Abstract
Preeclampsia (PE) is a leading cause of maternal and neonatal morbidity and mortality worldwide. Defects in trophoblast invasion, differentiation of extravillous trophoblasts and spiral artery remodeling are key factors in PE development. Currently there are no predictive biomarkers clinically available for PE. Recent technological advancements empowered transcriptome exploration and led to the discovery of numerous non-coding RNA species of which microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are the most investigated. They are implicated in the regulation of numerous cellular functions, and as such are being extensively explored as potential biomarkers for various diseases. Altered expression of numerous lncRNAs and miRNAs in placenta has been related to pathophysiological processes that occur in preeclampsia. In the following text we offer summary of the latest knowledge of the molecular mechanism by which lnRNAs and miRNAs (focusing on the chromosome 19 miRNA cluster (C19MC)) contribute to pathophysiology of PE development and their potential utility as biomarkers of PE, with special focus on sample selection and techniques for the quantification of lncRNAs and miRNAs in maternal circulation.
1. Introduction
Hypertensive pregnancy complications, including preeclampsia (PE), are one of the most common direct causes of maternal and fetal morbidity and mortality [1]. PE is defined as a new-onset hypertension, diagnosed after the 20th week of gestation, which solves postpartum, followed by proteinuria or some other form of end-organ damage: thrombocytopenia, impaired liver function, development of renal insufficiency, pulmonary edema, or cerebral and visual disturbances [2]. PE is a two-stage syndrome [3]. The first stage begins early in pregnancy, and is associated with impaired trophoblast differentiation, invasion and ultimately remodeling of the spiral arteries, resulting in an inadequate transition of the blood vessel phenotype from small-diameter and high-resistance blood vessels into blood vessels of high capacity and low resistance [3,4,5]. In healthy pregnancy, remodeling of the spiral arteries ensures adequate blood flow to the uteroplacental unit [4,5]. In PE, trophoblast invasion is limited only to spiral arteries in the superficial decidua, resulting in hemodynamic disturbances, leading to poor uterine-placental blood flow and placental hypoxia [4,6]. It should be emphasized that difference in the etiopathogenesis of early-onset (EO, developed before the 34th week of gestation) and late-onset (LO, developed after the 34th week of gestation) PE exists [7]. EO-PE is referred to as a fetal disorder associated with placental dysfunction (previously described as incomplete trophoblast invasion). On the other side, the LO syndrome is thought to be a disorder predisposed by specific maternal risk factors, such as obesity, diabetes, or chronic hypertension [7,8,9], where limited blood flow between the chorionic villi during placental maturation may lead to placental hypoxia [9].
In both cases, resulting hypoxia and ischemia/reperfusion lead to an increased production of humoral factors from trophoblast cells together with the sequestration of the particles from the surface of the injured syncytium of the human placenta, and their subsequent release into the uteroplacental circulation [10]. In healthy pregnancy, trophoblast derived extracellular vesicles (EV) are packed with specific cargo of humoral factors and miRNAs, which can influence endothelial cell function and maternal immune cells (monocytes, granulocytes, T cells and natural killer (NK) cells), having in that way a major effect on immune tolerance to the developing fetus and on the development and function of the placenta [11,12,13,14,15]. In PE, the composition of the cargo carried in the vesicles is altered, with the number of the vesicles being increased, which significantly contributes to the pathophysiology of the disease [16,17,18,19,20,21,22]. Maternal endothelium may be vulnerable to the products of trophoblast cells, which further stimulate cytokine production, adhesion molecules and neutrophils migration into blood vessels walls and generate systemic inflammatory response which results with clinical manifestations of the disease (the second stage of PE) [23,24]
The biggest challenge regarding PE is clear understanding of the disease etiopathogenesis, which entails limited therapeutic approach and lack of effective clinical and biochemical multi-marker algorithms for preeclampsia prediction [4].
Recent technological advancements empowered transcriptome exploration and led to the discovery of numerous non-coding RNA species (small nuclear RNA, small nucleolar RNAs, telomerase RNA, tRNA-derived fragments, tRNA halves, microRNA, small interfering RNA, piwi-interacting RNA, enhancer RNA, long non-coding RNA, and circular RNA), of which microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are the most investigated [25].
A growing body of evidence supports the involvement of placenta expressed miRNAs and lncRNAs in the regulation of crucial processes of placenta development and function. The complete biological roles of the most miRNAs and lncRNA in placenta remain scarce, nevertheless, it has been unequivocally shown that altered expression of numerous miRNAs as well as lncRNAs in placenta, is related to pathophysiological processes that occur in PE [25,26,27,28,29,30].
The aim of this review is to summarize the latest knowledge of the molecular mechanisms by which miRNAs (focusing on the chromosome 19 miRNA cluster (C19MC)) and lnRNAs contribute to pathophysiology of PE and to review the latest evidence of their potential utility as biomarkers of PE.
2. MicroRNAs
MiRNAs are small, about 22–24 nucleotides long, noncoding RNAs that regulate gene expression at post transcriptional level. Half of all currently identified miRNAs are transcribed mostly from introns and few exons of protein coding genes, while the remaining are intergenic, transcribed independently of a host gene [31,32]. Their biogenesis starts with the processing of RNA polymerase II/III transcripts post- or co-transcriptionally [33]. In the canonical, dominant pathway of miRNA biogenesis, primary miRNAs (pri-miRNAs) are transcribed from their genes and processed into double-stranded precursors (pre-miRNAs), by the microprocessor complex: RNA binding protein DiGeorge Syndrome Critical Region 8 (DGCR8) and a ribonuclease III enzyme, Drosha [34]. DGCR8 recognizes an N6-methyladenylated GGAC and other motifs within the pri-miRNA [35], while Drosha cleaves the pri-miRNA duplex at the base of the characteristic hairpin structure of pri-miRNA. Pre-miRNAs are exported to the cytoplasm by an exportin 5 (XPO5)/RanGTP complex and further processed by the RNase III endonuclease Dicer [34,36], which results in the removal of the terminal loop, yielding a mature miRNA duplex [33,37]. This step is followed by unwinding of mature miRNA duplex and loading into the Argonaute2 (Ago2)-containing, RNA-induced silencing complex (RISC) [38,39], where the miRNAs repress mRNAs in a sequence-specific manner [40]. The directionality of the miRNA strand determines the name of the mature miRNA form (5p of 3p derived from the 5′ end and the 3′ end, respectively). Both strands derived from the mature miRNA duplex can be loaded into RISC [38,39], depending on their thermodynamic stability at the 5′ ends of the miRNA duplex or a 5′ U at nucleotide position 1 [39]. The loaded strand is called the guide strand, while the unloaded strand is called the passenger strand, which will be unwound from the guide strand through various mechanisms and cleaved by AGO2 and degraded by cellular machinery [33]. The miRNA biogenesis pathway is shown in Figure 1. In addition, multiple non-canonical miRNA biogenesis pathways exist, using different combinations of the proteins involved in the canonical pathway, mainly Drosha, Dicer, exportin 5, and AGO2 [39].
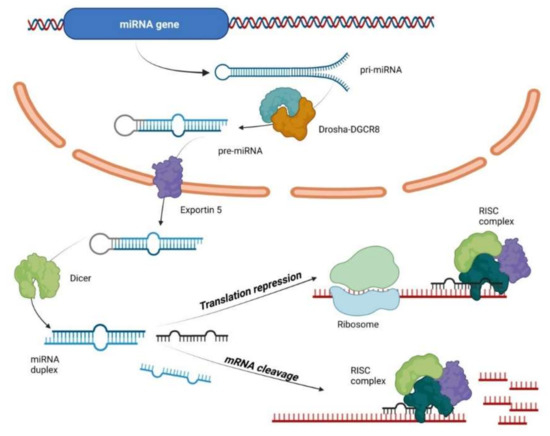
Figure 1.
MicroRNA biogenesis and mechanism of action. Primary miRNAs (pri-miRNAs) are transcribed from their genes and processed into double-stranded precursors (pre-miRNAs), by the microprocessor complex: RNA binding protein DiGeorge Syndrome Critical Region 8 (DGCR8) and Drosha. Pre-miRNAs are exported to the cytoplasm by an exportin 5 (XPO5)/RanGTP complex and processed by the RNase III endonuclease Dicer yielding a mature miRNA duplex. This step is followed by unwinding of mature miRNA duplex and loading into the Argonaute2 (Ago2)-containing, RNA-induced silencing complex (RISC) where the miRNAs repress mRNAs in a sequence-specific manner.
In most cases, miRNAs interact with the 3′ UTR of target RNAs to suppress expression [33,39], promoting their degradation or inhibiting translation; in others, interaction with 5′ UTR and coding part of target sequence and gene promoters occurs, while under certain conditions, miRNAs can even activate gene expression [39]. Recent studies have suggested that subcellular compartmentation of miRNAs has an effect on the rate of translation, and even transcription [41]. By these mechanisms, miRNAs regulate more than 30–60% of protein coding genes in the human genome [39,42]. MiRNAs are now recognized as pivotal posttranscriptional regulators in a broad range of biological processes [43]. During embryogenesis, critical periods are controlled by epigenetic modification of genes: gamete development, preimplantation embryo development, as well as placentation [44].
