Plant–Microbiome Crosstalk: Dawning from Composition and Assembly of Microbial Community to Improvement of Disease Resilience in Plants
Abstract
:1. Introduction
2. Structural Dynamics of Microbiome in Plant Life
3. Plant–Microbe Interplays: Recruiting Microbial Communities for Microbiome Assembly
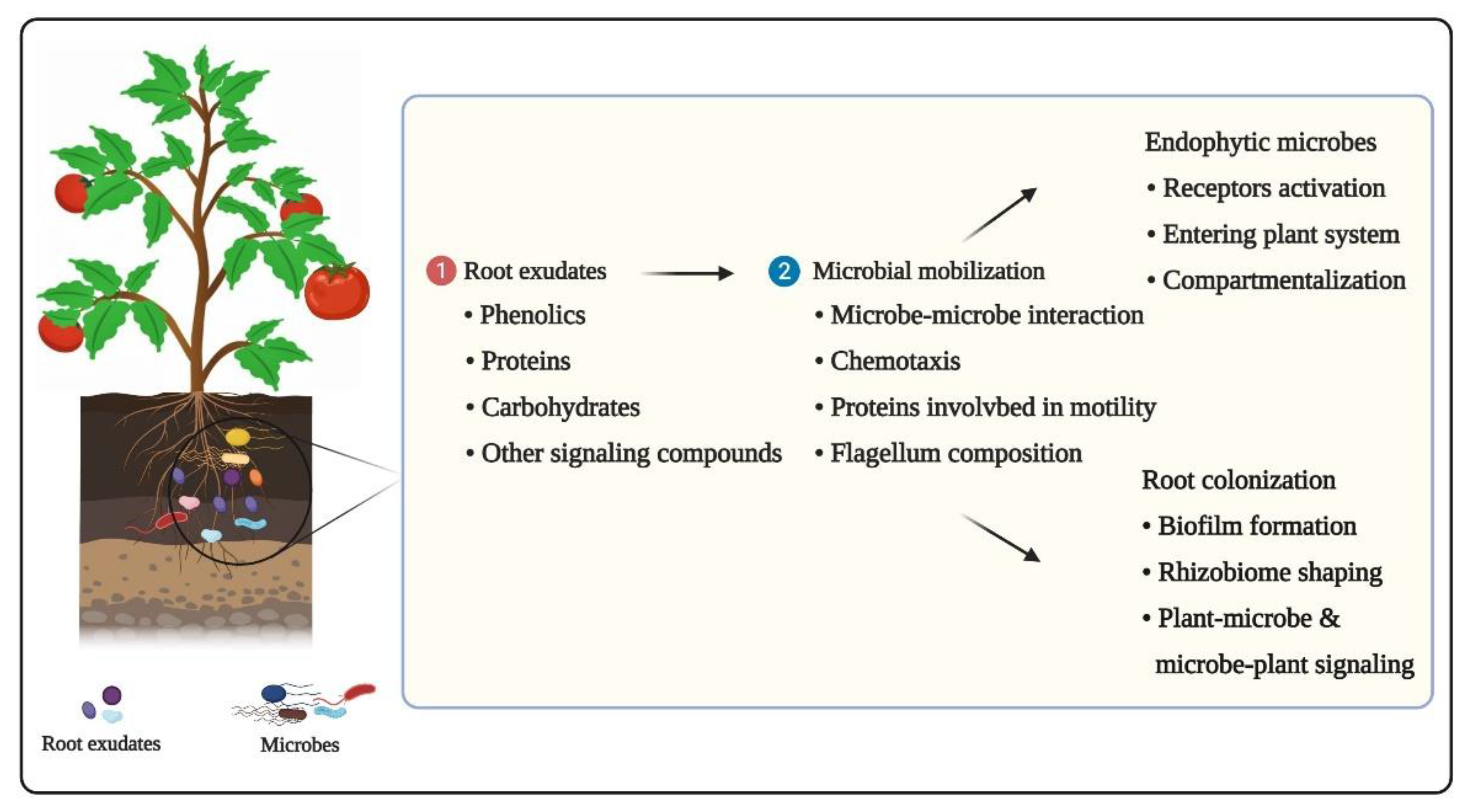
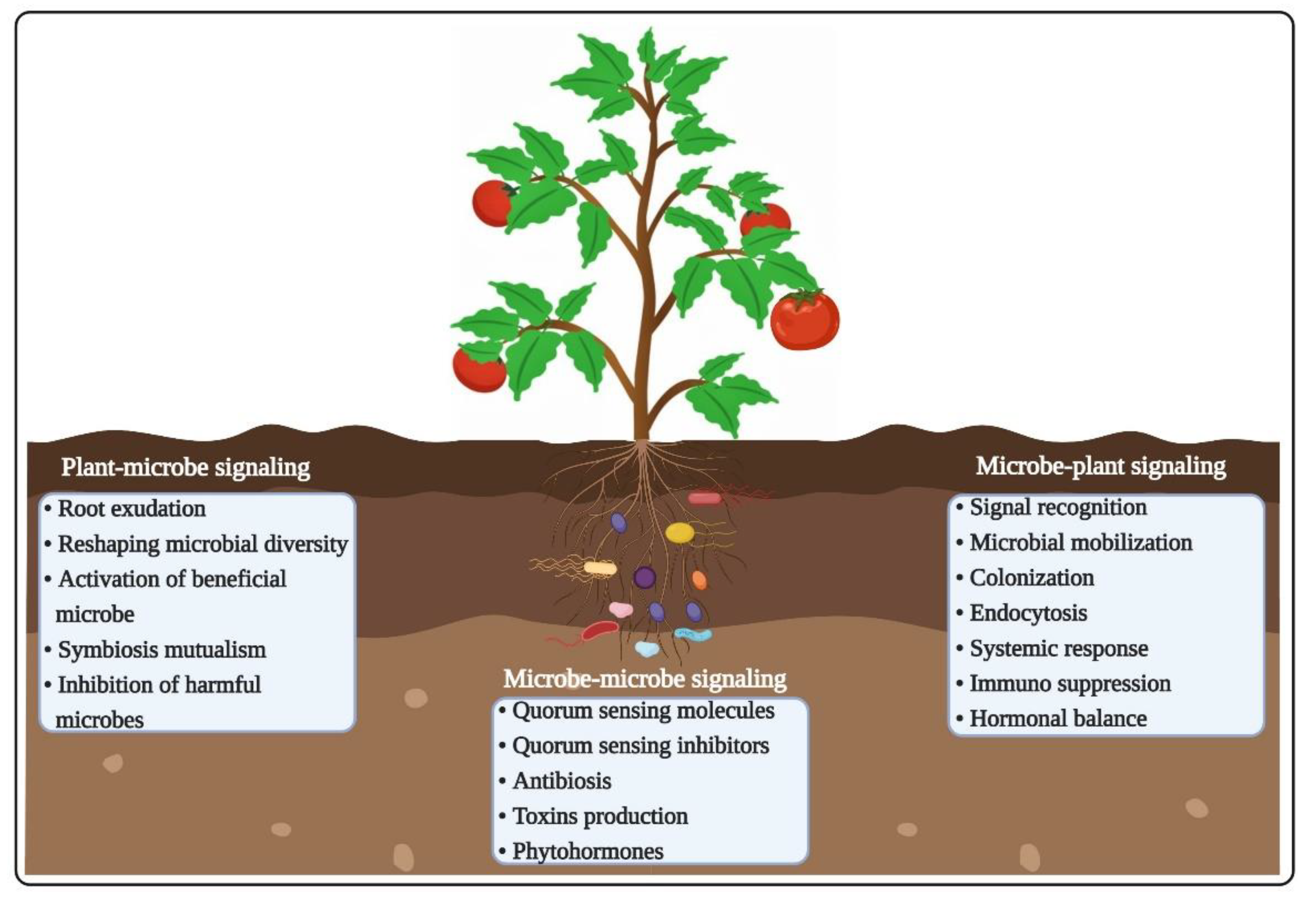
3.1. Root Exudates and Chemotaxis
3.2. Microbe–Microbe Interactions
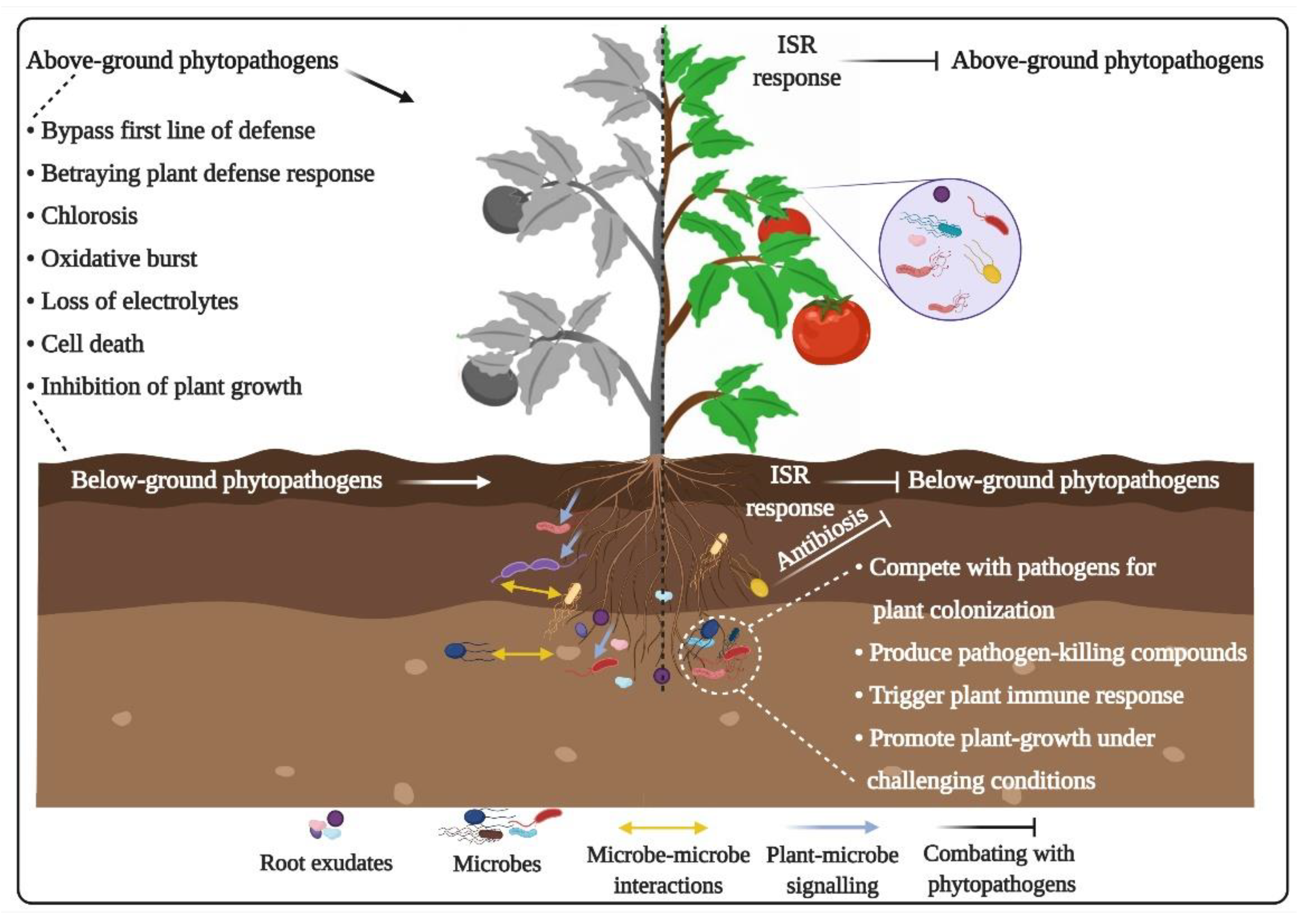
3.3. Plant-Pathogen Interactions
4. Functions of Microbiome in Plant Health
4.1. Roles in Direct Suppression of Plant Pathogens
4.2. Roles in Activation of Plant Immune Response
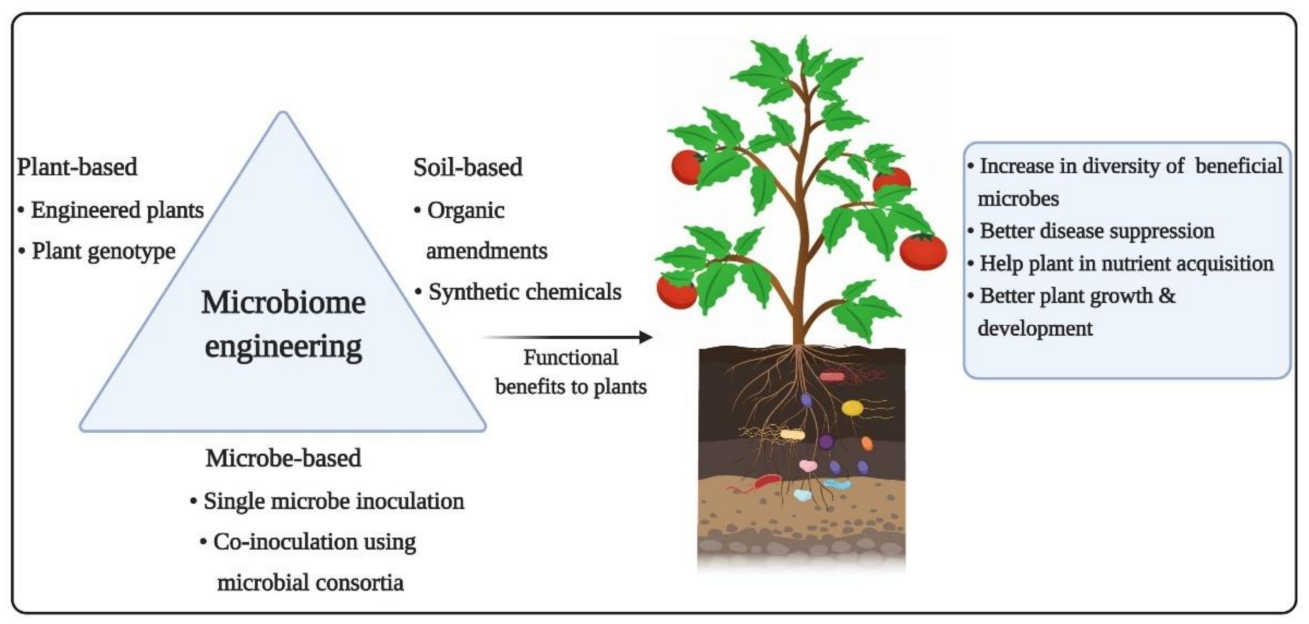
5. Microbiome Engineering: Plausible Functional Benefits on Plant Health
5.1. Traditional Soil Conditioning Using Organic and Chemical Amendments
5.2. Microbiome Engineering Using Artificial Microbial Consortia
5.3. Host Genotype-Dependent Microbiome Engineering
6. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef] [Green Version]
