Cysteine-Rich Hydrophobin Gene Family: Genome Wide Analysis, Phylogeny and Transcript Profiling in Cordyceps militaris
Abstract
1. Introduction
2. Results
2.1. Domain Structure, Hydropathy Pattern and Homology Modeling of Hydrophobins in Cordyceps militaris
2.2. Genomic Organization of the Hydrophobin Genes
2.3. Distribution of Hydrophobins in Ascomycota Fungi with Different Lifestyles
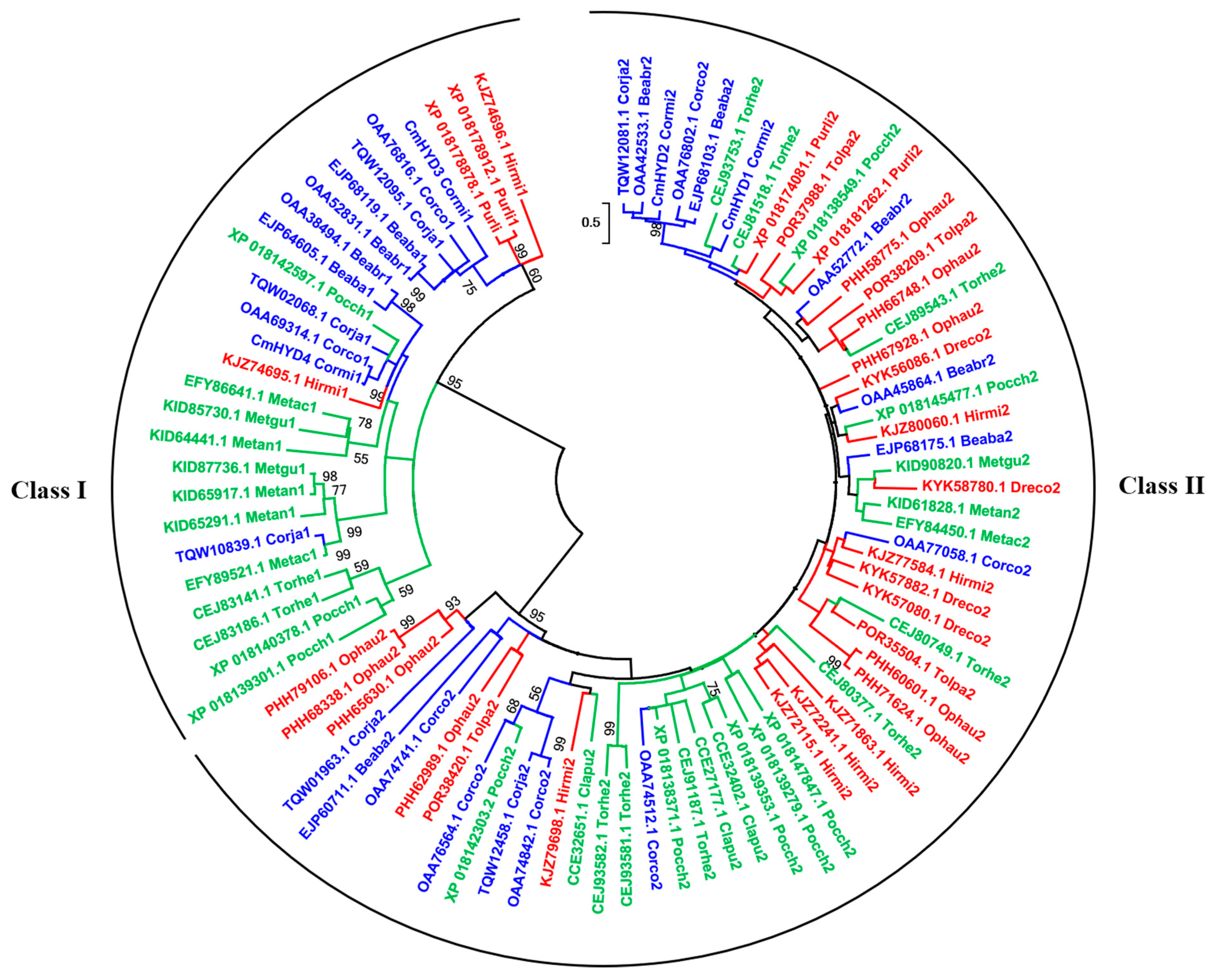
2.4. Phylogeny Analysis Based on the Hydrophobin Proteins of Cordyceps s.l.
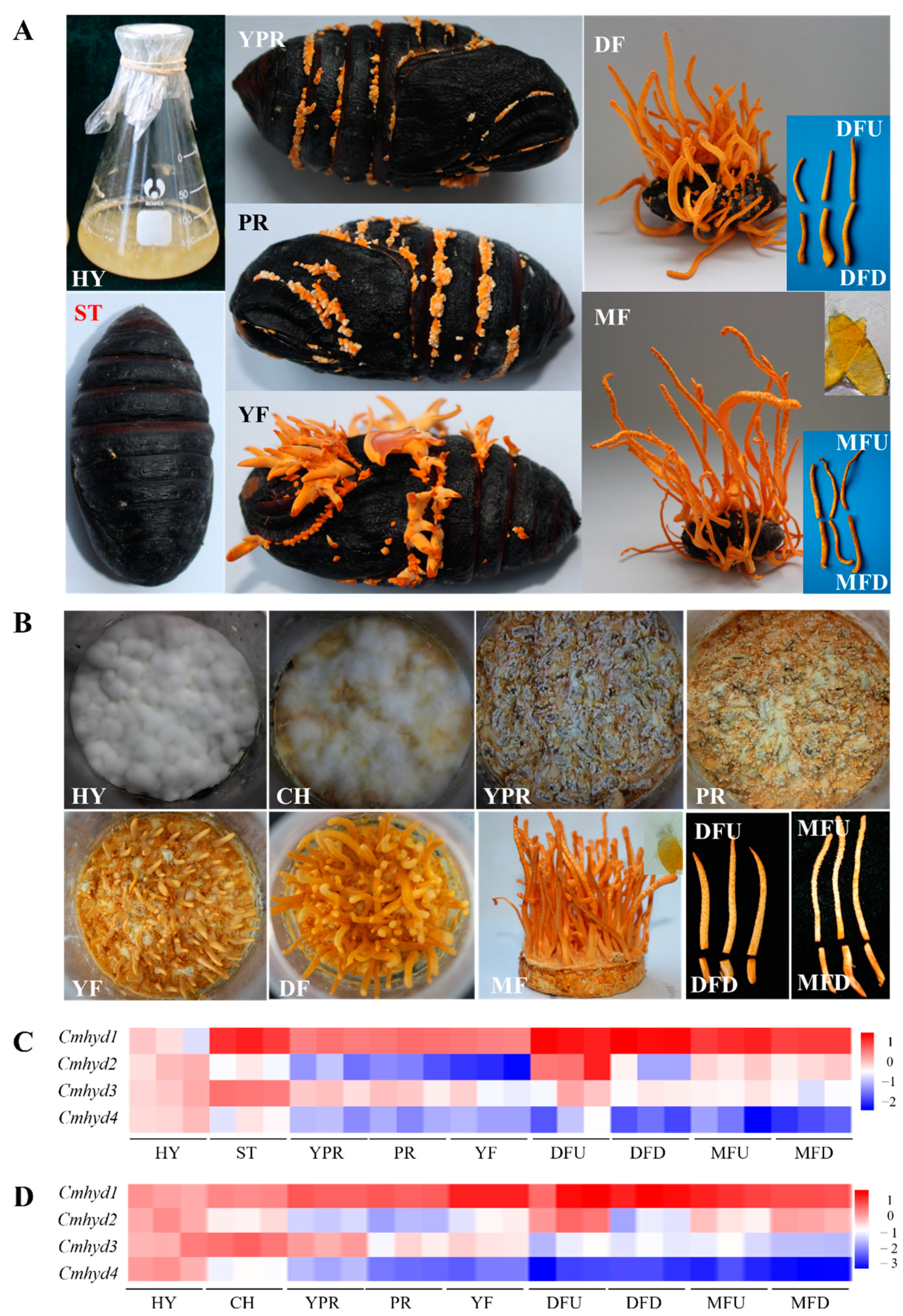
2.5. Transcript Patterns of Hydrophobin Genes during the Fruiting Body Development
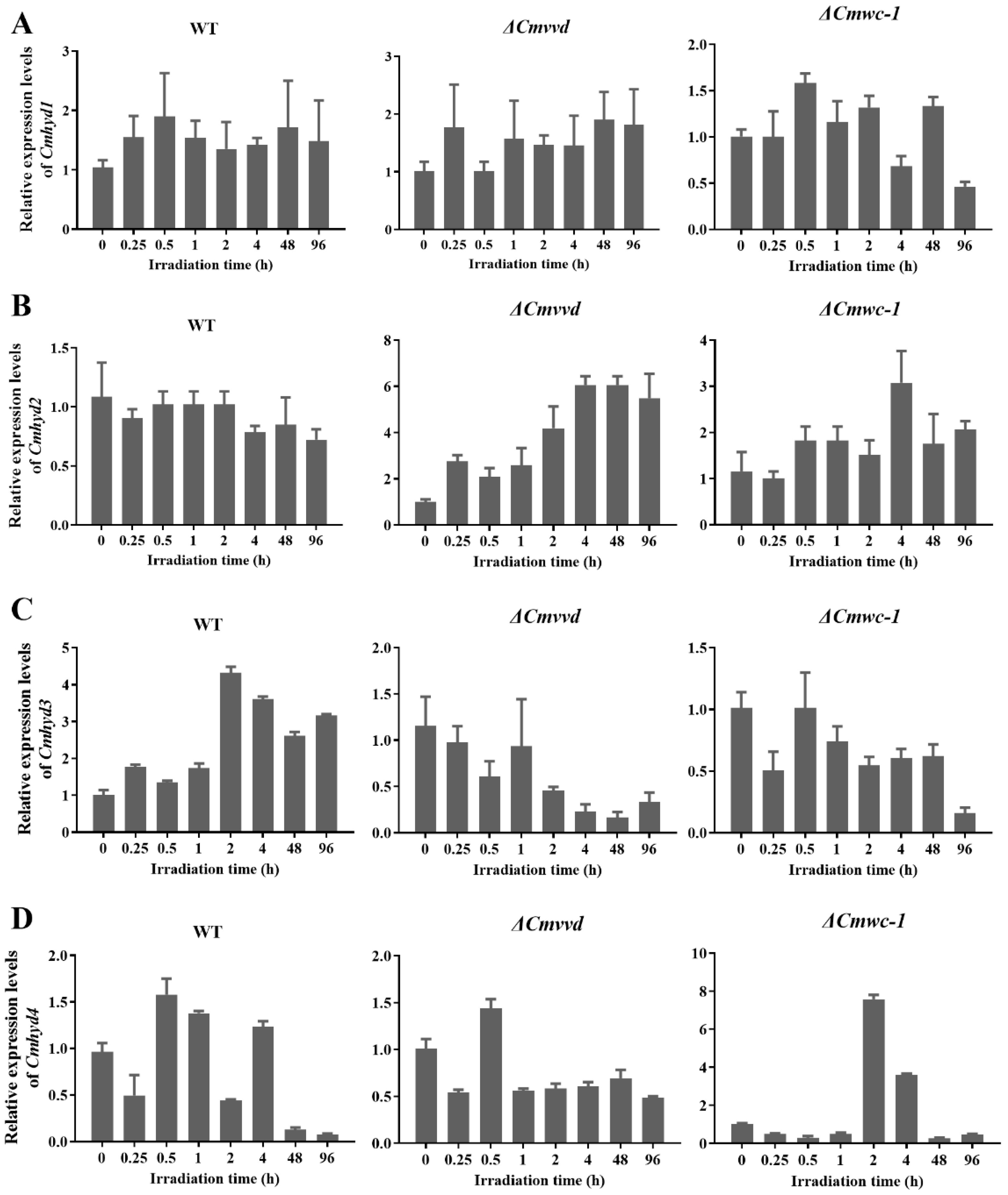
2.6. Transcript of Hydrophobin Genes Respond to Light Irradiation
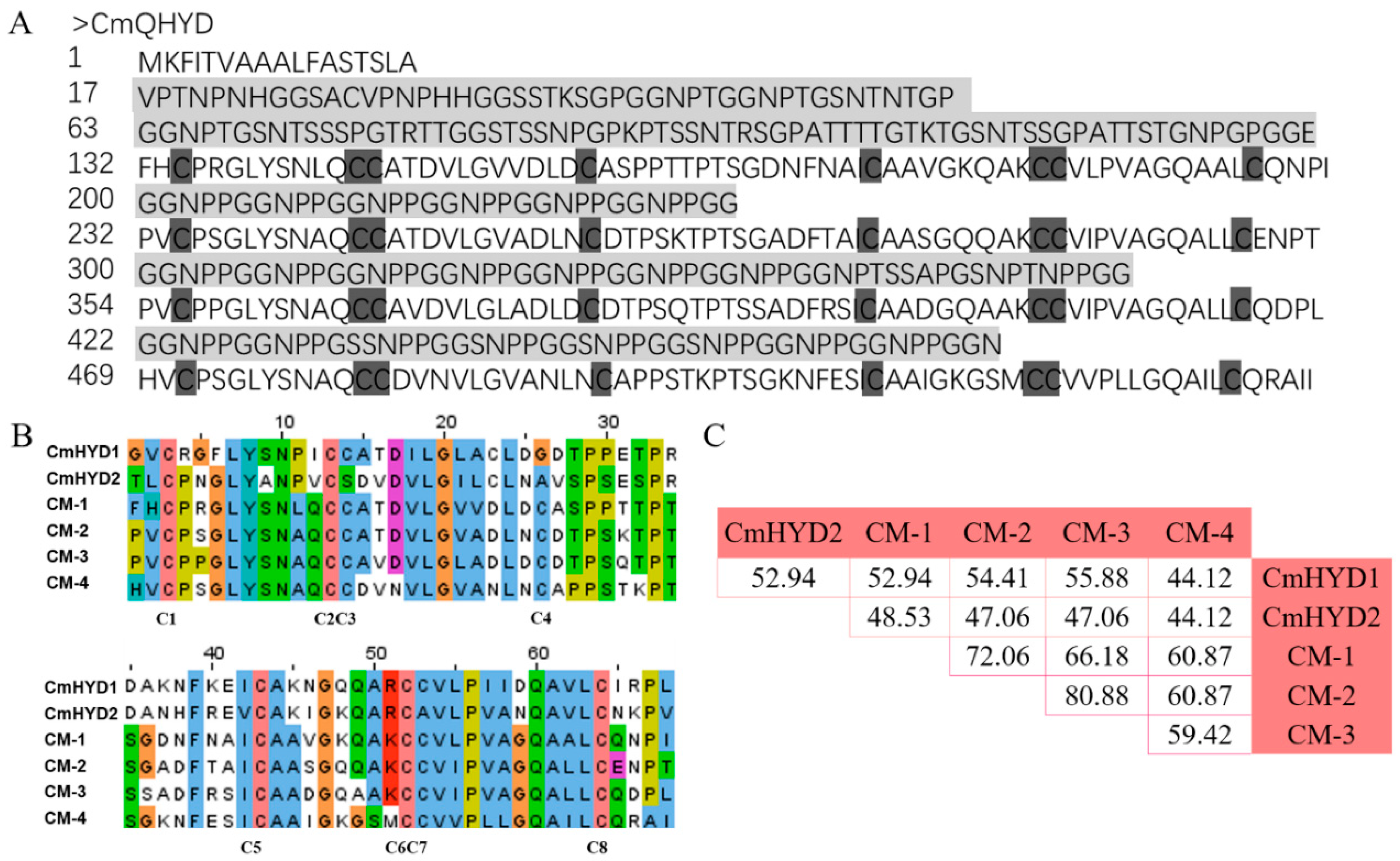
2.7. Multi-Domain in Hydrophobin Proteins
3. Discussion
4. Materials and Methods
4.1. Strains, Media and Growth Conditions
4.2. Sequence Search and Annotation of Hydrophobin Genes in Cordyceps militaris
4.3. Homology Modeling of Hydrophobins
4.4. Distribution of Hydrophobins in Ascomycota Fungi with Different Life Styles
4.5. Phylogenetic Analysis of Hydrophobins in Cordyceps Sensu Lato
4.6. Transcript Analysis of Hydrophobin Genes during the Fruiting Body Development
4.7. Transcript Analysis of Hydrophobin Genes after Light Exposure for Different Times
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elliot, M.A.; Talbot, N.J. Building filaments in the air: Aerial morphogenesis in bacteria and fungi. Curr. Opin. Microbiol. 2004, 7, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Linder, M.B.; Szilvay, G.R.; Nakari-Setälä, T.; Penttilä, M.E. Hydrophobins: The protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 2005, 29, 877–896. [Google Scholar] [CrossRef] [PubMed]
- Wösten, H.A.; Wessels, J.G. Hydrophobins, from molecular structure to multiple functions in fungal development. Mycoscience 1997, 38, 363–374. [Google Scholar] [CrossRef]