2.1. Placental MiRNAs
Key molecules involved in miRNA biogenesis, such as Drosha, Exportin 5, Dicer, Ago2 and DEAD-box helicase protein (DP103), have been identified in placental trophoblast cells [45,46]. In Ago2 mutant mouse miRNA machinery is disabled which causes malformation of placental labyrinthine layer with a greatly reduced thickness and death of the mutant embryos at midgestation, highlighting the essential role that miRNAs have in placental development and function [47].
So far, more than 600 miRNAs have been identified in human placenta, including miRNAs originating from imprinted gene clusters, many of which are predominantly or even solely expressed in placenta [48,49]. The imprinted genes are usually activated at critical developmental stages and are involved in cell differentiation, embryonic and placental growth and regulation of nutritional requirements [15,49]. The chromosome 19 microRNA cluster (C19MC) is located at the imprinted, paternally inherited allele on the human chromosome Chr19q13 site and represents one of the largest human miRNA clusters. It is composed of 46 pre-miRNA genes, yielding 59 mature miRNAs [49,50]. C19MC is expressed almost exclusively in placenta, with miRNAs originating from C19MC being the most abundant miRNAs expressed in human term trophoblasts [51]. High C19MC expression is now even considered as one of the most robust markers to define primary first-trimester trophoblast cells [52], while in situ hybridization revealed that the main source of these miRNAs is the trophoblast layer with dominant signals in the syncytiotrophoblast [53,54]. The expression of certain miRNAs belonging to C19MC, at far lower levels than placenta, is also detected in the germ cells, embryonic stem cells (ESC) and certain tumors [39,40,55]. The C19MC is found only in primates and is highly enriched in Alu elements [50]. The expression of C19MC is controlled by methylation at the upstream CpG rich promoter region [50]. Fothi et al. described an epigenetically regulated placental tissue-specific promoter, which is silenced in stem cells. This promoter is particularly efficient in attracting the transcription protein complex that optimizes cluster expression [56]. C19MC miRNAs are transcribed by RNA polymerase II and are processed from introns of a poorly characterized transcript, C19MC-HG (host gene), composed of many repeated non-coding exons [57].
Chromosome 14 microRNA cluster (C14MC), located at the imprinted, maternally inherited allele at chromosome 14q32 site, is composed of 52 miRNA genes, most of them originating from two genomic regions: the miR-127/miR-136 cluster and the miR-379-miR-410 cluster [39,40,49]. C14MC is regulated by methylation of a distal Intergenic Germline derived Differentially Methylated Region [58]. Members of C14MC are not placenta specific but are predominantly expressed in placenta and epithelial tissues [59], with high levels of expression in the adult brain [49].
The miR-371-3 cluster is located on chromosome 19 within a 1050 bp region approximately 20 kb downstream of the C19MC [60]. It is predominantly expressed in the placenta [61] however; some members of the miR-371-3 cluster are highly expressed in human ESCs [62]. The miR-371-3 cluster in humans is composed of hsa-miR-371a-3p, hsa-miR-372 and hsa-miR-373-3p, which all share the same seed sequence “AAG UGC”. Along with them, hsa-miR-371-5p and hsa-miR-373-5p, synthesized from the opposite site of the pre-microRNA, and hsa-miR-371b-3p, are also a part of the miR-371-3 cluster [40]. The miR-371-3 cluster appears to be important for maintaining cell cycle progression, regulation of proliferation, and controlling apoptosis [63].
The expression of the C19MC cluster is detected as early as 5 weeks of pregnancy, and markedly increases in placental trophoblasts from the first to the third trimester [40,64,65]. In contrast, the expression levels of the C14MC are the highest in the first trimester placenta, and decrease during the course of pregnancy [40,64,65]. The expression of the 371–373 cluster only slightly changes during pregnancy [40,65].
Along with these three clusters, many other miRNAs are found to be expressed in placenta and influence the development and function of the organ, however, they are abundantly expressed in many other human cells and therefore are not placenta specific [48,49]. In further text, we will focus on the roles of the largest, placenta specific C19MC in PE.
2.2. C19MC MiRNAs and Trophoblast Differentiation, Invasion and Angiogenesis
Several in vitro studies indicated that C19CM is implicated in crucial processes required for adequate placentation, and that their dysregulation has an impact on trophoblast differentiation, invasion and angiogenesis. It was shown that C19MC miRNAs selectively attenuate migration of human trophoblasts by regulating target transcripts related to cellular movement. In particular, miR-519d regulated the extravillous trophoblast (EVT) invasive phenotype by targeting CXCL6, NR4A2 and FOXL2 transcripts through a 3′UTR miRNA-responsive element [66]. MiR-519d-3p directly targeted the 3‘UTR of matrix metalloproteinase-2 (MMP-2), consequently suppressing trophoblast invasion and migration [67]. Another member of the C19MC, miR-520g also targeted MMP2, and suppressed migration and invasion of HTR-8/SVneo cells [68]. Mir515-5p was upregulated in PE placentas and its overexpression led to an inhibition of syncytiotrophoblast differentiation, targeting key molecules in the process [69]. Fu et al. showed that miR-517-5p is highly expressed in placenta samples of PE pregnancies, which decreased cell proliferative and invasive abilities of human choriocarcinoma cell line by inhibiting extracellular signal regulated kinase/MMP-2 pathway [70]. Liu et al. showed that miR-518b can promote trophoblast cell proliferation via Rap1b–Ras–MAPK pathway, and the aberrant upregulation of miR-518b in preeclamptic placenta may contribute to the excessive trophoblast proliferation [71]. Mong et al. have shown that C19MC miRNAs have a crucial role in regulating epithelial-to-mesenchymal transition (EMT) genes in villous trophoblasts and maintaining their stem-like epithelial cell phenotype. The hypoxic condition during early placentation reduced C19MC expression and released the inhibition of EMT genes leading to the acquisition of migratory and invasive characteristics of EVTs. The authors conclude that maintaining optimal expression levels of C19MC is critical for EVT differentiation and invasion, while dysregulation of C19MC may result in impaired invasion associated with either the shallow placentation of PE or the exuberant invasion of placenta accrete [72].
MiR-517a/b or miR-517c were upregulated in PE placentas, and their upregulation in first trimester primary EVTs resulted in decreased trophoblast invasion and increased release of the anti-angiogenic protein soluble fms-like tyrosine kinase 1 (sFlt1) [73]. sFlt1 binds circulating angiogenic factors and consequently blocks their ability to induce angiogenesis, disturb normal signaling in maternal endothelium, in that way having a major impact in PE pathogenesis [74,75,76]. Upregulation of both miRNAs in first trimester primary EVTs also led to upregulation of tumor necrosis factor superfamily15 (TNFSF15), a cytokine involved in Flt1 splicing [73]. Additionally, it was demonstrated that syncytiotrophoblast-derived EVs directly transferred functional placental miRNA belonging to C19MC (mir-517a, mir-517c, mir-519a) to primary human endothelial cells, which may directly affect the maternal and fetal endothelial function [77]. Interestingly, Strub et al. showed that C19MC miRNA members could be involved in the aberrant angiogenesis in infantile hemangioma, which goes in line with the possible role that C19MC miRNA members may have in angiogenesis during pregnancy [78].
These results clearly indicate that C19MC miRNAs are important factors in controlling differentiation, migration and invasion of trophoblast cells as well as angiogenesis. Their dysregulation could lead to the impaired function of the trophoblast cells, contributing to impaired remodeling of the spiral arteries and consequently PE development.
2.3. Placenta Derived Extracellular MiRNAs and Immunomodulation
Placental miRNAs are released into the extracellular space and reach maternal circulation, where they are found in a very stable form of microparticles [53]. The main source of these circulating miRNAs are trophoblast cells [40,51,53]. These molecules can be released as microvesicles, exosomes, apoptotic bodies and as protein-bound miRNAs, (miRNAs complexed with Ago2, nucleophosmin1, or high-density lipoproteins (HDL)) [79,80,81]. Microvesicles originate from direct shedding of the plasma membrane; while on the other hand, release of exosomes is a regulated process, composed out of several steps which ultimately define the composition of the cargo within the vesicles. The release of miRNAs from cells in nonvesicular form is presumably an ATP-dependent process which also involves cell necrosis [40,81]. It is important to underline that extracellular miRNAs are not merely a simple measure of waste of cellular metabolism, apoptosis or necrosis; they act as a close or distant cell to cell communication toll. The extracellular miRNAs can be delivered to target cells and act as autocrine, paracrine, and/or endocrine regulators in that way potentially having a great impact on the function of the recipient cells [39,82]. Exosomes carrying specific miRNA cargo can be taken up by recipient cells via endocytic pathways or via direct fusing of the exosome particle with the cell membrane, with the release of their cargo into the cytoplasm [40,83] while on the other hand, miRNAs bound to proteins could interact with recipient cells via ligand–receptor interactions. Extracellular miRNAs can bind to Toll-like receptors and activate downstream signaling events, in that way regulating cell-to-cell communications [84]. MiRNAs that are carried in the circulation bound to HDL can be delivered to recipient cells via the HDL receptor scavenger receptor B type I [80].
