Abstract
Many vector-borne viruses possess the ability to manipulate vector behaviors to facilitate their transmission. There is evidence that the mechanism of this phenomenon has been described in part as direct manipulation through regulating vector chemosensation. Rice stripe virus (RSV) is transmitted by the small brown planthopper, Laodelphax striatellus (Fallen), in a persistent, circulative–propagative manner. The effect of RSV infection on the olfactory system of L. striatellus has not been fully elucidated. Here, we employed transcriptomic sequencing to analyze gene expression profiles in antennae, legs and heads (without antennae) from L. striatellus females and males with/without RSV infection. Comparisons of the differentially expressed genes (DEGs) among antennae, legs and heads indicated that tissue-specific changes in the gene expression profile were greater than sex-specific changes. A total of 17 olfactory related genes were differentially expressed in viruliferous antennae as compared to nonviruliferous antennae, including LstrOBP4/9, LstrCSP1/2/5, LstrGR28a/43a/43a-1, LstrIR1/2/NMDA1, LstrOR67/85e/56a/94 and LstrSNMP2/2-2. There are 23 olfactory related DEGs between viruliferous and nonviruliferous legs, including LstrOBP2/3/4/12/13, LstrCSP13/5/10, LstrIR1/2/Delta2/Delta2-1/kainate2/NMDA2, LstrOR12/21/31/68 and LstrORco. A low number of olfactory related DEGs were found between viruliferous and nonviruliferous heads, including LstrCSP1, LstrOBP2, LstrOR67 and LstrSNMP2-2. Among these DEGs, the expression patterns of LstrOBP2, LstrOBP3 and LstrOBP9 in three tissues was validated by quantitative real-time PCR. The demonstration of overall changes in the genes in L. striatellus’ chemoreception organs in response to RSV infection would not only improve our understanding of the effect of RSV on the olfactory related genes of insect vectors but also provide insights into developing approaches to control the plant virus transmission and spread as well as pest management in the future.
1. Introduction
Vector-borne viruses contribute to a substantial portion of the global plant disease burden, accounting for more than 70% of all known plant viruses [1,2]. The horizontal transmission and spread of vector-borne viruses relies on the movement of insect vectors, which could be manipulated by vector-borne viruses [3,4]. The mechanism by which viruses manipulate vector behaviors has largely been described as indirect manipulation through modifications of the quality, color, odor and associated plant traits. For instance, cucumber mosaic virus (CMV) accelerates the emigration of aphid vectors by reducing host plant quality, and increased the attractiveness of infected plants to aphid vectors by inducing elevated emissions of a volatile blend [3]. Potato leafroll virus (PLV) infection changes the volatile blend of potato to attract vector aphids, thereby increasing the probability of virus acquisition [5]. Furthermore, some vector-borne viruses could manipulate vector behavior via directly affecting the nervous and olfactory system. Tomato yellow leaf curl virus (TYLCV) reduces whitefly preference to viruliferous plant by disabling the nervous system of vectors [6]. Southern rice black streaked dwarf virus (SRBDV) and tomato chlorosis virus (TCV) are able to regulate the gene expression of odorant-binding protein (OBP) 2 and OBP3 in their vectors, Sogatella furcifera and Bemisia tabaci, respectively, and to reverse the preferences of nonviruliferous/viruliferous vectors, finally affecting the virus transmission [7,8]. Therefore, various vector-borne viruses have developed the ability to utilize a direct or indirect manipulation mechanism to induce behavioral changes of insect vectors, to benefit their spread. However, the effect of vector-borne virus infection on the olfactory system of insect vectors has not been fully elucidated.
The critical roles of the insect’s olfactory system in their locomotion, host seeking, probing and feeding are mainly executed via detecting semiochemicals by sensilla on the main periphery, such as the main chemoreception organ, the antennae [9,10]. A number of protein families located in these peripheral sensory organs are responsible for the recognition of semiochemicals, which include two classes of carrier proteins: OBPs and chemosensory proteins (CSPs), and four classes of membrane proteins: ionotropic (IR), gustatory (GRs), membrane-bound odorant receptors (ORs)/olfactory co-receptor receptor (ORco) and sensory neuron membrane proteins (SNMPs) [11,12,13,14,15,16]. When environmental chemicals penetrate the pores of sensilla, OBPs and CSPs as carriers bind these chemicals in the sensilla lymph and transport the chemicals to ORs or IRs located on the olfactory sensory neurons. Then, chemical signals are converted into electrical signals, and conveyed to the insect’s brain to eventually modulate the insect’s behavior [14,17]. Thus far, the role of olfactory-related proteins in virus transmission remains unclear.
Rice stripe virus (RSV), a typical member of the genus Tenuivirus, has inflicted serious yield losses of rice by causing the notorious rice stripe disease in Eastern Asia [18,19]. It is transmitted by the small brown planthopper, Laodelphax striatellus (Fallen), in a persistent, circulative–propagative manner [20,21,22]. RSV particles enter L. striatellus via the alimentary canal during feeding and initially establish infection in the midgut epithelium, propagating in midgut visceral muscles. After transferring into the hemolymph, these virus invade several key organs via lymph circulation and, ultimately, arrive at the salivary gland, from which RSV particles are inoculated back to the plant hosts along with saliva. RSV transmission in L. striatellus requires specialized interactions between components of the virus and vector, evidenced by the transcriptome analyses of the expression changes of overall mRNA genes and the regulation of RSV-derived siRNA in L. striatellus after RSV infection [23,24,25]. In addition, the organ-specific transcriptomes of L. striatellus’ alimentary canal and salivary gland characterize the responses of two organs in confronting RSV infection [26]. However, the molecular mechanism underlying the interactions between RSV and chemoreception organs remains unknown at the omic level.
