Sci. 2017, 8, 652. [Google Scholar] [CrossRef] [PubMed]
- Labate, J.A.; Grandillo, S.; Fulton, T.; Muños, S.; Caicedo, A.L.; Peralta, I.; Ji, Y.; Shetelat, R.T.; Scott, J.W.; Gonzalo, M.J. Tomato. In Genome Mapping and Molecular Breeding in Plants; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2007; Volume 5. [Google Scholar]
- Just, D.; Garcia, V.; Fernandez, L.; Bres, C.; Mauxion, J.-P.; Petit, J.; Jorly, J.; Assali, J.; Bournonville, C.; Ferrand, C. Micro-Tom mutants for functional analysis of target genes and discovery of new alleles in tomato. Plant Biotechnol. 2013, 30. [Google Scholar] [CrossRef]
- Meissner, R.; Chague, V.; Zhu, Q.; Emmanuel, E.; Elkind, Y.; Levy, A.A. A high throughput system for transposon tagging and promoter trapping in tomato. Plant J. 2000, 22, 265–274. [Google Scholar] [CrossRef]
- Menda, N.; Semel, Y.; Peled, D.; Eshed, Y.; Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 2004, 38, 861–872. [Google Scholar] [CrossRef]
- Rothan, C.; Diouf, I.; Causse, M. Trait discovery and editing in tomato. Plant J. 2019, 97, 73–90. [Google Scholar] [CrossRef]
- Ariizumi, T.; Kishimoto, S.; Kakami, R.; Maoka, T.; Hirakawa, H.; Suzuki, Y.; Ozeki, Y.; Shirasawa, K.; Bernillon, S.; Okabe, Y. Identification of the carotenoid modifying gene PALE YELLOW PETAL 1 as an essential factor in xanthophyll esterification and yellow flower pigmentation in tomato (S olanum lycopersicum). Plant J. 2014, 79, 453–465. [Google Scholar] [CrossRef]
- Chaudhary, J.; Alisha, A.; Bhatt, V.; Chandanshive, S.; Kumar, N.; Mir, Z.; Kumar, A.; Yadav, S.K.; Shivaraj, S.; Sonah, H. Mutation breeding in tomato: Advances, applicability and challenges. Plants 2019, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Ariizumi, T.; Ezura, H. SEXUAL STERILITY is essential for both male and female gametogenesis in tomato. Plant Cell Physiol. 2017, 58, 22–34. [Google Scholar]
- Isaacson, T.; Ronen, G.; Zamir, D.; Hirschberg, J. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 2002, 14, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Minoia, S.; Petrozza, A.; D’Onofrio, O.; Piron, F.; Mosca, G.; Sozio, G.; Cellini, F.; Bendahmane, A.; Carriero, F. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 2010, 3, 69. [Google Scholar] [CrossRef] [PubMed]
- Okabe, Y.; Asamizu, E.; Saito, T.; Matsukura, C.; Ariizumi, T.; Brès, C.; Rothan, C.; Mizoguchi, T.; Ezura, H. Tomato TILLING technology: Development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 2011, 52, 1994–2005. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Jiang, K.; Tal, L.; Yichie, Y.; Gar, O.; Zamir, D.; Eshed, Y.; Lippman, Z.B. Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 2014, 46, 1337–1342. [Google Scholar] [CrossRef] [PubMed]
- Petit, J.; Bres, C.; Just, D.; Garcia, V.; Mauxion, J.-P.; Marion, D.; Bakan, B.; Joubès, J.; Domergue, F.; Rothan, C. Analyses of tomato fruit brightness mutants uncover both cutin-deficient and cutin-abundant mutants and a new hypomorphic allele of GDSL lipase. Plant Physiol. 2014, 164, 888–906. [Google Scholar] [CrossRef]
- Petit, J.; Bres, C.; Mauxion, J.-P.; Tai, F.W.J.; Martin, L.B.; Fich, E.A.; Joubès, J.; Rose, J.K.; Domergue, F.; Rothan, C. The glycerol-3-phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol. 2016, 171, 894–913. [Google Scholar] [CrossRef]
- Pulungan, S.I.; Yano, R.; Okabe, Y.; Ichino, T.; Kojima, M.; Takebayashi, Y.; Sakakibara, H.; Ariizumi, T.; Ezura, H. SlLAX1 is required for normal leaf development mediated by balanced adaxial and abaxial pavement cell growth in tomato. Plant Cell Physiol. 2018, 59, 1170–1186. [Google Scholar] [CrossRef]
- Shi, J.X.; Adato, A.; Alkan, N.; He, Y.; Lashbrooke, J.; Matas, A.J.; Meir, S.; Malitsky, S.; Isaacson, T.; Prusky, D. The tomato S l SHINE 3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytol. 2013, 197, 468–480. [Google Scholar] [CrossRef]
- Xu, J.; Wolters-Arts, M.; Mariani, C.; Huber, H.; Rieu, I. Heat stress affects vegetative and reproductive performance and trait correlations in tomato (Solanum lycopersicum). Euphytica 2017, 213, 156. [Google Scholar] [CrossRef]
- Yeats, T.H.; Buda, G.J.; Wang, Z.; Chehanovsky, N.; Moyle, L.C.; Jetter, R.; Schaffer, A.A.; Rose, J.K. The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J. 2012, 69, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Kissoudis, C.; Yan, Z.; Visser, R.G.; van der Linden, G. Plant behaviour under combined stress: Tomato responses to combined salinity and pathogen stress. Plant J. 2018, 93, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Foolad, M.; Lin, G.; Chen, F. Comparison of QTLs for seed germination under non-stress, cold stress and salt stress in tomato. Plant Breed. 1999, 118, 167–173. [Google Scholar] [CrossRef]
- Wen, J.; Jiang, F.; Weng, Y.; Sun, M.; Shi, X.; Zhou, Y.; Yu, L.; Wu, Z. Identification of heat-tolerance QTLs and high-temperature stress-responsive genes through conventional QTL mapping, QTL-seq and RNA-seq in tomato. BMC Plant Biol. 2019, 19, 398. [Google Scholar] [CrossRef] [PubMed]
- Causse, M.; Duffe, P.; Gomez, M.; Buret, M.; Damidaux, R.; Zamir, D.; Gur, A.; Chevalier, C.; Lemaire-Chamley, M.; Rothan, C. A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. J. Exp. Bot. 2004, 55, 1671–1685. [Google Scholar] [CrossRef]
- Liu, Y.S.; Gur, A.; Ronen, G.; Causse, M.; Damidaux, R.; Buret, M.; Hirschberg, J.; Zamir, D. There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol. J. 2003, 1, 195–207. [Google Scholar] [CrossRef]
- Stevens, R.; Page, D.; Gouble, B.; Garchery, C.; Zamir, D.; Causse, M. Tomato fruit ascorbic acid content is linked with monodehydroascorbate reductase activity and tolerance to chilling stress. Plant Cell Environ. 2008, 31, 1086–1096. [Google Scholar] [CrossRef]
- Bauchet, G.; Grenier, S.; Samson, N.; Bonnet, J.; Grivet, L.; Causse, M. Use of modern tomato breeding germplasm for deciphering the genetic control of agronomical traits by Genome Wide Association study. Theor. Appl. Genet. 2017, 130, 875–889. [Google Scholar] [CrossRef]
- Shirasawa, K.; Fukuoka, H.; Matsunaga, H.; Kobayashi, Y.; Kobayashi, I.; Hirakawa, H.; Isobe, S.; Tabata, S. Genome-wide association studies using single nucleotide polymorphism markers developed by re-sequencing of the genomes of cultivated tomato. DNA Res. 2013, 20, 593–603. [Google Scholar] [CrossRef]
- Tieman, D.; Zhu, G.; Resende, M.F.; Lin, T.; Nguyen, C.; Bies, D.; Rambla, J.L.; Beltran, K.S.O.; Taylor, M.; Zhang, B. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, C.; Segura, V.; Bauchet, G.; Stevens, R.; Do, P.T.; Nikoloski, Z.; Fernie, A.R.; Causse, M. Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol. 2014, 165, 1120–1132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, J.; Xu, Y.; Liang, J.; Chang, P.; Yan, F.; Li, M.; Liang, Y.; Zou, Z. Genome-wide association mapping for tomato volatiles positively contributing to tomato flavor. Front. Plant Sci. 2015, 6, 1042. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sauvage, C.; Zhao, J.; Bitton, F.; Bauchet, G.; Liu, D.; Huang, S.; Tieman, D.M.; Klee, H.J.; Causse, M. Meta-analysis of genome-wide association studies provides insights into genetic control of tomato flavor. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Sun, Y.; Liang, Y.; Wu, J.; Li, Y.; Cui, X.; Qin, L. Dynamic QTL analysis for fruit lycopene content and total soluble solid content in a Solanum lycopersicum x S. pimpinellifolium cross. Genet. Mol. Res. 2012, 11, 3696–3710. [Google Scholar] [CrossRef]
- Bolger, A.; Scossa, F.; Bolger, M.E.; Lanz, C.; Maumus, F.; Tohge, T.; Quesneville, H.; Alseekh, S.; Sørensen, I.; Lichtenstein, G. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014, 46, 1034–1038. [Google Scholar] [CrossRef]
- Consortium, T.G. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar] [CrossRef]
- Consortium, T.G.S.; Aflitos, S.; Schijlen, E.; de Jong, H.; de Ridder, D.; Smit, S.; Finkers, R.; Wang, J.; Zhang, G.; Li, N. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 2014, 80, 136–148. [Google Scholar]
- Gao, L.; Gonda, I.; Sun, H.; Ma, Q.; Bao, K.; Tieman, D.M.; Burzynski-Chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Razali, R.; Bougouffa, S.; Morton, M.J.; Lightfoot, D.J.; Alam, I.; Essack, M.; Arold, S.T.; Kamau, A.A.; Schmöckel, S.M.; Pailles, Y. The genome sequence of the wild tomato Solanum pimpinellifolium provides insights into salinity tolerance. Front. Plant Sci. 2018, 9, 1402. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Wang, S.; Huang, Z.; Zhang, S.; Liao, Q.; Zhang, C.; Lin, T.; Qin, M.; Peng, M.; Yang, C. Rewiring of the fruit metabolome in tomato breeding. Cell 2018, 172, 249–261.e212. [Google Scholar] [CrossRef] [PubMed]