Exosome vesicles, secreted from primary human trophoblasts (PHT) cells are packed with miRNAs, resembling the profile of trophoblastic cellular miRNA. The most abundant of them originate from C19MC [51]. It has been shown by several studies that C19MC derived miRNAs in these vesicles, can influence maternal immune cells and have a major impact in immunomodulation during pregnancy and defense against viral infection. Delorme-Axford et al. have provided evidence that PHT are highly resistant to infection by a number of viruses and more interestingly that they have an antiviral effect on other, non-placental cell types, by exosome-mediated delivery of specific miRNAs of the C19MC. They have shown that ectopically expressing the entire C19MC fragment in cells or transfecting a high level of certain C19MC members (miR517-3p, miR-512-3p, or 516b-5p) allocated viral resistance to the recipient non-placental cells by induction of autophagy [85]. Bayer et al. also demonstrated that PHT cells exert antiviral activity by at least two independent mechanisms, mediated by C19MC miRNA and by type III interferons [86], while Krawczynski et al. have found that miR-517a targets unc-13 homolog D (autophagy-related gene), suppressing in that way replication of vesicular stomatitis virus [87]. Furthermore, miR-517a-3p carried by placental exosomes regulates activation and proliferation of maternal T cells and NK cells, targeting the PRKG1 gene involved in activation of nitric oxide/cGMP signaling pathway [88]. Chaiwangyen et al. showed that miR-519d-3p released via extracellular vesicles from the trophoblast, can be taken up by other trophoblast cells and maternal immune cells and regulate their proliferation and migration, which may contribute to the immune tolerance in pregnancy [89]. All this evidence supports the specific role of miRNAs belonging to C19MC in regulation of the maternal immune system during pregnancy. The major part in pathogenesis of PE is related to immune response disorders and inflammation, triggered by humoral factors released from placenta and whole particles sequestered from the surface of the syncytium of the human placenta [10]. Therefore, it is possible that trophoblast derived exosomes carrying a potentially aberrant miRNA repertoire could severely influence maternal immune cells, leading to their dysfunction and in that way contribute to maternal systemic inflammatory response and the development and progression of PE [19,85,86,87,88]. The importance of the effect of miRNA cargo released from trophoblast cells in microparticles was underlined in the recent study by Hiu et al. [90]. The authors showed that trophoblastic small EV (sEV) are internalized into placental fibroblasts or uterine endothelial cells by macropinocytosis and clathrin-mediated endocytosis and demonstrated the trafficking of sEVs through the endosome-lysosome system and the delivery of sEV miRNA cargo to P-bodies. This represents the first data to suggest the delivery of sEV miRNAs to the RISC complex proteins, cellular site for miRNA-dependent silencing in the target cells [90] and strengthens the evidence of the impact of the microparticle miRNA cargo on cells participating in PE pathogenesis, both maternal and fetal origin.
2.4. Circulating MiRNAs as Biomarkers of PE
As already stated above, a specific placental miRNA pattern dynamically changes during pregnancy and those changes are reflected in maternal circulation [40,91,92]. Placenta derived EV are detected as early as six weeks of gestation, while after termination of pregnancy, plasma levels of C19MC associated miRNAs decrease significantly [93,94]. In contrary to cellular RNA species, the presence of miRNAs in vesicles or with accompanying proteins protects extracellular miRNAs from degradation and increases their stability in the extracellular milieu [39,95], making them a good biomarker candidate for the pathology of placenta. The additional fact in favor of this is that the release of microRNAs from the placenta mainly occurs from the villous trophoblast cells, indicating that circulating microRNAs could serve as unique markers for monitoring trophoblast and placental function [40,51,53]. Given the nature of miRNAs belonging to the C19MC cluster, their exclusive placenta-specific pattern of expression, high levels of expression in trophoblasts and presence in mother circulation, in vitro evidence for influencing crucial steps in PE pathogenesis, they have attracted great interest in PE biomarker research, both as prediction and diagnostic markers of the disease [45,53,85,94].
Many studies have shown abnormal levels of C19MC derived miRNAs in maternal circulation in pregnancies affected by PE, in different stages of the disease [64,68,70,96,97,98,99,100,101,102]. Several excellent reviews and a meta-analysis were written, giving us a useful overview of the potential of miRNA as prognostic and diagnostic tools in PE [29,30,103]. Recently, the miRNA analysis of plasma exosomes, either from placenta-specific isolated exosomes, or from whole plasma exosomes, has attracted much attention [17,19,20,104]. This approach could provide better sensitivity for the prediction of PE, however, cost effectiveness is a major issue. Even a decade later, great discrepancies still exist between the studies, with minimal overlap between the individual miRNAs identified as potential biomarkers Table 1 [17,29,30,96,97,98,99,100,101,102,103,104].

Table 1.
C19MC miRNA differential expression in maternal circulation of PE patients.
Nevertheless, in vitro studies along with the measurements of C19MC miRNAs in maternal blood provide evidence for the dysregulation of C19MC miRNAs during PE and their involvement in the regulation of crucial processes required for adequate placentation and immunomodulation during pregnancy. Their dysregulation could have a major effect in both stages of the disease.
2.5. Preanalytics and Technology
There are several major preanalytical and analytical points that should be considered when quantifying circulating miRNAs, which could be a cause of inconsistencies between the studies. When analyzing placenta derived non-specific miRNAs (which originate from other cells as well) in maternal circulation, different processing of the samples (time frame form venipuncture to the sample preparation and freezing) can have a major impact on the sample miRNA profile. The study of Mitchell et al. showed that freeze–thaw cycles, to a great extent, influence plasma miRNA profile, if not prepared as platelet poor plasma [105]. The reason for this is that non-specific placental miRNAs could be released from the residual blood cells, and in that way contaminate the sample [106]. These issues should also be considered in exosomal analysis of the plasma. Furthermore, serum and plasma have a different spectrum of miRNAs, due to the coagulation process itself, and the subsequent miRNA release, the process that we certainly cannot control in any sample [107]. Indeed, analyte which takes part in the coagulation process or changes during it is never quantified in serum samples, but in plasma instead. The logic here should be the same.
Regarding both placenta specific and nonspecific miRNAs, other causes of discrepancies could be due to cohort characteristics, gestational age (given that changes within placenta trough gestation are reflected in mothers’ circulation), PE form (EO or LO) [7,40,91,92]. Additionally, a great source of variation could be due to the use of different profiling high-throughput technologies (different microarray and next generation sequencing (NGS) platforms) [108,109,110,111,112,113,114,115].
As a first step in biomarker discovery, it is always wise to use some of the screening technologies. Earlier, some form of array technology was used: whether it was microarray [108] or qPCR arrays [109]. Microarrays were a good starting point. With time, more dense microarrays were designed, which allowed better target coverage. Its main challenge was normalization and buffering the potential bias between the samples and measurement variations which sometimes resulted in bad reproducibility of the results [110]. qPCR-arrays were later quite often used as a technique of screening, but still only a limited number of targets could be measured with this technique, since the qPCR-arrays are often thematic and cover dedicated signaling pathways [109]. With the development of new generation sequencing (NGS), or sometimes called deep sequencing, a new high-throughput technology emerged that allowed miRNA high-sensitive expression profiling without previous target selection. This technique enabled identification of novel miRNAs: revealing multiple miRNA variants with heterogeneous ends, lengths and expression levels [111,112]. In comparison to qPCR-arrays, NGS provides a cost-effective method in relation to high capacity. However, it requires a high input amount of the sample which could represent a technical challenge when circulating miRNAs are considered as potential biomarkers. There are available low-input miRNA library preparation kits, but the choice should be carefully evaluated for each sample type [113]. In addition, special attention must be paid to deal with biases associated with library preparation: adapter dimer formation, size selection of small RNA species, etc. [114,115]. In a study by Hui et al., of four maternal plasma exosome miRNAseq libraries, C19MC miRNAs were reliably detected in only one library, while numerous C19MC miRNAs were detected when qPCR-based TaqMan cards were used, questioning weather sequencing is sensitive enough for the detection of C19MC in plasma [19].
All these issues complicate the identification of a reliable biomarker for PE. In our research, we have used the digital droplet PCR (ddPCR) method for circulating miRNA quantification and shown that placenta specific miR-518b could serve as a potential biomarker for discriminating preeclampsia and healthy pregnancy [102]. DdPCR is a sensitive technique that enables absolute nucleic acid quantification, therefore represents a right tool for quantification of circulating miRNAs from low abundant samples. This technique is suitable for measuring a limited number of targets and is mostly used as a confirmation technology for the identified targets detected with screening techniques. It has many potential advantages over real-time PCR: obtaining higher precision and sensitivity, absolute quantification without the use of external references, with the advantage of removal of PCR efficiency bias [116,117]. It would be also the method of choice for the diagnostic use of miRNA biomarkers, since quantification by ddPCR in comparison to real-time PCR revealed greater precision (coefficients of variation decreased 37–86%) and improved day-to-day reproducibility of ddPCR [117]. More importantly, when applied to microRNA detection in clinical serum samples, ddPCR showed superior diagnostic performance compared to real-time PCR [117]. By comparing plasma exosome and whole plasma analysis of miRNAs in PE patients vs. controls, only one out of seven differentially expressed miRNAs in exosomes was detected in whole plasma samples [19]. This could be a consequence of the overall dilution of exosomes carrying placental miRNA in plasma. In this case, perhaps the application of ultra-sensitive ddPCR technology could be useful in overcoming these issues.