- Syed-Ab-Rahman, S.F.; Xiao, Y.; Carvalhais, L.C.; Ferguson, B.J.; Schenk, P.M. Suppression of Phytophthora capsici infection and promotion of tomato growth by soil bacteria. Rhizosphere 2019, 9, 72–75. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef] [Green Version]
- Hamonts, K.; Trivedi, P.; Garg, A.; Janitz, C.; Grinyer, J.; Holford, P.; Botha, F.C.; Anderson, I.C.; Singh, B.K. Field study reveals core plant microbiota and relative importance of their drivers. Environ. Microbiol. 2018, 20, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, Y.; Zhang, P.; Trivedi, P.; Riera, N.; Wang, Y.; Liu, X.; Fan, G.; Tang, J.; Coletta-Filho, H.D. The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 2018, 9, 4894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergelson, J.; Mittelstrass, J.; Horton, M.W. Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci. Rep. 2019, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roman-Reyna, V.; Pinili, D.; Borjaa, F.N.; Quibod, I.; Groen, S.C.; Mulyaningsih, E.S.; Rachmat, A.; Slamet-Loedin, I.H.; Alexandrov, N.; Mauleon, R. The rice leaf microbiome has a conserved community structure controlled by complex host-microbe interactions. bioRxiv 2019. [Google Scholar] [CrossRef]
