Abstract
Legumes are a better source of proteins and are richer in diverse micronutrients over the nutritional profile of widely consumed cereals. However, when exposed to a diverse range of abiotic stresses, their overall productivity and quality are hugely impacted. Our limited understanding of genetic determinants and novel variants associated with the abiotic stress response in food legume crops restricts its amelioration. Therefore, it is imperative to understand different molecular approaches in food legume crops that can be utilized in crop improvement programs to minimize the economic loss. ‘Omics’-based molecular breeding provides better opportunities over conventional breeding for diversifying the natural germplasm together with improving yield and quality parameters. Due to molecular advancements, the technique is now equipped with novel ‘omics’ approaches such as ionomics, epigenomics, fluxomics, RNomics, glycomics, glycoproteomics, phosphoproteomics, lipidomics, regulomics, and secretomics. Pan-omics—which utilizes the molecular bases of the stress response to identify genes (genomics), mRNAs (transcriptomics), proteins (proteomics), and biomolecules (metabolomics) associated with stress regulation—has been widely used for abiotic stress amelioration in food legume crops. Integration of pan-omics with novel omics approaches will fast-track legume breeding programs. Moreover, artificial intelligence (AI)-based algorithms can be utilized for simulating crop yield under changing environments, which can help in predicting the genetic gain beforehand. Application of machine learning (ML) in quantitative trait loci (QTL) mining will further help in determining the genetic determinants of abiotic stress tolerance in pulses.
1. Introduction
1.1. Rationale
Legumes belonging to the Fabaceae family are consumed globally and are the second most important food crop after cereals, which are best complemented with the latter to constitute a balanced diet [1]. In some regions of the world, legumes are also utilized as fodder for cattle. Legumes are rich in proteins, vitamins, and minerals and provide bulk to the diet [2,3,4]. Due to their rich nutritional profile, their daily intake can help in reducing micronutrient deficiencies among people in developing countries, who are predominantly impacted by this hidden hunger [5]. Thus, legumes contribute in meeting global food security requirements. Legumes also serve a prospective role in conservative agriculture because of their capability to fix atmospheric nitrogen (N), which improves soil fertility. Early on, most of the legumes were considered to be orphans; however, recent decoding of major food legumes, such as mungbean [6], chickpea [7], common bean [8], soybean [9], pigeonpea [10], cowpea [11], and pea [12], has turned them into rich genomic resources.
Climate change is an unavoidable predicament aggravating abiotic stresses, ultimately threatening global food security by reducing crop yields by around 70% [13,14]. Abiotic stresses, e.g., water stress (e.g., floods and drought), extreme temperature conditions (e.g., heat, cold, and frost), salinity, acidic soils, and heavy metal toxicity, severely affect legume production. To thrive in such harsh conditions, plants counter with strong stress responses. Generally, plants’ stress tolerance is dynamic, involving signal transduction pathways at different regulatory levels to adjust metabolic changes [15,16,17]. These pathways are controlled by several genes, proteins, and post-translational modifications [18,19]. Drought, salinity, and temperature stresses are the major factors that reduce the yield of leguminous crops. These stresses have been aggravated due to the climatic changes over the last few decades [20]. Apart from the most prominent and commonly studied abiotic stresses, there are a few stresses that are more prominent in temperate latitudes that affect phenological abiotic mismatches, which restrict gene flow due to small-scale heterogeneity and affect plant variability. Some phenological mismatches are common in alpine and arctic tundra ecosystems [21]. A restricted gene flow was observed in the long-lived dwarf shrub Salix herbacea L. due to variation in the snowmelt timing [22]. Frost stress due to variability in snow cover duration and elevation affected the size and the vulnerability of alpine dwarf shrubs [23,24]. Additionally, variation in altitudinal gradients affected the distribution of Espeletia taxa [25] and S. herbacea [26]. Flooding in habitats of these mountainous terrains showed variability along with plant traits, such as plant height, plant pubescence, and the presence of aerenchyma, that provided adaptations to variability in alpine environmental conditions [27]. Further, variations in nutrient availability also affect different microhabitats. For example, in the case of S. herbacea, there was differential accumulation of nutrients due to plant–soil interactions [28]. This was due to the novel microbial communities that participated in biotic interactions with plants [29]. It is essential to understand the response mechanisms and their regulatory factors to improve pulse production in extremely harsh environments. Since the pathways are regulated at each stage of the central dogma, it is crucial to deploy integrated advanced genomic approaches together with gene editing/transgenic approaches. The former can be best exemplified in the form of ‘pan-omics’, which collaboratively utilizes metabolomic, proteomic, genomic, and transcriptomic data to uncover the precise mechanisms behind stress regulation.
With the emergence of techniques such as next-generation sequencing (NGS) and high-throughput genotyping [30], it is now possible to interpret the precise roles of proteins, genes, and metabolites in legumes. These technologies have also helped in the sequencing and assembly of genomic drafts of major legumes. The details of the genomic drafts of these major legumes are listed in Table 1. High-throughput genomics studies utilizing techniques such as genome-wide association studies (GWAS), genome skimming, genotyping by sequencing (GBS), single nucleotide polymorphism (SNP) chip genotyping, and whole-genome resequencing (WGRS) have been employed in many crops, including legumes, to elucidate the role of stress-responsive genes [31,32]. Further, targeted genome editing is also evolving over time for the development of elite cultivars. Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) as well as other site-directed nucleases, such as transcription activator-like effectors (TALEs) and zinc fingers (ZFs), have emerged as new tools for next-generation breeding [33].

Table 1.
List of published reference genomes of major legumes.
Pan-omics and genome editing for the production of climate-smart pulse crops are still new concepts because of the limited availability of genomic information for most of the legumes. Accelerating the development of pulse pan-genomes is therefore, needed for future applications. Based on the advancement of basic omics approaches, the development of some novel omics techniques is gaining momentum. Analysis of metabolic fluxes in the metabolome of an organism is progressing in the form of fluxomics, whereas regulomics is associated with the evaluation of regulatory factors, such as transcription factors (TFs), proteins, and regulatory genes, which are involved in the regulation of gene responses to various abiotic stresses. Likewise, ionomics, glycomics, glycoproteomics, phosphoproteomics, lipidomics, and secretomics represent the advanced omics techniques for studying abiotic stress physiology in different forms [37]. The advent of artificial intelligence (AI) and computer programming for simulations has introduced smart farming as a new facet of climate-resilient crop breeding. Advanced machine learning (ML) algorithms are now being used for crop modeling to obtain maximum yields. Combinations of GWAS and ML algorithms are now being used to detect genetic variants associated with complex abiotic stress tolerance traits [38]. Integration of ML with novel omics techniques will definitely benefit future pulse breeding programs. The overall integration of omics technologies and artificial intelligence pipelines for the molecular functional prediction of abiotic stress tolerance is shown in Figure 1.
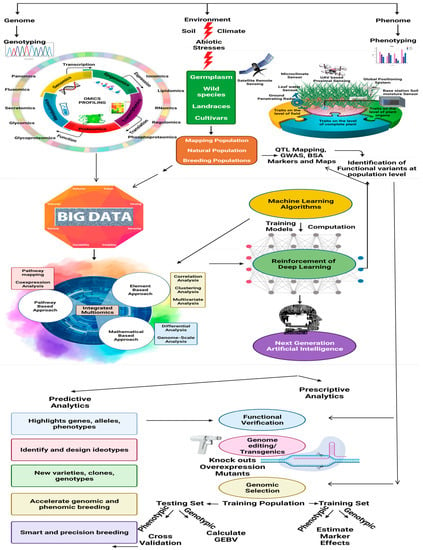
Figure 1.
Overall representation of different combinations of omics technologies and artificial intelligence pipelines to predict molecular function-induced abiotic stress. This image was generated by BioRender.com.
1.2. Objectives
The present review highlights the opportunities associated with: (i) novel ‘omics’ approaches; (ii) pan-omics approaches; (iii) multi-omics integration; and (iv) AI for smart farming that can handle the climate exigency and its adverse effects on legume production. This will help in the generation of simulation models for future legume breeding and in sustainable agri-production.
2. Methods
A systematic review was designed to understand the role of novel omics approaches independently or in association with artificial intelligence in the amelioration of abiotic stresses in legumes and for devising future strategies. The checklist reported in Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) was followed for an organized assembly of relevant data and information [39]. A comprehensive literature search was performed to identify relevant research articles. More specifically, the papers published until the end of February 2021 in scientific journals were included in this systematic review. Four hundred and twenty seven journals were sorted and added to the list of master journals. We used web search engines such as Google Scholar and Pubmed and, in some cases, websites such as FAO and Knowpulse to search for the information pertaining to legumes’ genomes and production. We searched for the terms “Legumes OR Fabaceae” AND “Omics OR Artificial Intelligence” in titles, abstracts, and/or keywords, which were restricted to articles in the English language, and no date restrictions were imposed. In Google Scholar, articles were sorted by relevance, which included citations, to provide 250 search results. Pubmed yielded 399 results with full-text availability and ‘randomized control trial’ and ‘review’ as the article types. No other relevant article types, such as books and documents, meta-analyses, and systematic reviews, provided any search results. Some studies on plants other than legumes were discarded, although introductory studies on other crops for the development of a particular technology (such as a novel omics technology) were included where necessary. The last search was run on 28 February 2021.
Information on the articles, including the title, abstract, keywords, names of authors, affiliations, journal name, and year of publication, was exported to MS excel. Highly relevant titles and abstracts were then filtered by two independent authors. Thereafter, full-text screenings of these articles for specificity towards the current topic were performed by two reviewers independently. Suggestions, disagreements, and information made by the reviewers to enhance the quality of the present review article were taken into consideration and added to or removed from the main body of the manuscript. The views of both the reviewers were taken into consideration to achieve a consensus. We included all scientific papers that used novel omics approaches or advanced scientific innovations together with any application of artificial intelligence in basic or applied studies on legumes.
3. Novel ‘Omics’ Approaches for Future Pulse Breeding Programs
3.1. Ionomics
The concept of an ‘ionome’ was first defined by Lahner et al. [40] as the metals, non-metals, and metalloids present in an organism. Later, the term ‘ionome’ was extended to ‘metallome’ [41] to refer to a collection of biologically important non-metals, such as N, phosphorus (P), and sulfur (S). Ionomics is the study of the complete ionome of a tissue/an organism, involving quantification of all elemental constituents in reaction to physiological processes or changes [42]. Ions have a substantial role in the maintenance of a plant’s homeostasis under different environmental conditions. Similarly, ion transporters are important for proper functioning of metabolic pathways as well as in stress regulation. The gene regulatory networks involved in the synthesis of these ions will surely help in furthering our knowledge about the role of ionome in the stress response. An extensive analysis of the Arabidopsis genome revealed that around 25,000 genes are engaged in regulating its ionome [40]. Plant ionomics has been extensively reviewed by Baxter (2010) [43], Huang and Salt [44]. A searchable database of more than 22,000 plants mutagenized with fast neutrons or Transfer-Deoxyribo nucleic acid (T-DNA) insertional lines is available at http://hort.agriculture.purdue.edu/Ionomics/database.asp (accessed on 12 January 2021). Similarly, ionome data of 975 soybean lines mutagenized using Nitroso-N-Methylurea (NMU) can be obtained from http://www.ionomicshub.org/home/PiiMS/dataexchange [45]. The Arabidopsis ionome project (http://www.ionomicshub.org, accessed on 3 August 2021)with the leaf ionome of more than 125,000 plants is the largest ionomic database till date [43].
The ionomics approach has been extensively used in model legumes such as Lotus japonicus [46] and food legumes such as soybean [45,47,48] when compared with other pulse crops. Utilizing this approach, mutants with an altered seed composition were identified in field-grown soybean [30]. Thereafter, they performed GWAS of ionomics traits in the soybean germplasm [47]. A set of 1653 soybean accessions were analyzed for the concentration of 20 elements in the seeds along with their weight. GWAS using oySNP50k chip data and 21 phenotypes showed a multilocus mixed model containing 29 SNPs for iron in one of the three Urbana locations in the year 2009 [47]. Similarly, seed ionome variation in 90 diverse soybean lines was also analyzed [48]. Recent developments in ionomics have provided novel ways to obtain a detailed account of the micro- and macronutrients as well as the elemental composition of legume grains in a rapid and cost-effective manner. The ionome data, thus, can be utilized for studies pertaining to the bioavailability of micronutrients in staple pulses. This way, ionomics can be used to achieve global food security and also to reduce the ‘hidden’ hunger associated with micronutrient deficiencies. The utilization of ionomics for the evaluation of abiotic-stress-responsive ion transporters, genes, ions, and elements requires extensive knowledge of the gene regulatory networks involved in ion homeostasis. Amalgamation of ionomics with other pan-omics approaches, such as proteomics and metabolomics, would increase the opportunities for studying the effects of abiotic stresses in legumes and their applications in producing climate-resilient legumes.
3.2. Epigenomics
Epigenomics is gaining importance as an alternate tool for germplasm enhancement. Epigenetic changes that are heritable in nature and affect the cellular processes of an organism form the basis of this tool. This includes modifications such as (de)methylation and (de)acetylation of histones or DNA that do not affect the actual DNA sequence but profoundly affect the gene’s functions [49]. Effects of abiotic stresses on the methylome of many pulse crops have been studied. For e.g., drought stress increased DNA methylation of drought-responsive genes in faba bean and pea [50,51]. Rakei et al. studied the effects of prolonged cold stress on chickpea, which induced DNA demethylation in cold-tolerant genotypes [52]. Similarly, Song et al. reported the consequent effects of salt stress on the epigenome of soybean and found changes in DNA methylation patterns together with histone modifications in salt-stress-responsive transcription factor genes [53]. Liang et al. found that, under continuous cropping stress, DNA demethylation occurred in tolerant soybean genotypes that was consistent with increased expression of demeter-like (DML) and repressor of silencing 1 (ROS1) genes [54]. Wu et al. reported that salinity induced crosstalk between histone methylation and histone acetylation in soybean [55]. In chickpea, salt and drought stresses activated CaHDZ12, a homeodomain leucine zipper (HD-Zip) TF, with acetylation of H3K9ac in the promoter region [56]. Awana et al. found hypermethylation of stress-responsive genes under salinity stress leading to upregulation of salinity-responsive genes in pigeonpea [57]. On the other hand, Chen et al. found hypermethylation of long non-coding ribonucleic acids (lncRNAs) leading to salinity stress tolerance in soybean [58]. Contrary to these studies, increased salinity was found to inactivate some stress-responsive genes in soybean, which was caused by increased deposition of H3K27me3 [59].