Small size and large number of closely related family members of miRNAs with highly similar sequences, makes it challenging to measure each miRNAs species separately accurately. Furthermore, several miRNAs at C19MC (e.g., the miR-520 and miR-519 families) have the similar “AAGUGC” seed sequence which is also found in members of the 371/miR-373 cluster and other miRNAs and nearly identical to that of the miR-17-92 cluster (also referred to as OncomiR1) [50]. Malnou et al. raised the question whether some miRNAs of C19MC sharing the same seed sequence may have related mRNA targets (or families thereof), perhaps underlying similar functions [49]. However, this question should also reflect the field of biomarker research, focusing perhaps on quantification of a panel of miRNAs with the same seed sequence or targets, especially taking into account that miRNAs from the C19MC cluster originate from the one large pre-miRNA transcript, and that the main regulation is its transcription [50,56,57].
3. Long Non-Coding RNAs
Long non-coding RNAs (lncRNAs) represent structurally and functionally a very diverse group of RNAs defined as transcripts over 200 nucleotides long characterized by poor conservation of primary sequences across species, low abundance in cells, high level of tissue specificity, and affinity to form secondary structural domains crucial for their functions [118,119]. Depending on the genomic loci of their origin, lncRNAs are classified as long intergenic, intronic, sense, antisense, promotor-associated, enhancer, and bidirectional [120]. Most lncRNAs are synthesized similarly to mRNAs through the activity of RNA polymerase II (RNAP II) [121]. In addition, RNAP III catalyzes the transcription of natural antisense transcripts from the antisense strand of exons, enhancer lncRNAs without a poly-A tale and intronic lncRNAs in combination with spRNAP IV [120,122]. LncRNAs also undergo intensive post-transcriptional modifications including 5’-capping, the addition of the poly-A tail, RNA editing and alternative splicing [120].
LncRNAs can regulate cellular processes through a variety of mechanisms. In the nucleus, lncRNAs can act through chromatin modifications, transcriptional regulations or interaction with enhancers. In the cytoplasm, lncRNAs act as miRNA sponges, influence mRNA stability and level of translation [123]. LncRNAs also play a role in regulating the expression of genes by acting as signals (participating in transmission of specific signaling pathways as a signal transduction molecule), scaffolds (downstream effectors can bind to the same lncRNA molecule to achieve information exchange and integration between different signaling pathways), guides (lncRNA can recruit chromatin-modifying enzymes to target genes) and decoys (lncRNA binds directly to proteins thus blocking the role of the molecule and signaling pathways) [124]. The biogenesis and functions of lncRNAs are described in Figure 2. Interestingly, some of the latest data suggested the existence of small open reading frames within lncRNAs with the potential to encode micropeptides, thus questioning the non-coding nature of lncRNAs [125]. Although the full biological roles of most lncRNA remain unknown, their roles in regulation of epigenetic and transcriptional mechanisms linked to PE are evident.
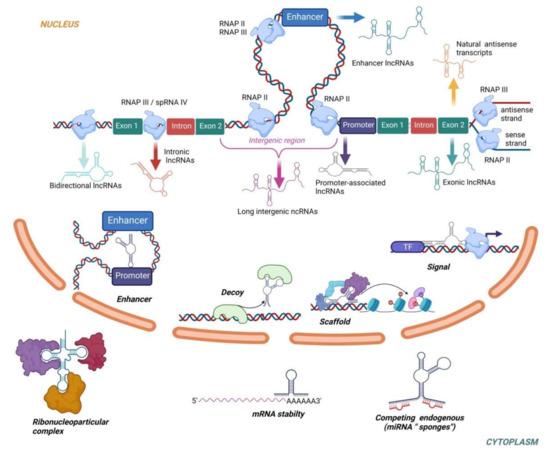
Figure 2.
The biogenesis and functions of lncRNAs. Based on the transcriptional origin, lncRNAs are classified as exonic, intronic, antisense, enhancer, intergenic, promoter-associated and bidirectional. The vast majority of lncRNAs is transcribed by RNAP II. RNAP III catalyzes transcription of natural antisense transcripts from the antisense strand of exons, enhancer lncRNAs without poly-A tale and intronic lncRNAs in combination with spRNAP IV. In the nucleus, lncRNAs regulate gene expression by acting as transcriptional signals, scaffolds, decoys or enhancers. In the cytoplasm, lncRNAs can increase/decrease mRNA stability, act as miRNAs “sponges” or interact with proteins to form ribonucleoprotein complexes.
3.1. LncRNAs in PE
Several studies have used a transcriptomic approach to identify lncRNAs involved with PE. Through microarray analysis and RNA profiling, He et al. identified 738 out of 28,443 differentially expressed lncRNAs in the placentas of PE in comparison to normal pregnancies [126]. Additionally, they have examined the expression of three lncRNAs (LOC391533, LOC284100, CEACAMP8) in 40 preeclampsia placenta tissues and 40-matched control placenta tissues using qPCR and proposed their involvement in PE through the regulation of angiogenesis and vasculogenesis (via vascular endothelial growth factor receptor 1) and lipid metabolism (via lipoprotein lipase) [126]. In another study, Liu S. et al. using combined microarray and RNA-seq approach identified several differentially expressed lncRNAs linked to PE and JAK-STAT signaling pathway activation implicated in the progression of PE [127]. By performing RNA-seq on decidual samples, Tong et al. discovered that 32 lncRNAs were differentially expressed between normal and early-onset PE, 53 lncRNAs were differentially expressed between normal and late-onset PE, and 32 differentially expressed lncRNAs between early-onset PE and late-onset PE, suggesting that different pathophysiological mechanisms are driving early-onset and late-onset PE [128]. In the study focusing on the lncRNAs role in the development of early-onset PE, Wang et al. identified 15,646 upregulated and 13,178 downregulated lncRNAs by microarray in the placenta of EOPE patients compared to the preterm controls. Through additional GO analysis, the authors showed that pathways overrepresented in the EOPE patients were related to cell migration and cell motility [129].
So far, particular roles of several lncRNAs in PE development and progression have been described. Upregulation and downregulation of specific lncRNAs can impact critical mechanisms in the development of PE and lead to changes in trophoblast proliferation, invasion, migration and apoptosis Table 2 [130,131,132,133,134,135,136,137,138,139,140,141,142,143,144].

Table 2.
Dysregulation of specific lncRNAs in PE and proposed mechanisms of action.
3.2. Circulating LncRNAs in PE
So far, several lncRNA in the circulation have been explored as potential biomarkers for PE. Sun et al. showed that levels of lncRNA BC030099 in whole blood had a good ability to discriminate preeclamptic from healthy pregnant women (AUC = 0.713). Furthermore, lncRNAs NR_026824.1, AK055151.1, NR_027457, and NR_024178 were highly upregulated in PE placentas but did not demonstrate good biomarker potential for PE (AUC < 0.6) [145]. Luo et al. suggested that levels of three lncRNAs in serum AF085938, G36948, and AK002210 can also serve as potential diagnostic biomarkers in preeclampsia (AUC = 0.7673, 0.7956, and 0.7575, respectively) [146]. Dai et al. have identified seven lncRNA in early pregnancy as potential biomarkers for the prediction of pregnancy-induced hypertension and preeclampsia. However, in ROC analysis, these seven lncRNAs (NR_002187, ENST00000398554, ENST00000586560, TCONS_00008014, ENST00000546789, ENST00000610270, and ENST00000527727,) showed only modest discriminative power (AUC between 0.6 and 0.7) [147].
Currently, the main problems in the quantification of circulating lncRNAs are related to their low abundance in the circulation and unstandardized strategies in normalization approaches. Many authors utilize mRNA levels of housekeeping genes as a reference in normalization. However, a significant difference in the stability of circulating mRNAs compared with lncRNAs questions the validity of this approach [148]. In the study by Dong et al., β-actin was shown to be the most stable in comparison to GAPDH, HPRT, 18S RNA, CYC, and GUSB in the serum of healthy and cancer patients [149]. The other approach to normalization includes the use of artificial spike-ins that enable good quality control of analytical variations (extraction, efficacy of reverse transcriptions and qPCR). On the other hand, the spike-ins are synthetic oligomers externally added to the sample, and as such have limited capacity to control preanalytical factors like sampling, hemolysis, etc. [150]. It seems important to reach a consensus regarding current dilemmas to exploit the full biomarker potential of circulating lncRNAs.
4. Conclusions
PE is characterized by extensive dysfunction of the placenta, caused by dysregulation of trophoblast differentiation, invasion and ultimately remodeling of the spiral arteries [4,5,6]. The disturbances in placentation have major consequences later on in pregnancy, causing extensive systemic inflammatory response which has a major impact on maternal and fetal health [7,8,9]. The results from many in vitro studies clearly indicate that miRNAs belonging to C19MC are important factors in controlling crucial processes required for adequate placentation, and that dysregulation of C19MC could lead to the impaired function of the trophoblast cells, impaired placentation and consequently PE development [66,67,68,69,70,71,72,73,74,75,76,77,78]. In addition, upregulation and downregulation of specific lncRNAs can also impact trophoblast proliferation, invasion, migration as well as apoptosis [130,131,132,133,134,135,136,137,138,139,140,141,142,143,144].