Our preliminary antennae-specific transcriptome analysis identified 14 OBPs, 12 CSPs, 7 SNMPs and 95 ORs in L. striatellus [27]. Meanwhile, viruliferous L. striatellus might have a stronger olfactory and seeking ability for rice than nonviruliferous insect [28]. Thus, the current study aims to further investigate the host preference mechanism of viruliferous L. striatellus and the role of the olfactory system in RSV transmission. We compared the response of the antennae, legs and heads (without antennae) from both L. striatellus females and males to RSV infection using next-generation deep-sequencing techniques. Furthermore, olfactory related genes from differentially expressed genes (DEGs) of chemoreception organ transcriptomes were enriched between viruliferous and nonviruliferous organs. Lastly, a qPCR was performed to confirm the expression changes of three OBPs between viruliferous and nonviruliferous tissues. Our results would not only improve our understanding on the role of the olfactory chemoreception organs system in RSV infection and transmission but also provide new insights into the symbiosis interaction between virus, insect and plant. A deeper understanding of this interaction provides new avenues for controlling plant viruses and their vectors.
2. Results
2.1. Illumina Sequencing and Assembly of the Chemoreception Organs of Female and Male Laodelphax striatellus
Twelve cDNA libraries were obtained from dissected antennae, heads and legs of male and female L. striatellus with or without RSV. The transcriptomes of twelve libraries (Viruliferous female antennae, VFA; Viruliferous male antennae, VMA; Viruliferous female legs, VFL; Viruliferous male legs, VML; Viruliferous female heads, VFH; Viruliferous male heads, VMH; Nonviruliferous female antennae, NFA; Nonviruliferous male antennae, NMA; Nonviruliferous female legs, NFL; Nonviruliferous male legs, NML; Nonviruliferous female heads, NFH; Nonviruliferous male heads, NMH) were sequenced using the Illumina HiSeq™ 2000 platform, and generated 663,707,248 clean sequence reads, containing 99.55 G bp clean bases (Table S1). GC percentages (%) of sequence reads from the twelve libraries were all approximately 40%, and Q20 (%) were more than 95.88%, which showed that the accuracy and quality of the sequencing data met the standard for further analysis (Table S1). Of the reads from each transcriptome, 58.06~68.94% paired-end clean reads were mapped to the genome of L. striatellus (Table S1) and assembled into 20,409 genes with function annotation. Additionally, the normalized read count with Fragments per Kilobase of transcript per Million mapped reads (FPKM) of these genes were calculated.
Principal component analysis (PCA) was used to compare the twelve transcriptomes from viruliferous and nonviruliferous organs of females and males. Overall, the principal component axis 1 and 2 (PC1 and PC2, accounting for 47.24% and 27.05% of the observed gene expression variation, respectively), separated these libraries into three groups: Antennae libraries (VMA, VFA, NMA and NFA), Legs libraries (VML, VFL, NML and NFL) and Heads libraries (VMH, VFH, NMH and NFH) (Figure 1A). The comparison between the three tissues demonstrated the relative most number of DEGs between heads and legs (especially in VMH versus (vs.) VML), modest in antennae vs. heads, and least in antennae vs. legs. However, the comparison between female tissues and male tissues showed fewer DEGs (Figure 1B). These results demonstrated that organ-specific and sex-specific DEGs profile in L. striatellus might be involved in the host preference of viruliferous L. striatellus.
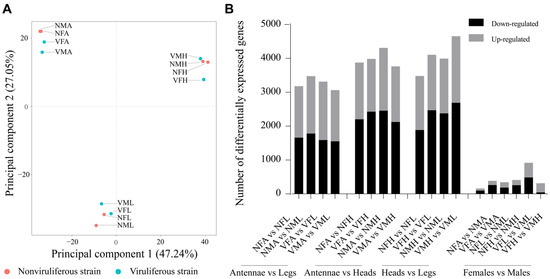
Figure 1.
The transcript abundance in antennae, legs and heads of female and male L. striatellus. (A) Principal component analysis of the transcriptomes from the three organs of viruliferous/nonviruliferous L. striatellus females and males. Viruliferous transcriptomes are denoted by green dots, and nonviruliferous transcriptomes are denoted by pink dots. (B) The total number of DEGs from comparisons among three organs or between females and males. The number of down-regulated DEGs is denoted by black, and the number of up-regulated DEGs shown by gray.
2.2. Gene Ontology Classification of Differentially Expressed Genes among the Chemoreception Organs of Females and Males
The DEGs among the antennae, legs and heads were functionally annotated and classified into different molecular function categories by gene ontology (GO) analysis (Figure 2). Eight pairwise comparisons between antennae and other tissues revealed a total of 1628, 1234, 1510, 1267, 1451, 1107, 1514 and 1409 DEGs with GO analysis. There were approximately 37 significantly enriched GO categories concerning molecular function seen in the DEGs of eight comparisons, of which the top three significant GO terms were protein binding (16.42~21.77%), DNA/RNA/chromatin binding (6.63~9.77%) and odorant-binding/olfactory receptor activity (6.94~10.12%) (Figure 2A,B,D,E and Figure S1A,B,D,E). In the DEGs’ molecular function of heads vs. legs, approximately 35 significantly enriched GO categories were identified with the top three GO terms involved in protein binding, DNA/RNA/chromatin binding and G-protein coupled receptor activity or peptidase activity (Figure 2C,F and Figure S1C,F).