Plants gain an epigenetic memory as a result of environmental interactions and pass it on to the next generation. The trans-generational inheritance of epimarks can thus be exploited for crop improvement programs. This involves the use of epialleles, recombinant inbred lines (RILs), and epigenetic quantitative trait loci (epiQTLs) to breed for abiotic stress resistance [60]. Schmitz et al. [61] exploited the epigenetic inheritance of local methyl quantitative trait loci (QTLs) in a soybean RIL population and utilized them to study methyl variations contributing to phenotypic variations over generations. In the same crop, Raju et al. [62] devised an epigenetic breeding strategy utilizing isogenic memory lines crossed to the wild type. The study exploited the amenability of MutS HOMOLOG1 (MSH1), which is responsible for developmental changes such as modulation of defense, the abiotic stress response, and the production of phytohormones, for inducing agronomically important epigenetic variations in soybean. The derived epi-populations of soybean also showed reduced epitype-by-environment (e × E) interactions, representing improved yield stability under changing environmental conditions. Such epigenetic breeding programs can be exploited in other pulse crops for enhancing yield under changing environments.
Comparative epigenomics is an emerging field that provides insights into gene and genome evolution in a similar manner to comparative genomics. Epigenetic mechanisms of gene regulation under abiotic stress may differ between species or may be conserved. Comparative epigenetics is used to understand the evolutionary conservation of the epigenetic regulation of biological functions by comparing epimarks between species [63]. This technique was used to compare the epigenomes of two closely related legumes, namely pigeonpea and soybean. The two genera diverged ~23 million years ago (mya) accompanied by a whole-genome duplication in the latter [64]. The study exploited gene body methylation (GbM) and gene expression patterns to reveal the conservation of nitrogen-metabolism-related genes in the two legumes. Similarly, in another study, methylomes of soybean and common bean were compared to add to the epigenetic resources for leguminous crops [65]. These two legumes share a whole-genome duplication event at around 56.5 mya followed by a genus-specific (Glycine) polyploidy event at around 10 mya. Studies on the application of epigenetic breeding in legumes are sparse due to the non-availability of genomic resources. Exploitation of naturally occurring epialleles will fast-track the development of alternate germplasms in orphan legumes with limited genomic information.
3.3. Fluxomics
Gathering information on genetic and metabolic regulation through pan-omics has become much easier; however, linking the gathered information to obtain a meaningful crux is difficult. Therefore, combined studies of fluxes through major metabolic pathways controlling the stress response are essential. This necessity has given rise to the study of metabolome-wide fluxes, called fluxomics. This novel omics approach provides the functional output of the cellular machinery involved in stress regulation. Fluxomics can be performed in various ways, including metabolic flux analysis (MFA) and flux balance analysis. The former is concerned with understanding metabolism at the system level under the influence of the environment, whereas the latter is a mathematical model of the metabolism in genomic-scale rebuilding of metabolic networks. MFA can generate metabolic maps that provide details about the metabolic networks involved in the environmental response and represent detailed metabolic phenotypes. Iyer et al. [66] prepared a metabolic flux map from soybean cotyledons to study the consequences of temperature variation for oil and protein biosynthesis using 13 Carbon (C) MFA. The knowledge obtained from metabolic networks can be utilized in the preparation of kinetic models for predicting the effects of environmental factors on genetic changes. Predictive modeling based on fluxomics has been successfully employed in crops such as maize [67] and Brassica napus [68]; however, studies on legumes are limited [69]. Moreira et al. developed a metabolic model highlighting metabolic fluxes in soybean seedlings during germination [70]. Similarly, Kannan et al. predicted the cumulative effects of an increase in atmospheric carbon dioxide (CO2) on the photosynthesis of soybean using a metabolic model based on gene regulatory networks and metabolic pathways [71]. Fluxomics delineates the key metabolic steps and processes by which fluxes are affected by environmental stresses. Therefore, fluxomics can also be employed to reconstruct metabolic networks in plants for metabolic engineering applications.
3.4. RNomics
RNomics is a new omics approach that involves the study of non-coding RNAs, e.g., micro ribonucleic acids (miRNAs) and lncRNAs. MiRNAs are believed to be engaged in stress response regulation in plants. Using NGS, four legume-specific miRNAs (miR5213, miR5232, miR2111, and miR2118) were discovered in chickpea libraries constructed and sequenced for fungal infection, salt treatment, and control conditions [72]. Multiple miRNAs responded under both biotic and abiotic stresses, suggesting the presence of crosstalk between stress-responsive pathways [72]. Barrera-Figueroa et al. [73] reported miRNAs that might have played significant roles in drought tolerance. Likewise, using a homology-based search, Kohli et al. [72] identified various conserved and new miRNAs associated with gene regulation under salt and wilt stress in chickpea.
LncRNAs make up a substantial proportion of non-coding RNAs and are engaged in a variety of biological operations. In one study, PLncPRO, a novel tool, was utilized for predicting lncRNAs in plants using transcriptome data, which revealed a total of 3714 (for drought) and 3457 (for salinity) high-confidence lncRNAs in chickpea [74]. This tool is based on ML and utilizes random forest algorithms to classify coding and long non-coding transcripts. The tool is suitable for plants and has better prediction accuracy compared with existing tools.
3.5. Glycomics, Glycoproteomics, and Phosphoproteomics
Glycomics is a comprehensive and developing scientific field that is based on defining the functional and structural roles of glycans in biological systems. Comprehensive knowledge of glycomes is important for understanding biological pathways as glycan modifications are critical to these pathways. The shocking complexity of the glycome, loosely defined as the collection of glycans expressed in a cell/an organism, has resulted in various challenges that must be overcome [75]. Recent advances in mass spectrometry as well as cell and molecular biology tools have helped us address the challenges posed by glycomics. Glycan microarrays are useful in the identification of glycan recognition determinants of glycan protein binding in a system. It is also useful in understanding the functions of glycans and their signaling in a cell or an organism. Moller et al. [76] profiled cell wall glycans in Arabidopsis by utilizing a novel technique based on microarrays called comprehensive microarray polymer profiling (CoMPP).
Accurate and high-resolution glycomes can allow for the assignment of an individual glycan molecule that is expressed on a particular glycoprotein. The study of such glycoproteins is called glycoproteomics [77]. The larger the number of glycosylation sites on a protein, the more complex and time consuming the analysis is. In addition, it will require a large amount of sampling material. Advanced techniques for the fragmentation and identification of glycans, such as electron capture dissociation (ECD), ion-trap mass spectrometry (MS), and collision-induced dissociation (CID), have increased the accuracy and feasibility of allocating glycans to specific amino acid sites in a collection of glyopeptides [78]. However, techniques for allocating glycans to specific amino acid sites remain understudied in plants.
Glycoproteomics can unveil the role of protein glycosylation in pulses under stress conditions. In the case of soybean, it was revealed that flood stress negatively impacted the N-glycosylation of functional proteins involved in stress regulation. In contrast, glycoproteins involved in glycolysis were found to be activated [79]. Protein phosphorylation is a key signaling mechanism in the plant abiotic stress response. Phosphoproteomics and glycoproteomics were exploited to study changes under stress conditions in chickpea and soybean [80,81]. Apart from novel molecular techniques, bioinformatics tools focused on glycomics are gaining importance as a new scientific discipline called glyco-bioinformatics. Glyco-bioinformatics utilizes algorithms to study and identify glycans together with their regulation and functions in a system. Recently, Showalter et al. developed a program called BIO OHIO 2.0 to detect hydroxyproline-rich glycoproteins (HRGPs) in the poplar cell wall as well as repeating amino acid sequences, signal peptide sequences, HRGPs, and glycosylphosphatidylinositol lipid anchor addition sequences in other plant species [81]. Similar tools can also be utilized to develop screening platforms for pulses under different stress conditions. Therefore, it is imperative to develop techniques to study protein modifications by glycans in plant cells in order to develop alternate strategies for breeding programs for the enhancement of stress tolerance.
3.6. Lipidomics, Regulomics, and Secretomics
Apart from proteins, lipids also play a significant role in stress regulation by maintaining cell wall dynamics under changing environmental conditions. The lipidome, which comprises the lipids expressed in a system, is studied as a subcategory of the metabolome, but its immense importance to cell regulation has made it an emerging scientific discipline [82]. On the other hand, a regulome can be defined as the whole set of the regulatory components present in an organism, including transcription factors, proteins, and mRNAs, which are known to be involved in stress response generation in plants. A few searchable databases are available for analyzing plant regulomes, including Plant Regulomics Portal (PRP) [83] and Plant Regulomics [84], which provide detailed information on transcription factors, small ribonucleic acids (sRNAs), DNA methylation, regulatory elements, gene networks, etc.
Similarly, a plant’s secretome is composed of a group of proteins released into the extracellular matrix that represents the plant’s interaction with its environment [85]. The plant secretome can reveal significant information regarding stress regulation, protein–protein interactions, and defense response generation in a changing environment. Apart from proteins released into the extracellular matrix, protein modifications under abiotic stress also reveal the cellular machinery and cell-to-cell communication in a changing environment. Some of the novel omics technologies described above have been utilized for the enhancement of tabiotic stress tolerance in some legume crops as presented in Table 2.

Table 2.
New omics technologies for pulse breeding.
4. Pan-Omics Approaches
Modern biotechnological tools, such as mutagenic breeding, marker-assisted breeding, and transgenic breeding, help in combating the bottlenecks of conventional plant breeding strategies, such as the non-availability of natural resistance and sexual incompatibility in some crops. These can also be utilized to understand the molecular mechanisms of the adaptive response towards abiotic stress(es) in legumes. Genome sequence information is invaluable to the application of next-generation breeding tools in any organism, but it cannot answer some queries related to the gene functions, biochemical pathways, and gene regulatory networks activated during the stress response. Therefore, a more comprehensive approach is required to study the intricate mechanism of the stress response in plants, which should include qualitative and quantitative analyses of gene functions. The knowledge obtained by studying the complex regulatory pathways can be applied in marker-assisted selection (MAS) and transgenic breeding programs for ameliorating the stress tolerance in legumes. Pan-omics integrates the complex omics datasets arising from different omics platforms that can facilitate the improvement of abiotic stress tolerance in crops via precision breeding. The recent progress in pan-omics approaches has remarkably contributed to an enhanced comprehension of the genetic and molecular bases of abiotic stress response generation in many leguminous plants [92].
4.1. Genomics
Genomics can be defined as the study of structural, functional, and evolutionary aspects of an organism’s genome. It includes determination of the whole DNA sequence and in-depth genome mapping of an organism. With the advent of NGS and other molecular biology techniques, a large amount of genomics data is available for legumes. Genome sequencing of legume species such as Lotus japonicus, Glycine max, and Medicago truncatula has already been accomplished [93]. Comparative genomics of these legume crops has revealed key regulatory networks of genes involved in adaptation to stress and crop productivity [94]. Abiotic-stress-related productivity losses in orphan legumes can be managed well using genomic data from the sequenced model legumes. The genomics approach can be linked to marker-assisted backcrossing (MAB) programs for easy manipulation of QTLs associated with stress tolerance and yield parameters. Molecular markers identified using genomics can thus be used in genomics-assisted breeding (GAB) programs, which have higher accuracy than conventional breeding practices [95]. Some of the QTLs identified for various abiotic stresses in legumes are presented in Table 3. Functional genomics techniques, such as insertional mutagenesis, gene overexpression studies, targeted induced local lesions in genomes (TILLING), and gene silencing, play an important role in developing an understanding of the complex gene regulatory networks associated with stress response generation, stress tolerance, and adaptation towards stress in plants. Functional validation of the large amounts of genomics data generated from experiments can be achieved by utilizing reverse genetics and gene silencing approaches such as RNA interference (RNAi), TILLING, and virus-induced gene silencing (VIGS) [96].

Table 3.
QTLs identified for various abiotic stresses in legumes.
4.2. Transgenomics
Transgenomics, also known as transgenic technology, is a popular, targeted gene-based technique that provides valuable insights into gene regulation under stress conditions. Foreign genes coding for important agronomic traits from different sources such as plants, animals, and microbes are transferred to the targeted organism’s germline. Many novel phenotypes are developed using transgenomics [126,127]. Transgenic technologies have been employed to elucidate the function of stress-responsive genes in many legumes, such as chickpea [128,129] and soybean [130]. Orphan legumes with limited genetic resources are often utilized in transgenomics for delineating the roles of unknown genes by expressing them in other crops (Table 4).

Table 4.
Genes and transcription factors (TFs) from different pulse crops overexpressed to generate improved traits or abiotic-stress-tolerant transgenic plants.
Several transgenic pulse crops with varying responses to different abiotic stresses have been developed. In transgenic chickpea, miR408 was overexpressed, which resulted in miRNA (miR4080)-induced gene regulation that improved its drought tolerance [150]. The transgenic approach was utilized to develop salinity-tolerant lentils expressing the transgenic DREB1 gene [151] and mung bean expressing the Arabidopsis antiporter (NHX1) gene [152]. Additionally, by co-expressing the Arabidopsis antiporter (NHX1) and bar genes in mung bean, Kumar et al. developed salinity-, herbicide-, and oxidative- stress-resistant lines [153]. Many studies have utilized the expression of Arabidopsis genes in soybean, such as the AtMYB44 gene, which resulted in improved drought and salinity tolerance [154] and AtΔKinase gene, which resulted in improved salt tolerance [155]. When the mung bean antiporter gene VrNHX1 was overexpressed in transgenic cowpea, it delivered increased salinity tolerance [156]. Several studies exploited stress-responsive genes from other food crops, such as cereals and vegetables, to improve the overall productivity of legume crops. Kwapata et al. [157] created a drought-tolerant common bean crop using Hordeum vulgare’s late embryogenesis abundant (LEA) protein HVA1. Likewise, Singh et al. utilized the rice DNA helicase (OsRuvB) gene to confer salinity tolerance in pigeonpea [158]. Similarly, Hanafy et al. heterologously expressed the potato gene PR10a in faba bean to enhance its salinity and drought tolerance [159]. Transgenic approaches hold a great deal of potential in the development of climate-smart crops, but the lack of proper legislation and the lack of their application in commercial breeding are holding them back from conquering these applications.