- Hakanpää, J.; Paananen, A.; Askolin, S.; Nakari-Setälä, T.; Parkkinen, T.; Penttilä, M.; Linder, M.B.; Rouvinen, J. Atomic resolution structure of the HFBII Hydrophobin, a Self-assembling Amphiphile. J. Biol. Chem. 2004, 279, 534–539. [Google Scholar] [CrossRef]
- Kallio, J.M.; Linder, M.; Rouvinen, J. Crystal structures of hydrophobin HFBII in the presence of detergent implicate the formation of fibrils and monolayer films. J. Biol. Chem. 2007, 282, 28733–28739. [Google Scholar] [CrossRef]
- Wösten, H.A.B. Hydrophobins: multipurpose proteins. Annu. Rev. Microbiol. 2001, 55, 625–646. [Google Scholar] [CrossRef]
- Hakanpää, J.; Szilvay, G.R.; Kaljunen, H.; Maksimainen, M.; Linder, M.; Rouvinen, J. Two crystal structures of Trichoderma reesei hydrophobin HFBI−The structure of a protein amphiphile with and without detergent interaction. Protein Sci. 2006, 15, 2129–2140. [Google Scholar] [CrossRef]
- Mgbeahuruike, A.C.; Kovalchuk, A.; Asiegbu, F. Comparative genomics and evolutionary analysis of hydrophobins from three species of wood-degrading fungi. Mycologia 2013, 105, 1471–1478. [Google Scholar] [CrossRef]
- Wessels, J.G.H. Developmental regulation of fungal cell wall formation. Annu. Rev. Phytopathol. 1994, 32, 413–437. [Google Scholar] [CrossRef]
- Gandier, J.-A.; Langelaan, D.N.; Won, A.; O’Donnell, K.; Grondin, J.L.; Spencer, H.L.; Wong, P.; Tillier, E.; Yip, C.; Smith, S.P.; et al. Characterization of a Basidiomycota hydrophobin reveals the structural basis for a high-similarity Class I subdivision. Sci. Rep. 2017, 7, srep45863. [Google Scholar] [CrossRef]
- Banerjee, G.; Robertson, D.L.; Leonard, T.J. Hydrophobins Sc3 and Sc4 gene expression in mounds, fruiting bodies and vegetative hyphae of Schizophyllum commune. Fungal Genet. Biol. 2008, 45, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Chen, R.; Yan, J.; Long, Y.; Tong, Z.; Song, H.; Xie, B. A hydrophobin gene, Hyd9, plays an important role in the formation of aerial hyphae and primordia in Flammulina filiformis. Gene 2019, 706, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Sevim, A.; Donzelli, B.G.G.; Wu, D.; Demirbag, Z.; Gibson, D.M.; Turgeon, B.G. Hydrophobin genes of the entomopathogenic fungus, Metarhizium brunneum, are differentially expressed and corresponding mutants are decreased in virulence. Curr. Genet. 