- Mondini, L.; Noorani, A.; Pagnotta, M.A. Assessing plant genetic diversity by molecular tools. Diversity 2009, 1, 19–35. [Google Scholar] [CrossRef]
- Wenzel, G. Molecular plant breeding: Achievements in green biotechnology and future perspectives. Appl. Microbiol. Biotechnol. 2006, 70, 642–650. [Google Scholar] [CrossRef]
- Hidema, J.; Teranishi, M.; Iwamatsu, Y.; Hirouchi, T.; Ueda, T.; Sato, T.; Burr, B.; Sutherland, B.M.; Yamamoto, K.; Kumagai, T. Spontaneously occurring mutations in the cyclobutane pyrimidine dimer photolyase gene cause different sensitivities to ultraviolet-B in rice. Plant J. 2005, 43, 57–67. [Google Scholar] [CrossRef]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar] [CrossRef]
- Brock, R. The role of induced mutations in plant improvement. Radiat. Bot. 1971, 11, 181–196. [Google Scholar] [CrossRef]
- Brock, R. Prospects and perspectives in mutation breeding. In Genetic Diversity in Plants; Springer: Boston, MA, USA, 1977; pp. 117–132. [Google Scholar]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.-P.; Guyon-Debast, A.; Chauvin, J.-E.; Nogué, F.; Mazier, M. Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef]
- Puchta, H. Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 2017, 36, 1–8. [Google Scholar] [CrossRef]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 1–5. [Google Scholar]
- Vats, S.; Kumawat, S.; Kumar, V.; Patil, G.B.; Joshi, T.; Sonah, H.; Sharma, T.R.; Deshmukh, R. Genome editing in plants: Exploration of technological advancements and challenges. Cells 2019, 8, 1386. [Google Scholar] [CrossRef]
- Yoder, J.I. Rapid proliferation of the maize transposable element Activator in transgenic tomato. Plant Cell 1990, 2, 723–730. [Google Scholar] [PubMed]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef]
- Gupta, P.; Reddaiah, B.; Salava, H.; Upadhyaya, P.; Tyagi, K.; Sarma, S.; Datta, S.; Malhotra, B.; Thomas, S.; Sunkum, A. Next-generation sequencing (NGS)-based identification of induced mutations in a doubly mutagenized tomato (Solanum lycopersicum) population. Plant J. 2017, 92, 495–508. [Google Scholar] [CrossRef]
- David-Schwartz, R.; Badani, H.; Smadar, W.; Levy, A.A.; Galili, G.; Kapulnik, Y. Identification of a novel genetically controlled step in mycorrhizal colonization: Plant resistance to infection by fungal spores but not extra-radical hyphae. Plant J. 2001, 27, 561–569. [Google Scholar] [CrossRef]
- Negi, S.; Santisree, P.; Kharshiing, E.V.; Sharma, R. Inhibition of the ubiquitin—Proteasome pathway alters cellular levels of nitric oxide in tomato seedlings. Mol. Plant 2010, 3, 854–869. [Google Scholar] [CrossRef]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef]
- Liu, R.; How-Kit, A.; Stammitti, L.; Teyssier, E.; Rolin, D.; Mortain-Bertrand, A.; Halle, S.; Liu, M.; Kong, J.; Wu, C. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. USA 2015, 112, 10804–10809. [Google Scholar] [CrossRef]
- Stadler, L.J. Genetic effects of X-rays in maize. Proc. Natl. Acad. Sci. USA 1928, 14, 69. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.H. Radiation in the production of useful mutations. Bot. Rev. 1958, 24, 1–24. [Google Scholar] [CrossRef]
- Naito, K.; Kusaba, M.; Shikazono, N.; Takano, T.; Tanaka, A.; Tanisaka, T.; Nishimura, M. Transmissible and nontransmissible mutations induced by irradiating Arabidopsis thaliana pollen with γ-rays and carbon ions. Genetics 2005, 169, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Okamura, M.; Yasuno, N.; Ohtsuka, M.; Tanaka, A.; Shikazono, N.; Hase, Y. Wide variety of flower-color and -shape mutants regenerated from leaf cultures irradiated with ion beams. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2003, 206, 574–578. [Google Scholar] [CrossRef]
- Shikazono, N.; Suzuki, C.; Kitamura, S.; Watanabe, H.; Tano, S.; Tanaka, A. Analysis of mutations induced by carbon ions in Arabidopsis thaliana. J. Exp. Bot. 2005, 56, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Ryuto, H.; Fukunishi, N. Ion beam radiation mutagenesis. Plant Mutat. Breed. Biotechnol. 2012, 99–106. [Google Scholar] [CrossRef]
- Magori, S.; Tanaka, A.; Kawaguchi, M. Physically induced mutation: Ion beam mutagenesis. In The Handbook of Plant Mutation Screening; WILEY-VCH Verlag: Weinheim, Germany, 2010; pp. 3–16. [Google Scholar]
- Tanaka, A.; Shikazono, N.; Hase, Y. Studies on biological effects of ion beams on lethality, molecular nature of mutation, mutation rate, and spectrum of mutation phenotype for mutation breeding in higher plants. J. Radiat. Res. 2010, 51, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Mba, C. Induced mutations unleash the potentials of plant genetic resources for food and agriculture. Agronomy 2013, 3, 200–231. [Google Scholar] [CrossRef]
- Kazama, Y.; Hirano, T.; Nishihara, K.; Ohbu, S.; Shirakawa, Y.; Abe, T. Effect of high-LET Fe-ion beam irradiation on mutation induction in Arabidopsis thaliana. Genes Genet. Syst. 2013, 88, 189–197. [Google Scholar] [CrossRef]
- Kazama, Y.; Hirano, T.; Saito, H.; Liu, Y.; Ohbu, S.; Hayashi, Y.; Abe, T. Characterization of highly efficient heavy-ion mutagenesis in Arabidopsis thaliana. BMC Plant Biol. 2011, 11, 161. [Google Scholar] [CrossRef]
- Kazama, Y.; Ishii, K.; Hirano, T.; Wakana, T.; Yamada, M.; Ohbu, S.; Abe, T. Different mutational function of low-and high-linear energy transfer heavy-ion irradiation demonstrated by whole-genome resequencing of Arabidopsis mutants. Plant J. 2017, 92, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Kazama, Y.; Ma, L.; Hirano, T.; Ohbu, S.; Shirakawa, Y.; Hatakeyama, S.; Tanaka, S.; Abe, T. Rapid evaluation of effective linear energy transfer in heavy-ion mutagenesis of Arabidopsis thaliana. Plant Biotechnol. 2012, 29, 441–445. [Google Scholar] [CrossRef]
- Cheema, A.A.; Atta, B.M. Radiosensitivity studies in basmati rice. Pak. J. Bot 2003, 35, 197–207. [Google Scholar]
- Sato, Y.; Shirasawa, K.; Takahashi, Y.; Nishimura, M.; Nishio, T. Mutant selection from progeny of gamma-ray-irradiated rice by DNA heteroduplex cleavage using Brassica petiole extract. Breed. Sci. 2006, 56, 179–183. [Google Scholar] [CrossRef]
- Matsukura, C.; Yamaguchi, I.; Inamura, M.; Ban, Y.; Kobayashi, Y.; Yin, Y.-G.; Saito, T.; Kuwata, C.; Imanishi, S.; Nishimura, S. Generation of gamma irradiation-induced mutant lines of the miniature tomato (Solanum lycopersicum L.) cultivar ‘Micro-Tom’. Plant Biotechnol. 2007, 24, 39–44. [Google Scholar] [CrossRef]
- Melamed, S. Chemical and Physical Mutagenesis in the Tomato Cultivar Micro-Tom; Hebrew University of Jerusalem: Jerusalem, Israel, 1999. [Google Scholar]
- Leitão, J. Chemical mutagenesis. In Plant Mutat. Breed. Biotechnol; Shu, Q.Y., Forster, B.P., Nakagawa, H., Eds.; CABI Publishing: Wallingford, UK, 2012; pp. 135–158. [Google Scholar]
- Heslot, H. Review of main mutagenic compounds. In Manual on Mutation Breeding; Technical Report Series; Iaea Vienna Second: IAEA, Vienna, 1977; pp. 51–58. [Google Scholar]
- Saito, T.; Ariizumi, T.; Okabe, Y.; Asamizu, E.; Hiwasa-Tanase, K.; Fukuda, N.; Mizoguchi, T.; Yamazaki, Y.; Aoki, K.; Ezura, H. TOMATOMA: A novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 2011, 52, 283–296. [Google Scholar] [CrossRef]
- Van Harten, A.M. Mutation Breeding: Theory and Practical Applications; Cambridge University Press: Cambridge, UK, 1998; p. 353. [Google Scholar]
- Till, B.J.; Cooper, J.; Tai, T.H.; Colowit, P.; Greene, E.A.; Henikoff, S.; Comai, L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 2007, 7, 19. [Google Scholar] [CrossRef]
- Szarejko, I.; Maluszynski, M. Translocations and inversions in barley induced by fast neutrons and N-nitroso-N-methylurea (MNUA)[Mutagenesis]. Barley Genet. Newsl. 1980, 10, 67–69. [Google Scholar]
- Maluszynski, M.; Szarejko, I.; Maluszynska, J. Encyclopedia of Applied Plant Sciences; Academic Press: New York, NY, USA, 2003; pp. 186–201. [Google Scholar]
- Wienholds, E.; Van Eeden, F.; Kosters, M.; Mudde, J.; Plasterk, R.H.; Cuppen, E. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003, 13, 2700–2707. [Google Scholar] [CrossRef]
- Wisman, E.; Koornneef, M.; Chase, T.; Lifshytz, E.; Ramanna, M.; Zabel, P. Genetic and molecular characterization of an Adh-1 null mutant in tomato. Mol. Gen. Genet. MGG 1991, 226, 120–128. [Google Scholar] [CrossRef]
- Piron, F.; Nicolaï, M.; Minoïa, S.; Piednoir, E.; Moretti, A.; Salgues, A.; Zamir, D.; Caranta, C.; Bendahmane, A. An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 2010, 5, e11313. [Google Scholar] [CrossRef] [PubMed]
- Kostov, K.; Batchvarova, R.; Slavov, S. Application of chemical mutagenesis to increase the resistance of tomato to Orobanche ramosa L. Bulg. J. Agric. Sci. 2007, 13, 505–513. [Google Scholar]