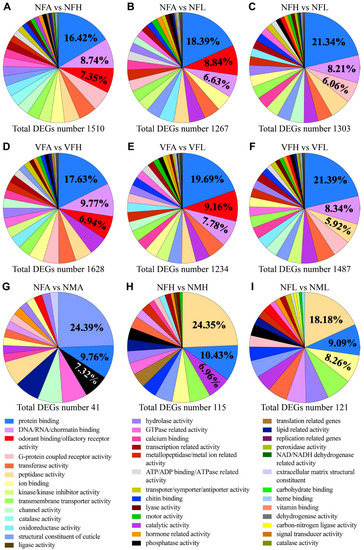
Figure 2.
Proportions of DEGs among (A–F) three organs or (G–I) between two sexes classified by a level 1 molecular function gene ontology. The legend indicates the top three listed GO (Gene Ontology) terms in at least one pairwise comparison.
When comparing the transcriptomes between females and males, only 152, 93, 286, 41, 115 and 121 genes were differentially expressed between VFA vs. VMA, VFL vs. VML, VFH vs. VMH, NFA vs. NMA, NFL vs. NML, NFH vs. NMH, respectively (Figure 2G–I and Figure S1G–I). The enriched GO terms for these sex-specific DEGs among the three tissues include protein binding, structural constituent of the ribosome, phosphatase activity, peptidase activity, and so on.
Taken together, these results demonstrated that a large number of genes involved in the regulation of cellular environment and peripheral chemosensory were more or less expressed in chemoreception organs, antennae and legs, but a low number of DEGs were found between females and males. It suggested that these organ- and sex-specific DEGs might participate in the host preference of L. striatellus on rice, not considering whether the samples are from viruliferous or nonviruliferous L. striatellus.
2.3. Differentially Expressed Genes Induced by RSV Infection
Thus, we next inspected the gene expression profile of viruliferous and nonviruliferous organs through a comparative heatmap. The gene expression patterns for the same organ clustered together. Among these clustered samples, the viruliferous samples were outgrouped from the nonviruliferous samples (Figure 3). According to the Venn diagram, the comparison between viruliferous and nonviruliferous female tissues revealed 439 (262 Up, 177 Down), 980 (521 Up, 459 Down) and 370 (274 Up, 96 Down) DEGs in antennae, legs and heads, respectively. Among them, there were 62 DEGs shared to responses to RSV infection (with 29 up-regulated and 20 down-regulated in all three tissues) (Figure 4A). Similarly, comparisons between viruliferous and nonviruliferous male tissues identified 659, 579 and 255 DEGs, respectively. The three tissues of males shared 79 DEGs in response to RSV infection (Figure 4B).
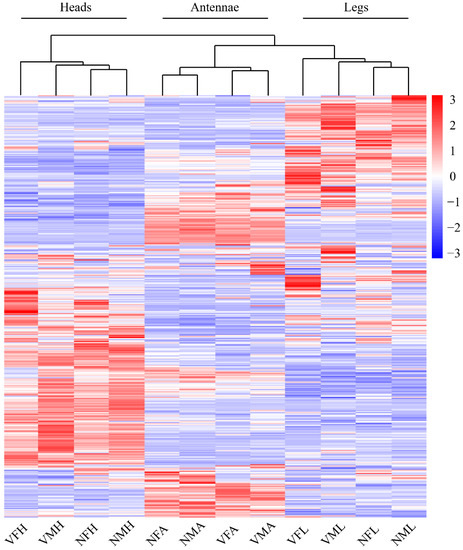
Figure 3.
The expression profiles of DEGs between viruliferous and nonviruliferous organs from female and male L. striatellus. All DEGs expression were performed in each organ in both males and females L. striatellus with or without RSV infection by heatmap. The expression profiles of each gene were calculated based on log2FPKM.
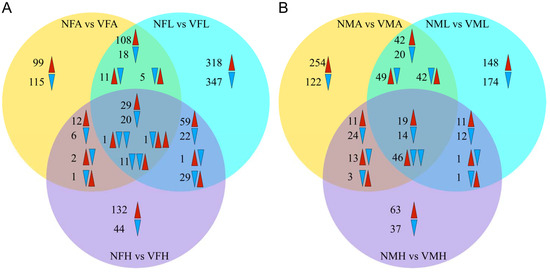
Figure 4.
The changes in DEGs expression between viruliferous and nonviruliferous organs from female and male L. striatellus. (A) Venn diagram summary of DEGs comparing VFA vs. NFA, VFL vs. NFL, VFH vs. NFH. (B) Venn diagram summary of DEGs comparing VMA vs. NMA, VML vs. NML, VMH vs. NMH. The up-regulated DEGs are denoted by red arrows, and the down-regulated DEGs shown as blue arrows.