2012, 58, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Ruocco, M.; Lanzuise, S.; Lombardi, N.; Woo, S.L.; Vinale, F.; Marra, R.; Varlese, R.; Manganiello, G.; Pascale, A.; Scala, V.; et al. Multiple roles and effects of a novel trichoderma Hydrophobin. Mol. Plant-Microbe Interact. 2015, 28, 167–179. [Google Scholar] [CrossRef]
- Moonjely, S.; Keyhani, N.O.; Bidochka, M.J. Hydrophobins contribute to root colonization and stress responses in the rhizosphere-competent insect pathogenic fungus Beauveria bassiana. Microbiology 2018, 164, 517–528. [Google Scholar] [CrossRef]
- Van Wetter, M.-A.; Schuren, F.H.; Schuurs, T.A.; Wessels, J.G. Targeted mutation of the SC3 hydrophobin gene of Schizophyllum commune affects formation of aerial hyphae. FEMS Microbiol. Lett. 1996, 140, 265–269. [Google Scholar] [CrossRef]
- Sammer, D.; Krause, K.; Gube, M.; Wagner, K.; Kothe, E. Hydrophobins in the life cycle of the ectomycorrhizal basidiomycete Tricholoma vaccinum. PLoS ONE 2016, 11, e0167773. [Google Scholar] [CrossRef]
- Kim, H.-I.; Lee, C.-S.; Park, Y.-J. Further characterization of hydrophobin genes in genome of Flammulina velutipes. Mycoscience 2016, 57, 320–325. [Google Scholar] [CrossRef]
- Mikus, M.; Hatvani, L.; Neuhof, T.; Komoń-Zelazowska, M.; Dieckmann, R.; Schwecke, T.; Druzhinina, I.S.; Von Döhren, H.; Kubicek, C.P. Differential regulation and posttranslational processing of the class ii hydrophobin genes from the biocontrol fungus Hypocrea atroviridis. Appl. Environ. Microbiol. 2009, 75, 3222–3229. [Google Scholar] [CrossRef]
- Ohm, R.A.; Aerts, D.; Wösten, H.A.B.; Lugones, L.G. The blue light receptor complex WC-1/2 of Schizophyllum communeis involved in mushroom formation and protection against phototoxicity. Environ. Microbiol. 2013, 15, 943–955. [Google Scholar] [CrossRef]