- Mazzucato, A.; Cellini, F.; Bouzayen, M.; Zouine, M.; Mila, I.; Minoia, S.; Petrozza, A.; Picarella, M.E.; Ruiu, F.; Carriero, F. A TILLING allele of the tomato Aux/IAA9 gene offers new insights into fruit set mechanisms and perspectives for breeding seedless tomatoes. Mol. Breed. 2015, 35, 22. [Google Scholar] [CrossRef]
- Maghuly, F.; Jankowicz-Cieslak, J.; Till, B.J.; Laimer, M. The Use of Ecotilling for the Genetic Improvement of Jatropha curcas L. In Jatropha, Challenges for a New Energy Crop; Springer: New York, NY, USA, 2013; pp. 335–349. [Google Scholar]
- Maghuly, F.; Laimer, M. Forward and reverse genetics for the improvement of Jatropha. In The Jatropha Genome; Springer: Cham, Switzerland, 2017; pp. 131–148. [Google Scholar]
- Greene, E.A.; Codomo, C.A.; Taylor, N.E.; Henikoff, J.G.; Till, B.J.; Reynolds, S.H.; Enns, L.C.; Burtner, C.; Johnson, J.E.; Odden, A.R. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 2003, 164, 731–740. [Google Scholar] [PubMed]
- Till, B.J.; Reynolds, S.H.; Weil, C.; Springer, N.; Burtner, C.; Young, K.; Bowers, E.; Codomo, C.A.; Enns, L.C.; Odden, A.R. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004, 4, 12. [Google Scholar] [CrossRef]
- Caldwell, D.G.; McCallum, N.; Shaw, P.; Muehlbauer, G.J.; Marshall, D.F.; Waugh, R. A structured mutant population for forward and reverse genetics in Barley (Hordeum vulgare L.). Plant J. 2004, 40, 143–150. [Google Scholar] [CrossRef]
- Slade, A.J.; Fuerstenberg, S.I.; Loeffler, D.; Steine, M.N.; Facciotti, D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat. Biotechnol. 2005, 23, 75–81. [Google Scholar] [CrossRef]
- Perry, J.A.; Wang, T.L.; Welham, T.J.; Gardner, S.; Pike, J.M.; Yoshida, S.; Parniske, M. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 2003, 131, 866–871. [Google Scholar] [CrossRef]
- Marroni, F.; Pinosio, S.; Di Centa, E.; Jurman, I.; Boerjan, W.; Felice, N.; Cattonaro, F.; Morgante, M. Large-scale detection of rare variants via pooled multiplexed next-generation sequencing: Towards next-generation Ecotilling. Plant J. 2011, 67, 736–745. [Google Scholar] [CrossRef]
- Rigola, D.; van Oeveren, J.; Janssen, A.; Bonné, A.; Schneiders, H.; van der Poel, H.J.; van Orsouw, N.J.; Hogers, R.C.; de Both, M.T.; van Eijk, M.J. High-throughput detection of induced mutations and natural variation using KeyPoint™ technology. PLoS ONE 2009, 4, e4761. [Google Scholar] [CrossRef]
- Tsai, H.; Howell, T.; Nitcher, R.; Missirian, V.; Watson, B.; Ngo, K.J.; Lieberman, M.; Fass, J.; Uauy, C.; Tran, R.K. Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol. 2011, 156, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Smith, S.M.; Ayele, M.; Yang, L.; Jogi, A.; Chaluvadi, S.R.; Bennetzen, J.L. High-throughput discovery of mutations in tef semi-dwarfing genes by next-generation sequencing analysis. Genetics 2012, 192, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Bado, S. Advances in Plant Mutation Breeding. Ph.D. Thesis, University of Natural Resources and Life Sciences, Vienna, Austria, 2015. [Google Scholar]
- Sauer, N.J.; Mozoruk, J.; Miller, R.B.; Warburg, Z.J.; Walker, K.A.; Beetham, P.R.; Schöpke, C.R.; Gocal, G.F. Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol. J. 2016, 14, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Gocal, G.F.; Schöpke, C.; Beetham, P.R. Oligo-mediated targeted gene editing. In Advances in New Technology for Targeted Modification of Plant Genomes; Springer: New York, NY, USA, 2015; pp. 73–89. [Google Scholar]
- Cole-Strauss, A.; Yoon, K.; Xiang, Y.; Byrne, B.C.; Rice, M.C.; Gryn, J.; Holloman, W.K.; Kmiec, E.B. Correction of the mutation responsible for sickle cell anemia by an RNA-DNA oligonucleotide. Science 1996, 273, 1386–1389. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.; Cole-Strauss, A.; Kmiec, E.B. Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA. DNA oligonucleotide. Proc. Natl. Acad. Sci. USA 1996, 93, 2071–2076. [Google Scholar] [CrossRef] [PubMed]
- Beetham, P.R.; Kipp, P.B.; Sawycky, X.L.; Arntzen, C.J.; May, G.D. A tool for functional plant genomics: Chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc. Natl. Acad. Sci. USA 1999, 96, 8774–8778. [Google Scholar] [CrossRef]
- Zhu, T.; Peterson, D.J.; Tagliani, L.; Clair, G.S.; Baszczynski, C.L.; Bowen, B. Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc. Natl. Acad. Sci. USA 1999, 96, 8768–8773. [Google Scholar] [CrossRef]
- Kochevenko, A.; Willmitzer, L. Chimeric RNA/DNA oligonucleotide-based site-specific modification of the tobacco acetolactate syntase gene. Plant Physiol. 2003, 132, 174–184. [Google Scholar] [CrossRef]
- Okuzaki, A.; Toriyama, K. Chimeric RNA/DNA oligonucleotide-directed gene targeting in rice. Plant Cell Rep. 2004, 22, 509–512. [Google Scholar] [CrossRef]
- Dong, C.; Beetham, P.; Vincent, K.; Sharp, P. Oligonucleotide-directed gene repair in wheat using a transient plasmid gene repair assay system. Plant Cell Rep. 2006, 25, 457–465. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.-K. RNA-directed DNA methylation and demethylation in plants. Sci. China Ser. C Life Sci. 2009, 52, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bartolomé, J.; Liu, W.; Kuo, P.H.; Feng, S.; Ghoshal, B.; Gardiner, J.; Zhao, J.M.-C.; Park, S.Y.; Chory, J.; Jacobsen, S.E. Co-targeting RNA polymerases IV and V promotes efficient de novo DNA methylation in Arabidopsis. Cell 2019, 176, 1068–1082.e19. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.; Deleris, A.; Hale, C.J.; Liu, A.; Feng, S.; Jacobsen, S.E. Interplay between active chromatin marks and RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet 2013, 9, e1003946. [Google Scholar] [CrossRef]
- Zhong, X.; Du, J.; Hale, C.J.; Gallego-Bartolome, J.; Feng, S.; Vashisht, A.A.; Chory, J.; Wohlschlegel, J.A.; Patel, D.J.; Jacobsen, S.E. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 2014, 157, 1050–1060. [Google Scholar] [CrossRef]
- Xie, M.; Yu, B. siRNA-directed DNA methylation in plants. Curr. Genom. 2015, 16, 23–31. [Google Scholar] [CrossRef]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 1–8. [Google Scholar] [CrossRef]
- Haag, J.R.; Ream, T.S.; Marasco, M.; Nicora, C.D.; Norbeck, A.D.; Pasa-Tolic, L.; Pikaard, C.S. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell 2012, 48, 811–818. [Google Scholar] [CrossRef]
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004, 2, e104. [Google Scholar] [CrossRef]
- Pikaard, C. Cell biology of the Arabidopsis nuclear siRNA pathway for RNA-directed chromatin modification. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 473–480. [Google Scholar] [CrossRef]
- Brocklehurst, S.; Watson, M.; Carr, I.M.; Out, S.; Heidmann, I.; Meyer, P. Induction of epigenetic variation in Arabidopsis by over-expression of DNA METHYLTRANSFERASE1 (MET1). PLoS ONE 2018, 13, e0192170. [Google Scholar] [CrossRef] [PubMed]
- Habu, Y. Epigenetic silencing of endogenous repetitive sequences by MORPHEUS’MOLECULE1 in Arabidopsis thaliana. Epigenetics 2010, 5, 562–565. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ji, L.; Jordan, W.T.; Shi, X.; Hu, L.; He, C.; Schmitz, R.J. TET-mediated epimutagenesis of the Arabidopsis thaliana methylome. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Saze, H.; Kakutani, T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 2007, 26, 3641–3652. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Vaughn, M.; Borges, F.; Tanurdžić, M.; Becker, J.D.; Feijó, J.A.; Martienssen, R.A. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 2009, 136, 461–472. [Google Scholar] [CrossRef]
- Otagaki, S.; Kawai, M.; Masuta, C.; Kanazawa, A. Size and positional effects of promoter RNA segments on virus-induced RNA-directed DNA methylation and transcriptional gene silencing. Epigenetics 2011, 6, 681–691. [Google Scholar] [CrossRef]
- Li, Q.; Eichten, S.R.; Hermanson, P.J.; Zaunbrecher, V.M.; Song, J.; Wendt, J.; Rosenbaum, H.; Madzima, T.F.; Sloan, A.E.; Huang, J. Genetic perturbation of the maize methylome. Plant Cell 2014, 26, 4602–4616. [Google Scholar] [CrossRef]
- Kasai, A.; Bai, S.; Hojo, H.; Harada, T. Epigenome editing of potato by grafting using transgenic tobacco as siRNA donor. PLoS ONE 2016, 11, e0161729. [Google Scholar] [CrossRef]
- Hu, L.; Li, N.; Xu, C.; Zhong, S.; Lin, X.; Yang, J.; Zhou, T.; Yuliang, A.; Wu, Y.; Chen, Y.-R. Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. USA 2014, 111, 10642–10647. [Google Scholar] [CrossRef]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef]
- Zhong, S.; Fei, Z.; Chen, Y.-R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m 6 A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Trewick, S.C.; Henshaw, T.F.; Hausinger, R.P.; Lindahl, T.; Sedgwick, B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 2002, 419, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Iusem, N.D. Tomato (Lycopersicon esculentum) genomic clone homologous to a gene encoding an abscisic acid-induced protein. Plant Physiol. 1994, 104, 1073. [Google Scholar] [CrossRef]