Considering miRNAs belonging to C19MC are involved in the regulation of maternal immune system during pregnancy, it is possible that in PE, trophoblast derived exosomes carrying a potentially aberrant miRNA repertoire could severely influence maternal immune cells, contributing to maternal systemic inflammatory response and progression of PE [19,85,86,87,88]. All these data are supported by the evidence that miRNAs belonging to C19MC, secreted from trophoblast cells and carried by microparticles, are taken up recipient cells [40,85,88,89,90] and actually delivered to the RISC complex proteins of both maternal and fetal recipient cells [90].
Extensive involvement of miRNAs belonging to C19MC in PE pathogenesis [66,67,68,69,70,71,72,73,74,75,76,77,78,85,86,87,88], their detection in maternal circulation early in pregnancy and exclusive placenta-specific pattern of expression [40,49,93,94] marks them as promising prognostic and diagnostic tools for PE. However, major inconsistencies exist between the studies examining their diagnostic utility in PE analyzing miRNAs either from maternal serum/plasma or from plasma exosomes [68,70,96,97,98,99,100,101,102]. Circulating lncRNAs have also been considered as biomarkers of PE, however, their low abundance in the circulation and unstandardized strategies in normalization approaches also represent challenges for their clinical utility in PE [148,149]. Therefore, critical preanalytical (sample handling, sample type) and analytical issues (use of various quantification approaches with different sensitivity and specificity, normalization), regarding both miRNAs and lncRNA quantification, should be resolved prior to potential clinical application of these markers in PE assessment.
Author Contributions
Conceptualization, J.M., M.S., U.P.Z.; literature research, J.M., M.S., U.P.Z., A.S., A.N., I.J., T.A.; writing of the manuscript, J.M., M.S., U.P.Z., A.S., A.N., I.J.; writing—review, editing, and supervision, R.K., V.S.-K., J.M., M.S., U.P.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by ddPCR Grant challenge initiative from Labena Ltd., Verovškova 64, 1000 Ljubljana, Slovenia and by a grant from the Ministry of Education, Science and Technological Development, Serbia (project number 175035).
Institutional Review Board Statement
Not applicable.
Acknowledgments
Figures were created with BioRender.com.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Health Topics—Maternal Health. Available online: https://www.who.int/health-topics/maternal-health#tab=tab_1 (accessed on 25 July 2020).
- American College of Obstetricians and Gynecologists. Gestational hypertension and preeclampsia: ACOG Practice Bulletin, number 222. Obstet. Gynecol. 2020, 135, e237–e260. [Google Scholar] [CrossRef] [PubMed]
- Lyall, F.; Robson, S.C.; Bulmer, J.N. Spiral artery remodeling and trophoblast invasion in preeclampsia and fetal growth restriction: Relationship to clinical outcome. Hypertension 2013, 62, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, F.J.; Pérez-Sepúlveda, A.; Torres, M.J.; Correa, P.; Repetto, G.M.; Illanes, S.E. Pathogenesis of preeclampsia: The genetic component. J. Pregnancy 2012, 2012, 632732. [Google Scholar] [CrossRef] [PubMed]
- Damsky, C.H.; Fisher, S.J. Trophoblast pseudo-vasculogenesis: Faking it with endothelial adhesion receptors. Curr. Opin. Cell Biol. 1998, 10, 660–666. [Google Scholar] [CrossRef]
- American College of Obstetricians and Gynecologists. Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet. Gynecol. 2013, 122, 1122–1131. [Google Scholar] [CrossRef]
- Raymond, D.; Peterson, E. A critical review of early-onset and late-onset preeclampsia. Obstet. Gynecol. Surv. 2011, 66, 497–506. [Google Scholar] [CrossRef]
- Staff, A.C. The two-stage placental model of preeclampsia: An update. J. Reprod. Immunol. 2019, 134–135, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Redman, C.W.; Sargent, I.L.; Staff, A.C. IFPA Senior Award Lecture: Making sense of pre-eclampsia—Two placental causes of preeclampsia? Placenta 2014, 35, S20–S25. [Google Scholar] [CrossRef]
- Germain, S.J.; Sacks, G.P.; Sooranna, S.R.; Sargent, I.L.; Redman, C.W. Systemic inflammatory priming in normal pregnancy and preeclampsia: The role of circulating syncytiotrophoblast microparticles. J. Immunol. 2007, 178, 5949–5956, correction in 2007, 179, 1390. [Google Scholar] [CrossRef] [PubMed]
- Göhner, C.; Plösch, T.; Faas, M.M. Immune-modulatory effects of syncytiotrophoblast extracellular vesicles in pregnancy and preeclampsia. Placenta 2017, 60 (Suppl. 1), S41–S51. [Google Scholar] [CrossRef]
- Cockell, A.P.; Learmont, J.G.; Smárason, A.K.; Redman, C.W.; Sargent, I.L.; Poston, L. Human placental syncytiotrophoblast microvillous membranes impair maternal vascular endothelial function. Br. J. Obstet. Gynaecol. 1997, 104, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.C. The effect of placental syncytiotrophoblast microvillous membranes from normal and pre-eclamptic women on the growth of endothelial cells in vitro. Br. J. Obstet. Gynaecol. 1994, 101, 559. [Google Scholar] [CrossRef]
- Sargent, I.L.; Germain, S.J.; Sacks, G.P.; Kumar, S.; Redman, C.W. Trophoblast deportation and the maternal inflammatory response in pre-eclampsia. J. Reprod. Immunol. 2003, 59, 153–160. [Google Scholar] [CrossRef]
- Fu, G.; Brkić, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [PubMed]
- Motta-Mejia, C.; Kandzija, N.; Zhang, W.; Mhlomi, V.; Cerdeira, A.S.; Burdujan, A.; Tannetta, D.; Dragovic, R.; Sargent, I.L.; Redman, C.W.; et al. Placental Vesicles Carry Active Endothelial Nitric Oxide Synthase and Their Activity is Reduced in Preeclampsia. Hypertension 2017, 70, 372–381. [Google Scholar] [CrossRef]
- Buca, D.; Bologna, G.; D’Amico, A.; Cugini, S.; Musca, F.; Febbo, M.; D’Arcangelo, D.; Buca, D.; Simeone, P.; Liberati, M.; et al. Extracellular Vesicles in Feto-Maternal Crosstalk and Pregnancy Disorders. Int. J. Mol. Sci. 2020, 21, 2120. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Salomon, C. Extracellular vesicles and their immunomodulatory functions in pregnancy. Semin. Immunopathol. 2018, 40, 425–437. [Google Scholar] [CrossRef]
- Li, H.; Ouyang, Y.; Sadovsky, E.; Parks, W.T.; Chu, T.; Sadovsky, Y. Unique microRNA Signals in Plasma Exosomes from Pregnancies Complicated by Preeclampsia. Hypertension 2020, 75, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Salomon, C.; Guanzon, D.; Scholz-Romero, K.; Longo, S.; Correa, P.; Illanes, S.E.; Rice, G.E. Placental Exosomes as Early Biomarker of Preeclampsia: Potential Role of Exosomal MicroRNAs across Gestation. J. Clin. Endocrinol. Metab. 2017, 102, 3182–3194. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Yao, J.; He, Q.; Liu, M.; Duan, T.; Wang, K. Exosomes from Women with Preeclampsia Induced Vascular Dysfunction by Delivering sFlt (Soluble Fms-like Tyrosine Kinase)-1 and sEng (Soluble Endoglin) to Endothelial Cells. Hypertension 2018, 72, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Czernek, L.; Düchler, M. Exosomes as Messengers between Mother and Fetus in Pregnancy. Int. J. Mol. Sci. 2020, 21, 4264. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.M.; Gammill, H.S. Preeclampsia: Recent insights. Hypertension 2005, 46, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- LaMarca, B.D.; Gilbert, J.; Granger, J.P. Recent progress toward the understanding of the pathophysiology of hypertension during preeclampsia. Hypertension 2008, 51, 982–988. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Qin, S.; Zhang, L.; Liu, S. Roles of noncoding RNAs in preeclampsia. Reprod. Biol. Endocrinol. 2021, 19, 100. [Google Scholar] [CrossRef]
- Chen, A.; Yu, R.; Jiang, S.; Xia, Y.; Chen, Y. Recent Advances of MicroRNAs, Long Non-Coding RNAs, and Circular RNAs in Preeclampsia. Front. Physiol. 2021, 12, 659638. [Google Scholar] [CrossRef]
- Yang, X.; Meng, T. Long Noncoding RNA in Preeclampsia: Transcriptional Noise or Innovative Indicators? BioMed Res. Int. 2019, 2019, 5437621. [Google Scholar] [CrossRef] [PubMed]