- Stappler, E.; Dattenböck, C.; Tisch, D.; Schmoll, M. Analysis of light- and carbon-specific transcriptomes implicates a class of G-protein-coupled receptors in cellulose sensing. Msphere 2017, 2, e00089-17. [Google Scholar] [CrossRef] [PubMed]
- Bell-Pedersen, D.; Dunlap, J.C.; Loros, J.J. The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 1992, 6, 2382–2394. [Google Scholar] [CrossRef] [PubMed]
- Casas-Flores, S.; Rios-Momberg, M.; Bibbins, M.; Ponce-Noyola, P.; Herrera-Estrella, A. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology 2004, 150, 3561–3569. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Saavedra, T.; Esquivel-Naranjo, E.U.; Casas-Flores, S.; Martínez-Hernández, P.; Ibarra-Laclette, E.; Cortes-Penagos, C.; Herrera-Estrella, A. Novel light-regulated genes in Trichoderma atroviride: A dissection by cDNA microarrays. Microbiology 2006, 152, 3305–3317. [Google Scholar] [CrossRef] [PubMed]
- Lauter, F.R.; Russo, V.E.; Yanofsky, C. Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev. 1992, 6, 2373–2381. [Google Scholar] [CrossRef]
- Liu, F.; Zhu, Z.; Sun, X.; Gao, H.; Zhang, Y. The preparation of three selenium-containing Cordyceps militaris polysaccharides: characterization and anti-tumor activities. Int. J. Biol. Macromol. 2017, 99, 196–204. [Google Scholar] [CrossRef]
- Lee, H.H.; Park, H.; Sung, G.-H.; Lee, K.; Lee, T.; Lee, I.; Park, M.-S.; Jung, Y.W.; Shin, Y.S.; Kang, H.; et al. Anti-influenza effect of Cordyceps militaris through immunomodulation in a DBA/2 mouse model. J. Microbiol. 2014, 52, 696–701. [Google Scholar] [CrossRef]
- Jeong, M.H.; Park, Y.-S.; Jeong, D.-H.; Lee, C.G.; Kim, J.-S.; Oh, S.-J.; Jeong, S.-K.; Yang, K.; Jo, W.-S. In vitro evaluation of Cordyceps militaris as a potential radioprotective agent. Int. J. Mol. Med. 2014, 34, 1349–1357. [Google Scholar] [CrossRef]