- Frankel, N.; Hasson, E.; Iusem, N.D.; Rossi, M.S. Adaptive evolution of the water stress-induced gene Asr2 in Lycopersicon species dwelling in arid habitats. Mol. Biol. Evol. 2003, 20, 1955–1962. [Google Scholar] [CrossRef]
- González, R.M.; Ricardi, M.M.; Iusem, N.D. Epigenetic marks in an adaptive water stress-responsive gene in tomato roots under normal and drought conditions. Epigenetics 2013, 8, 864–872. [Google Scholar] [CrossRef]
- Voinnet, O. RNA silencing as a plant immune system against viruses. Trends Genet. 2001, 17, 449–459. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Kørner, C.J.; Pitzalis, N.; Peña, E.J.; Erhardt, M.; Vazquez, F.; Heinlein, M. Crosstalk between PTGS and TGS pathways in natural antiviral immunity and disease recovery. Nat. Plants 2018, 4, 157–164. [Google Scholar] [CrossRef]
- Sijen, T.; Vijn, I.; Rebocho, A.; van Blokland, R.; Roelofs, D.; Mol, J.N.; Kooter, J.M. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 2001, 11, 436–440. [Google Scholar] [CrossRef]
- Baulcombe, D.C.; English, J.J. Ectopic pairing of homologous DNA and post-transcriptional gene silencing in transgenic plants. Curr. Opin. Biotechnol. 1996, 7, 173–180. [Google Scholar] [CrossRef]
- Chuang, C.-F.; Meyerowitz, E.M. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 4985–4990. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, P.M.; Graham, M.W.; Wang, M.-B. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 1998, 95, 13959–13964. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D. Viruses and gene silencing in plants. In 100 Years of Virology; Springer: Vienna, Austria, 1999; pp. 189–201. [Google Scholar]
- Burch-Smith, T.M.; Schiff, M.; Liu, Y.; Dinesh-Kumar, S.P. Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 2006, 142, 21–27. [Google Scholar] [CrossRef]
- Lange, M.; Yellina, A.L.; Orashakova, S.; Becker, A. Virus-Induced Gene Silencing (VIGS) in Plants: An Overview of Target Species and the Virus-Derived Vector Systems. In Advanced Structural Safety Studies; Springer Science and Business Media LLC.: Berlin/Heidelberg, Germany, 2013; Volume 975, pp. 1–14. [Google Scholar]
- Lu, R.; Martin-Hernandez, A.M.; Peart, J.R.; Malcuit, I.; Baulcombe, D.C. Virus-induced gene silencing in plants. Methods 2003, 30, 296–303. [Google Scholar] [CrossRef]
- Van Kammen, A. Virus-induced gene silencing in infected and transgenic plants. Trends Plant Sci. 1997, 2, 409–411. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Hutvágner, G.; Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 2002, 297, 2056–2060. [Google Scholar] [CrossRef]
- Kawamata, T.; Tomari, Y. Making risc. Trends Biochem. Sci. 2010, 35, 368–376. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef]
- Kumagai, M.H.; Donson, J.; Della-Cioppa, G.; Harvey, D.; Hanley, K.; Grill, L. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 1995, 92, 1679–1683. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002, 30, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.; Martin-Hernandez, A.M.; Baulcombe, D.C. Technical advance: Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001, 25, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Ekengren, S.K.; Liu, Y.; Schiff, M.; Dinesh-Kumar, S.; Martin, G.B. Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 2003, 36, 905–917. [Google Scholar] [CrossRef]
- Ramegowda, V.; Mysore, K.S.; Senthil-Kumar, M. Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front. Plant Sci. 2014, 5, 323. [Google Scholar] [CrossRef]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef]
- Iida, S.; Terada, R. Modification of endogenous natural genes by gene targeting in rice and other higher plants. Plant Mol. Biol. 2005, 59, 205–219. [Google Scholar] [CrossRef]
- Pereira, J.F.; Ryan, P.R. The role of transposable elements in the evolution of aluminium resistance in plants. J. Exp. Bot. 2019, 70, 41–54. [Google Scholar] [CrossRef]
- Flavell, A.J.; Pearce, S.R.; Kumar, A. Plant transposable elements and the genome. Curr. Opin. Genet. Dev. 1994, 4, 838–844. [Google Scholar] [CrossRef]
- Seidl, M.F.; Thomma, B.P. Transposable elements direct the coevolution between plants and microbes. Trends Genet. 2017, 33, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Gray, Y.H. It takes two transposons to tango: Transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000, 16, 461–468. [Google Scholar] [CrossRef]
- Thieme, M.; Bucher, E. Transposable Elements as Tool for Crop Improvement. Adv. Bota. Res. 2018, 88, 165–202. [Google Scholar] [CrossRef]
- McClintock, B. Carnegie Inst. Wash. Year Book 1948, 47, 155–169. [Google Scholar]
- McClintock, B. Nobel lecture. The significance of response of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Yoshida, T.; Tsukahara, S.; Kawabe, A. Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene 2013, 518, 256–261. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Grzebelus, D. The Functional Impact of Transposable Elements on the Diversity of Plant Genomes. Diversity 2018, 10, 18. [Google Scholar] [CrossRef]
- Sangwan, R.S.; Ochatt, S.; Nava-Saucedo, J.-E.; Sangwan-Norreel, B. T-DNA insertion mutagenesis. In Plant Mutat. Breed. Biotechnol; Shu, Q.Y., Forster, B.P., Nakagawa, H., Eds.; CABI Publishing: Wallingford, UK, 2012; pp. 489–506. [Google Scholar]
- Belzile, F.; Yoder, J.I. Pattern of somatic transposition in a high copy Ac tomato line. Plant J. 1992, 2, 173–179. [Google Scholar]
- Briza, J.; Carroll, B.J.; Klimyuk, V.I.; Thomas, C.M.; Jones, D.A.; Jones, J. Distribution of unlinked transpositions of a Ds element from a T-DNA locus on tomato chromosome 4. Genetics 1995, 141, 383–390. [Google Scholar]
- Goldsbrough, A.P.; Lastrella, C.N.; Yoder, J.I. Transposition mediated re–positioning and subsequent elimination of marker genes from transgenic tomato. Bio/technology 1993, 11, 1286–1292. [Google Scholar] [CrossRef]
- Osborne, B.I.; Corr, C.A.; Prince, J.P.; Hehl, R.; Tanksley, S.D.; McCormick, S.; Baker, B. Ac transposition from a T-DNA can generate linked and unlinked clusters of insertions in the tomato genome. Genetics 1991, 129, 833–844. [Google Scholar] [PubMed]
- Peterson, P.W.; Yoder, J.I. Amplification of Ac in tomato is correlated with high Ac transposition activity. Genome 1995, 38, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Cooley, M.B.; Yoder, J.; Goldsbrough, A.; Still, D. Site-selected insertional mutagenesis of tomato with maizeAc andDs elements. Mol. Gen. Genet. MGG 1996, 252, 184–194. [Google Scholar] [CrossRef]
- Atarés, A.; Moyano, E.; Morales, B.; Schleicher, P.; García-Abellán, J.O.; Antón, T.; García-Sogo, B.; Perez-Martin, F.; Lozano, R.; Flores, F.B. An insertional mutagenesis programme with an enhancer trap for the identification and tagging of genes involved in abiotic stress tolerance in the tomato wild-related species Solanum pennellii. Plant Cell Rep. 2011, 30, 1865. [Google Scholar] [CrossRef]
- Van der Biezen, E.A.; Brandwagt, B.F.; van Leeuwen, W.; Nijkamp, H.J.J.; Hille, J. Identification and isolation of theFEEBLY gene from tomato by transposon tagging. Mol. Gen. Genet. MGG 1996, 251, 267–280. [Google Scholar] [CrossRef][Green Version]
- Bibikova, M.; Golic, M.; Golic, K.G.; Carroll, D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 2002, 161, 1169–1175. [Google Scholar]
- Osakabe, K.; Osakabe, Y.; Toki, S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 12034–12039. [Google Scholar] [CrossRef]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef]
- Persikov, A.V.; Wetzel, J.L.; Rowland, E.F.; Oakes, B.L.; Xu, D.J.; Singh, M.; Noyes, M.B. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 2015, 43, 1965–1984. [Google Scholar] [CrossRef]
- Bitinaite, J.; Wah, D.A.; Aggarwal, A.K.; Schildkraut, I. FokI dimerization is required for DNA cleavage. Proc. Natl. Acad. Sci. USA 1998, 95, 10570–10575. [Google Scholar] [CrossRef] [PubMed]
- Hiroyuki, S.; Susumu, K. New restriction endonucleases from Flavobacterium okeanokoites (FokI) and Micrococcus luteus (MluI). Gene 1981, 16, 73–78. [Google Scholar] [CrossRef]
- Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Petolino, J.F. Genome editing in plants via designed zinc finger nucleases. Vitr. Cell. Dev. Biol. Plant 2015, 51, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Bashir, T.; Hashem, A.; Abd_Allah, E.F.; Bae, H. Genome editing tools in plants. Genes 2017, 8, 399. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef]
- Townsend, J.A.; Wright, D.A.; Winfrey, R.J.; Fu, F.; Maeder, M.L.; Joung, J.K.; Voytas, D.F. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 2009, 459, 442–445. [Google Scholar] [CrossRef]