2.4. Gene Ontology Classification of Differentially Expressed Genes Induced by RSV Infection
Hundreds of DEGs between viruliferous and nonviruliferous tissues were functionally annotated by GO analysis and were classified into 28~33 significantly enriched categories between viruliferous and nonviruliferous organs (Figure 5A–F). The top ten GO categories included protein binding, peptidase activity, structural constituent of cuticle, DNA/RNA/nucleotide/chromatin binding, transferase activity, transmembrane transporter activity, hydrolase activity, catalytic activity, ion binding and odorant-binding/olfactory receptor activity (Figure 5A,B,D,E). There were only 18 and 24 significantly enriched GO terms from nonviruliferous vs. viruliferous male and female heads, respectively (Figure 5C,F). In particular, a large number of olfactory-related genes (7.14%, 4.51%, 2.14% and 4.17%) were found to be more or less enriched in viruliferous chemoreception organs (antennae and legs) as compared to nonviruliferous chemoreception organs (Figure 5A,B,D,E). The data indicated a link of the olfactory related genes to the difference between the viruliferous and nonviruliferous chemoreception organs, however, deep analysis of these genes may shed light on the interaction between RSV and the vector’s chemoreception organs.
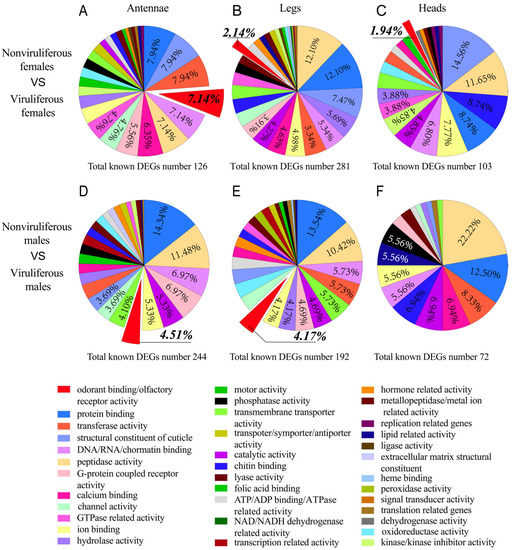
Figure 5.
GO analysis of DEGs between viruliferous and nonviruliferous organs. Proportions of DEGs between viruliferous and nonviruliferous (A,D) antennae/(B,E) legs/(C,F) heads classified by a level 1 molecular function gene ontology. The legend indicates the top ten listed GO terms in at least one pairwise comparison.
2.5. Changes in the Expression of Olfactory-Related Genes in the Chemoreception Organs Infected by RSV
In order to validate the effect of RSV infection on the olfactory system, a more in-depth expression profiling the olfactory-related chemosensation and olfaction DEGs was performed. A total of 17 olfactory-related genes were differentially expressed in viruliferous antennae compared to nonviruliferous antennae (Figure 6 left). A cluster of olfactory genes, including LstrOBP4, LstrCSP2/5, LstrGR28a, LstrIR1/NMDA1, LstrOR67/85e and LstrSNMP2/2-2, showed higher abundance after infection in both male and female antennae (Figure 6 left). The expression level of six other olfactory genes (LstrOBP9, LstrGR43a/43a-1, LstrIR2 and LstrOR56a/94) decreased in both male and female viruliferous antennae (Figure 6 left). Between viruliferous and nonviruliferous legs, a total of 23 olfactory-related genes were found to be differentially expressed (down-regulated: LstrCSP1, LstrIR kainate2/NMDA2/Delta2-1, LstrOR21/31/68, LstrORco and LstrSNMP1-4/2/2-2; up-regulated: LstrCSP3/5/10, LstrIR1/2/Delta2, LstrOBP3/4/12/13 and LstrOR12) (Figure 6 middle). Comparative gene expression profiling of viruliferous vs. nonviruliferous heads revealed only four DEGs (LstrCSP1, LstrOBP2, LstrOR67 and LstrSNMP2-2) (Figure 6 right). Interestingly, three olfactory genes of these DEGs (LstrCSP1, LstrOBP2, LstrSNMP2-2) appeared to show opposite changes between female and male organs after RSV infection (Figure 6). Consistently, the influence of RSV infection on the expression of olfactory-related genes in antennae and legs was greater than that in heads (without antennae).
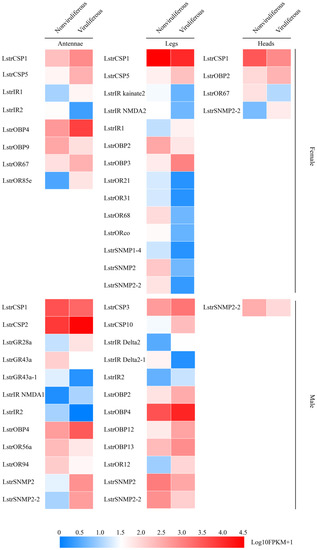
Figure 6.
The expression profiles of differentially abundant olfactory-related genes between viruliferous and nonviruliferous female/male organs. The expression profiles of each gene were calculated based on log10FPKM+1.