4.3. Transcriptomics
Transcriptomics is a powerful tool used to quantify gene expression and can provide a precise depiction of the gene expression in a target cell or tissue. Transcriptomics can reveal the gene regulatory networks and candidate genes engaged in abiotic stress response generation, which can be utilized for legume breeding. With the discovery of high-throughput technologies, the deduction of comprehensive transcriptomic data can be executed using serial analysis of gene expression (SAGE) and microarrays. Differential expression of genes (DEGs) can be determined using ribonucleic acid sequencing (RNA-seq) data. A recently developed technique called digital gene expression (DGE) for quantitative estimation of gene expression can also be used. RNA-seq analysis is a cost-effective, high-throughput sequencing technique that makes it possible to analyze large amounts of transcriptomic data. This technique offers several advantages over microarray technology as it does not require genomic information for designing probe sets and can identify novel transcripts [160]. Many studies have exploited this technique for elucidating the gene regulatory networks involved in abiotic stress tolerance in pulse crops (Table 5). Utilizing the NGS approach, a transcriptome atlas has been developed for soybean under drought-stressed conditions [161]. Comparative transcriptomic analysis has described the transcriptional changes in both drought-tolerant and drought-sensitive varieties of soybean [162,163]. Diverse sets of common bean genotypes that were resistant to biotic and abiotic stresses, such as aluminum toxicity, heat, drought, and low phosphorous, were assessed for parental polymorphisms, genetic diversity, and genetic and genomic association mapping using single nucleotide polymorphisms (SNPs) as a marker system, which were derived from Sanger sequencing and Illumina’s GoldenGate technology [164,165,166,167]. Das et al. used metabolomic profiling to reveal that sugar metabolism, nitrogen metabolism, and phytochemical metabolism are of prime significance under water deficit conditions in soybean [168]. From a transcriptomic analysis, Singh et al. identified putative candidate genes expressed under drought stress at the seedling stage in lentil [169], whereas dehydration-responsive proteins were identified by Pandey et al. in chickpea [170]. Molina et al. investigated transcriptomes of chickpea under drought stress using SuperSAGE and deep SuperSAGE and identified 80,238 tags representing 17,493 unique transcripts [171]. Root transcriptome analysis of oxylipin synthesis genes in chickpea unveiled the expeditious induction of jasmonate in roots under drought conditions [172]. Application of RNA-seq for understanding the genes expressed during the stress response will benefit future pulse breeding programs.

Table 5.
RNA-Seq for transcriptome profiling of pulse crops under abiotic stress(es).
The RNA-seq data or microarray data extracted from transcriptome analyses of various crops are used to make high-resolution gene expression atlases (GEAs). GEAs provide information regarding the expression of mRNAs and other important proteins involved in certain biological functions. They act as a valuable resource for studying the expression of genes and proteins engaged in developmental functions as well as in the abiotic stress response. Several GEAs have been developed in pulse crops (Table 6). Apart from GEAs, many transcriptome databases have also been made available for different pulse crops; for example, SoySeq (http://soybase.org/), SoyPLEX (http://www.plexdb.org/plex.php?database=Soybean), and the Chickpea Transcriptome Database (CTDB) (http://www.nipgr.res.in/ctdb.html, accessed on 3 August 2021) [187,188,189]. These extensive transcriptome databases can be used to retrieve data regarding the genes expressed in different tissues in different biological processes under different conditions.

Table 6.
High-resolution gene expression atlases (GEAs) for different pulse crops.
4.4. Proteomics and Metabolomics in Abiotic Stress Mitigation
Apart from changes in genes and mRNAs during abiotic stress, plants’ metabolomes and proteomes are also greatly impacted due to these stresses since they are actively involved in defense mechanisms against different stresses [196]. The proteome of an organism, which acts as a bridge between the transcriptome and the metabolome, reflects the actual state of the cellular response better than the DNA markers. The cellular mRNA levels represented by the transcriptome are not accurate depiction of the protein expression as proteins generally undergo post-translational modifications that influence the actual function of proteins [197]. These proteins are of significance to signal transduction pathways and are involved in stress adaptation processes, stress repair mechanisms, etc. Thus, they assist the plant with its recovery from a stress injury and help with its survival under stress [198]. On the other hand, metabolites are a reflection of the gene expression and interactions responsible for gene regulation under stress conditions and have close relations to the phenotype rather than the mRNA or proteins [199]. Of all the different omics technologies, metabolomics is the most cross-functional and reflects most of the processes as they are [196]. Furthermore, metabolic pathways are usually involved in highly complex networks and never function alone, which implies that interrupting a single metabolic pathway, could have adverse effects on other pathways, resulting in damaging traits in the modified plant. Hence, comprehensive analyses that elucidate the metabolic networks involved in the growth and development of plants under varying environmental conditions are very important. The molecular phenotypes of legumes under abiotic stresses have been studied by using proteomics and metabolomics as presented in Table 7.

Table 7.
Application of proteomics and metabolomics in abiotic stress mitigation in pulse crops.
5. Multi-Omics Integration (MOI) for Future Pulse Breeding
Across all disciplines of biology, the rapid development of high-throughput data generation techniques has allowed us to conduct multi-omics-based systems biology research. The data generated from transcriptomics, metabolomics, and proteomics can provide insights regarding the expression of transcripts, metabolites, and proteins, respectively. However, systematic multi-omics integration (MOI) of such data can comprehensively annotate, assimilate, and model these large datasets to provide meaningful, detailed information. Integration of omics data from various platforms together with novel omics approaches can help in bridging the genome-to-phenome gap in crop plants and ultimately help in identifying the right phenotype based on the genetic contribution for breeding purposes [211,212]. The integration of different omics techniques for improving abiotic stress tolerance in legumes is presented in Figure 2.
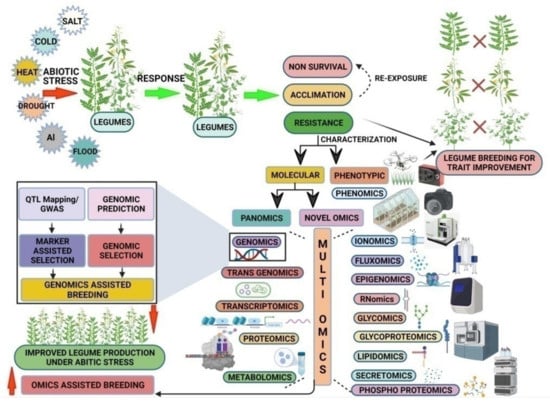
Figure 2.
Integrated omics approaches for improving abiotic stress tolerance in legumes. QTL, Quantitative Trait Loci; GWAS, Genome-Wide Association Study. This figure was created by Biorender.com.
Large NGS-derived genomic datasets and MOI approaches have substantially contributed towards increasing our knowledge of living organisms at the molecular level. Furthermore, translational genomics (TG) can be used to bridge the information gap between model systems and relatively understudied crop plants. The paramount aim of crop breeding is to achieve the maximum genetic gain of desirable traits in crop genomes in a cost- and time-effective manner. The TG technique has recently been utilized in some of the major legume crops [213].
Recently, GWAS analysis has gained immense popularity due to its ability to find genes, genomic loci, and SNP/InDels in genomes that are associated with beneficial crop traits [214]. Sequencing and/or array-based GWAS tools are making it possible to accurately predict/identify the alleles that are directly linked to particular phenotypic features, which is beyond the reach of map-based QTL analyses. WGRS can reveal genome-wide nucleotide variations, which can be further used for GWAS analyses. Moreover, the development of a high-throughput phenotyping system (HTPS) is imperative for phenotype-associated genomic analyses. Based on their syntenic relationships, the information derived from HTPS can be used for closely related plant genomes. These multi-dimensional and omics-driven techniques can assist with deriving useful information from multi-species phenotypic annotations linked to complex traits. Multi-omics platforms have been integrated together in some legumes to improve abiotic stress tolerance as presented in Table 8.

Table 8.
Multi-omics integration for improved abiotic stress tolerance in pulse crops.
6. Smart Farming: Artificial Intelligence (AI)-Based Pulse Breeding for Climate Resilience
The selection of cultivars with the best traits, especially under stress conditions, requires the modeling of genomics and phenomics data in such a manner that can provide the best output with the minimum cost and effort. MOI data are multidimensional, heterogeneous, and complex data that require advanced solutions for their application in plant breeding technologies. With the advancement of AI technologies, the development of climate-smart crop varieties with enhanced yield can enhance the tolerance/resistance to multiple abiotic stresses and can produce higher genetic gains in less time [222]. A combination of phenotypic, genotypic, and environmental data can reflect a plant’s stress response profile thoroughly; however, due to the complexity of the phenotypic plasticity in changing environments, obtaining meaningful information from integrated data is difficult as it is burdened by the genotype-to-phenotype (GP) gap. Intensive phenotyping involving concurrent comparative phenotypic measurements under changing environmental conditions is required to compensate for unapproachable factors such as the creation of identical growth conditions that are impossible to repeat. The coupling of such measured data with next-generation AI tools will diminish the bias arising from the GP gap. Negin and Moshelion [223] devised a strategy for screening drought-tolerant crops based on the use of a physiology-based high-throughput functional phenotyping system (HFPS) in combination with the soil–plant–atmosphere-continuum (SPAC), which can be used to measure the plant’s response to continuous and fluctuating environmental conditions. The use of a HFPS along with GWAS can result in a better understanding of gene characteristics under changing environments as well as in the development of novel genetic resources for pulse breeding. High-throughput phenotyping in changing environments has also been adopted successfully in certain legumes as presented in Table 9.

Table 9.
Automated phenotyping platforms for screening pulse crops in changing environments.
6.1. Machine learning (ML)-Enabled Genomic Selection, QTL Mining, GWAS, and Functional Prediction for Pulse Breeding
Over the years, GWAS have identified thousands of important genes associated with the stress response. However, due to the complex nature of stress response mechanisms in plants, these responses have been reattributed to multiple interacting genetic variants that are usually ignored in GWAS. ML algorithms can be used to detect these genetic variants. Zhang et al. [38] used a ML-facilitated image phenotyping approach to study the genetic basis of abiotic-stress-related iron deficiency chlorosis (IDC) in soybean. The generated data were subsequently utilized in genomic prediction and GWAS analyses to identify a previously described locus and a new locus containing a gene homolog engaged in iron acquisition. In another study, Naik et al. [230] reported an end-to-end phenotyping approach for soybean stress severity phenotyping that emphasizes IDC-severity-indifferent field plots. The high-throughput framework helped with the digital analysis of stress traits in real-time, identified markers, helped with genomic selection (GS)-based prediction, and increased the rate of genetic gain, which has stress scouting applications in plant breeding as illustrated in the figures of previous works reported by Libbrecht and Noble [231], Schrider and Kern [232], and Cortés et al. [233]. Liu et al. [234] used deep learning technology to predict quantitative phenotypes and to discover markers associated with them. The deep learning framework was based on convolutional neural networks (CNNs), which were used to predict the quantitative traits from SNPs and achieved more accurate results. Similarly, artificial neural networks (ANNs) have been employed for GS-based prediction modeling in common bean [235]. The genotype Aporé, which was studied using ANNs, was recommended for use in unfavorable environments because of its grain yield and high phenotypic stability even under unfavorable conditions. Examples of the use of different machine learning approaches, such as convolutional neural networks, deep belief networks, multivariate Poisson deep learning, multilayer perceptrons, probabilistic neural networks, and radial basis function neural networks, to improve the prediction of tolerance to abiotic stresses, such as heat and drought, in different crop plants have been listed by Cortés and López-Hernández [236]. ML is also a promising tool for QTL mining in crops. Falk et al. [237] developed a computer vision and ML-enabled high-throughput root phenotyping platform for soybean. Using this ML-enabled root phenotyping platform, they studied the genetic variability of root system architecture (RSA) traits in different soybean accessions. The combination of predictive and machine learning algorithms that support genome-wide marker-assisted breeding with innovative methodologies for adaptation to a changing climate together with thermal adaptation has been thoroughly reviewed by Cortés et al. [233,238].
ML systems are cost-effective, non-destructive, and high-throughput tools for the assessment of root growth and development for genomics and phenomics studies. Thus, ML can be efficiently utilized in plant breeding technologies for characterizing the genetic variants controlling complex traits associated with abiotic stress tolerance.
6.2. Artificial Intelligence (AI)-Enabled Genome Editing
Genome editing has evolved as an advanced technique to remove deleterious genes from the genome of an organism. Interestingly, the removal of deleterious alleles is one of the important components of plant breeding. Linkage drag can be avoided by the introduction of beneficial alleles into elite cultivars utilizing a genome editing technique rather than backcrossing with other donor parents carrying deleterious alleles at linked loci [239,240]. The utilization of the CRISPR/Cas9 system as a genome-editing tool has opened new avenues in understanding the functional roles of many important regulatory genes. The efficiency of the CRISPR system relies on a specifically designed single-guide RNA (sgRNA) that is complementary to the specific genomic regions under study. However, off-target deletions could result from the binding of sgRNA to off-target sites. AI-enabled identification of target prediction is currently being exploited for designing sgRNAs with increased specificity and improved efficiency. Abadi et al. [241] have designed a computer algorithm using a ML framework called CRISPR Target Assessment (CRISTA) for predicting the target in the genome. The predictions made with CRISTA were found to be more accurate and precise compared with other available methodologies. Most of the existing off-target binding prediction tools are based on the calculation of a mismatch score; thus, they cannot be scaled up with the rapidly increasing amount of experimental data generated through the CRISPR/Cas9 technique [242]. To address this issue, Lin et al. [242] designed two algorithms using deep neural networks, i.e., deep CNNs and deep feed-forward neural networks, to predict off-target mutations in CRISPR/Cas9-based gene editing. The models were evaluated for performance using off-target datasets, such as the CRISPOR dataset (http://crispor.org, accessed on 3 August 2021) and datasets discovered by Genome-wide, Unbiased Identification of DSBs Enabled by Sequencing (GUIDE-seq). The deep neural network-based models were further compared to advanced off-target prediction methods (CCTop, Convolutional Neural Networks, CROP-IT, and MIT) and three conventional ML models (gradient boosting trees, logistic regression, and random forest) in both datasets. The deep neural network-based algorithm made more precise predictions than the conventionally used models. Such ML- and deep-learning-based models can also be utilized in pulse crops for the prediction of off-target binding and, thus, gene editing can also be easily achieved in pulse crops.
7. Challenges and Opportunities for Future Pulse Crop Breeding
Legumes share important taxon-specific data opportunities that must be fully explored to improve their abiotic stress resilience. At the individual legume species level, assimilation of novel or unique data is a challenge that can be addressed by integrating different omics approaches and the coupling of phenotyping data with next-generation AI tools. Predictive modeling based on a novel omics approach, such as fluxomics—which can predict the effects of environmental factors on genetic changes—has not yet been explored in case of legumes and should be given attention. In addition, allocating glycans to specific amino acid sites remains understudied in legumes, as tools and algorithms have not yet reached the level of automation required, which needs to be addressed promptly. Furthermore, as large amounts of genomic data on members of the Leguminosae family are becoming available, the creation of comprehensive resource atlases is required. GEAs will be useful for generating markers that are associated with specific productivity- and tolerance-related traits that can be employed in pulse crop breeding. However, many important pulse crops still have a limited number of genetic resources available for the development of databases, which limits the application of epigenetic breeding in legumes. For such pulse crops, the construction of pan-genomes will help us to develop a comprehensive understanding of stress response mechanisms.