- Zhang, F.; Maeder, M.L.; Unger-Wallace, E.; Hoshaw, J.P.; Reyon, D.; Christian, M.; Li, X.; Pierick, C.J.; Dobbs, D.; Peterson, T. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 12028–12033. [Google Scholar] [CrossRef]
- Curtin, S.J.; Zhang, F.; Sander, J.D.; Haun, W.J.; Starker, C.; Baltes, N.J.; Reyon, D.; Dahlborg, E.J.; Goodwin, M.J.; Coffman, A.P. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 2011, 156, 466–473. [Google Scholar] [CrossRef]
- Mushtaq, M.; Bhat, J.A.; Mir, Z.A.; Sakina, A.; Ali, S.; Singh, A.K.; Tyagi, A.; Salgotra, R.K.; Dar, A.A.; Bhat, R. CRISPR/Cas approach: A new way of looking at plant-abiotic interactions. J. Plant Physiol. 2018, 224, 156–162. [Google Scholar] [CrossRef]
- Gaj, T.; Sirk, S.J.; Shui, S.-l.; Liu, J. Genome-editing technologies: Principles and applications. Cold Spring Harb. Perspect. Biol. 2016, 8, a023754. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.M.; Musunuru, K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Investig. 2014, 124, 4154–4161. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, C.L.; Foley, J.E.; Wright, D.A.; Müller-Lerch, F.; Rahman, S.H.; Cornu, T.I.; Winfrey, R.J.; Sander, J.D.; Fu, F.; Townsend, J.A. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods 2008, 5, 374–375. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Zhao, H. Transcription activator-like effector nucleases (TALENs): A highly efficient and versatile tool for genome editing. Biotechnol. Bioeng. 2013, 110, 1811–1821. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Moscou, M.J.; Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef]
- Ding, Q.; Regan, S.N.; Xia, Y.; Oostrom, L.A.; Cowan, C.A.; Musunuru, K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 2013, 12, 393. [Google Scholar] [CrossRef]
- Mussolino, C.; Morbitzer, R.; Lütge, F.; Dannemann, N.; Lahaye, T.; Cathomen, T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011, 39, 9283–9293. [Google Scholar] [CrossRef]
- Reyon, D.; Tsai, S.Q.; Khayter, C.; Foden, J.A.; Sander, J.D.; Joung, J.K. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012, 30, 460. [Google Scholar] [CrossRef]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Qi, Y.; Zhang, Y.; Voytas, D.F. Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3 Genes Genomes Genet. 2013, 3, 1697–1705. [Google Scholar]
- Mahfouz, M.M.; Li, L.; Shamimuzzaman, M.; Wibowo, A.; Fang, X.; Zhu, J.-K. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl. Acad. Sci. USA 2011, 108, 2623–2628. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, F.; Li, X.; Baller, J.A.; Qi, Y.; Starker, C.G.; Bogdanove, A.J.; Voytas, D.F. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2013, 161, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390. [Google Scholar] [CrossRef]
- Shah, S.A.; Erdmann, S.; Mojica, F.J.; Garrett, R.A. Protospacer recognition motifs: Mixed identities and functional diversity. RNA Biol. 2013, 10, 891–899. [Google Scholar] [CrossRef]
- Wendt, T.; Holm, P.B.; Starker, C.G.; Christian, M.; Voytas, D.F.; Brinch-Pedersen, H.; Holme, I.B. TAL effector nucleases induce mutations at a pre-selected location in the genome of primary barley transformants. Plant Mol. Biol. 2013, 83, 279–285. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Chen, K.; Liang, Z.; Li, J.; Zhang, Y.; Zhang, K.; Liu, J.; Voytas, D.F.; Zheng, X. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant 2013, 6, 1365–1368. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Zhao, H.; Sheng, G.; Wang, M.; Yin, M.; Wang, Y. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 2015, 163, 840–853. [Google Scholar] [CrossRef]
- Pennisi, E. The CRISPR craze. Science 2013, 341, 833–836. [Google Scholar] [CrossRef] [PubMed]
- Segal, D.J.; Meckler, J.F. Genome engineering at the dawn of the golden age. Annu. Rev. Genom. Hum. Genet. 2013, 14, 135–158. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Dagdas, Y.S.; Chen, J.S.; Sternberg, S.H.; Doudna, J.A.; Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 2017, 3, eaao0027. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef]
- Knott, G.J.; Doudna, J.A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef] [PubMed]
- Freudhofmaier, M. Gene Editing of Commercially Important Genes of Jatropha curcas L. CRISPR/Cas9 Mediated Gene Knock-Out. Master’s Thesis, University of Natural Resources and Life Sciences (BOKU), Vienna, Austria, 2018. [Google Scholar]
- Peterson, B.A.; Haak, D.C.; Nishimura, M.T.; Teixeira, P.J.; James, S.R.; Dangl, J.L.; Nimchuk, Z.L. Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS ONE 2016, 11, e0162169. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Gasiunas, G.; Young, J.; Bigelyte, G.; Silanskas, A.; Cigan, M.; Siksnys, V. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.M.; Cradick, T.J.; Bao, G. The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol. Ther. 2016, 24, 645–654. [Google Scholar] [CrossRef]
- Müller, M.; Lee, C.M.; Gasiunas, G.; Davis, T.H.; Cradick, T.J.; Siksnys, V.; Bao, G.; Cathomen, T.; Mussolino, C. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol. Ther. 2016, 24, 636–644. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for crop improvement: An update review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef] [PubMed]
- Damalas, C.A.; Eleftherohorinos, I.G. Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Public Health 2011, 8, 1402–1419. [Google Scholar] [CrossRef] [PubMed]
- Tilman, D.; Cassman, K.G.; Matson, P.A.; Naylor, R.; Polasky, S. Agricultural sustainability and intensive production practices. Nature 2002, 418, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M. Transgenic approaches to crop improvement. J. Exp. Bot. 2000, 51, 487–496. [Google Scholar] [CrossRef]
- Yin, K.; Qiu, J.-L. Genome editing for plant disease resistance: Applications and perspectives. Philos. Trans. R. Soc. B 2019, 374, 20180322. [Google Scholar] [CrossRef]
- Buxdorf, K.; Rubinsky, G.; Barda, O.; Burdman, S.; Aharoni, A.; Levy, M. The transcription factor SlSHINE3 modulates defense responses in tomato plants. Plant Mol. Biol. 2014, 84, 37–47. [Google Scholar] [CrossRef]
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of Sl PL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 2017, 15, 1544–1555. [Google Scholar] [CrossRef]
- Ye, J.; Liu, G.; Chen, W.; Zhang, F.; Li, H.; Ye, Z.; Zhang, Y. Knockdown of SlNL33 accumulates ascorbate, enhances disease and oxidative stress tolerance in tomato (Solanum lycopersicum). Plant Growth Regul. 2019, 89, 49–58. [Google Scholar] [CrossRef]
- Di Pietro, A.; García-Maceira, F.I.; Méglecz, E.; Roncero, M.I.G. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 2001, 39, 1140–1152. [Google Scholar] [CrossRef]
- Pareek, M.; Rajam, M.V. RNAi-mediated silencing of MAP kinase signalling genes (Fmk1, Hog1, and Pbs2) in Fusarium oxysporum reduces pathogenesis on tomato plants. Fungal Biol. 2017, 121, 775–784. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Zhao, R.; Yu, W.; Li, R.; Li, Y.; Sheng, J.; Shen, L. Knockout of SlMAPK3 reduced disease resistance to Botrytis cinerea in tomato plants. J. Agric. Food Chem. 2018, 66, 8949–8956. [Google Scholar] [CrossRef]
- Prihatna, C.; Barbetti, M.J.; Barker, S.J. A novel tomato fusarium wilt tolerance gene. Front. Microbiol. 2018, 9, 1226. [Google Scholar] [CrossRef]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 1–6. [Google Scholar] [CrossRef]
- Koseoglou, E. The Study of SlPMR4 CRISPR/Cas9-Mediated Tomato Allelic Series for Resistance Against Powdery Mildew. Master’s Thesis, Wageningen University and Research, Wageningen, The Netherlands, 2017. [Google Scholar]
- Huibers, R.P.; Loonen, A.E.; Gao, D.; Van den Ackerveken, G.; Visser, R.G.; Bai, Y. Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS ONE 2013, 8, e67467. [Google Scholar] [CrossRef]
- Tashkandi, M.; Ali, Z.; Aljedaani, F.; Shami, A.; Mahfouz, M.M. Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 2018, 13, e1525996. [Google Scholar] [CrossRef]
- Fuentes, A.; Ramos, P.L.; Fiallo, E.; Callard, D.; Sánchez, Y.; Peral, R.; Rodríguez, R.; Pujol, M. Intron–hairpin RNA derived from replication associated protein C1 gene confers immunity to Tomato yellow leaf curl virus infection in transgenic tomato plants. Transgenic Res. 2006, 15, 291–304. [Google Scholar] [CrossRef]
- Ramesh, S.; Mishra, A.; Praveen, S. Hairpin RNA-mediated strategies for silencing of tomato leaf curl virus AC1 and AC4 genes for effective resistance in plants. Oligonucleotides 2007, 17, 251–257. [Google Scholar] [CrossRef]