- Bounds, K.R.; Chiasson, V.L.; Pan, L.J.; Gupta, S.; Chatterjee, P. MicroRNAs: New Players in the Pathobiology of Preeclampsia. Front. Cardiovasc. Med. 2017, 4, 60. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Kolluru, G.K.; Ahmed, A. Small Molecule, Big Prospects: MicroRNA in Pregnancy and Its Complications. J. Pregnancy 2017, 2017, 6972732. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.M.; Small, H.Y.; Currie, G.; Delles, C. Systematic Review of Micro-RNA Expression in Pre-Eclampsia Identifies a Number of Common Pathways Associated with the Disease. PLoS ONE 2016, 11, e0160808. [Google Scholar] [CrossRef] [PubMed]
- De Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Mouillet, J.F.; Coyne, C.B.; Sadovsky, Y. Review: Placenta-specific microRNAs in exosomes—Good things come in nano-packages. Placenta 2014, 35, S69–S73. [Google Scholar] [CrossRef] [PubMed]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Huang, L.; Cao, J.; Zen, K.; Chen, X.; Zhang, C.Y. Regulation of mammalian gene expression by exogenous microRNAs. Wiley Interdiscip. Rev. RNA 2012, 3, 733–742. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Wilkins-Haug, L. Epigenetics and assisted reproduction. Curr. Opin. Obstet. Gynecol. 2009, 21, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Donker, R.B.; Mouillet, J.F.; Nelson, D.M.; Sadovsky, Y. The expression of Argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol. Hum. Reprod. 2007, 13, 273–279. [Google Scholar] [CrossRef]
- Forbes, K.; Farrokhnia, F.; Aplin, J.D.; Westwood, M. Dicer-dependent miRNAs provide an endogenous restraint on cytotrophoblast proliferation. Placenta 2012, 33, 581–585. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef]
- Chen, D.B.; Wang, W. Human placental microRNAs and preeclampsia. Biol. Reprod. 2013, 88, 130. [Google Scholar] [CrossRef] [PubMed]
- Malnou, E.C.; Umlauf, D.; Mouysset, M.; Cavaillé, J. Imprinted MicroRNA Gene Clusters in the Evolution, Development, and Functions of Mammalian Placenta. Front. Genet. 2019, 9, 706. [Google Scholar] [CrossRef]
- Noguer-Dance, M.; Abu-Amero, S.; Al-Khtib, M.; Lefèvre, A.; Coullin, P.; Moore, G.E.; Cavaillé, J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum. Mol. Genet. 2010, 19, 3566–3582. [Google Scholar] [CrossRef]
- Donker, R.B.; Mouillet, J.F.; Chu, T.; Hubel, C.A.; Stolz, D.B.; Morelli, A.E.; Sadovsky, Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol. Hum. Reprod. 2012, 18, 417–424. [Google Scholar] [CrossRef]
- Lee, C.Q.; Gardner, L.; Turco, M.; Zhao, N.; Murray, M.J.; Coleman, N.; Rossant, J.; Hemberger, M.; Moffett, A. What Is Trophoblast? A Combination of Criteria Define Human First-Trimester Trophoblast. Stem Cell Rep. 2016, 6, 257–272. [Google Scholar] [CrossRef]
- Luo, S.S.; Ishibashi, O.; Ishikawa, G.; Ishikawa, T.; Katayama, A.; Mishima, T.; Takizawa, T.; Shigihara, T.; Goto, T.; Izumi, A.; et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009, 81, 717–729. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.Q.; Su, B. Molecular evolution of a primate-specific microRNA family. Mol. Biol. Evol. 2008, 25, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Morales-Prieto, D.M.; Ospina-Prieto, S.; Chaiwangyen, W.; Schoenleben, M.; Markert, U.R. Pregnancy-associated miRNA-clusters. J. Reprod. Immunol. 2013, 97, 51–61. [Google Scholar] [CrossRef]
- Fóthi, Á.; Biró, O.; Erdei, Z.; Apáti, Á.; Orbán, T.I. Tissue-specific and transcription-dependent mechanisms regulate primary microRNA processing efficiency of the human chromosome 19 MicroRNA cluster. RNA Biol. 2021, 18, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Bortolin-Cavaillé, M.L.; Dance, M.; Weber, M.; Cavaillé, J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009, 37, 3464–3473. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.; Royo, H.; Bortolin, M.L.; Lin, S.P.; Ferguson-Smith, A.C.; Cavaillé, J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004, 14, 1741–1748. [Google Scholar] [CrossRef]
- Liang, Y.; Ridzon, D.; Wong, L.; Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007, 8, 166. [Google Scholar] [CrossRef]
- Suh, M.R.; Lee, Y.; Kim, J.Y.; Kim, S.K.; Moon, S.H.; Lee, J.Y.; Cha, K.Y.; Chung, H.M.; Yoon, H.S.; Moon, S.Y.; et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004, 270, 488–498. [Google Scholar] [CrossRef]
- Bentwich, I.; Avniel, A.; Karov, Y.; Aharonov, R.; Gilad, S.; Barad, O.; Barzilai, A.; Einat, P.; Einav, U.; Meiri, E.; et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005, 37, 766–770. [Google Scholar] [CrossRef]
- Laurent, L.C.; Chen, J.; Ulitsky, I.; Mueller, F.J.; Lu, C.; Shamir, R.; Fan, J.B.; Loring, J.F. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells 2008, 26, 1506–1516. [Google Scholar] [CrossRef]
- Jairajpuri, D.S.; Almawi, W.Y. MicroRNA expression pattern in pre-eclampsia (Review). Mol. Med. Rep. 2016, 13, 2351–2358. [Google Scholar] [CrossRef] [PubMed]
- Dumont, T.M.F.; Mouillet, J.F.; Bayer, A.; Gardner, C.L.; Klimstra, W.B.; Wolf, D.G.; Yagel, S.; Balmir, F.; Binstock, A.; Sanfilippo, J.S.; et al. The expression level of C19MC miRNAs in early pregnancy and in response to viral infection. Placenta 2017, 53, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Sun, J.; Groome, L.J.; Wang, Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Endocrinol. Metab. 2013, 304, e836–e843. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Mouillet, J.F.; Chu, T.; Parks, W.T.; Sadovsky, E.; Knöfler, M.; Sadovsky, Y. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology 2014, 155, 4975–4985. [Google Scholar] [CrossRef]
- Ding, J.; Huang, F.; Wu, G.; Han, T.; Xu, F.; Weng, D.; Wu, C.; Zhang, X.; Yao, Y.; Zhu, X. MiR-519d-3p suppresses invasion and migration of trophoblast cells via targeting MMP-2. PLoS ONE 2015, 10, e0120321. [Google Scholar] [CrossRef]
- Jiang, L.; Long, A.; Tan, L.; Hong, M.; Wu, J.; Cai, L.; Li, Q. Elevated microRNA-520g in pre-eclampsia inhibits migration and invasion of trophoblasts. Placenta 2017, 51, 70–75. [Google Scholar] [CrossRef]
- Zhang, M.; Muralimanoharan, S.; Wortman, A.C.; Mendelson, C.R. Primate-specific miR-515 family members inhibit key genes in human trophoblast differentiation and are upregulated in preeclampsia. Proc. Natl. Acad. Sci. USA 2016, 113, e7069–e7076. [Google Scholar] [CrossRef]
- Fu, J.Y.; Xiao, Y.P.; Ren, C.L.; Guo, Y.W.; Qu, D.H.; Zhang, J.H.; Zhu, Y.J. Up-regulation of miR-517-5p inhibits ERK/MMP-2 pathway: Potential role in preeclampsia. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6599–6608. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Y.; Lu, H.; Wang, H.; Shi, X.; Shao, X.; Li, Y.X.; Zhao, Y.; Wang, Y.L. miR-518b Enhances Human Trophoblast Cell Proliferation Through Targeting Rap1b and Activating Ras-MAPK Signal. Front. Endocrinol. 2018, 9, 100. [Google Scholar] [CrossRef]
- Mong, E.F.; Yang, Y.; Akat, K.M.; Canfield, J.; VanWye, J.; Lockhart, J.; Tsibris, J.; Schatz, F.; Lockwood, C.J.; Tuschl, T.; et al. Chromosome 19 microRNA cluster enhances cell reprogramming by inhibiting epithelial-to-mesenchymal transition. Sci. Rep. 2020, 10, 3029. [Google Scholar] [CrossRef] [PubMed]
- Anton, L.; Olarerin-George, A.O.; Hogenesch, J.B.; Elovitz, M.A. Placental expression of miR-517a/b and miR-517c contributes to trophoblast dysfunction and preeclampsia. PLoS ONE 2015, 10, e0122707. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Rana, S.; Karumanchi, S.A. Preeclampsia: The role of angiogenic factors in its pathogenesis. Physiology 2009, 24, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Mutter, W.P.; Karumanchi, S.A. Molecular mechanisms of preeclampsia. Microvasc. Res. 2008, 75, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Leaños-Miranda, A.; Navarro-Romero, C.S.; Sillas-Pardo, L.J.; Ramírez-Valenzuela, K.L.; Isordia-Salas, I.; Jiménez-Trejo, L.M. Soluble Endoglin As a Marker for Preeclampsia, Its Severity, and the Occurrence of Adverse Outcomes. Hypertension 2019, 74, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Cronqvist, T.; Tannetta, D.; Mörgelin, M.; Belting, M.; Sargent, I.; Familari, M.; Hansson, S.R. Syncytiotrophoblast derived extracellular vesicles transfer functional placental miRNAs to primary human endothelial cells. Sci. Rep. 2017, 7, 4558. [Google Scholar] [CrossRef]