Integration of pan-omics platforms with novel omics tools and AI will further assist with the discovery of target genes and pathways controlled by complex mechanisms, which will allow ‘speed cum precision breeding’ to develop climate-resilient, high-yield legumes. However, the MOI approach is often hampered by variations in the data output, data structure, and unwanted noise between the different technological platforms used for data collection. MOI can also be problematic for datasets that are irreproducible, qualitative, contain false positive/negative values, and lack metadata. Therefore, for productive integration and comparison, data management and sharing standards need to be updated. There is a desire to include consistent metadata and ontologies in properly maintained repositories to facilitate their use. Further, genome editing has significantly accelerated livestock breeding; however, genome editing is difficult to achieve in the case of legumes due to the complexity of allelic effects and the GP gap. Computer-simulated, environment-specific models generated from ML- and deep-learning-based models can alleviate the problems associated with genome editing in legumes in changing environments. ML can enable better genomic selection, QTL mining, and genome-wide association studies in orphan legumes. It can also be employed to predict a plant’s response to an abiotic stress by utilizing the miRNA expression in the plant, which to date has only been exemplified in the case of Arabidopsis [243]. Similar approaches can also be employed in economically important pulse crops to uncover the role of various stress-responsive miRNAs.
The adjustment of legumes towards changes in climatic scenario and molecular breeding of legumes for resilience to abiotic stresses have conventionally been aided by QTL mapping, marker-assisted selection, and GWAS [244]. Recently, extensive augmentation in the area of predictive breeding has helped us accelerate the selection from natural origins and within the breeding succession by abbreviating the generation intervals and escalating the selection fidelity ahead of field trials. Therefore, predictive breeding has enormous potential for complex polygenic adaptive traits such as abiotic stress tolerance. Lenz et al. have hitherto recognized and talked about refinement in this area, such as multi-trait GP models together with integrative selection scores [245]. These models can describe multi-scale trait–environment inter-relations in legumes. Machine learning provides a predictive method competent at amalgamating GWAS, GEA, and GP approaches. For genetic and genomic datasets, ML algorithms can be dissected into supervised, semi-supervised, and unsupervised methods. Abiotic stress tolerance amelioration requires various ML methods based on the aim of expounding the output model or elucidating the predictive power. Generative models are great for interpretability, whereas discriminative models are suitable for predictive power [231].
Genomic selection also depends upon progress in ML and the accessibility of genotypic data to predict stress-related phenotypic traits. Further scrutiny of the association between mechanistic models that permit the simulation of phenotypes under abiotic stresses and ML models that can incorporate marker data holds potential to solve the problem of model transferability among environments [246]. ML has traditionally been employed in functional genomics [247]. Currently, it is metamorphosing into GWAS coupled to MAS [247] and GP [248,249]. Creative advancements in ML will further assist with precise predictions by agglutinating environmental variables and phenotypic and genotypic diversity [238]. In conclusion, leveraging tools from various scientific disciplines together with “omics” and advanced breeding technologies is crucial to sustaining legume productivity under changing climatic conditions.
Author Contributions
Conceptualization, D.S. (Dharmendra Singh) and M.P.; methodology, D.S. (Dharmendra Singh) and M.P.; formal analysis, P.C., J.T., C.K.S. and D.S. (Deepti Singh); data curation, P.C., J.T., C.K.S. and D.S. (Deepti Singh); writing—original draft preparation, P.C., J.T. and C.K.S.; writing—review and editing, R.S.S.T., M.A., N.S.K., R.S.R., S.S., R.S.S. and R.K.Y.; visualization, P.C. and J.T.; supervision, D.S. (Dharmendra Singh) and M.P.; funding acquisition, D.S. (Dharmendra Singh) and M.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by a grant received from the Indian Agricultural Research Institute, New Delhi (Project no- JAN 09/16). The funding bodies had no role in the design of the review and the writing of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank the director of the Indian Council of Agricultural Research (ICAR) and the Indian Agricultural Research Institute (IARI), New Delhi and the head of the Division of Genetics, ICAR- IARI, New Delhi for healthy discussions during the preparation of this article.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Maphosa, Y.; Jideani, V.A. The Role of Legumes in Human Nutrition, Functional Food. In Improve Health Through Adequate Food; Hueda, M.C., Ed.; IntechOpen: Rijeka, Croatia, 2017. [Google Scholar] [CrossRef]
- Bohra, A.; Sahrawat, K.L.; Kumar, S.; Joshi, R.; Parihar, A.K.; Singh, U.; Singh, D.; Singh, N.P. Genetics and genomics-based interventions for nutritional enhancement of grain legume crops: Status and outlook. J. Appl. Genet. 2015, 56, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Lam, H.M.; Nguyen, H.T.; Siddique, K.H.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M.; et al. Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2016, 2, 16112. [Google Scholar] [CrossRef]
- Considine, M.J.; Siddique, K.H.M.; Foyer, C.H. Nature’s pulse power: Legumes, food security and climate change. J. Expt. Bot. 2017, 68, 1815–1818. [Google Scholar] [CrossRef] [PubMed]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant. Physiol. 2003. 131, 872–877. [CrossRef]
- Kang, Y.J.; Kim, S.K.; Kim, M.Y.; Lestari, P.; Kim, K.H.; Ha, B.-K.; Jun, T.H.; Hwang, W.J.; Lee, T.; Lee, J.; et al. Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Gaur, P.M.; Chamarthi, S.K.; Krishnamurthy, L.; Tripathi, S.; Kashiwagi, J.; Samineni, S.; Singh, V.K.; Thudi, M.; Jaganathan, D. Fast-track introgression of “QTL-hotspot” for root traits and other drought tolerance traits in JG 11, an elite and leading variety of chickpea. Plant Genome 2013, 6, 3. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Chen, W.; Li, Y.; Bharti, A.K.; Saxena, R.K.; Schlueter, J.A.; Donoghue, M.T.; Azam, S.; Fan, G.; Whaley, A.M.; et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat. Biotechnol. 2012, 30, 83. [Google Scholar] [CrossRef]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.C.; et al. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Gen. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Jewell, M.C.; Campbell, B.C.; Godwin, I.D. Transgenic Plants for Abiotic Stress Resistance. In Transgenic Crop Plants; Springer: Berlin/Heidelberg, Germany, 2010; pp. 67–132. [Google Scholar]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef]
- Le, B.H.; Wagmaister, J.A.; Kawashima, T.; Bui, A.Q.; Harada, J.J.; Goldberg, R.B. Using genomics to study legume seed development. Plant. Physiol. 2007, 144, 562–574. [Google Scholar] [CrossRef]
- Saito, K.; Matsuda, F. Metabolomics for functional genomics, systems biology and biotechnology. Annu. Rev. Plant. Biol. 2010, 61, 463–489. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant. Sci. 2016, 7, 67. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signalling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Varshney, R.K.; Roorkiwal, M.; Nguyen, T. Legume genomics: From genomic resources to molecular breeding. Plant. Genome 2013, 6, 1–7. [Google Scholar] [CrossRef]
- Wheeler, J.A.; Cortes, A.J.; Sedlacek, J.; Karrenberg, S.; van Kleunen, M.; Wipf, S.; Hoch, G.; Bossdorf, O.; Rixen, C. The snow and the willows: Earlier spring snowmelt reduces performance in the low-lying alpine shrub Salix herbacea. J. Ecol. 2016, 104, 1041–1050. [Google Scholar] [CrossRef]
- Cortés, A.J.; Waeber, S.; Lexer, C.; Sedlacek, J.; Wheeler, J.A.; van Kleunen, M.; Boßdorf, O.; Hoch, G.; Rixen, C.; Wipf, S.; et al. Small-scale patterns in snowmelt timing affect gene flow and the distribution of genetic diversity in the alpine dwarf shrub Salix herbacea. Heredity 2014, 113, 233–239. [Google Scholar] [CrossRef]
- Wheeler, J.A.; Hoch, G.; Cortés, A.J.; Sedlacek, J.; Wipf, S.; Rixen, C. Increased spring freezing vulnerability for alpine shrubs under early snowmelt. Oecologia 2014, 175, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, J.A.; Schnider, F.; Sedlacek, J.; Cortés, A.J.; Wipf, S.; Hoch, G.; Rixen, C. With a little help from my friends: Community facilitation increases performance in the dwarf shrub Salix herbacea. Basic Appl. Ecol. 2015, 16, 202–209. [Google Scholar] [CrossRef]
- Valencia, J.B.; Mesa, J.; León, J.G.; Madriñán, S.; Cortés, A.J. Climate vulnerability assessment of the espeletia complex on Páramo Sky Islands in the Northern Andes. Front. Ecol. Evol. 2020, 8, 309. [Google Scholar] [CrossRef]
- Sedlacek, J.; Cortés, A.J.; Wheeler, J.; Bossdorf, O.; Hoch, G.; Klápště, J.; Lexer, C.; Rixen, C.; Wipf, S.; Karrenberg, S.; et al. Evolutionary potential in the Alpine: Trait heritabilities and performance variation of the dwarf willow Salix herbacea from different elevations and microhabitats. Ecol. Evol. 2016, 6, 3940–3952. [Google Scholar] [CrossRef]
- Cortés, A.J.; Garzón, L.N.; Valencia, J.B.; Madriñán, S. On the causes of rapid diversification in the páramos: Isolation by ecology and genomic divergence in espeletia. Front. Plant. Sci. 2018, 9, 1700. [Google Scholar] [CrossRef] [PubMed]
- Little, C.J.; Wheeler, J.A.; Sedlacek, J.; Cortés, A.J.; Rixen, C. Small-scale drivers: The importance of nutrient availability and snowmelt timing on performance of the alpine shrub Salix herbacea. Oecologia 2016, 180, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Sedlacek, J.F.; Bossdorf, O.; Cortés, A.J.; Wheeler, J.A.; van Kleunen, M. What role do plant–soil interactions play in the habitat suitability and potential range expansion of the alpine dwarf shrub Salix herbacea? Basic Appl. Ecol. 2014, 15, 305–315. [Google Scholar] [CrossRef]
- Varshney, R.K. Application of Next Generation Sequencing and Genotyping Technologies to Develop Large-Scale Genomic Resources in SAT Legume Crops. In Genomics and Crop Improvement: Relevance and Reservations; Muralidharan, K., Siddiq., E.A., Acharya, N.G., Eds.; Ranga Agricultural University: Hyderabad, India, 2011; pp. 1–10. [Google Scholar]
- Varshney, R.K.; Singh, V.K.; Kumar, A.; Powell, W.; Sorrells, M.E. Can genomics deliver climate-change ready crops? Curr. Opin. Plant. Biol. 2018, 45, 205–211. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Jogaiah, S.; Burritt, D.J.; Tran, L.S.P. Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant. Cell Env. 2018, 41, 1972–1983. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K. Genome editing with engineered nucleases in plants. Plant. Cell Physiol. 2015, 56, 389–400. [Google Scholar] [CrossRef]
- Knowpulse website. Available online: http://knowpulse.usask.ca/ (accessed on 15 January 2021).