- Praveen, S.; Mishra, A.K.; Dasgupta, A. Antisense suppression of replicase gene expression recovers tomato plants from leaf curl virus infection. Plant Sci. 2005, 168, 1011–1014. [Google Scholar] [CrossRef]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of Sl JAZ 2. Plant Biotechnol. J. 2019, 17, 665–673. [Google Scholar] [CrossRef]
- Almirón, M.; Link, A.J.; Furlong, D.; Kolter, R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992, 6, 2646–2654. [Google Scholar] [CrossRef]
- Ceci, P.; Ilari, A.; Falvo, E.; Chiancone, E. The Dps Protein of Agrobacterium tumefaciens Does Not Bind to DNA but Protects It toward Oxidative Cleavage X-RAY crystal structure, iron binding, and hydroxyl-radical scavenging properties. J. Biol. Chem. 2003, 278, 20319–20326. [Google Scholar] [CrossRef]
- Choi, S.H.; Baumler, D.J.; Kaspar, C.W. Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157: H7. Appl. Environ. Microbiol. 2000, 66, 3911–3916. [Google Scholar] [CrossRef]
- Halsey, T.A.; Vazquez-Torres, A.; Gravdahl, D.J.; Fang, F.C.; Libby, S.J. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect. Immun. 2004, 72, 1155–1158. [Google Scholar] [CrossRef]
- Martinez, A.; Kolter, R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 1997, 179, 5188–5194. [Google Scholar] [CrossRef]
- Nair, S.; Finkel, S.E. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 2004, 186, 4192–4198. [Google Scholar] [CrossRef]
- Saumaa, S.; Tover, A.; Tark, M.; Tegova, R.; Kivisaar, M. Oxidative DNA damage defense systems in avoidance of stationary-phase mutagenesis in Pseudomonas putida. J. Bacteriol. 2007, 189, 5504–5514. [Google Scholar] [CrossRef]
- Colburn-Clifford, J.M.; Scherf, J.M.; Allen, C. Ralstonia solanacearum Dps contributes to oxidative stress tolerance and to colonization of and virulence on tomato plants. Appl. Environ. Microbiol. 2010, 76, 7392–7399. [Google Scholar] [CrossRef]
- Hu, T.; Ye, J.; Tao, P.; Li, H.; Zhang, J.; Zhang, Y.; Ye, Z. The tomato HD-Zip I transcription factor Sl HZ 24 modulates ascorbate accumulation through positive regulation of the d-mannose/l-galactose pathway. Plant J. 2016, 85, 16–29. [Google Scholar] [CrossRef]
- De Toledo Thomazella, D.P.; Brail, Q.; Dahlbeck, D.; Staskawicz, B. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. BioRxiv 2016, 064824. [Google Scholar] [CrossRef]
- Shu, P.; Li, Z.; Min, D.; Zhang, X.; Ai, W.; Li, J.; Zhou, J.; Li, Z.; Li, F.; Li, X. CRISPR/Cas9-Mediated SlMYC2 Mutagenesis Adverse to Tomato Plant Growth and MeJA-Induced Fruit Resistance to Botrytis cinerea. J. Agric. Food Chem. 2020, 68, 5529–5538. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Guo, L.; Wang, Z.; Zhang, X.; Zhao, B.; Guo, Y.-D.; Zhang, N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a Lateral Organ Boundaries Domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020, 301, 110683. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.I.S.; Bracuto, V.; Koseoglou, E.; Appiano, M.; Jacobsen, E.; Visser, R.G.; Wolters, A.-M.A.; Bai, Y. CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 2020, 20, 284. [Google Scholar]
- Song, L.X.; Xu, X.C.; Wang, F.N.; Wang, Y.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Zhou, J.; Yu, J.Q. Brassinosteroids act as a positive regulator for resistance against root-knot nematode involving RESPIRATORY BURST OXIDASE HOMOLOG-dependent activation of MAPKs in tomato. Plant Cell Environ. 2018, 41, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.; Rajam, M. Targeting chitinase gene of Helicoverpa armigera by host-induced RNA interference confers insect resistance in tobacco and tomato. Plant Mol. Biol. 2016, 90, 281–292. [Google Scholar]
- Wang, T.; Deng, Z.; Zhang, X.; Wang, H.; Wang, Y.; Liu, X.; Liu, S.; Xu, F.; Li, T.; Fu, D. Tomato DCL2b is required for the biosynthesis of 22-nt small RNAs, the resulting secondary siRNAs, and the host defense against ToMV. Hortic. Res. 2018, 5, 1–14. [Google Scholar] [CrossRef]
- Wang, Z.; Hardcastle, T.J.; Pastor, A.C.; Yip, W.H.; Tang, S.; Baulcombe, D.C. A novel DCL2-dependent miRNA pathway in tomato affects susceptibility to RNA viruses. Genes Dev. 2018, 32, 1155–1160. [Google Scholar] [CrossRef]
- Schwind, N.; Zwiebel, M.; Itaya, A.; Ding, B.; WANG, M.B.; Krczal, G.; Wassenegger, M. RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct. Mol. Plant Pathol. 2009, 10, 459–469. [Google Scholar] [CrossRef]
- Li, Y.; Qin, L.; Zhao, J.; Muhammad, T.; Cao, H.; Li, H.; Zhang, Y.; Liang, Y. SlMAPK3 enhances tolerance to tomato yellow leaf curl virus (TYLCV) by regulating salicylic acid and jasmonic acid signaling in tomato (Solanum lycopersicum). PLoS ONE 2017, 12, e0172466. [Google Scholar] [CrossRef]
- Srivastava, R.; Kumar, R. The expanding roles of APETALA2/Ethylene Responsive Factors and their potential applications in crop improvement. Brief. Funct. Genom. 2019, 18, 240–254. [Google Scholar] [CrossRef]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, H.; Quan, R.; Wang, X.-C.; Huang, R. Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 2009, 150, 365–377. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, R. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 2010, 73, 241–249. [Google Scholar] [CrossRef]
- Jang, J.-C.; Sheen, J. Sugar sensing in higher plants. Plant Cell 1994, 6, 1665–1679. [Google Scholar]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef]
- Smeekens, S. Sugar-induced signal transduction in plants. Annu. Rev. Plant Biol. 2000, 51, 49–81. [Google Scholar] [CrossRef] [PubMed]
- Smeekens, S.; Rook, F. Sugar sensing and sugar-mediated signal transduction in plants. Plant Physiol. 1997, 115, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-X.; Hu, Q.; Yang, W.-N.; Jin, Y. The roles of cell wall invertase inhibitor in regulating chilling tolerance in tomato. BMC Plant Biol. 2017, 17, 195. [Google Scholar] [CrossRef]
- Cai, B.; Li, Q.; Xu, Y.; Yang, L.; Bi, H.; Ai, X. Genome-wide analysis of the fructose 1, 6-bisphosphate aldolase (FBA) gene family and functional characterization of FBA7 in tomato. Plant Physiol. Biochem. 2016, 108, 251–265. [Google Scholar] [CrossRef]
- Michelis, R.; Gepstein, S. Identification and characterization of a heat-induced isoform of aldolase in oat chloroplast. Plant Mol. Biol. 2000, 44, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Purev, M.; Kim, M.K.; Samdan, N.; Yang, D.-C. Isolation of a novel fructose-1, 6-bisphosphate aldolase gene from Codonopsis lanceolata and analysis of the response of this gene to abiotic stresses. Mol. Biol. 2008, 42, 179. [Google Scholar] [CrossRef]
- Cai, B.; Li, Q.; Liu, F.; Bi, H.; Ai, X. Decreasing fructose-1, 6-bisphosphate aldolase activity reduces plant growth and tolerance to chilling stress in tomato seedlings. Physiol. Plant. 2018, 163, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Tubiello, F.N.; Soussana, J.-F.; Howden, S.M. Crop and pasture response to climate change. Proc. Natl. Acad. Sci. USA 2007, 104, 19686–19690. [Google Scholar] [CrossRef]
- Link, V.; Sinha, A.K.; Vashista, P.; Hofmann, M.G.; Proels, R.K.; Ehness, R.; Roitsch, T. A heat-activated MAP kinase in tomato: A possible regulator of the heat stress response. FEBS Lett. 2002, 531, 179–183. [Google Scholar] [CrossRef]
- Evrard, A.; Kumar, M.; Lecourieux, D.; Lucks, J.; von Koskull-Döring, P.; Hirt, H. Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ 2013, 1, e59. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; He, J.; Wu, Y.; Wu, X.; Ge, C.; Wang, Y.; Zhong, S.; Peiter, E.; Liang, J.; Xu, W. The tomato mitogen-activated protein kinase slmpk1 is as a negative regulator of the high-temperature stress response. Plant Physiol. 2018, 177, 633–651. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.-D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef]
- Liu, D.; Kumar, R.; Claus, L.A.; Johnson, A.J.; Siao, W.; Vanhoutte, I.; Wang, P.; Bender, K.W.; Yperman, K.; Martins, S. Endocytosis of BRASSINOSTEROID INSENSITIVE1 Is Partly Driven by a Canonical Tyr-Based Motif. Plant Cell 2020, 32, 3598–3612. [Google Scholar] [CrossRef]
- Divi, U.K.; Krishna, P. Brassinosteroid: A biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol. 2009, 26, 131–136. [Google Scholar] [CrossRef]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef]
- Nakashita, H.; Yasuda, M.; Nitta, T.; Asami, T.; Fujioka, S.; Arai, Y.; Sekimata, K.; Takatsuto, S.; Yamaguchi, I.; Yoshida, S. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003, 33, 887–898. [Google Scholar] [CrossRef]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase-mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 2018, 59, 2239–2254. [Google Scholar] [CrossRef]