- Leach, J.E.; Triplett, L.R.; Argueso, C.T.; Trivedi, P. Communication in the phytobiome. Cell 2017, 169, 587–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzpatrick, C.R.; Salas-González, I.; Conway, J.M.; Finkel, O.M.; Gilbert, S.; Russ, D.; Teixeira, P.J.P.L.; Dangl, J.L. The plant microbiome: From ecology to reductionism and beyond. Annu. Rev. Microbiol. 2020, 74, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Arif, I.; Batool, M.; Schenk, P.M. Plant microbiome engineering: Expected benefits for improved crop growth and resilience. Trends Biotechnol. 2020, 38, 1385–1396. [Google Scholar] [CrossRef]
- Trivedi, P.; Delgado-Baquerizo, M.; Trivedi, C.; Hamonts, K.; Anderson, I.C.; Singh, B.K. Keystone microbial taxa regulate the invasion of a fungal pathogen in agro-ecosystems. Soil Biol. Biochem. 2017, 111, 10–14. [Google Scholar] [CrossRef]
- Xiong, C.; Zhu, Y.G.; Wang, J.T.; Singh, B.; Han, L.L.; Shen, J.P.; Li, P.P.; Wang, G.B.; Wu, C.F.; Ge, A.H. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2020, 229, 1091–1104. [Google Scholar] [CrossRef]