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Kato, T.; Nakao, M.; Sasamoto, S.; Watanabe, A.; Ono, A.; Kawashima, K.; et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008, 15, 227–239. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nature Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Tripathi, P.; Rabara, R.C.; Rushton, P.J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 2014, 239, 255–266. [Google Scholar] [CrossRef]
- Zhang, J.; Naik, H.S.; Assefa, T.; Sarkar, S.; Reddy, R.C.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer vision and machine learning for robust phenotyping in genome-wide studies. Sci. Rep. 2017, 7, 44048. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clinical Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef]
- Lahner, B.; Gong, J.; Mahmoudian, M.; Smith, E.L.; Abid, K.B.; Rogers, E.E.; Guerinot, M.L.; Harper, J.F.; Ward, J.M.; McIntyre, L.; et al. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 1215–1221. [Google Scholar] [CrossRef]
- Szpunar, J. Metallomics: A new frontier in analytical chemistry. Anal. Bioanal Chem. 2004, 378, 54–56. [Google Scholar] [CrossRef]
- Salt, D.E.; Baxter, I.; Lahner, B. Ionomics and the study of the plant ionome. Annu. Rev. Plant. Biol. 2008, 59, 709–733. [Google Scholar] [CrossRef]
- Baxter, I. Ionomics: The functional genomics of elements. Brief. Funct. Genom. 2010, 9, 14956. [Google Scholar] [CrossRef]
- Huang, X.Y.; Salt, D.E. Plant ionomics: From elemental profiling to environmental adaptation. Mol. Plant 2016, 9, 787–797. [Google Scholar] [CrossRef]
- Ziegler, G.; Terauchi, A.; Becker, A.; Armstrong, P.; Hudson, K.; Baxter, I. Ionomic screening of field-grown soybean identifies mutants with altered seed elemental composition. Plant. Genome. 2013, 6. [Google Scholar] [CrossRef]
- Chen, Z.; Watanabe, T.; Shinano, T.; Okazaki, K.; Osaki, M. Rapid characterization of plant mutants with an altered ion-profile: A case study using Lotus japonicus. New Phytol. 2009, 181, 795–801. [Google Scholar] [CrossRef]
- Ziegler, G.; Nelson, R.; Granada, S.; Krishnan, H.B.; Gillman, J.D.; Baxter, I. Genome wide association study of ionomic traits on diverse soybean populations from germplasm collections. Plant. Direct. 2018, 15, e00033. [Google Scholar] [CrossRef]
- Hacisalihoglu, G.; Settles, A. Quantification of seed ionome variation in 90 diverse soybean (Glycine max) lines. J. Plant. Nutr. 2017, 40, 2808–2817. [Google Scholar] [CrossRef]
- Springer, N.M. Epigenetics and crop improvement. Trends Genet. 2013, 29, 241–247. [Google Scholar] [CrossRef]
- Labra, M.; Ghiani, A.; Citterio, S.; Sgorbati, S.; Sala, F.; Vannini, C.; Ruffini-Castiglione, M.; Bracale, M. Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant. Biol. 2002, 4, 694–699. [Google Scholar] [CrossRef]
- Abid, G.; Mingeot, D.; Muhovski, Y.; Mergeai, G.; Aouida, M.; Abdelkarim, S.; Aroua, I.; El Ayed, M.; M’hamdi, M.; Sassi, K.; et al. Analysis of DNA methylation patterns associated with drought stress response in faba bean (Vicia faba L.) using methylation-sensitive amplification polymorphism (MSAP). Environ. Exp. Bot. 2017, 142, 34–44. [Google Scholar] [CrossRef]
- Rakei, A.; Maali-Amiri, R.; Zeinali, H.; Ranjbar, M. DNA methylation and physio-biochemical analysis of chickpea in response to cold stress. Protoplasma. 2015, 253, 61–76. [Google Scholar] [CrossRef]
- Song, Y.; Ji, D.; Li, S.; Wang, P.; Li, Q.; Xiang, F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 2012, 7, e41274. [Google Scholar] [CrossRef]
- Liang, X.; Hou, X.; Li, J.; Han, Y.; Zhang, Y.; Feng, N.; Du, J.; Zhang, W.; Zheng, D.; Fang, S. High-resolution DNA methylome reveals that demethylation enhances adaptability to continuous cropping comprehensive stress in soybean. BMC Plant. Biol. 2019, 19, 79. [Google Scholar] [CrossRef]
- Wu, T.; Pi, E.X.; Tsai, S.N.; Lam, H.M.; Sun, S.M.; Kwan, Y.W.; Ngai, S.M. GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant. Biol. 2011, 11, 178. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Chakraborty, J.; Ghosh, P.; Basu, D.; Das, S. Chickpea WRKY70 regulates the expression of a homeodomain-leucine zipper (HD-Zip) I transcription factor CaHDZ12, which confers abiotic stress tolerance in transgenic tobacco and chickpea. Plant. Cell Physiol. 2017, 58, 1934–1952. [Google Scholar] [CrossRef] [PubMed]
- Awana, M.; Yadav, K.; Rani, K.; Gaikwad, K.; Praveen, S.; Kumar, S.; Singh, A. Insights into salt stress-induced biochemical, molecular and epigenetic regulation of spatial responses in pigeonpea (Cajanus cajan L.). J. Plant Growth Regul. 2019, 38, 1545–1561. [Google Scholar] [CrossRef]
- Chen, R.; Li, M.; Zhang, H.; Duan, L.; Sun, X.; Jiang, Q.; Zhang, H.; Hu, Z. Continuous salt stress-induced long non-coding RNAs and DNA methylation patterns in soybean roots. BMC Genomic. 2019, 20, 730. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Song, G.; Guo, W.; Wang, W.; Zhao, H.; Gao, T.; Lv, Q.; Yang, X.; Xu, F.; Dong, Y.; et al. Dynamic changes in genome-wide histone3 lysine27 trimethylation and gene expression of soybean roots in response to salt stress. Front. Plant. Sci. 2019, 10, 1031. [Google Scholar] [CrossRef] [PubMed]
- Gahlaut, V.; Zinta, G.; Jaiswal, V.; Kumar, S. Quantitative Epigenetics: A new avenue for crop improvement. Epigenomes 2020, 4, 25. [Google Scholar] [CrossRef]
- Schmitz, R.J.; He, Y.; Valdés-López, O.; Khan, S.M.; Joshi, T.; Urich, M.A.; Nery, J.R.; Diers, B.; Xu, D.; Stacey, G.; et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013, 23, 1663–1674. [Google Scholar] [CrossRef]
- Raju, S.K.K.; Shao, M.R.; Sanchez, R.; Xu, Y.Z.; Sandhu, A.; Graef, G.; Mackenzie, S. An epigenetic breeding system in soybean for increased yield and stability. Plant. Biotechnol. J. 2018, 16, 1836–1847. [Google Scholar] [CrossRef]
- Zhong, X. Comparative epigenomics: A powerful tool to understand the evolution of DNA methylation. New Phytol. 2016, 210, 76–80. [Google Scholar] [CrossRef]
- Junaid, A.; Singh, N.; Gaikwad, K. Patterns of gene-body-methylation conservation and its divergent association with gene expression in pigeonpea and soybean. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kim, K.D.; El Baidouri, M.; Abernathy, B.; Iwata-Otsubo, A.; Chavarro, C.; Gonzales, M.; Libault, M.; Grimwood, J.; Jackson, S.A. A comparative epigenomic analysis of polyploidy-derived genes in soybean and common bean. Plant. Physiol. 2015, 168, 1433–1447. [Google Scholar] [CrossRef]
- Iyer, V.V.; Sriram, G.; Fulton, D.B.; Zhou, R.; Westgate, M.E.; Shanks, J.V. Metabolic flux maps comparing the effect of temperature on protein and oil biosynthesis in developing soybean cotyledons. Plant. Cell Environ. 2008, 31, 506–517. [Google Scholar] [CrossRef]
- Cocuron, J.C.; Koubaa, M.; Kimmelfield, R.; Ross, Z.; Alonso, A.P. A combined metabolomics and fluxomics analysis identifies steps limiting oil synthesis in maize embryos. Plant. Physiol. 2019, 181, 961–975. [Google Scholar] [CrossRef]
- Schwender, J.; Hay, J.O. Predictive modeling of biomass component tradeoffs in Brassica napus developing oilseeds based on in silico manipulation of storage metabolism. Plant. Physiol. 2012, 160, 1218–1236. [Google Scholar] [CrossRef]
- Salon, C.; Avice, J.C.; Colombié, S.; Dieuaide-Noubhani, M.; Gallardo, K.; Jeudy, C.; Ourry, A.; Prudent, M.; Voisin, A.S.; Rolin, D. Fluxomics links cellular functional analyses to whole-plant phenotyping. J. Expt. Bot. 2017, 68, 2083–2098. [Google Scholar] [CrossRef]
- Moreira, T.B.; Shaw, R.; Luo, X.; Ganguly, O.; Kim, H.S.; Coelho, L.G.F.; Cheung, C.Y.M.; Rhys Williams, T.C. A genome-scale metabolic model of soybean (Glycine max) highlights metabolic fluxes in seedlings. Plant. Physiol. 2019, 180, 1912–1929. [Google Scholar] [CrossRef]
- Kannan, K.; Wang, Y.; Lang, M.; Challa, G.S.; Long, S.P.; Marshall-Colon, A. Combining gene network, metabolic and leaf-level models shows means to future-proof soybean photosynthesis under rising CO2. In Silico Plants 2019, 1, diz008. [Google Scholar] [CrossRef]
- Kohli, D.; Joshi, G.; Deokar, A.A.; Bhardwaj, A.R.; Agarwal, M.; Katiyar-Agarwal, S.; Srinivasan, R.; Jain, P.K. Identification and characterization of Wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PLoS ONE 2014, 9, e108851. [Google Scholar] [CrossRef]
- Barrera-Figueroa, B.E.; Gao, L.; Diop, N.N.; Wu, Z.; Ehlers, J.D.; Roberts, P.A.; Close, T.J.; Zhu, J.K.; Liu, R. Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant. Biol. 2011, 11, 127. [Google Scholar] [CrossRef]
- Singh, U.; Khemka, N.; Rajkumar, M.S.; Garg, R.; Jain, M. PLncPRO for prediction of long non-coding RNAs (lncRNAs) in plants and its application for discovery of abiotic stress-responsive lncRNAs in rice and chickpea. Nucleic Acids Res. 2017, 45, e183. [Google Scholar] [CrossRef]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.E.; Pettolino, F.A.; Hart, C.; Lampugnani, E.R.; Willats, W.G.T.; Bacic, A. Glycan profiling of plant cell wall polymers using microarrays. J. Vis. Exp. 2012, 70, 4238. [Google Scholar] [CrossRef]
- Cummings, R.D.; Pierce, J.M. The challenge and promise of glycomics: Chem. Biol. 2014, 21, 1–15. [Google Scholar] [CrossRef]
- Halim, A.; Nilsson, J.; Rüetschi, U.; Hesse, C.; Larson, G. Human urinary glycoproteomics attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol. Cell Proteomics. 2012, 11. [Google Scholar] [CrossRef]
- Mustafa, G.; Komatsu, S. Quantitative proteomics reveals the effect of protein glycosylation in soybean root under flooding stress. Front. Plant. Sci. 2014, 5, 627. [Google Scholar] [CrossRef]
- Subba, P.; Barua, P.; Kumar, R.; Datta, A.; Soni, K.K.; Chakraborty, S.; Chakraborty, N. Phosphoproteomic dynamics of chickpea (Cicer arietinum L.) reveals shared and distinct components of dehydration response. J. Proteome Res. 2013, 12, 5025–5047. [Google Scholar] [CrossRef]
- Subba, P.; Barua, P.; Kumar, R.; Datta, A.; Soni, K.K.; Chakraborty, S.; Chakraborty, N. Bioinformatic identification and analysis of hydroxyproline-rich glycoproteins in Populustrichocarpa. BMC Plant. Biol. 2016, 16, 229. [Google Scholar] [CrossRef]
- Balkir, P.; Kemahlioglu, K.; Yucel, U. Foodomics: A new approach in food quality and safety. Trends Food Sci. Technol. 2021, 108, 49–57. [Google Scholar] [CrossRef]
- Panzade, G.; Gangwar, I.; Awasthi, S.; Sharma, N.; Shankar, R. Plant Regulomics Portal (PRP): A comprehensive integrated regulatory information and analysis portal for plant genomes. Database 2019, 2019, baz130. [Google Scholar] [CrossRef]
- Ran, X.; Zhao, F.; Wang, Y.; Liu, J.; Zhuang, Y.; Ye, L.; Qi, M.; Cheng, J.; Zhang, Y. Plant Regulomics: A data-driven interface for retrieving upstream regulators from plant multi-omics data. Plant. J. Cell Mol. Biol. 2020, 101, 237–248. [Google Scholar] [CrossRef]
- Tanveer, T.; Shaheen, K.; Parveen, S.; Kazi, A.G.; Ahmad, P. Plant secretomics: Identification, isolation, and biological significance under environmental stress. Plant. Signal. Behav. 2014, 9, e29426. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Wardhan, V.; Kumar, A.; Rathi, D.; Pandey, A.; Chakraborty, S.; Chakraborty, N. Secretome analysis of chickpea reveals dynamic extracellular remodeling and identifies a Bet v1-like protein, CaRRP1 that participates in stress response. Sci. Rep. 2015, 5, 18427. [Google Scholar] [CrossRef] [PubMed]
- Parveen, S.; Gupta, D.B.; Dass, S.; Kumar, A.; Pandey, A.; Chakraborty, S.; Chakraborty, N. Chickpea ferritin CaFer1 participates in oxidative stress response, and promotes growth and development. Sci. Rep. 2016, 6, 31218. [Google Scholar] [CrossRef]
- Fernandes, C.; Figueira, E.; Tauler, R. Exposure to chlorpyrifos induces morphometric, biochemical and lipidomic alterations in green beans (Phaseolus vulgaris). Ecotoxicol. Environ. Saf. 2018, 156, 25–33. [Google Scholar] [CrossRef]
- Narayanan, S.; Zoong-Lwe, Z.S.; Gandhi, N.; Welti, R.; Fallen, B.; Smith, J.R.; Rustgi, S. Comparative lipidomic analysis reveals heat stress responses of two soybean genotypes differing in temperature sensitivity. Plants 2020, 9, 457. [Google Scholar] [CrossRef]
- Okazaki, Y.; Takano, K.; Saito, K. Lipidomic analysis of soybean leaves revealed tissue-dependent difference in lipid remodeling under phosphorus-limited growth conditions. Plant. Biotechnol. 2017, 34, 57–63. [Google Scholar] [CrossRef]
- Yin, X.; Sakata, K.; Komatsu, S. Phosphoproteomics reveals the effect of ethylene in soybean root under flooding stress. J. Proteome Res. 2014, 13, 5618–5634. [Google Scholar] [CrossRef]
- Razzaq, M.K.; Aleem, M.; Mansoor, S.; Khan, M.A.; Rauf, S.; Iqbal, S.; Siddique, K.H.M. Omics and CRISPR-Cas9 approaches for molecular insight, functional gene analysis, and stress tolerance development in crops. Int. J. Mol. Sci. 2021, 22, 1292. [Google Scholar] [CrossRef]