- Strub, G.M.; Kirsh, A.L.; Whipple, M.E.; Kuo, W.P.; Keller, R.B.; Kapur, R.P.; Majesky, M.W.; Perkins, J.A. Endothelial and circulating C19MC microRNAs are biomarkers of infantile hemangioma. JCI Insight 2016, 1, e88856. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433, correction in Nat. Cell Biol. 2015, 17, 104. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef]
- Iftikhar, H.; Carney, G.E. Evidence and potential in vivo functions for biofluid miRNAs: From expression profiling to functional testing: Potential roles of extracellular miRNAs as indicators of physiological change and as agents of intercellular information exchange. Bioessays 2016, 38, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Filipazzi, P.; Bürdek, M.; Villa, A.; Rivoltini, L.; Huber, V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin. Cancer Biol. 2012, 22, 342–349. [Google Scholar] [CrossRef]
- Fabbri, M. MicroRNAs and miRceptors: A new mechanism of action for intercellular communication. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20160486. [Google Scholar] [CrossRef] [PubMed]
- Delorme-Axford, E.; Donker, R.B.; Mouillet, J.F.; Chu, T.; Bayer, A.; Ouyang, Y.; Wang, T.; Stolz, D.B.; Sarkar, S.N.; Morelli, A.E.; et al. Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl. Acad. Sci. USA 2013, 110, 12048–12053. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.; Lennemann, N.J.; Ouyang, Y.; Sadovsky, E.; Sheridan, M.A.; Roberts, R.M.; Coyne, C.B.; Sadovsky, Y. Chromosome 19 microRNAs exert antiviral activity independent from type III interferon signaling. Placenta 2018, 61, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Krawczynski, K.; Ouyang, Y.; Mouillet, J.F.; Chu, T.; Coyne, C.B.; Sadovsky, Y. Unc-13 homolog D mediates an antiviral effect of the chromosome 19 microRNA cluster miR-517a. J. Cell Sci. 2020, 134, jcs246769. [Google Scholar] [CrossRef] [PubMed]
- Kambe, S.; Yoshitake, H.; Yuge, K.; Ishida, Y.; Ali, M.M.; Takizawa, T.; Kuwata, T.; Ohkuchi, A.; Matsubara, S.; Suzuki, M.; et al. Human exosomal placenta-associated miR-517a-3p modulates the expression of PRKG1 mRNA in Jurkat cells. Biol. Reprod. 2014, 91, 129. [Google Scholar] [CrossRef] [PubMed]
- Chaiwangyen, W.; Murrieta-Coxca, J.M.; Favaro, R.R.; Photini, S.M.; Gutiérrez-Samudio, R.N.; Schleussner, E.; Markert, U.R.; Morales-Prieto, D.M. MiR-519d-3p in Trophoblastic Cells: Effects, Targets and Transfer to Allogeneic Immune Cells via Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 3458. [Google Scholar] [CrossRef]
- Li, H.; Pinilla-Macua, I.; Ouyang, Y.; Sadovsky, E.; Kajiwara, K.; Sorkin, A.; Sadovsky, Y. Internalization of trophoblastic small extracellular vesicles and detection of their miRNA cargo in P-bodies. J. Extracell. Vesicles 2020, 9, 181226. [Google Scholar] [CrossRef]
- Morales-Prieto, D.M.; Chaiwangyen, W.; Ospina-Prieto, S.; Schneider, U.; Herrmann, J.; Gruhn, B.; Markert, U.R. MicroRNA expression profiles of trophoblastic cells. Placenta 2012, 33, 725–734. [Google Scholar] [CrossRef]
- Chim, S.S.; Shing, T.K.; Hung, E.C.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.; Lo, Y.M. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef]
- Miura, K.; Miura, S.; Yamasaki, K.; Higashijima, A.; Kinoshita, A.; Yoshiura, K.; Masuzaki, H. Identification of pregnancy-associated microRNAs in maternal plasma. Clin. Chem. 2010, 56, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Kotlabova, K.; Doucha, J.; Hromadnikova, I. Placental-specific microRNA in maternal circulation—Identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J. Reprod. Immunol. 2011, 89, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Hromadnikova, I.; Kotlabova, K.; Ondrackova, M.; Kestlerova, A.; Novotna, V.; Hympanova, L.; Doucha, J.; Krofta, L. Circulating C19MC microRNAs in preeclampsia, gestational hypertension, and fetal growth restriction. Mediat. Inflamm. 2013, 2013, 186041. [Google Scholar] [CrossRef]
- Yang, S.; Li, H.; Ge, Q.; Guo, L.; Chen, F. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Mol. Med. Rep. 2015, 12, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Higashijima, A.; Murakami, Y.; Tsukamoto, O.; Hasegawa, Y.; Abe, S.; Fuchi, N.; Miura, S.; Kaneuchi, M.; Masuzaki, H. Circulating chromosome 19 miRNA cluster microRNAs in pregnant women with severe pre-eclampsia. J. Obstet. Gynaecol. Res. 2015, 41, 1526–1532. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Kotlabova, K.; Ivankova, K.; Krofta, L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS ONE 2017, 12, e0171756. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fierro, M.L.; Garza-Veloz, I.; Gutierrez-Arteaga, C.; Delgado-Enciso, I.; Barbosa-Cisneros, O.Y.; Flores-Morales, V.; Hernandez-Delgadillo, G.P.; Rocha-Pizaña, M.R.; Rodriguez-Sanchez, I.P.; Badillo-Almaraz, J.I.; et al. Circulating levels of specific members of chromosome 19 microRNA cluster are associated with preeclampsia development. Arch. Gynecol. Obstet. 2018, 297, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Hromadnikova, I.; Dvorakova, L.; Kotlabova, K.; Krofta, L. The Prediction of Gestational Hypertension, Preeclampsia and Fetal Growth Restriction via the First Trimester Screening of Plasma Exosomal C19MC microRNAs. Int. J. Mol. Sci. 2019, 20, 2972. [Google Scholar] [CrossRef] [PubMed]
- Munjas, J.; Sopić, M.; Joksić, I.; Zmrzljak, U.P.; Karadžov-Orlić, N.; Košir, R.; Egić, A.; Miković, Ž.; Ninić, A.; Spasojević-Kalimanovska, V. Placenta-specific plasma miR518b is a potential biomarker for preeclampsia. Clin. Biochem. 2020, 79, 28–33, correction in 2020, 85, 57. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, M.; Yu, H.; Zhang, J.; Zhou, R. Circulating microRNAs as biomarkers for diagnosis and prediction of preeclampsia: A systematic review and meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 253, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Matsubara, Y.; Uchikura, Y.; Sugiyama, T. Pathophysiology of Preeclampsia: The Role of Exosomes. Int. J. Mol. Sci. 2021, 22, 2572. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.J.; Gray, W.D.; Hayek, S.S. Platelets confound the measurement of extracellular miRNA in archived plasma. Sci. Rep. 2016, 6, 32651. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuan, Y.; Cho, J.H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef] [PubMed]
- Founds, S.A.; Dorman, J.S.; Conley, Y.P. Microarray technology applied to the complex disorder of preeclampsia. J. Obstet. Gynecol. Neonatal Nurs. 2008, 37, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Cronqvist, T.; Erlandsson, L.; Tannetta, D.; Hansson, S.R. Placental syncytiotrophoblast extracellular vesicles enter primary endothelial cells through clathrin-mediated endocytosis. Placenta 2020, 100, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yao, C.; Guo, Z.; Zou, J.; Zhang, L.; Xiao, H.; Wang, D.; Yang, D.; Gong, X.; Zhu, J.; et al. Apparently low reproducibility of true differential expression discoveries in microarray studies. Bioinformatics 2008, 24, 2057–2063. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Meder, B.; Hart, M.; Ludwig, N.; Leidinger, P.; Vogel, B.; Galata, V.; Roth, P.; Menegatti, J.; Grässer, F.; et al. Prioritizing and selecting likely novel miRNAs from NGS data. Nucleic Acids Res. 2016, 44, e53. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Heinicke, F.; Zhong, X.; Zucknick, M.; Breidenbach, J.; Sundaram, A.; Flåm, S.T.; Leithaug, M.; Dalland, M.; Farmer, A.; Henderson, J.M.; et al. Systematic assessment of commercially available low-input miRNA library preparation kits. RNA Biol. 2020, 17, 75–86. [Google Scholar] [CrossRef]
- Raabe, C.A.; Tang, T.H.; Brosius, J.; Rozhdestvensky, T.S. Biases in small RNA deep sequencing data. Nucleic Acids Res. 2014, 42, 1414–1426. [Google Scholar] [CrossRef]