- Shrestha, B.; Zhang, W.; Zhang, Y.; Liu, X.-Z. The medicinal fungus Cordyceps militaris: Research and development. Mycol. Prog. 2012, 11, 599–614. [Google Scholar] [CrossRef]
- Yang, T.; Dong, C. Photo morphogenesis and photo response of the blue-light receptor geneCmwc-1in different strains of Cordyceps militaris. FEMS Microbiol. Lett. 2014, 352, 190–197. [Google Scholar] [CrossRef][Green Version]
- Yang, T.; Guo, M.; Yang, H.; Guo, S.; Dong, C. The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 2015, 100, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Song, X.; Dong, X.; Zhang, J.; Dong, C. DASH-type cryptochromes regulate fruiting body development and secondary metabolism differently than CmWC-1 in the fungus Cordyceps militaris. Appl. Microbiol. Biotechnol. 2017, 101, 4645–4657. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, Q.; Zhang, J.; Liu, K.; Li, K.; Liu, G.; Dong, C. Comparative transcriptome analysis between a spontaneous albino mutant and its sibling strain of Cordyceps militaris in response to light stress. Front. Microbiol. 2018, 9, 1237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, F.; Yang, Y.; Wang, Y.; Dong, C. CmVVD is involved in fruiting body development and carotenoid production and the transcriptional linkage among three blue-light receptors in edible fungus Cordyceps militaris. Environ. Microbiol. 2019, 22, 466–482. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Baker, S.E.; Gamauf, C.; Kenerley, C.M.; Druzhinina, I.S. Purifying selection and birth-and-death evolution in the class II hydrophobin gene families of the ascomycete Trichoderma/Hypocrea. BMC Evol. Biol. 2008, 8, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Kramer, G.; Nodwell, J.R. Chromosome level assembly and secondary metabolite potential of the parasitic fungus Cordyceps militaris. BMC Genom. 2017, 18, 912. [Google Scholar] [CrossRef]