- Young, N.D.; Cannon, S.B.; Sato, S.; Kim, D.; Cook, D.R.; Town, C.D.; Roe, B.A.; Tabata, S. Sequencing the gene spaces of Medicago truncatula and Lotus japonicus. Plant. Physiol. 2005, 137, 1174–1181. [Google Scholar] [CrossRef]
- Young, N.D.; Bharti, A.K. Genome-enabled insights into legume biology. Annu. Rev. Plant. Biol. 2012, 63, 283–305. [Google Scholar] [CrossRef]
- Bohra, A.; Pandey, M.K.; Jha, U.C.; Singh, B.; Singh, I.P.; Datta, D.; Chaturvedi, S.K.; Nadarajan, N.; Varshney, R.K. Genomics assisted breeding in four major pulse crops of developing countries: Present status and prospects. Theor. Appl. Genet. 2014, 127, 1263–1291. [Google Scholar] [CrossRef]
- Gilchrist, E.; Haughn, G. Reverse genetics techniques: Engineering loss and gain of gene function in plants. Brief. Funct. Genom. 2010, 9, 103–110. [Google Scholar] [CrossRef]
- Rehman, A.U.; Malhotra, R.S.; Bett, K.; Tar’An, B.; Bueckert, R.; Warkentin, T.D. Mapping QTL associated with traits affecting grain yield in chickpea (Cicer arietinum L.) under terminal drought stress. Crop. Sci. 2011, 51, 450–463. [Google Scholar] [CrossRef]
- Muchero, W.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Mapping QTL for drought stress-induced premature senescence and maturity in cowpea [Vigna unguiculata (L.) Walp.]. Theor. Appl. Genet. 2009, 118, 849–863. [Google Scholar] [CrossRef]
- Asfaw, A.; Blair, M.W.; Struik, P.C. Multienvironment quantitative trait loci analysis for photosynthate acquisition, accumulation, and remobilization traits in common bean under drought stress. G3 2012, 2, 579–595. [Google Scholar] [CrossRef]
- Nabateregga, M.; Mukankusi, C.; Raatz, B. Quantitative trait loci (QTL) mapping for intermittent drought tolerance in BRB 191 × SEQ 1027 Andean Intra-gene cross recombinant inbred line population of common bean (Phaseolus vulgaris L. African J. Biotechnol. 2019, 18, 452–461. [Google Scholar]
- y Teran, J.C.B.M.; Konzen, E.R.; Palkovic, A.; Tsai, S.M.; Rao, I.M.; Beebe, S.; Gepts, P. Effect of drought stress on the genetic architecture of photosynthate allocation and remobilization in pods of common bean (Phaseolus vulgaris L.), a key species for food security. BMC Plant. Biol. 2019, 19, 171. [Google Scholar] [CrossRef]
- Diaz, L.M.; Ricaurte, J.; Tovar, E.; Cajiao, C.; Teran, H.; Grajales, M.; Polania, J.; Rao, I.; Beebe, S.; Raatz, B. QTL analyses for tolerance to abiotic stresses in a common bean (Phaseolus vulgaris L.) population. PLoS ONE 2018, 13, e0202342. [Google Scholar] [CrossRef]
- Diaz, S.; Ariza-Suarez, D.; Izquierdo, P.; Lobaton, J.D.; de la Hoz, J.F.; Acevedo, F.; Duitama, J.; Guerrero, A.F.; Cajiao, C.; Mayor, V. Genetic mapping for agronomic traits in a MAGIC population of common bean (Phaseolus vulgaris L.) under drought conditions. BMC Genomics. 2020, 21, 799. [Google Scholar] [CrossRef]
- Idrissi, O.; Udupa, S.M.; De Keyser, E.; McGee, R.J.; Coyne, C.J.; Saha, G.C.; Muehlbauer, F.J.; Van Damme, P.; De Riek, J. Identification of quantitative trait loci controlling root and shoot traits associated with drought tolerance in a lentil (Lens culinaris Medik.) recombinant inbred line population. Front. Plant. Sci. 2016, 7, 1174. [Google Scholar] [CrossRef]
- Specht, J.E.; Chase, K.; Macrander, M.; Graef, G.L.; Chung, J.; Markwell, J.P.; Germann, M.; Orf, J.H.; Lark, K.G. Soybean response to water: A QTL analysis of drought tolerance. Crop. Sci. 2001, 41, 493–509. [Google Scholar] [CrossRef]
- Ye, H.; Song, L.; Schapaugh, W.T.; Ali, M.L.; Sinclair, T.R.; Riar, M.K.; Raymond, R.N.; Li, Y.; Vuong, T.; Valliyodan, B.; et al. The importance of slow canopy wilting in drought tolerance in soybean. J. Expt. Bot. 2020, 71, 642–652. [Google Scholar] [CrossRef]
- Sholihin, H.D.M. Molecular mapping of drought resistance in mungbean (Vigna radiata): 1. Linkage map in mungbean using AFLP markers. J.B. Pertanian. 2002, 7, 17–24. [Google Scholar]
- Liu, C.; Wu, J.; Wang, L. Quantitative trait locus mapping under irrigated and drought treatments based on a novel genetic linkage map in mungbean (Vigna radiata L.). Theor. Appl. Genet. 2017, 130, 2375–2393. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-García, R.; Prats, E.; Fondevilla, S.; Satovic, Z.; Rubiales, D. Quantitative trait loci associated to drought adaptation in pea (Pisum sativum L.). Plant. Mol. Biol. Rep. 2015, 33, 1768–1778. [Google Scholar] [CrossRef]
- Paul, P.J.; Samineni, S.; Thudi, M.; Sajja, S.B.; Rathore, A.; Das, R.R.; Khan, A.W.; Chaturvedi, S.K.; Lavanya, G.R.; Varshney, R. Molecular mapping of QTLs for heat tolerance in chickpea. Int. J. Mol. Sci. 2018, 19, 2166. [Google Scholar] [CrossRef] [PubMed]
- Lucas, M.R.; Ehlers, J.D.; Huynh, B.L.; Diop, N.N.; Roberts, P.A.; Close, T.J. Markers for breeding heat-tolerant cowpea. Mol. Breed. 2013, 31, 529–536. [Google Scholar] [CrossRef]
- Singh, D.; Singh, C.K.; Singh Tomar, R.S.; Pal, M. Genetics and molecular mapping of heat tolerance for seedling survival and pod set in lentil. Crop. Sci. 2017, 57, 3059–3067. [Google Scholar] [CrossRef]
- Mugabe, D.; Coyne, C.J.; Piaskowski, J.; Zheng, P.; Ma, Y.; Landry, E.; McGee, R.; Main, D.; Vandemark, G.; Zhang, H.; et al. Quantitative trait loci for cold tolerance in chickpea. Crop. Sci. 2019, 59, 573–582. [Google Scholar] [CrossRef]
- Sallam, A.; Arbaoui, M.; El-Esawi, M.; Abshire, N.; Martsch, R. Identification and verification of QTL associated with frost tolerance using linkage mapping and GWAS in winter faba bean. Front. Plant. Sci. 2016, 7, 1098. [Google Scholar] [CrossRef]
- Kahraman, A.; Kusmenoglu, I.; Aydin, N.; Aydogan, A.; Erskine, W.; Muehlbauer, F.J. QTL mapping of winter hardiness genes in Lentil. Crop. Sci. 2004, 44, 13. [Google Scholar] [CrossRef]
- Dumont, E.; Fontaine, V.; Vuylsteker, C.; Sellier, H.; Bodèle, S.; Voedts, N.; Devaux, R.; Frise, M.; Avia, K.; Hilbert, J.L. Association of sugar content QTL and PQL with physiological traits relevant to frost damage resistance in pea under field and controlled conditions. Theor. Appl. Genet. 2009, 118, 1561–1571. [Google Scholar] [CrossRef]
- Jähne, F.; Balko, C.; Hahn, V.; Würschum, T.; Leiser, W.L. Cold stress tolerance of soybeans during flowering: QTL mapping and efficient selection strategies under controlled conditions. Plant. Breed. 2019, 138, 708–720. [Google Scholar] [CrossRef]
- Vadez, V.; Krishnamurthy, L.; Thudi, M.; Anuradha, C.; Colmer, T.D.; Turner, N.C.; Siddique, K.H.; Gaur, P.M.; Varshney, R.K. Assessment of ICCV 2 × JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTL for seed yield and yield components. Mol. Breed. 2012, 30, 9–21. [Google Scholar] [CrossRef]
- Pushpavalli, R.; Krishnamurthy, L.; Thudi, M.; Gaur, P.M.; Rao, M.V.; Siddique, K.H.; Colmer, T.D.; Turner, N.C.; Varshney, R.K.; Vadez, V. Two key genomic regions harbour QTLs for salinity tolerance in ICCV 2 × JG 11 derived chickpea (Cicer arietinum L.) recombinant inbred lines. BMC Plant. Biol. 2015, 15, 124. [Google Scholar] [CrossRef]
- Chankaew, S.; Isemura, T.; Naito, K.; Ogiso-Tanaka, E.; Tomooka, N.; Somta, P.; Kaga, A.; Vaughan, D.A.; Srinives, P. QTL mapping for salt tolerance and domestication-related traits in Vigna marina subsp. oblonga, a halophytic species. Theor. Appl. Genet. 2013, 127, 691–702. [Google Scholar] [CrossRef]
- Leonforte, A.; Sudheesh, S.; Cogan, N.O.; Salisbury, P.A.; Nicolas, M.E.; Materne, M.; Forster, J.W.; Kaur, S. SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum sativum L.). BMC Plant. Biol. 2013, 13, 161. [Google Scholar] [CrossRef]
- Lee, G.J.; Boerma, H.R.; Villagarcia, M.R.; Zhou, X.; Carter, T.E.; Li, Z.; Gibbs, M.O. A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars. Theor. Appl. Genet. 2004, 109, 1610–1619. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, Y.; Yang, C.; Yang, C.; Mu, Y.; Xia, Q.; Ma, Q. QTL mapping for aluminum tolerance in RIL population of soybean (Glycine max L.) by RAD sequencing. PLoS ONE 2019, 14, e0223674. [Google Scholar] [CrossRef]
- Korir, P.C.; Zhang, J.; Wu, K.; Zhao, T.; Gai, J. Association mapping combined with linkage analysis for aluminum tolerance among soybean cultivars released in Yellow and Changjiang River Valleys in China. Theor. Appl. Genet. 2013, 126, 1659–1675. [Google Scholar] [CrossRef]
- Sharma, A.D.; Sharma, H.; Lightfoot, D.A. The genetic control of tolerance to aluminum toxicity in the ‘Essex’ by ‘Forrest’ recombinant inbred line population. Theor. Appl. Genet. 2011, 122, 687–694. [Google Scholar] [CrossRef]
- Correa, R.; Stanga, J.; Larget, B.; Roznowski, A.; Shu, G.; Dilkes, B.; Baum, D.A. An assessment of transgenomics as a tool for identifying genes involved in the evolutionary differentiation of closely related plant species. New Phytol. 2012, 193, 494–503. [Google Scholar] [CrossRef]
- Tzfira, T.; Weinthal, D.; Marton, I.; Zeevi, V.; Zuker, A.; Vainstein, A. Genome modifications in plant cells by custom-made restriction enzymes. Plant. Biotechnol. J. 2012, 10, 373–389. [Google Scholar] [CrossRef]
- Das Bhowmik, S.S.; Cheng, A.Y.; Long, H.; Tan, G.; Hoang, T.; Karbaschi, M.R.; Williams, B.; Higgins, T.; Mundree, S.G. Robust genetic transformation system to obtain non-chimeric transgenic chickpea. Front. Plant. Sci. 2019, 10, 524. [Google Scholar] [CrossRef]
- Das, A.; Basu, P.S.; Kumar, M.; Ansari, J.; Shukla, A.; Thakur, S.; Singh, P.; Datta, S.; Chaturvedi, S.K.; Sheshshayee, M.S.; et al. Transgenic chickpea (Cicer arietinum L.) harbouring AtDREB1a are physiologically better adapted to water deficit. BMC Plant. Biol. 2021, 21, 39. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Vu, L.T.K.; Nguyen, L.T.N.; Pham, N.T.T.; Nguyen, Y.T.H.; Van Le, S.; Chu, M.H. Overexpression of the GmDREB6 gene enhances proline accumulation and salt tolerance in genetically modified soybean plants. Sci. Rep. 2019, 9, 196630. [Google Scholar] [CrossRef]
- Meena, M.K.; Ghawana, S.; Dwivedi, V.; Roy, A.; Chattopadhyay, D. Expression of chickpea CIPK25 enhances root growth and tolerance to dehydration and salt stress in transgenic tobacco. Front. Plant. Sci. 2015, 6, 683. [Google Scholar] [CrossRef]
- Shukla, R.K.; Raha, S.; Tripathi, V.; Chattopadhyay, D. Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant. Physiol. 2006, 142, 113–123. [Google Scholar] [CrossRef]
- Jain, D.; Chattopadhyay, D. Promoter of CaZF, a chickpea gene that positively regulates growth and stress tolerance, Is activated by an AP2-family transcription factor CAP2. PLoS ONE 2013, 8, e56737. [Google Scholar] [CrossRef]
- Kumar, M.; Yusuf, M.A.; Yadav, P.; Narayan, S. Overexpression of chickpea defensin gene gonfers tolerance to water-deficit stress in Arabidopsis thaliana. Front. Plant. Sci. 2019, 10, 290. [Google Scholar] [CrossRef]
- Yu, X.; Peng, H.; Liu, Y.; Zhang, Y.; Shu, Y.; Chen, Q.; Shi, S.; Ma, L.; Ma, H.; Zhang, H. CarNAC2, a novel NAC transcription factor in chickpea (Cicer arietinum L.), is associated with drought-response and various developmental processes in transgenic Arabidopsis. J. Plant. Biol. 2014, 57, 55–66. [Google Scholar] [CrossRef]
- Figueroa-Balderas, R.E.; García-Ponce, B.; Rocha-Sosa, M. Hormonal and stress induction of the gene encoding common bean acetyl-coenzyme A carboxylase. Plant. Physiol. 2006, 142, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Niron, H.; Türet, M. A putative common bean Chalcone O-Methyltransferase improves salt tolerance in transgenic Arabidopsis thaliana. J. Plant. Growth Regul. 2020, 39, 957–969. [Google Scholar] [CrossRef]
- Chung, E.; Cho, C.W.; So, H.A.; Kang, J.S.; Chung, Y.S.; Lee, J.H. Overexpression of VrUBC1, a mungbean E2 ubiquitin-conjugating enzyme, enhances osmotic stress tolerance in Arabidopsis. PLoS ONE 2013, 8, e66056. [Google Scholar] [CrossRef]
- Banu, M.S.A.; Huda, K.M.K.; Sahoo, R.K.; Garg, B.; Tula, S.; Islam, S.S.; Tuteja, R.; Tuteja, N. Pea p68 imparts salinity stress tolerance in rice by scavenging of ROS-mediated H2O2 and interacts with argonaute. Plant. Mol. Biol. Rep. 2015, 33, 221–238. [Google Scholar] [CrossRef]
- Tuteja, N.; Banu, M.S.A.; Huda, K.M.K.; Gill, S.S.; Jain, P.; Pham, X.H.; Tuteja, R. Pea p68, a DEAD-Box helicase, provides salinity stress tolerance in transgenic tobacco by reducing oxidative stress and improving photosynthesis machinery. PLoS ONE 2014, 9, e98287. [Google Scholar] [CrossRef]
- Sahoo, R.K.; Gill, S.S.; Tuteja, N. Pea DNA helicase 45 promotes salinity stress tolerance in IR64 rice with improved yield. Plant. Signal. Behav. 2012, 7, 1042–1046. [Google Scholar] [CrossRef]
- Srivastava, S.; Rahman, M.H.; Shah, S.; Kav, N.N. Constitutive expression of the pea ABA-responsive 17 (ABR17) cDNA confers multiple stress tolerance in Arabidopsis thaliana. Plant. Biotechnol. J. 2006, 4, 529–549. [Google Scholar] [CrossRef]