2.6. Validation of the OBPs Expressions in RSV-Infected Chemoreception Organs by Quantitative Real-Time PCR
To confirm the above transcriptome results, we selected a significant differentially expression OBP from each organ transcriptome for qPCR examination. The expression of selected LstrOBP9 gene in viruliferous female antennae was significantly down-regulated by 33.1% compared to that in nonviruliferous female antennae, while it exhibited no significant change in viruliferous vs. nonviruliferous male antennae (Figure 7A). On the contrary, the expression level LstrOBP3 in viruliferous female legs was about 4-fold of that in nonviruliferous female legs (Figure 7B). Similarly, the LstrOBP2 was also highly expressed in the heads of viruliferous female, but no changes were found between viruliferous and nonviruliferous male heads (Figure 7C). In general, the expression trends of the three LstrOBPs were consistent with our transcriptome data, supporting the involvement of these organ- and sex-specific olfactory-related LstrOBPs in mediating the host preference of L. striatellus on rice plant behavior as well as RSV transmission.
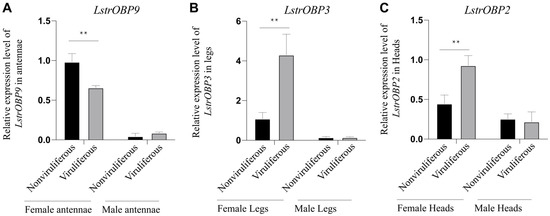
Figure 7.
Relative expression pattern of three selected olfactory-related genes in viruliferous/nonviruliferous female and male organs. (A) Expression changes of LstrOBP9 between viruliferous and nonviruliferous antennae. (B) Expression changes of LstrOBP3 between viruliferous and nonviruliferous legs. (C) Expression changes of LstrOBP2 between viruliferous and nonviruliferous heads. ** indicated p < 0.01.
3. Discussion
The small brown planthopper, as one of the most destructive rice pests, is notorious for causing serious yield loss by feeding damage as well as transmitting RSV [29]. Before the release of the L. striatellus genome sequence in 2017 [30], several previous pyrosequencing-based transcriptome studies have characterized the whole body or organ transcriptomic response [23,26]. In the current study, based on a high-quality genome assembly and annotation of L. striatellus, we performed transcriptomic RNA sequencing to analyze gene expression profiles in three tissues (antennae, legs and heads) from L. striatellus females and males with/without RSV infection. The identification of these DEGs would provide insights into the host preference of L. striatellus on rice plant behavior as well as RSV transmission.
The GO analysis for these DEGs found that the top listed GO terms included protein binding, DNA/RNA/chromatin binding, odorant-binding/olfactory receptor activity, G-protein coupled receptor activity and peptidase activity, suggesting a tissue- or sex- specific signaling reaction. Moreover, the comparison of DEGs between antennae vs. legs/heads was more enriched in the regulation of the cell cycle, cellular environment and peripheral chemosensory than those of heads vs. legs. Since most olfactory sensilla of insects are distributed on the surface of primary chemoreception organs, the antennae, while insect legs and heads are secondary chemoreception organs and olfactory signal processing organs, respectively [31], it is reasonable that olfactory-related genes were higher expressed in the antennae than that in the legs and heads. Thus, the increase or decrease in olfactory-related genes in antennae after virus infection has dire implications for vectorial capacity.
To the best of our knowledge, there has been no systematic study on the effect of virus infection on the olfactory genes expression of insect vector’s antennae. Our study demonstrated overall changes in insect chemoreception organs in response to infection with plant virus RSV. Transcriptomic comparisons of viruliferous vs. nonviruliferous chemoreception organs indicated that a large number of olfactory related genes were differentially expressed in viruliferous antennae and legs but not in the heads. A previous report also found that dengue virus could infect the mosquito’s antennae and result in changes in the transcript abundance of two AgOBPs [32]. The SRBDV infection led to a decrease in SfOBP2 and SfOBP11 in S. furcifera [7]. However, the TCV infection increased the expression of BtOBP3 in B. tabaci [8]. Li and his colleagues found that the LstrORco was stimulated by RSV in the head (with antennae) [28]. However, our data showed that RSV infection down-regulated the LstrORco expression in female L. striatellus legs and did not affect the LstrORco expression in antennae and heads (without antennae). These different results may be due to different sampling and detection methods. The qPCR method was used to detect the LstrORco expression in mixed samples of heads and antennae in Li and his colleagues’ study, while transcriptome sequencing was used to detect the LstrORco expression in antennae and heads (without antennae) in our study. The protein level expression and the location of LstrORco in chemoreception organs need to be further verified by Western blot and in situ hybridization.
Previous reports have demonstrated that plant viruses could regulate olfactory genes to affect the behavior of insect vectors and their associations with host plants. SRBDV and TCV reversed the preferences of insect vectors by regulating OBPs [7,8]. Li and his colleagues reported that silencing of the LstrORco expression inhibited the host seeking behavior and increased the ‘no response’ percent and the response time of L. striatellus [28]. Our transcriptome analysis revealed 17 olfactory-related genes differentially expressed in viruliferous antennae (including two LstrOBPs, three LstrCSPs, three LstrGRs, three LstrIRs, four LstrORs and two LstrSNMPs) as well as five LstrOBPs, three LstrCSPs, six LstrIRs, four LstrORs and LstrORco differentially expressed in viruliferous legs. Additionally, only four olfactory genes were differentially abundant in viruliferous heads. During insect chemical perception, OBPs and CSPs can selectively transport specific odorants to four classes of membrane proteins for odorant recognition [14]. It is possible that the expression changes in olfactory-related genes alter the plant’s volatile organic compound (VOC) recognition and affect insect behavioral responses. Taken together, it was implied that the RSV induction of olfactory related genes, including LstrORco, in antennae and legs could alter the feeding behavior of L. striatellus on rice with the host preference and, thus, at least theoretically, affect virus transmission. In the next step, the plant VOCs recognized by differentially expressed olfactory genes can be screened using gas chromatography–electroantennographic detection (GC–EAD). The two electrodes voltage clamp technique and field trapping experiments can be conducted to validate the interaction between differentially expressed olfactory genes and plant VOCs. Through these techniques, we will further reveal the role of olfactory genes in RSV-induced behavior.