- Mgbeahuruike, A.C.; Kovalchuk, A.; Chen, H.; Ubhayasekera, W.; Asiegbu, F. Evolutionary analysis of hydrophobin gene family in two wood-degrading basidiomycetes, Phlebia brevispora and Heterobasidion annosum s.l. BMC Evol. Biol. 2013, 13, 240. [Google Scholar] [CrossRef]
- Kershaw, M.J.; Thornton, C.R.; Wakley, G.E.; Talbot, N.J. Four conserved intramolecular disulphide linkages are required for secretion and cell wall localization of a hydrophobin during fungal morphogenesis. Mol. Microbiol. 2005, 56, 117–125. [Google Scholar] [CrossRef]
- Sunde, M.; Kwan, A.H.; Templeton, M.D.; Beever, R.E.; Mackay, J.P. Structural analysis of hydrophobins. Micron 2008, 39, 773–784. [Google Scholar] [CrossRef]
- Lobo, I.; Shaw, K. Thomas Hunt Morgan, genetic recombination, and gene mapping. Nat. Educ. 2008, 1, 205. [Google Scholar]
- Adomas, A.; Eklund, M.; Johansson, M.; Asiegbu, F. Identification and analysis of differentially expressed cDNAs during nonself-competitive interaction between Phlebiopsis gigantea and Heterobasidion parviporum. FEMS Microbiol. Ecol. 2006, 57, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Whiteford, J.R.; Spanu, P.D. Hydrophobins and the interactions between fungi and plants. Mol. Plant Pathol. 2002, 3, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Arntz, C.; Tudzynski, P. Identification of genes induced in alkaloid-producing cultures of Claviceps sp. Curr. Genet. 1997, 31, 357–360. [Google Scholar] [CrossRef] [PubMed]
- De Vries, O.M.H.; Moore, S.; Arntz, C.; Wessels, J.G.H.; Tudzynski, P. Identification and characterization of a tri-partite hydrophobin from Claviceps fusiformis: A novel type of class II hydrophobin. JBIC J. Biol. Inorg. Chem. 1999, 262, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Mey, G.; Correia, T.; Oeser, B.; Kershaw, M.J.; Garre, V.; Arntz, C.; Talbot, N.J.; Tudzynski, P. Structural and functional analysis of an oligomeric hydrophobin gene from Claviceps purpurea. Mol. Plant Pathol. 2002, 4, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S.; et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2018, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Gasteiger, E. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Malda, J.; Woodfield, T.; Van Der Vloodt, F.; Wilson, C.; Martens, D.; Tramper, J.; Van Blitterswijk, C.; Riesle, J. The effect of PEGT/PBT scaffold architecture on the composition of tissue engineered cartilage. Biomaterials 2005, 26, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Rineau, F.; Lmalem, H.; Ahren, D.; Shah, F.; Johansson, T.; Coninx, L.; Ruytinx, J.; Nguyen, H.; Grigoriev, I.; Kuo, A.; et al. Comparative genomics and expression levels of hydrophobins from eight mycorrhizal genomes. Mycorrhiza 2017, 27, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Guo, M.; Guo, S.; Huaijun, Y.; Bu, N.; Dong, C. Comparison of major bioactive compounds of the caterpillar medicinal mushroom, Cordyceps militaris (Ascomycetes), fruiting bodies cultured on wheat substrate and pupae. Int. J. Med. Mushrooms 2016, 18, 327–336. [Google Scholar] [CrossRef]
- Lian, T.; Yang, T.; Liu, G.; Sun, J.; Dong, C. Reliable reference gene selection for Cordyceps militaris gene expression studies under different developmental stages and media. FEMS Microbiol. Lett. 2014, 356, 97–104. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Name | Protein ID * | Chr. ** | Genomic Location *** | AA **** | MW ***** (kDa) | GRAVY ****** | Class | Signal Peptide |
|---|---|---|---|---|---|---|---|---|
| Cmhyd1 | CCM_03537/ A9K55_005453 | 6 | 356807–357251 | 105 | 10.57 | 0.37 | II | Yes |
| Cmhyd2 | CCM_06854/ A9K55_008053 | 7 | 3749538–3749938 | 99 | 10.42 | 0.412 | II | Yes |
| Cmhyd3 | CCM_06862/ A9K55_008043 | 7 | 3715724–3716248 | 138 | 13.48 | 0.583 | Ι | Yes |
| Cmhyd4 | CCM_07964/ A9K55_007066 | 7 | 398330–398703 | 104 | 10.35 | 0.478 | Ι | Yes |
| Species | No. MH * | GenBank ID | No. H II ** Domain | AA | MW (kDa) | GRAVY *** | pI **** |
|---|---|---|---|---|---|---|---|
| Cordycipitaceae | |||||||
| Beauveria bassiana | 3 | PMB64239.1 | 4 | 734 | 71.74 | −0.48 | 6.1 |