- Priyanka, B.; Sekhar, K.; Sunita, T.; Reddy, V.D.; Rao, K.V. Characterization of expressed sequence tags (ESTs) of pigeonpea (Cajanus cajan L.) and functional validation of selected genes for abiotic stress tolerance in Arabidopsis thaliana. Mol. Genet. Genomics 2010, 283, 273–287. [Google Scholar] [CrossRef]
- Tamirisa, S.; Vudem, D.R.; Khareedu, V.R. Overexpression of pigeonpea stress-induced cold and drought regulatory gene (CcCDR) confers drought, salt, and cold tolerance in Arabidopsis. J. Exp. Bot. 2014, 65, 4769–4781. [Google Scholar] [CrossRef]
- Sekhar, K.; Priyanka, B.; Reddy, V.D.; Rao, K.V. Isolation and characterization of a pigeonpeacyclophilin (CcCYP) gene, and its over-expression in Arabidopsis confers multiple abiotic stress tolerance. Plant. Cell Environ. 2010, 33, 1324–1338. [Google Scholar] [CrossRef]
- Sunitha, M.; Srinath, T.; Reddy, V.D.; Rao, K.V. Expression of cold and drought regulatory protein (CcCDR) of pigeonpea imparts enhanced tolerance to major abiotic stresses in transgenic rice plants. Planta 2017, 245, 1137–1148. [Google Scholar] [CrossRef]
- Rodrigues, S.M.; Andrade, M.O.; Gomes, A.P.S.; DaMatta, F.M.; Baracat-Pereira, M.C.; Fontes, E.P. Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J. Expt. Bot. 2006, 57, 1909–1918. [Google Scholar] [CrossRef]
- Chen, M.; Wang, Q.Y.; Cheng, X.G.; Xu, Z.S.; Li, L.C.; Ye, X.G.; Xia, L.Q.; Ma, Y.Z. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007, 353, 299–305. [Google Scholar] [CrossRef]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. miR408 overexpression causes increased drought tolerance in chickpea. Gene. 2015, 555, 186–193. [Google Scholar] [CrossRef]
- Khatib, F.; Makris, A.; Yamaguchi-Shinozaki, K.; Kumar, S.; Sarker, A.; Erskine, W.; Baum, M. Expression of the DREB1A gene in lentil (Lens culinaris Medik. subsp. culinaris) transformed with the Agrobacterium system. Crop. Pasture Sci. 2011, 62, 488–495. [Google Scholar] [CrossRef]
- Sahoo, D.P.; Kumar, S.; Mishra, S.; Kobayashi, Y.; Panda, S.K.; Sahoo, L. Enhanced salinity tolerance in transgenic mungbean overexpressing Arabidopsis antiporter (NHX1) gene. Mol. Breed. 2016, 36, 144. [Google Scholar] [CrossRef]
- Kumar, S.; Kalita, A.; Srivastava, R.; Sahoo, L. Co-expression of Arabidopsis NHX1 and bar improves the tolerance to salinity, oxidative stress, and herbicide in transgenic mungbean. Front. Plant. Sci. 2017, 8, 1896. [Google Scholar] [CrossRef]
- Seo, J.S.; Sohn, H.B.; Noh, K.; Jung, C.; An, J.H.; Donovan, C.M.; Somers, D.A.; Kim, D.I.; Jeong, S.C.; Kim, C.G.; et al. Expression of the Arabidopsis AtMYB44 gene confers drought/salt-stress tolerance in transgenic soybean. Mol. Breed. 2011, 29, 601–608. [Google Scholar] [CrossRef]
- Shanmugam, S.; Zhao, S.; Nandy, S.; Srivastava, V. and Khodakovskaya, M.;. Modification of soybean growth and abiotic stress tolerance by expression of truncated ERECTA Protein from Arabidopsis Thaliana. PLoS ONE 2020, 15, e0233383. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Behura, R.; Awasthi, J.P. Ectopic overexpression of a mungbean vacuolar Na+/H+ antiporter gene (VrNHX1) leads to increased salinity stress tolerance in transgenic Vigna unguiculata L. Walp. Mol. Breed. 2014, 34, 1345–1359. [Google Scholar] [CrossRef]
- Kwapata, K.; Nguyen, T.; Sticklen, M. Genetic transformation of common bean (Phaseolus vulgaris L.) with the guscolor Marker, the Bar herbicide resistance, and the barley (Hordeum vulgare) HVA1 drought tolerance genes. Int. J. Agron 2012, 2012. [Google Scholar] [CrossRef][Green Version]
- Singh, R.; Sharma, S.; Kharb, P.; Saifi, S.; Tuteja, N. OsRuvB transgene induces salt tolerance in pigeon pea. J. Plant. Interactions. 2020, 15, 17–26. [Google Scholar] [CrossRef]
- Hanafy, M.S.; El-Banna, A.; Schumacher, H.M.; Jacobsen, H.-J.; Hassan, F.S. Enhanced tolerance to drought and salt stresses in transgenic faba bean (Vicia faba L.) plants by heterologous expression of the PR10a gene from potato. Plant. Cell Rep. 2013, 32, 663–674. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant. J. 2010, 63, 86–99. [Google Scholar] [CrossRef]
- Prince, S.J.; Joshi, T.; Mutava, R.N.; Syed, N.; Vitor, M.D.S.J.; Patil, G.; Song, L.; Wang, J.; Lin, L.; Chen, W.; et al. Comparative analysis of the drought-responsive transcriptome in soybean lines contrasting for canopy wilting. Plant. Sci. 2015, 240, 65–78. [Google Scholar] [CrossRef]
- Wang, L.; Dong, S.; Liu, L.; Ma, Y.; Li, S.; Zu, W. Transcriptome profiling reveals PEG-simulated drought, heat and combined stress response mechanisms in soybean. Comput. Biol. Chem. 2018, 77, 413–419. [Google Scholar] [CrossRef]
- Cortés, A.J.; Chavarro, M.C.; Blair, M.W. SNP marker diversity in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2011, 123, 827. [Google Scholar] [CrossRef]
- Blair, M.W.; Soler, A.; Cortes, A.J. Diversification and population structure in common beans (Phaseolus vulgaris L.). PLoS ONE 2012, 7, e49488. [Google Scholar] [CrossRef]
- Blair, M.W.; Cortés, A.J.; Penmetsa, R.V.; Farmer, A.; Carrasquilla-Garcia, N.; Cook, D.R. A high-throughput SNP marker system for parental polymorphism screening, and diversity analysis in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2013, 126, 535–548. [Google Scholar] [CrossRef]
- Galeano, C.H.; Cortés, A.J.; Fernández, A.C.; Soler, Á.; Franco-Herrera, N.; Makunde, G.; Vanderleyden, J.; Blair, M.W. Gene-based single nucleotide polymorphism markers for genetic and association mapping in common bean. BMC Genetics. 2012, 13, 1–11. [Google Scholar] [CrossRef]
- Das, A.; Rushton, P.J.; Rohila, J.S. Metabolomic profiling of soybeans (Glycine max L.) reveals the importance of sugar and nitrogen metabolism under drought and heat stress. Plants 2017, 6, 21. [Google Scholar] [CrossRef]
- Singh, D.; Singh, C.K.; Taunk, J.; Tomar, R.S.S.; Chaturvedi, A.K.; Gaikwad, K.; Pal, M. Transcriptome analysis of lentil (Lens culinaris Medikus) in response to seedling drought stress. BMC Genomics. 2017, 18, 206. [Google Scholar] [CrossRef]
- Pandey, A.; Chakraborty, S.; Datta, A.; Chakraborty, N. Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.). Mol. Cell Proteomics. 2008, 7, 88–107. [Google Scholar] [CrossRef]
- Molina, C.; Rotter, B.; Horres, R.; Udupa, S.M.; Besser, B.; Bellarmino, L.; Baum, M.; Matsumura, H.; Terauchi, R.; Kahl, G.; et al. SuperSAGE: The drought stress-responsive transcriptome of chickpea roots. BMC Genomics 2008, 9, 553. [Google Scholar] [CrossRef]
- De Domenico, S.; Bonsegna, S.; Horres, R.; Pastor, V.; Taurino, M.; Poltronieri, P.; Imtiaz, M.; Kahl, G.; Flors, V.; Winter, P. Transcriptomic analysis of oxylipin biosynthesis genes and chemical profiling reveal an early induction of jasmonates in chickpea roots under drought stress. Plant. Physiol. Biochem. 2012, 61, 115–122. [Google Scholar] [CrossRef]
- Mahdavi Mashaki, K.; Garg, V.; Nasrollahnezhad Ghomi, A.A.; Kudapa, H.; Chitikineni, A.; Zaynali Nezhad, K.; Yamchi, A.; Soltanloo, H.; Varshney, R.K.; Thudi, M. RNA-Seq analysis revealed genes associated with drought stress response in kabuli chickpea (Cicer arietinum L.). PLoS ONE 2018, 13, e0199774. [Google Scholar] [CrossRef]
- Badhan, S.; Kole, P.; Ball, A.; Mantri, N. RNA sequencing of leaf tissues from two contrasting chickpea genotypes reveals mechanisms for drought tolerance. Plant. Physiol. Biochem. 2018, 129, 295–304. [Google Scholar] [CrossRef]
- Khandal, H.; Parween, S.; Roy, R.; Meena, M.K.; Chattopadhyay, D. MicroRNA profiling provides insights into post-transcriptional regulation of gene expression in chickpea root apex under salinity and water deficiency. Sci. Rep. 2017, 7, 4632. [Google Scholar] [CrossRef]
- Wu, J.; Wang, L.; Li, L.; Wang, S. De novo assembly of the common bean transcriptome using short reads for the discovery of drought-responsive genes. PLoS ONE 2014, 9, e109262. [Google Scholar] [CrossRef]
- Pereira, W.J.; Melo, A.T.D.O.; Coelho, A.S.G.; Rodrigues, F.A.; Mamidi, S.; Alencar, S.A.D.; Lanna, A.C.; Valdisser, P.A.M.R.; Brondani, C.; Nascimento-Júnior, I.R.D.; et al. Genome-wide analysis of the transcriptional response to drought stress in root and leaf of common bean. Genet. Mol. Biol. 2020, 43, e20180259. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, W.J.; Yin, Z.G.; Li, W.J.; Zhao, H.H.; Zhang, S.; Zhuang, L.; Wang, Y.X.; Zhang, W.H.; Du, J.D. Genome—And transcriptome-wide identification of C3Hs in common bean (Phaseolus vulgaris L.) and structural and expression-based analyses of their functions during the sprout stage under salt-stress conditions. Front. Genet. 2020, 11, 564607. [Google Scholar] [CrossRef]
- Hiz, M.C.; Canher, B.; Niron, H.; Turet, M. Transcriptome analysis of salt tolerant common bean (Phaseolus vulgaris L.) under saline conditions. PLoS ONE 2014, 9, e92598. [Google Scholar] [CrossRef]
- Coetzer, N.; Gazendam, I.; Oelofse, D.; Berger, D.K. SSHscreen and SSHdb, generic software for microarray-based gene discovery: Application to the stress response in cowpea. Plant. Methods 2010, 6, 10. [Google Scholar] [CrossRef]
- Zuo, J.; Wang, Y.; Zhu, B.; Luo, Y.; Wang, Q.; Gao, L. sRNAome and transcriptome analysis provide insight into chilling response of cowpea pods. Gene 2018, 671. [Google Scholar] [CrossRef]
- Khan, M.A.; Alghamdi, S.S.; Ammar, M.H.; Sun, Q.; Teng, F.; Migdadi, H.M.; Al-Faifi, S.A. Transcriptome profiling of faba bean (Viciafaba L.) drought-tolerant variety hassawi-2 under drought stress using RNA sequencing. Electron. J. Biotechnol. 2019, 39, 15–29. [Google Scholar] [CrossRef]
- Alghamdi, S.S.; Khan, M.A.; Ammar, M.H.; Sun, Q.; Huang, L.; Migdadi, H.M.; El-Harty, E.H.; Al-Faifi, S.A. Characterization of drought stress-responsive root transcriptome of faba bean (Viciafaba L.) using RNA sequencing. 3 Biotech. 2018, 8, 502. [Google Scholar] [CrossRef]
- Yang, F.; Chen, H.; Liu, C.; Li, L.; Liu, L.; Han, X.; Wan, Z.; Sha, A. Transcriptome profile analysis of two Vicia faba cultivars with contrasting salinity tolerance during seed germination. Sci. Rep. 2020, 10, 7250. [Google Scholar] [CrossRef]
- Singh, D.; Singh, C.K.; Taunk, J.; Jadon, V.; Pal, M.; Gaikwad, K. Genome wide transcriptome analysis reveals vital role of heat responsive genes in regulatory mechanisms of lentil (Lens culinaris Medikus). Sci. Rep. 2019, 9, 12976. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, S.; Liu, Y.; Liu, X. Transcriptomic profiling reveals metabolic and regulatory pathways in the desiccation tolerance of mungbean (Vigna radiata [L.] R. Wilczek). Front. Plant. Sci. 2016, 7, 1921. [Google Scholar] [CrossRef]
- Severin, A.J.; Woody, J.L.; Bolon, Y.T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant. Biol. 2010, 10, 160. [Google Scholar] [CrossRef]
- Dash, S.; Van Hemert, J.; Hong, L.; Wise, R.P.; Dickerson, J.A. PLEXdb: Gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2012, 40, D1194–D1201. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Patel, R.K.; Jhanwar, S.; Priya, P.; Bhattacharjee, A.; Yadav, G.; Bhatia, S.; Chattopadhyay, D.; Tyagi, A.K.; Jain, M. Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant. Physiol. 2011, 156, 1661–1678. [Google Scholar] [CrossRef] [PubMed]
- Kudapa, H.; Garg, V.; Chitikineni, A.; Varshney, R.K. The RNA-Seq-based high resolution gene expression atlas of chickpea (Cicer arietinum L.) reveals dynamic spatio-temporal changes associated with growth and development. Plant. Cell Environ. 2018, 41, 2209–2225. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.A.; Iniguez, L.P.; Fu, F.; Bucciarelli, B.; Miller, S.S.; Jackson, S.A.; McClean, P.E.; Li, J.; Dai, X.; Zhao, P.X.; et al. An RNA-Seq based gene expression atlas of the common bean. BMC Genomics 2014, 15, 866. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Jiang, C.; Huang, Z.; Torres-Jerez, I.; Chang, J.; Zhang, H.; Udvardi, M.; Liu, R.; Verdier, J. The Vigna unguiculata Gene expression atlas (VuGEA) from de novo assembly and quantification of RNA-seq data provides insights into seed maturation mechanisms. Plant. J. 2016, 88, 318–327. [Google Scholar] [CrossRef]
- Alves-Carvalho, S.; Aubert, G.; Carrere, S.; Cruaud, C.; Brochot, A.L.; Jacquin, F.; Klein, A.; Martin, C.; Boucherot, K.; Kreplak, J.; et al. Full-length de novo assembly of RNA-seq data in pea (Pisumsativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant. J. 2015, 84, 1–19. [Google Scholar] [CrossRef]
- Pazhamala, L.T.; Purohit, S.; Saxena, R.K.; Garg, V.; Krishnamurthy, L.; Verdier, J.; Varshney, R.K. Gene expression atlas of pigeonpea and its application to gain insights into genes associated with pollen fertility implicated in seed formation. J. Exp. Bot. 2017, 68, 2037–2054. [Google Scholar] [CrossRef]