4. Materials and Methods
4.1. Nonviruliferous and Viruliferous L. striatellus Rearing
Nonviruliferous and viruliferous strains of L. striatellus were a gift from Prof. Yijun Zhou’s laboratory of Jiangsu Academy of Agricultural Sciences, Jiangsu province, China, and were reared independently on seedlings of rice cv. Wuyujing 3 in a growth incubator at 25 ± 1 °C, with 80% ± 5% RH and a 12-h light–dark photoperiod.
To ensure that insects were viruliferous, individual female insects were allowed to feed independently. The presence of RSV in their offspring were collected and detected via Dot-ELISA with the monoclonal anti-CP antibody [33]. Highly viruliferous colonies were screened and prepared for subsequent studies.
4.2. Samples Preparation and Transcriptomic Sequencing
The antennae, legs and heads (without antennae) of more than 3000 L. striatellus adults were dissected independently. Viruliferous female/male and nonviruliferous female/male samples were collected and stored at −80 °C for each RNA library. TRIzol method was used to extract total RNA from organ samples as recommended by the manufacturer (Ambion, Austin, TX, USA). RNA purity and concentration were checked using the NanoPhotometer® spectrophotometer (IMPLEN, Westlake Village, CA, USA) and RNA assay kit in Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Twelve paired-end RNA-seq libraries were established from the extracted RNA using the NEBNext® Ultra™ RNA Library Prep Kit (New England BioLabs, Ipswich, MA, USA). First and second strand cDNA were synthesized according to the methods described previously [27]. After the adapters had been attached, PCR was performed with the adapter-ligated cDNA as templates, phusion high-fidelity DNA polymerase, universal PCR primers and index (X) Primer to generate cDNA libraries. These libraries were sequenced with Illumina HiSeq 2000 sequencer (Illumina, San Diego, CA, USA) and 125–150 bp paired-end reads were generated.
4.3. Transcriptomic Assembly and GO Annotation
After sequencing, FastQC was employed to check the quality distribution of the raw data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc, accessed on 20 December 2018). The clean reads were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw reads. The Q20, Q30 and GC content of the clean data were calculated. All paired-end clean reads were aligned to a reference genome of L. striatellus using HISAT2 v2.0.5 (http://www.ccb.jhu.edu/software/hisat, accessed on 25 December 2018) and assembled by StringTie software [34] to generate mapped unigenes. The reference genome of L. striatellus (GigaDB RRID: SCR004002) and gene model annotation files were released by Zhu’s lab in the GigaScience repository [30]. All mapped unigenes were annotated by Gene Ontology (GO) enrichment analysis. GO terms with corrected p-value less than 0.05 were considered significantly enriched.
4.4. Analysis of Differentially Expressed Genes
Read counts mapped to each unigene were calculated by featureCounts v1.5.0-p3 [35]. FPKM of each unigene was obtained based on the length of the gene and reads count mapped to this gene. Unigene expression profiles were performed by heatmapping and Heml software [36]. DEGs between two treatments were identified using the edgeR R package [37]. Corrected p-value of 0.05 and absolute fold change of 2 were set as the threshold for significantly differential expression according to Benjamini and Hochberg method [38].
4.5. Identification and Comparative Expression Profiles of Olfactory Related Genes
Based on GO term described as odorant-binding/olfactory receptor activity, all unigenes were manually retrieved by keywords (OBP, CSP, OR, IR. GR SNMP, chemosensory protein, olfactory protein). Then, retrieved olfactory-related genes were matched with 128 olfactory genes in previous description [27] and identified using a BLASTx algorithm-based search in NCBI website. Expression profiles of these olfactory-related genes were displayed using Heml software according to FPKM of each gene [36].
4.6. Quantitative Real-Time PCR Analysis
Total RNA was isolated from 600 antennae, 100 legs and 50 heads of viruliferous and nonviruliferous adults using the TRIzol Total RNA Isolation Kit (Takara, Dalian, China). First-strand cDNA was synthesized using One Step SYBR PrimeScript RT-PCR kit (Takara, Dalian, China). Primers for LstrOBPs and LstrActin (control) (Table S2) were designed as previous description [27]. The qPCR was conducted with SYBR Premix Ex Taq (Takara) and a CFX96™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) as follows: denaturation for 3 min at 95 °C, followed by 40 cycles at 95 °C for 10 s, and 60 °C for 30 s. Relative expression levels for triplicate samples were calculated using the 2−ΔΔCt method [39], and expression levels of target genes were normalized to the LstrActin gene. Three technical repeats were performed for each of the three biological replicates. The T-test method was used to analyze the significance of relative expression level from various samples [40].