- Goldman, D.; Domschke, K. Making sense of deep sequencing. Int. J. Neuropsychopharmacol. 2014, 17, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.F.; Cowen, S.; Foy, C.A. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin. Chem. 2015, 61, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef] [PubMed]
- Garratt, H.; Ashburn, R.; Sopić, M.; Nogara, A.; Caporali, A.; Mitić, T. Long Non-Coding RNA Regulation of Epigenetics in Vascular Cells. Non-Coding RNA 2021, 7, 62. [Google Scholar] [CrossRef]
- Lanzillotti, C.; De Mattei, M.; Mazziotta, C.; Taraballi, F.; Rotondo, J.C.; Tognon, M.; Martini, F. Long Non-coding RNAs and MicroRNAs Interplay in Osteogenic Differentiation of Mesenchymal Stem Cells. Front. Cell Dev. Biol. 2021, 9, 646032. [Google Scholar] [CrossRef]
- Khandelwal, A.; Bacolla, A.; Vasquez, K.M.; Jain, A. Long non-coding RNA: A new paradigm for lung cancer. Mol. Carcinog. 2015, 54, 1235–1251. [Google Scholar] [CrossRef]
- Nie, L.; Wu, H.J.; Hsu, J.M.; Chang, S.S.; Labaff, A.M.; Li, C.W.; Wang, Y.; Hsu, J.L.; Hung, M.C. Long non-coding RNAs: Versatile master regulators of gene expression and crucial players in cancer. Am. J. Transl. Res. 2012, 4, 127–150. [Google Scholar]
- Louro, R.; Smirnova, A.S.; Verjovski-Almeida, S. Long intronic noncoding RNA transcription: Expression noise or expression choice? Genomics 2009, 93, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Graf, J.; Kretz, M. From structure to function: Route to understanding lncRNA mechanism. Bioessays 2020, 42, e2000027. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.; Liu, R.; Zhang, Y.W.; Zhang, Y.; Zhou, R.; Sun, J.; Lv, X.B.; Zhang, Z. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed. Pharmacother. 2019, 115, 108912. [Google Scholar] [CrossRef] [PubMed]
- Calviello, L.; Mukherjee, N.; Wyler, E.; Zauber, H.; Hirsekorn, A.; Selbach, M.; Landthaler, M.; Obermayer, B.; Ohler, U. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods 2015, 13, 165–170. [Google Scholar] [CrossRef] [PubMed]
- He, X.; He, Y.; Xi, B.; Dai, X.; Xue, X.; Lu, Y.; Shen, R.; Li, J.; Li, J.; Ding, H. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS ONE 2013, 8, e81437. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xie, X.; Lei, H.; Zou, B.; Xie, L. Identification of Key circRNAs/lncRNAs/miRNAs/mRNAs and Pathways in Preeclampsia Using Bioinformatics Analysis. Med. Sci. Monit. 2019, 25, 1679–1693. [Google Scholar] [CrossRef]
- Tong, J.; Zhao, W.; Lv, H.; Li, W.P.; Chen, Z.J.; Zhang, C. Transcriptomic Profiling in Human Decidua of Severe Preeclampsia Detected by RNA Sequencing. J. Cell. Biochem. 2018, 119, 607–615. [Google Scholar] [CrossRef]
- Long, W.; Rui, C.; Song, X.; Dai, X.; Xue, X.; Lu, Y.; Shen, R.; Li, J.; Li, J.; Ding, H. Distinct expression profiles of lncRNAs between early-onset preeclampsia and preterm controls. Clin. Chim. Acta 2016, 463, 193–199. [Google Scholar] [CrossRef]
- Cao, C.; Li, J.; Li, J.; Liu, L.; Cheng, X.; Jia, R. Long Non-Coding RNA Uc.187 Is Upregulated in Preeclampsia and Modulates Proliferation, Apoptosis, and Invasion of HTR-8/SVneo Trophoblast Cells. J. Cell. Biochem. 2017, 118, 1462–1470. [Google Scholar] [CrossRef]
- Zuo, Q.; Huang, S.; Zou, Y.; Xu, Y.; Jiang, Z.; Zou, S.; Xu, H.; Sun, L. The Lnc RNA SPRY4-IT1 Modulates Trophoblast Cell Invasion and Migration by Affecting the Epithelial-Mesenchymal Transition. Sci. Rep. 2016, 6, 37183. [Google Scholar] [CrossRef]
- Song, X.; Rui, C.; Meng, L.; Zhang, R.; Shen, R.; Ding, H.; Li, J.; Li, J.; Long, W. Long non-coding RNA RPAIN regulates the invasion and apoptosis of trophoblast cell lines via complement protein C1q. Oncotarget 2017, 8, 7637–7646. [Google Scholar] [CrossRef]
- Li, J.L.; Li, R.; Gao, Y.; Guo, W.C.; Shi, P.X.; Li, M. LncRNA CCAT1 promotes the progression of preeclampsia by regulating CDK4. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1216–1223. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Liu, Y.L.; Fei, K.L.; Li, P. Long non-coding RNA HOTAIR modulates the progression of preeclampsia through inhibiting miR-106 in an EZH2-dependent manner. Life Sci. 2020, 253, 117668. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, Y.; Ding, Y. Lnc-DC mediates the over-maturation of decidual dendritic cells and induces the increase in Th1 cells in preeclampsia. Am. J. Reprod. Immunol. 2017, 77, e12647. [Google Scholar] [CrossRef]
- Gao, W.L.; Liu, M.; Yang, Y.; Yang, H.; Liao, Q.; Bai, Y.; Li, Y.X.; Li, D.; Peng, C.; Wang, Y.L. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1). RNA Biol. 2012, 9, 1002–1010. [Google Scholar] [CrossRef]
- Xu, J.; Xia, Y.; Zhang, H.; Guo, H.; Feng, K.; Zhang, C. Overexpression of long non-coding RNA H19 promotes invasion and autophagy via the PI3K/AKT/mTOR pathways in trophoblast cells. Biomed. Pharmacother. 2018, 101, 691–697. [Google Scholar] [CrossRef]
- Liu, X.; Chen, H.; Kong, W.; Zhang, Y.; Cao, L.; Gao, L.; Zhou, R. Down-regulated long non-coding RNA-ATB in preeclampsia and its effect on suppressing migration, proliferation, and tube formation of trophoblast cells. Placenta 2017, 49, 80–87, correction in 2020, 91, 67. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, X.; Li, C.; Zhang, W.; Lv, Y.; Wang, L.; Wu, L.; Meng, L.; Fan, Y.; Ding, H.; et al. Down-regulated long non-coding RNA PVT1 contributes to gestational diabetes mellitus and preeclampsia via regulation of human trophoblast cells. Biomed. Pharmacother. 2019, 120, 109501. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lian, Y.; Zhang, Y.; Huang, S.; Zuo, Q.; Yang, N.; Chen, Y.; Wu, D.; Sun, L. The long non-coding RNA PVT1 represses ANGPTL4 transcription through binding with EZH2 in trophoblast cell. J. Cell. Mol. Med. 2018, 22, 1272–1282. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ge, Z.; Zhang, E.; Zuo, Q.; Huang, S.; Yang, N.; Wu, D.; Zhang, Y.; Chen, Y.; Xu, H.; et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017, 8, e3104. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, J.; Su, D.M.; Guan, L.N.; Mu, W.H.; Yu, M.; Ma, X.; Yang, R.J. lncRNA TUG1 modulates proliferation, apoptosis, invasion, and angiogenesis via targeting miR-29b in trophoblast cells. Hum. Genom. 2019, 13, 50. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, N.; Zhang, X.J. Long noncoding TUG1 promotes angiogenesis of HUVECs in PE via regulating the miR-29a-3p/VEGFA and Ang2/Tie2 pathways. Microvasc. Res. 2021, 139, 104231. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Li, Q.; Xu, Y.; Yu, X.; Zuo, Q.; Huang, S.; Chu, Y.; Jiang, Z.; Sun, L. Promotion of trophoblast invasion by lncRNA MVIH through inducing Jun-B. J. Cell. Mol. Med. 2018, 22, 1214–1223. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hou, Y.; Lv, N.; Liu, Q.; Lin, N.; Zhao, S.; Chu, X.; Chen, X.; Cheng, G.; Li, P. Circulating lncRNA BC030099 Increases in Preeclampsia Patients. Mol. Ther. Nucleic Acids 2019, 14, 562–566. [Google Scholar] [CrossRef]
- Luo, X.; Li, X. Long Non-Coding RNAs Serve as Diagnostic Biomarkers of Preeclampsia and Modulate Migration and Invasiveness of Trophoblast Cells. Med. Sci. Monit. 2018, 24, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Zhao, C.; Xu, M.; Sui, X.; Sun, L.; Liu, Y.; Su, M.; Wang, H.; Yuan, Y.; Zhang, S.; et al. Serum lncRNAs in early pregnancy as potential biomarkers for the prediction of pregnancy-induced hypertension, including preeclampsia. Mol. Ther. Nucleic Acids 2021, 24, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Guglas, K.; Bogaczyńska, M.; Kolenda, T.; Ryś, M.; Teresiak, A.; Bliźniak, R.; Łasińska, I.; Mackiewicz, J.; Lamperska, K. lncRNA in HNSCC: Challenges and potential. Contemp. Oncol. 2017, 21, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Qi, P.; Xu, M.D.; Ni, S.J.; Huang, D.; Xu, Q.H.; Weng, W.W.; Tan, C.; Sheng, W.Q.; Zhou, X.Y.; et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int. J. Cancer 2015, 137, 1128–1135. [Google Scholar] [CrossRef]
- Androvic, P.; Valihrach, L.; Elling, J.; Sjoback, R.; Kubista, M. Two-tailed RT-qPCR: A novel method for highly accurate miRNA quantification. Nucleic Acids Res. 2017, 45, e144. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).