- Arikit, S.; Xia, R.; Kakrana, A.; Huang, K.; Zhai, J.; Yan, Z.; Valdés-López, O.; Prince, S.; Musket, T.A.; Nguyen, H.T.; et al. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant. Cell 2014, 26, 4584–4601. [Google Scholar] [CrossRef]
- Arbona, V.; Manzi, M.; Ollas, C.D.; Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef]
- Jorrín, J.V.; Maldonado, A.M.; Castillejo, M.A. Plant proteome analysis: A 2006 update. Proteomics. 2007, 7, 2947–2962. [Google Scholar] [CrossRef]
- Hakeem, K.R.; Chandna, R.; Ahmad, P.; Iqbal, M.; Ozturk, M. Relevance of proteomic investigations in plant abiotic stress physiology. OMICS 2012, 16, 621–635. [Google Scholar] [CrossRef]
- Wienkoop, S.; Morgenthal, K.; Wolschin, F.; Scholz, M.; Selbig, J.; Weckwerth, W. Integration of metabolomic and proteomic phenotypes analysis of data covariance dissects starch and RFO metabolism from low and high temperature compensation response in Arabidopsis thaliana. Mol. Cell Proteomics. 2008, 7, 1725–1736. [Google Scholar] [CrossRef]
- Vessal, S.; Arefian, M.; Siddique, K.H.M. Proteomic responses to progressive dehydration stress in leaves of chickpea seedlings. BMC Genomics 2020, 21, 523. [Google Scholar] [CrossRef]
- Gupta, S.; Mishra, S.K.; Misra, S.; Pandey, V.; Agrawal, L.; Nautiyal, C.S.; Chauhan, P.S. Revealing the complexity of protein abundance in chickpea root under drought-stress using a comparative proteomics approach. Plant. Physiol. Biochem. 2020, 151, 88–102. [Google Scholar] [CrossRef]
- Cevik, S.; Akpinar, G.; Yildizli, A.; Kasap, M.; Karaosmanoğlu, K.; Ünyayar, S. Comparative physiological and leaf proteome analysis between drought-tolerant chickpea Cicer reticulatum and drought-sensitive chickpea C. arietinum. J. Biosci. 2019, 44, 20. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M.A. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci. Rep. 2019, 9, 2097. [Google Scholar] [CrossRef]
- Goufo, P.; Moutinho-Pereira, J.M.; Jorge, T.F.; Correia, C.M.; Oliveira, M.R.; Rosa, E.A.; António, C.; Trindade, H. Cowpea (Vigna unguiculata L. Walp.) metabolomics: Osmoprotection as a physiological strategy for drought stress resistance and improved yield. Front. Plant. Sci. 2017, 8, 586. [Google Scholar] [CrossRef]
- Li, Y.; Ruperao, P.; Batley, J.; Edwards, D.; Khan, T.; Colmer, T.D.; Pang, J.; Siddique, K.H.; Sutton, T. Investigating drought tolerance in chickpea using genome-wide association mapping and genomic selection based on whole-genome resequencing data. Front. Plant. Sci. 2018, 9, 190. [Google Scholar] [CrossRef]
- Arefian, M.; Vessal, S.; Malekzadeh-Shafaroudi, S.; Siddique, K.H.; Bagheri, A. Comparative proteomics and gene expression analyses revealed responsive proteins and mechanisms for salt tolerance in chickpea genotypes. BMC Plant. Biol. 2019, 19, 300. [Google Scholar] [CrossRef]
- Richter, J.A.; Behr, J.H.; Erban, A.; Kopka, J.; Zörb, C. Ion-dependent metabolic responses of Vicia faba L to salt stress. Plant Cell Environ. 2018, 42, 295–309. [Google Scholar] [CrossRef]
- Li, M.; Guo, R.; Jiao, Y.; Jin, X.; Zhang, H.; Shi, L. Comparison of salt tolerance in Soja based on metabolomics of seedling roots. Front. Plant. Sci. 2017, 8, 1101. [Google Scholar] [CrossRef] [PubMed]
- Parankusam, S.; Bhatnagar-Mathur, P.; Sharma, K.K. Heat responsive proteome changes reveal molecular mechanisms underlying heat tolerance in chickpea. Environ. Exp. Bot. 2017, 141, 132–144. [Google Scholar] [CrossRef]
- Duressa, D.; Soliman, K.; Taylor, R.; Senwo, Z. Proteomic analysis of soybean roots under aluminum stress. Int. J. Plant. Genomics 2011, 2011, 282531. [Google Scholar] [CrossRef] [PubMed]
- Langridge, P.; Fleury, D. Making the most of ‘omics’ for crop breeding. Trends Biotechnol. 2011, 29, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.K. Translational genomics and multi-omics integrated approaches as a useful strategy for crop breeding. Genes Genomics. 2019, 41, 133–146. [Google Scholar] [CrossRef]
- Varshney, R.K.; Kudapa, H.; Pazhamala, L.; Chitikineni, A.; Thudi, M.; Bohra, A.; Gaur, P.M.; Janila, P.; Fikre, A.; Kimurto, P.; et al. Translational genomics in agriculture: Some examples in grain legumes. Critical Rev. Plant. Sci. 2015, 34, 169–194. [Google Scholar] [CrossRef]
- Ma, Y.; Reif, J.C.; Jiang, Y.; Wen, Z.; Wang, D.; Liu, Z.; Guo, Y.; Wei, S.; Wang, S.; Yang, C.; et al. Potential of marker selection to increase prediction accuracy of genomic selection in soybean (Glycine max L.). Mol. Breed. 2016, 36, 113. [Google Scholar] [CrossRef]
- Sinha, A.; Haider, T.; Narula, K.; Ghosh, S.; Chakraborty, N.; Chakraborty, S. Integrated seed proteome and phosphoproteome analyses reveal interplay of nutrient dynamics, carbon-nitrogen partitioning, and oxidative signalling in chickpea. Proteomics 2020, 20, e1900267. [Google Scholar] [CrossRef]
- Yang, Z.B.; Eticha, D.; Führs, H.; Heintz, D.; Ayoub, D.; Van Dorsselaer, A.; Schlingmann, B.; Rao, I.M.; Braun, H.P.; Horst, W.J. Proteomic and phosphoproteomic analysis of polyethylene glycol-induced osmotic stress in root tips of common bean (Phaseolus vulgaris L.). J. Exp. Bot. 2013, 64, 5569–5586. [Google Scholar] [CrossRef]
- Tripathi, P.; Rabara, R.C.; Reese, R.N.; Miller, M.A.; Rohila, J.S.; Subramanian, S.; Shen, Q.J.; Morandi, D.; Bücking, H.; Shulaev, V. A toolbox of genes, proteins, metabolites and promoters for improving drought tolerance in soybean includes the metabolite coumestrol and stomatal development genes. BMC Genomics 2016, 17, 102. [Google Scholar] [CrossRef]
- Deshmukh, R.K.; Vivancos, J.; Guérin, V.; Sonah, H.; Labbé, C.; Belzile, F.; Bélanger, R.R. Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant. Mol. Biol. 2013, 83, 303–315. [Google Scholar] [CrossRef]
- Pi, E.; Zhu, C.; Fan, W.; Huang, Y.; Qu, L.; Li, Y.; Zhao, Q.; Ding, F.; Qiu, L.; Wang, H.; et al. Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt Stress. Mol. Cell Proteomics 2018, 17, 1209–1224. [Google Scholar] [CrossRef]
- Pi, E.; Qu, L.; Hu, J.; Huang, Y.; Qiu, L.; Lu, H.; Jiang, B.; Liu, C.; Peng, T.; Zhao, Y.; et al. Mechanisms of soybean roots’ tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol. Cell Proteomics 2016, 15, 266–288. [Google Scholar] [CrossRef]
- Valdés-López, O.; Batek, J.; Gomez-Hernandez, N.; Nguyen, C.T.; Isidra-Arellano, M.C.; Zhang, N.; Joshi, T.; Xu, D.; Hixson, K.K.; Weitz, K.K.; et al. Soybean roots grown under heat stress show global changes in their transcriptional and proteomic profiles. Front. Plant. Sci. 2016, 7, 517. [Google Scholar] [CrossRef]
- Harfouche, A.L.; Jacobson, D.A.; Kainer, D.; Romero, J.C.; Harfouche, A.H.; Mugnozza, G.S.; Moshelion, M.; Tuskan, G.A.; Keurentjes, J.J.; Altman, A. Accelerating climate resilient plant breeding by applying next-generation artificial intelligence. Trends Biotechnol. 2019, 37, 1217–1235. [Google Scholar] [CrossRef]
- Negin, B.; Moshelion, M. The advantages of functional phenotyping in pre-field screening for drought-tolerant crops. Funct. Plant. Biol. 2016, 44, 1–107. [Google Scholar] [CrossRef]
- Salter, W.T.; Shrestha, A.; Barbour, M.M. Open source 3D phenotyping of chickpea plant architecture across plant development. BioRxiv 2020. [Google Scholar] [CrossRef]
- Burridge, J.; Jochua, C.N.; Bucksch, A.; Lynch, J.P. Legume shovelomics: High—Throughput phenotyping of common bean (Phaseolus vulgaris L.) and cowpea (Vigna unguiculata subsp, unguiculata) root architecture in the field. Field Crops Res. 2016, 192, 21–32. [Google Scholar] [CrossRef]
- Nguyen, G.N.; Norton, S.L.; Rosewarne, G.M.; James, L.E.; Slater, A.T. Automated phenotyping for early vigour of field pea seedlings in controlled environment by colour imaging technology. PLoS ONE 2018, 13, e0207788. [Google Scholar] [CrossRef]
- Humplík, J.F.; Lazár, D.; Fürst, T.; Husičková, A.; Hýbl, M.; Spíchal, L. Automated integrative high-throughput phenotyping of plant shoots: A case study of the cold-tolerance of pea (Pisum sativum L.). Plant. Methods 2015, 11, 20. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, H.; Zhou, J.; Fu, X.; Ye, H.; Nguyen, H.T. Development of an automated phenotyping platform for quantifying soybean dynamic responses to salinity stress in greenhouse environment. Comput. Electron. Agr. 2018, 151, 319–330. [Google Scholar] [CrossRef]
- Peirone, L.S.; PereyraIrujo, G.A.; Bolton, A. Assessing the efficiency of phenotyping early traits in a greenhouse automated platform for predicting drought tolerance of soybean in the field. Front. Plant. Sci. 2018, 9, 587. [Google Scholar] [CrossRef] [PubMed]
- Naik, H.S.; Zhang, J.; Lofquist, A.; Assefa, T.; Sarkar, S.; Ackerman, D.; Singh, A.; Singh, A.K.; Ganapathysubramanian, B. A real-time phenotyping framework using machine learning for plant stress severity rating in soybean. Plant. Methods. 2017, 13, 23. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Schrider, D.R.; Kern, A.D. Supervised machine learning for population genetics: A new paradigm. Trends Genet. 2018, 343, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; Restrepo-Montoya, M.; Bedoya-Canas, L.E. Modern strategies to assess and breed forest tree adaptation to changing climate. Front. Plant. Sci. 2020, 11, 1606. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, D.; He, F.; Wang, J.; Joshi, T.; Xu, D. Phenotype prediction and genome-wide association study using deep convolutional neural network of soybean. Front. Genet. 2019, 10, 1091. [Google Scholar] [CrossRef]
- Corrêa, A.M.; Teodoro, P.E.; Gonçalves, M.C.; Barroso, L.M.A.; Nascimento, M.; Santos, A.; Torres, F.E. Artificial intelligence in the selection of common bean genotypes with high phenotypic stability. Genet. Mol. Res. 2016, 15, gmr-15028230. [Google Scholar] [CrossRef]
- Cortés, A.J.; López-Hernández, F. Harnessing Crop Wild Diversity for Climate Change Adaptation. Genes 2021, 12, 783. [Google Scholar] [CrossRef]
- Falk, K.G.; Jubery, T.Z.; Mirnezami, S.V.; Parmley, K.A.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer vision and machine learning enabled soybean root phenotyping pipeline. Plant. Methods. 2020, 16, 5. [Google Scholar] [CrossRef]
- Cortés, A.J.; López-Hernández, F.; Osorio-Rodriguez, D. Predicting thermal adaptation by looking into populations’ genomic past. Front. Genet. 2020, 11, 1093. [Google Scholar] [CrossRef]
- Jenko, J.; Gorjanc, G.; Cleveland, M.A.; Varshney, R.K.; Whitelaw, C.B.A.; Woolliams, J.A.; Hickey, J.M. Potential of promotion of alleles by genome editing to improve quantitative traits in livestock breeding programs. Genet. Sel. Evol. 2015, 47, 55. [Google Scholar] [CrossRef]
- Wang, H.; Cimen, E.; Singh, N.; Buckler, E. Deep learning for plant genomics and crop improvement. Curr. Opin. Plant. Biol. 2020, 54, 34–41. [Google Scholar] [CrossRef]
- Abadi, S.; Yan, W.X.; Amar, D.; Mayrose, I. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput. Biol. 2017, 13, e1005807. [Google Scholar] [CrossRef]
- Lin, J.; Wong, K.C. Off-target predictions in CRISPR-Cas9 gene editing using deep learning. Bioinformatics 2018, 34, i656–i663. [Google Scholar] [CrossRef]
- Vakilian, K.A. Machine learning improves our knowledge about miRNA functions towards plant abiotic stresses. Sci. Rep. 2020, 10, 1–10. [Google Scholar]
- Varshney, R.K.; Pandey, M.K.; Bohra, A.; Singh, V.K.; Thudi, M.; Saxena, R.K. Toward the sequence-based breeding in legumes in the post-genome sequencing era. Theor. Appl. Genet. 2019, 132, 797–816. [Google Scholar] [CrossRef]
- Lenz, P.R.; Nadeau, S.; Mottet, M.J.; Perron, M.; Isabel, N.; Beaulieu, J.; Bousquet, J. Multi-trait genomic selection for weevil resistance, growth, and wood quality in Norway Spruce. Evol. Appl. 2020, 13, 76–94. [Google Scholar] [CrossRef] [PubMed]
- Tong, H.; Nikoloski, Z. Machine learning approaches for crop improvement: Leveraging phenotypic and genotypic big data. J. Plant. Physiol. 2021, 257, 153354. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; Liu, X.; Sedlacek, J.; Wheeler, J.A.; Lexer, C.; Karrenberg, S. Maintenance of Female-Bias in a Polygenic Sex Determination System is Consistent with Genomic Conflict. In On the Big Challenges of a Small Shrub: Ecological Genetics of Salix Herbacea, L.; Acta Universitatis Upsaliensis: Uppsala, Sweden, 2015. [Google Scholar]
- Crossa, J.; Martini, J.W.; Gianola, D.; Pérez-Rodríguez, P.; Jarquin, D.; Juliana, P.; Montesinos-López, O.; Cuevas, J. Deep kernel and deep learning for genome-based prediction of single traits in multienvironment breeding trials. Front. Genet. 2019, 10, 1168. [Google Scholar] [CrossRef] [PubMed]
- Abdollahiarpanahi, R.; Gianola, D.; Peñagaricano, F. Deep learning versus parametric and ensemble methods for genomic prediction of complex phenotypes. Genet. Sel. Evol. 2020, 52, 12. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).