- Priest, D.M.; Ambrose, S.J.; Vaistij, F.E.; Elias, L.; Higgins, G.S.; Ross, A.R.; Abrams, S.R.; Bowles, D.J. Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 2006, 46, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ji, K.; Liang, B.; Du, Y.; Jiang, L.; Wang, J.; Kai, W.; Zhang, Y.; Zhai, X.; Chen, P. Suppressing ABA uridine diphosphate glucosyltransferase (Sl UGT 75C1) alters fruit ripening and the stress response in tomato. Plant J. 2017, 91, 574–589. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef]
- Zhu, M.; Meng, X.; Cai, J.; Li, G.; Dong, T.; Li, Z. Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 2018, 18, 83. [Google Scholar] [CrossRef]
- Guo, X.; Chen, G.; Naeem, M.; Yu, X.; Tang, B.; Li, A.; Hu, Z. The MADS-box gene SlMBP11 regulates plant architecture and affects reproductive development in tomato plants. Plant Sci. 2017, 258, 90–101. [Google Scholar] [CrossRef]
- Yin, W.; Hu, Z.; Cui, B.; Guo, X.; Hu, J.; Zhu, Z.; Chen, G. Suppression of the MADS-box gene SlMBP8 accelerates fruit ripening of tomato (Solanum lycopersicum). Plant Physiol. Biochem. 2017, 118, 235–244. [Google Scholar] [CrossRef]
- Li, Y.; Chu, Z.; Luo, J.; Zhou, Y.; Cai, Y.; Lu, Y.; Xia, J.; Kuang, H.; Ye, Z.; Ouyang, B. The C2H2 zinc-finger protein Sl ZF 3 regulates AsA synthesis and salt tolerance by interacting with CSN 5B. Plant Biotechnol. J. 2018, 16, 1201–1213. [Google Scholar] [CrossRef]
- Bao, H.; Chen, X.; Lv, S.; Jiang, P.; Feng, J.; Fan, P.; Nie, L.; Li, Y. Virus-induced gene silencing reveals control of reactive oxygen species accumulation and salt tolerance in tomato by γ-aminobutyric acid metabolic pathway. Plantcell Environ. 2015, 38, 600–613. [Google Scholar] [CrossRef]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012. [Google Scholar] [CrossRef]
- Yang, Y.; Tang, N.; Xian, Z.; Li, Z. Two SnRK2 protein kinases genes play a negative regulatory role in the osmotic stress response in tomato. Plant Cell Tissue Organ Cult. PCTOC 2015, 122, 421–434. [Google Scholar] [CrossRef]
- Borsani, O.; Cuartero, J.; Valpuesta, V.; Botella, M.A. Tomato tos1 mutation identifies a gene essential for osmotic tolerance and abscisic acid sensitivity. Plant J. 2002, 32, 905–914. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Reduction of tomato-plant chilling tolerance by CRISPR–Cas9-mediated SlCBF1 mutagenesis. J. Agric. Food Chem. 2018, 66, 9042–9051. [Google Scholar] [CrossRef]
- Hu, T.; Wang, Y.; Wang, Q.; Dang, N.; Wang, L.; Liu, C.; Zhu, J.; Zhan, X. The tomato 2-oxoglutarate-dependent dioxygenase gene SlF3HL is critical for chilling stress tolerance. Hortic. Res. 2019, 6, 1–12. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, X.; Qin, Y.; Feng, B.; Wu, Y.; He, Y.; Wang, A.; Zhu, J. The chilling tolerance divergence 1 protein confers cold stress tolerance in processing tomato. Plant Physiol. Biochem. 2020, 151, 34–46. [Google Scholar] [CrossRef]
- Zhuang, K.; Kong, F.; Zhang, S.; Meng, C.; Yang, M.; Liu, Z.; Wang, Y.; Ma, N.; Meng, Q. Whirly1 enhances tolerance to chilling stress in tomato via protection of photosystem II and regulation of starch degradation. New Phytol. 2019, 221, 1998–2012. [Google Scholar] [CrossRef]
- Zhuang, K.; Wang, J.; Jiao, B.; Chen, C.; Zhang, J.; Ma, N.; Meng, Q. SlWHIRLY1 maintains leaf photosynthetic capacity in tomato by regulating the expression of SlRbcS1 under chilling stress. J. Exp. Bot. 2020, 71, 3653–3663. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, Y.; Zhu, N.; Zhong, S.; Bouzayen, M.; Li, Z. SlGRAS4 mediates a novel regulatory pathway promoting chilling tolerance in tomato. Plant Biotechnol. J. 2020, 18, 1620–1633. [Google Scholar] [CrossRef]
- Li, H.; Jiang, X.; Lv, X.; Ahammed, G.J.; Guo, Z.; Qi, Z.; Yu, J.; Zhou, Y. Tomato GLR3. 3 and GLR3. 5 mediate cold acclimation-induced chilling tolerance by regulating apoplastic H2O2 production and redox homeostasis. Plantcell Environ. 2019, 42, 3326–3339. [Google Scholar]
- Guo, X.; Li, J.; Zhang, L.; Zhang, Z.; He, P.; Wang, W.; Wang, M.; Wang, A.; Zhu, J. Heterotrimeric G-protein α subunit (LeGPA1) confers cold stress tolerance to processing tomato plants (Lycopersicon esculentum Mill). BMC Plant Biol. 2020, 20, 394. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef]
- Li, R.; Liu, C.; Zhao, R.; Wang, L.; Chen, L.; Yu, W.; Zhang, S.; Sheng, J.; Shen, L. CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019, 19, 38. [Google Scholar]
- Zhang, Y.; Li, H.; Shu, W.; Zhang, C.; Zhang, W.; Ye, Z. Suppressed expression of ascorbate oxidase gene promotes ascorbic acid accumulation in tomato fruit. Plant Mol. Biol. Report. 2011, 29, 638–645. [Google Scholar] [CrossRef]
- Ewas, M.; Gao, Y.; Wang, S.; Liu, X.; Zhang, H.; Nishawy, E.M.; Ali, F.; Shahzad, R.; Ziaf, K.; Subthain, H. Manipulation of SlMXl for enhanced carotenoids accumulation and drought resistance in tomato. Sci. Bull. 2016, 61, 1413–1418. [Google Scholar] [CrossRef]
- Song, J.; Xing, Y.; Munir, S.; Yu, C.; Song, L.; Li, H.; Wang, T.; Ye, Z. An ATL78-Like RING-H2 finger protein confers abiotic stress tolerance through interacting with RAV2 and CSN5B in tomato. Front. Plant Sci. 2016, 7, 1305. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.; Jiang, L.; Kai, W.; Liang, B.; Wang, J.; Du, Y.; Zhai, X.; Wang, J.; Zhang, Y. Suppressing type 2C protein phosphatases alters fruit ripening and the stress response in tomato. Plant Cell Physiol. 2018, 59, 142–154. [Google Scholar] [CrossRef]
- Zhu, T.; Zou, L.; Li, Y.; Yao, X.; Xu, F.; Deng, X.; Zhang, D.; Lin, H. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 2063–2076. [Google Scholar] [CrossRef]
- Li, J.; Chen, C.; Wei, J.; Pan, Y.; Su, C.; Zhang, X. SpPKE1, a Multiple Stress-Responsive Gene Confers Salt Tolerance in Tomato and Tobacco. Int. J. Mol. Sci. 2019, 20, 2478. [Google Scholar] [CrossRef]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN 1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.; Salts, Y.; Barg, R. Tomato facultative parthenocarpy results from Sl AGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 2017, 15, 634–647. [Google Scholar] [CrossRef]
- Spicher, L.; Almeida, J.; Gutbrod, K.; Pipitone, R.; Dörmann, P.; Glauser, G.; Rossi, M.; Kessler, F. Essential role for phytol kinase and tocopherol in tolerance to combined light and temperature stress in tomato. J. Exp. Bot. 2017, 68, 5845–5856. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, S.; Lv, J.; Liu, Z.; Wang, Y.; Ma, N.; Meng, Q. SUMO E3 ligase SlSIZ1 facilitates heat tolerance in tomato. Plant Cell Physiol. 2018, 59, 58–71. [Google Scholar] [CrossRef]
- Zhuang, K.; Gao, Y.; Liu, Z.; Diao, P.; Sui, N.; Meng, Q.; Meng, C.; Kong, F. WHIRLY1 regulates HSP21. 5A expression to promote thermotolerance in tomato. Plant Cell Physiol. 2020, 61, 169–177. [Google Scholar] [CrossRef]
- Olias, R.; Eljakaoui, Z.; Li, J.; DE MORALES, P.A.; MARÍN-MANZANO, M.C.; Pardo, J.M.; Belver, A. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 2009, 32, 904–916. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, C.; Shu, W.; Ye, Z.; Li, H.; Zhang, Y. The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem. Biophys. Res. Commun. 2016, 474, 736–741. [Google Scholar] [CrossRef]
- Wang, L.; Hu, Z.; Zhu, M.; Zhu, Z.; Hu, J.; Qanmber, G.; Chen, G. The abiotic stress-responsive NAC transcription factor SlNAC11 is involved in drought and salt response in tomato (Solanum lycopersicum L.). Plant Celltissue Organ Cult. (PCTOC) 2017, 129, 161–174. [Google Scholar] [CrossRef]
- Campos, J.F.; Cara, B.; Pérez-Martín, F.; Pineda, B.; Egea, I.; Flores, F.B.; Fernandez-Garcia, N.; Capel, J.; Moreno, V.; Angosto, T. The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation. Plant Biotechnol. J. 2016, 14, 1345–1356. [Google Scholar] [CrossRef]
- Liu, B.; Ouyang, Z.; Zhang, Y.; Li, X.; Hong, Y.; Huang, L.; Liu, S.; Zhang, H.; Li, D.; Song, F. Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS ONE 2014, 9, e102067. [Google Scholar] [CrossRef]
- Vick, B.A.; Zimmerman, D.C. Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 1984, 75, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Heitz, T.; Bergey, D.R.; Ryan, C.A. A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol. 1997, 114, 1085–1093. [Google Scholar] [CrossRef]
- Hu, T.; Hu, Z.; Zeng, H.; Qv, X.; Chen, G. Tomato lipoxygenase D involved in the biosynthesis of jasmonic acid and tolerance to abiotic and biotic stress in tomato. Plant Biotechnol. Rep. 2015, 9, 37–45. [Google Scholar] [CrossRef]
- AbuQamar, S.; Luo, H.; Laluk, K.; Mickelbart, M.V.; Mengiste, T. Crosstalk between biotic and abiotic stress responses in tomato is mediated by the AIM1 transcription factor. Plant J. 2009, 58, 347–360. [Google Scholar] [CrossRef]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.; Poovaiah, B. Ca 2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef]
- Galon, Y.; Nave, R.; Boyce, J.M.; Nachmias, D.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008, 582, 943–948. [Google Scholar] [CrossRef]