5. Conclusions
In summary, our transcriptomic analysis revealed the expression profiles of genes in antennae, legs and heads of L. striatellus infected by RSV. RSV infection regulated the expression of multiple olfactory-related genes in the a chemoreception organs. In particular, qPCR confirmed the down-regulation of LstrOBP2, and increased LstrOBP3 and LstrOBP9 in the chemoreception organs of females L. striatellus post RSV infection. It is suggested that RSV may alter host preference of the insect vector by changing these olfactory-related genes, which are ultimately beneficial to its transmission. Therefore, it is promising to develop approaches (such as RNAi based gene-silencing technology) to interrupt the olfactory-related genes in chemoreception organs of L. striatellus to control the transmission and spread of RSV in the future. In this regard, this work is a starting point for research, which will eventually help the control of RSV and provide new strategies for the control of other vector-borne viruses.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms221910299/s1, Figure S1: GO analysis of DEGs among male organs (or between female and male viruliferous organs), Table S1. Summary statistics of Illumina sequence data, Table S2: The primers used in this study.
Author Contributions
The work presented here was carried out in collaboration among all the authors. Conceptualization, F.L. and Y.L.; methodology, Y.L. and Y.Z.; software, Y.L.; validation, Y.L., Y.Z., Y.X., D.C. and J.H.; formal analysis, Y.L. and Y.Z.; investigation, Y.L. and Y.Z.; resources, Y.Z.; data curation, Y.L.; writing—original draft preparation, Y.L.; writing—review and editing, F.L.; visualization, Y.L.; supervision, F.L. and Y.L.; project administration, F.L. and Y.L.; funding acquisition, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (No. 31801732), Jiangsu Agricultural Scientific Self-innovation Fund (No. CX[181003] and CX[18]3057).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Illumina sequencing reads from these transcriptomes were submitted to the NCBI Sequence Read Archive under accession number PRJNA589749.
Acknowledgments
We thank Yijun Zhou and his colleagues of Jiangsu Academy of Agricultural Sciences for L. striatellus strains and anti-CP antibody.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| DEGs | Differentially expressed genes |
| RSV | Rice stripe virus |
| CMV | Cucumber mosaic virus |
| PLV | Potato leafroll virus |
| TYLCV | Tomato yellow leaf curl virus |
| SRBDV | Southern rice black streaked dwarf virus |
| TCV | Tomato chlorosis virus |
| OBPs | Odorant-binding proteins |
| CSPs | Chemosensory proteins |
| IRs | Ionotropic receptors |
| GRs | Gustatory receptors |
| ORs | Odorant receptors |
| ORco | Olfactory co-receptor receptor |
| SNMPs | Sensory neuron membrane proteins |
| VFA | Viruliferous female antennae |
| VMA | Viruliferous male antennae |
| VFL | Viruliferous female legs |
| VML | Viruliferous male legs |
| VFH | Viruliferous female heads |
| VMH | Viruliferous male heads |
| NFA | Nonviruliferous female antennae |
| NMA | Nonviruliferous male antennae |
| NFL | Nonviruliferous female legs |
| NML | Nonviruliferous male legs |
| NFH | Nonviruliferous female heads |
| NMH | Nonviruliferous male heads |
| FPKM | Fragments per Kilobase of transcript per Million mapped reads |
| PCA | Principal component analysis |
| GO | Gene ontology |
| qPCR | Real-Time Quantitative PCR |
| VOC | volatile organic compounds |
| GC–EAD | gas chromatography–electroantennographic detection |
References
- Ng, J.C.K.; Falk, B.W. Virus-Vector Interactions Mediating Nonpersistent and Semipersistent Transmission of Plant Viruses. Annu. Rev. Phytopathol. 2006, 44, 183–212. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Ammar, E.-D.; Whitfield, A.E.; Redinbaugh, M.G. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef] [PubMed]
- Mauck, K.E.; De Moraes, C.M.; Mescher, M.C. Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc. Natl. Acad. Sci. USA 2010, 107, 3600–3605. [Google Scholar] [CrossRef] [PubMed]
- Keesey, I.W.; Koerte, S.; Khallaf, M.A.; Retzke, T.; Guillou, A.; Grosse-Wilde, E.; Buchon, N.; Knaden, M.; Hansson, B.S. Pathogenic bacteria enhance dispersal through alteration of Drosophila social communication. Nat. Commun. 2017, 8, 265. [Google Scholar] [CrossRef]
- Ngumbi, E.; Eigenbrode, S.D.; Bosque-Pérez, N.A.; Ding, H.; Rodriguez, A. Myzus persicae is arrested more by blends than by individual compounds elevated in headspace of plrv-infected potato. J. Chem. Ecol. 2007, 33, 1733–1747. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Guo, H.; Ge, F.; Sun, Y. Apoptotic neurodegeneration in whitefly promotes the spread of tylcv. Elife 2020, 9, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Yang, H.; Liu, S.; He, H.; Li, Y. Odorant-Binding Protein 2 is Involved in the Preference of Sogatella furcifera (Hemiptera: Delphacidae) for Rice Plants Infected with the Southern Rice Black-Streaked Dwarf Virus. Florida Entomol. 2019, 102, 353. [Google Scholar] [CrossRef]