| PQK10896.1 | 3 | 593 | 57.46 | −0.50 | 6.01 | ||
| KAF1736859.1 | 3 | 596 | 58.22 | −0.54 | 6.59 | ||
| Cordyceps militaris | 1 | ATY58761.1 | 4 | 537 | 51.00 | −0.36 | 6.16 |
| Cordyceps fumosorosea | 1 | XP_018708370.1 | 5 | 599 | 58.36 | −0.31 | 5.01 |
| Cordyceps javanica | 1 | TQV96986.1 | 2 | 264 | 25.91 | −0.16 | 5.15 |
| Cordyceps confragosa | 1 | OAA82275.1 | 4 | 605 | 57.51 | −0.44 | 5.77 |
| Clavicipitaceae | |||||||
| Torrubiella hemipterigena | 2 | CEJ88606.1 | 7 | 1187 | 111.18 | −0.58 | 4.06 |
| CEJ95278.1 | 3 | 719 | 67.70 | −0.53 | 4.23 | ||
| Claviceps fusiformis | 1 | CAB61236.1 | 3 | 394 | 36.81 | −0.53 | 4.70 |
| Claviceps purpurea | 1 | CAD10781.1 | 5 | 756 | 69.90 | −0.73 | 5.27 |
| Moelleriella libera | 1 | KZZ89374.1 | 4 | 508 | 49.50 | −0.59 | 4.74 |
| Pochonia chlamydosporia | 1 | RZR67753.1 | 3 | 402 | 38.21 | −0.62 | 7.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, F.; Xu, Y.; Liu, G.; Dong, C. Cysteine-Rich Hydrophobin Gene Family: Genome Wide Analysis, Phylogeny and Transcript Profiling in Cordyceps militaris. Int. J. Mol. Sci. 2021, 22, 643. https://doi.org/10.3390/ijms22020643
Li X, Wang F, Xu Y, Liu G, Dong C. Cysteine-Rich Hydrophobin Gene Family: Genome Wide Analysis, Phylogeny and Transcript Profiling in Cordyceps militaris. International Journal of Molecular Sciences. 2021; 22(2):643. https://doi.org/10.3390/ijms22020643
Chicago/Turabian StyleLi, Xiao, Fen Wang, Yanyan Xu, Guijun Liu, and Caihong Dong. 2021. "Cysteine-Rich Hydrophobin Gene Family: Genome Wide Analysis, Phylogeny and Transcript Profiling in Cordyceps militaris" International Journal of Molecular Sciences 22, no. 2: 643. https://doi.org/10.3390/ijms22020643
APA StyleLi, X., Wang, F., Xu, Y., Liu, G., & Dong, C. (2021). Cysteine-Rich Hydrophobin Gene Family: Genome Wide Analysis, Phylogeny and Transcript Profiling in Cordyceps militaris. International Journal of Molecular Sciences, 22(2), 643. https://doi.org/10.3390/ijms22020643