- Laluk, K.; Prasad, K.; Savchenko, T.; Celesnik, H.; Dehesh, K.; Levy, M.; Mitchell-Olds, T.; Reddy, A. The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol. 2012, 53, 2008–2015. [Google Scholar] [CrossRef]
- Nie, H.; Zhao, C.; Wu, G.; Wu, Y.; Chen, Y.; Tang, D. SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 2012, 158, 1847–1859. [Google Scholar] [CrossRef]
- Qiu, Y.; Xi, J.; Du, L.; Suttle, J.C.; Poovaiah, B. Coupling calcium/calmodulin-mediated signaling and herbivore-induced plant response through calmodulin-binding transcription factor AtSR1/CAMTA3. Plant Mol. Biol. 2012, 79, 89–99. [Google Scholar] [CrossRef]
- Doherty, C.J.; Van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar] [CrossRef]
- Kim, Y.; Park, S.; Gilmour, S.J.; Thomashow, M.F. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of A rabidopsis. Plant J. 2013, 75, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N.; Ranjan, A.; Pant, P.; Tripathi, R.K.; Ateek, F.; Pandey, H.P.; Patre, U.V.; Sawant, S.V. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genom. 2013, 14, 216. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Peng, H.; Whitaker, B.D.; Jurick, W.M. Differential expression of calcium/calmodulin-regulated SlSRs in response to abiotic and biotic stresses in tomato fruit. Physiol. Plant. 2013, 148, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Peng, H.; Whitaker, B.D.; Conway, W.S. Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol. 2012, 12, 19. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, L.; Zhang, Y.; Ouyang, Z.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 2014, 14, 286. [Google Scholar] [CrossRef] [PubMed]
- Priyanka, B.; Sekhar, K.; Reddy, V.D.; Rao, K.V. Expression of pigeonpea hybrid-proline-rich protein encoding gene (CcHyPRP) in yeast and Arabidopsis affords multiple abiotic stress tolerance. Plant Biotechnol. J. 2010, 8, 76–87. [Google Scholar] [CrossRef]
- Tan, J.; Zhuo, C.; Guo, Z. Nitric oxide mediates cold-and dehydration-induced expression of a novel MfHyPRP that confers tolerance to abiotic stress. Physiol. Plant. 2013, 149, 310–320. [Google Scholar]
- Xu, D.; Huang, X.; Xu, Z.-Q.; Schläppi, M. The HyPRP gene EARLI1 has an auxiliary role for germinability and early seedling development under low temperature and salt stress conditions in Arabidopsis thaliana. Planta 2011, 234, 565–577. [Google Scholar] [CrossRef]
- Yeom, S.I.; Seo, E.; Oh, S.K.; Kim, K.W.; Choi, D. A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant J. 2012, 69, 755–768. [Google Scholar] [CrossRef]
- Zhang, Y.; Schläppi, M. Cold responsive EARLI1 type HyPRPs improve freezing survival of yeast cells and form higher order complexes in plants. Planta 2007, 227, 233–243. [Google Scholar] [CrossRef]
- Li, J.; Ouyang, B.; Wang, T.; Luo, Z.; Yang, C.; Li, H.; Sima, W.; Zhang, J.; Ye, Z. HyPRP1 gene suppressed by multiple stresses plays a negative role in abiotic stress tolerance in tomato. Front. Plant Sci. 2016, 7, 967. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, S.; Tuori, R.P.; D’Ascenzo, M.D.; Fobert, P.R.; Després, C.; Martin, G.B. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 2003, 15, 3033–3050. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.-Q.; Wildermuth, M.C.; Chakravarthy, S.; Loh, Y.-T.; Yang, C.; He, X.; Han, Y.; Martin, G.B. Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 2002, 14, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Mysore, K.S.; Crasta, O.R.; Tuori, R.P.; Folkerts, O.; Swirsky, P.B.; Martin, G.B. Comprehensive transcript profiling of Pto-and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 2002, 32, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liang, B.; Wang, J.; Kai, W.; Chen, P.; Jiang, L.; Du, Y.; Leng, P. SlPti4 affects regulation of fruit ripening, seed germination and stress responses by modulating ABA signaling in tomato. Plant Cell Physiol. 2018, 59, 1956–1965. [Google Scholar] [CrossRef]
- Xu, C.; Liberatore, K.L.; MacAlister, C.A.; Huang, Z.; Chu, Y.-H.; Jiang, K.; Brooks, C.; Ogawa-Ohnishi, M.; Xiong, G.; Pauly, M. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015, 47, 784–792. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering quantitative trait variation for crop improvement by genome editing. Cell 2017, 171, 470–480.e478. [Google Scholar] [CrossRef]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef]
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Toki, S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 2015, 467, 76–82. [Google Scholar] [CrossRef]
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Shima, Y.; Nakamura, N.; Kotake-Nara, E.; Kawasaki, S.; Toki, S. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat. Plants 2017, 3, 866–874. [Google Scholar] [CrossRef]
- Ito, Y.; Sekiyama, Y.; Nakayama, H.; Nishizawa-Yokoi, A.; Endo, M.; Shima, Y.; Nakamura, N.; Kotake-Nara, E.; Kawasaki, S.; Hirose, S. Allelic Mutations in the Ripening-Inhibitor Locus Generate Extensive Variation in Tomato Ripening. Plant Physiol. 2020, 183, 80–95. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, G.; Li, R.; Yan, S.; Fu, D.; Zhu, B.; Tian, H.; Luo, Y.; Zhu, H. The RNA editing factor SlORRM4 is required for normal fruit ripening in tomato. Plant Physiol. 2017, 175, 1690–1702. [Google Scholar] [CrossRef]
- Yu, Q.-h.; Wang, B.; Li, N.; Tang, Y.; Yang, S.; Yang, T.; Xu, J.; Guo, C.; Yan, P.; Wang, Q. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Sravankumar, T.; Naik, N.; Kumar, R. A ripening-induced SlGH3-2 gene regulates fruit ripening via adjusting auxin-ethylene levels in tomato (Solanum lycopersicum L.). Plant Mol. Biol. 2018, 98, 455–469. [Google Scholar] [CrossRef]
- Ueta, R.; Abe, C.; Watanabe, T.; Sugano, S.S.; Ishihara, R.; Ezura, H.; Osakabe, Y.; Osakabe, K. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Chen, S.; Tian, H.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 2018, 9, 559. [Google Scholar] [CrossRef]
- Gago, C.; Drosou, V.; Paschalidis, K.; Guerreiro, A.; Miguel, G.; Antunes, D.; Hilioti, Z. Targeted gene disruption coupled with metabolic screen approach to uncover the LEAFY COTYLEDON1-LIKE4 (L1L4) function in tomato fruit metabolism. Plant Cell Rep. 2017, 36, 1065–1082. [Google Scholar] [CrossRef]
- Hilioti, Z.; Ganopoulos, I.; Ajith, S.; Bossis, I.; Tsaftaris, A. A novel arrangement of zinc finger nuclease system for in vivo targeted genome engineering: The tomato LEC1-LIKE4 gene case. Plant Cell Rep. 2016, 35, 2241–2255. [Google Scholar] [CrossRef]
- Li, R.; Li, R.; Li, X.; Fu, D.; Zhu, B.; Tian, H.; Luo, Y.; Zhu, H. Multiplexed CRISPR/Cas9-mediated metabolic engineering of γ-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Park, S.J.; Van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Ye, L.; Qin, L.; Liu, X.; He, Y.; Wang, J.; Chen, L.; Lu, G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 2016, 6, 24765. [Google Scholar] [CrossRef] [PubMed]
- Keddie, J.S.; Carroll, B.; Jones, J.; Gruissem, W. The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 1996, 15, 4208–4217. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Kajala, K.; Pauluzzi, G.; Wang, D.; Reynoso, M.A.; Zumstein, K.; Garcha, J.; Winte, S.; Masson, H.; Inagaki, S. Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 2014, 166, 455–469. [Google Scholar] [CrossRef]
- Lor, V.S.; Starker, C.G.; Voytas, D.F.; Weiss, D.; Olszewski, N.E. Targeted mutagenesis of the tomato PROCERA gene using transcription activator-like effector nucleases. Plant Physiol. 2014, 166, 1288–1291. [Google Scholar] [CrossRef]
- Hayut, S.F.; Bessudo, C.M.; Levy, A.A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 2017, 8, 1–9. [Google Scholar]
- Xie, K.; Zhang, J.; Yang, Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR–Cas9-mediated genome editing in model plants and major crops. Mol. Plant 2014, 7, 923–926. [Google Scholar] [CrossRef]
- Zaidi, S.S.A.; Mahfouz, M.M.; Mansoor, S. CRISPR-Cpf1: A new tool for plant genome editing. Trends Plant Sci. 2017, 22, 550–553. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Zhang, J.; Botella, J.R.; Ma, C.; Kong, F.; Liu, B.; Zhu, J.-K. Perspectives on the application of genome-editing technologies in crop breeding. Mol. Plant 2019, 12, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.L.; Zhang, P. Plant biotechnology for food security and bioeconomy. Plant Mol. Biol. 2013, 83, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Bhargava, A.; Srivastava, S. Participatory Plant Breeding: Concept and Applications; Springer: Singapore, 2019. [Google Scholar]
- Mann, C.C. Genetic engineers aim to soup up crop photosynthesis. Science 1999, 283, 314–316. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).