- Shi, X.B.; Wang, X.Z.; Zhang, D.Y.; Zhang, Z.H.; Zhang, Z.; Cheng, J.; Zheng, L.M.; Zhou, X.G.; Tan, X.Q.; Liu, Y. Silencing of odorant-binding protein gene OBP3 using RNA interference reduced virus transmission of tomato chlorosis virus. Int. J. Mol. Sci. 2019, 20, 4969. [Google Scholar] [CrossRef]
- Renou, M.; Guerrero, A. Insect Parapheromones in Olfaction Research and Semiochemical-Based Pest Control Strategies. Annu. Rev. Entomol. 2000, 45, 605–630. [Google Scholar] [CrossRef]
- Morita, H. Primary processes of insect chemoreception. Adv. Biophys. 1972, 3, 161. [Google Scholar]
- Benton, R.; Sachse, S.; Michnick, S.W.; Vosshall, L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006, 4, e20. [Google Scholar] [CrossRef] [PubMed]
- Benton, R.; Vannice, K.S.; Vosshall, L.B. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 2007, 450, 289. [Google Scholar] [CrossRef] [PubMed]
- Clyne, P.J.; Warr, C.G.; Carlson, J.R. Candidate taste receptors in Drosophila. Science 2000, 287, 1830–1834. [Google Scholar] [CrossRef]
- Leal, S.W. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 2018, 93, 184–200. [Google Scholar] [CrossRef]
- Pelosi, P.; Zhou, J.-J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. C 2006, 63, 1658–1676. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef]
- Toriyama, S. Rice stripe virus: Prototype of a new group of viruses that replicate in plants and insects. Microbiol. Sci. 1986, 3, 347–351. [Google Scholar]
- Falk, B.W.; Tsai, J.H. Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 1998, 36, 139–163. [Google Scholar] [CrossRef]
- Hibino, H. Biology and Epidemiology of Rice Viruses. Annu. Rev. Phytopathol. 1996, 34, 249–274. [Google Scholar] [CrossRef]
- Heong, K.L.; Cheng, J.; Escalada, M.M. Rice Planthoppers—Ecology, Management, Socio Economics and Policy; Springer: Dordrecht, The Netherlands, 2015; ISBN 9789401795340. [Google Scholar]
- Li, Y.; Chen, D.; Hu, J.; Zhang, K.; Liu, F. The α-tubulin of Laodelphax striatellus mediates the passage of rice stripe virus (RSV) and enhances horizontal transmission. PLoS Pathog. 2020, 16, e1008710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Guo, H.; Zheng, H.; Zhou, T.; Zhou, Y.; Wang, S.; Fang, R.; Qian, W.; Chen, X. Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV). BMC Genom. 2010, 11, 303. [Google Scholar] [CrossRef]
- Yang, M.; Xu, Z.; Zhao, W.; Liu, Q.; Li, Q.; Lu, L.; Liu, R.; Zhang, X.; Cui, F. Rice stripe virus-derived siRNAs play different regulatory roles in rice and in the insect vector Laodelphax striatellus. BMC Plant Biol. 2018, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Choi, J.Y.; Tao, X.Y.; Kim, J.S.; Kim, W.; Je, Y.H. Transcriptome analysis of the small brown planthopper, Laodelphax striatellus carrying Rice stripe virus. Plant Pathol. J. 2013, 29, 330–337. [Google Scholar] [CrossRef]
- Zhao, W.; Lu, L.; Yang, P.; Cui, N.; Kang, L.; Cui, F. Organ-specific transcriptome response of the small brown planthopper toward rice stripe virus. Insect Biochem. Mol. Biol. 2016, 70, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, J.; Xiang, Y.; Zhang, Y.; Liu, F. Identification and comparative expression profiles of chemosensory genes in major chemoreception organs of a notorious pests, Laodelphax striatellus. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 33, 100646. [Google Scholar] [CrossRef]
- Li, S.; Zhou, C.; Zhou, Y. Olfactory co-receptor Orco stimulated by Rice stripe virus is essential for host seeking behavior in small brown planthopper. Pest Manag. Sci. 2019, 75, 187–194. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, S.; Tao, X.; Zhou, X. Rice stripe virus: Exploring Molecular Weapons in the Arsenal of a Negative-Sense RNA Virus. Annu. Rev. Phytopathol. 2021, 59, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Jiang, F.; Wang, X.; Yang, P.; Bao, Y.; Zhao, W.; Wang, W.; Lu, H.; Wang, Q.; Cui, N. Genome sequence of the small brown planthopper, Laodelphax Striatellus. Gigascience 2017, 6, gix109. [Google Scholar] [CrossRef] [PubMed]
- Naters, W.; Carlson, J.R. Insects as chemosensors of humans and crops. Nature 2006, 444, 302–307. [Google Scholar] [CrossRef]
- Sim, S.; Ramirez, J.L.; Dimopoulos, G. Dengue virus infection of the aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012, 8, e1002631. [Google Scholar] [CrossRef]
- Wang, G.Z.; Zhou, Y.J.; Chen, Z.X.; University, Z. Hangzhou Production of monoclonal antibodies to Rice stripe virus and application in virus detection. Acta Phytopathol. Sin. 2004, 34, 302–306. [Google Scholar]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Smyth, G.K.; Wei, S. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A Toolkit for Illustrating Heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Haynes, W. Benjamini—Hochberg Method BT. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; p. 78. ISBN 978-1-4419-9863-7. [Google Scholar]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2−ΔΔCT method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).