- Shade, A.; Jacques, M.-A.; Barret, M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 2017, 37, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Torres-Cortés, G.; Bonneau, S.; Bouchez, O.; Genthon, C.; Briand, M.; Jacques, M.-A.; Barret, M. Functional microbial features driving community assembly during seed germination and emergence. Front. Plant. Sci. 2018, 9, 902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, J.A.; Santos-Medellín, C.M.; Liechty, Z.S.; Nguyen, B.; Lurie, E.; Eason, S.; Phillips, G.; Sundaresan, V. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol. 2018, 16, e2003862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Gu, Y.; Friman, V.P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [Green Version]
- De Souza, R.S.C.; Okura, V.K.; Armanhi, J.S.L.; Jorrín, B.; Lozano, N.; Da Silva, M.J.; González-Guerrero, M.; De Araújo, L.M.; Verza, N.C.; Bagheri, H.C. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci. Rep. 2016, 6, 28774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cregger, M.; Veach, A.; Yang, Z.; Crouch, M.; Vilgalys, R.; Tuskan, G.; Schadt, C. The Populus holobiont: Dissecting the effects of plant niches and genotype on the microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef]
- Tian, B.; Zhang, C.; Ye, Y.; Wen, J.; Wu, Y.; Wang, H.; Li, H.; Cai, S.; Cai, W.; Cheng, Z. Beneficial traits of bacterial endophytes belonging to the core communities of the tomato root microbiome. Agric. Ecosyst. Environ. 2017, 247, 149–156. [Google Scholar] [CrossRef]
- Soni, R.; Suyal, D.C.; Sahu, B.; Phulara, S.C. Metagenomics: An approach to unravel the plant microbiome and its function. In Phytomicrobiome Interactions and Sustainable Agriculture; Verma, A., Saini, J.K., Hesham, A.E.-L., Singh, H.B., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2021; pp. 32–44. [Google Scholar]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Schlaeppi, K.; Bulgarelli, D. The plant microbiome at work. MPMI 2015, 28, 212–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkel, O.M.; Salas-González, I.; Castrillo, G.; Spaepen, S.; Law, T.F.; Teixeira, P.J.P.L.; Jones, C.D.; Dangl, J.L. The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 2019, 17, e3000534. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Acuña, J.J.; Inostroza, N.G.; Mora, M.L.; Radic, S.; Sadowsky, M.J.; Jorquera, M.A. Endophytic bacterial communities associated with roots and leaves of plants growing in Chilean extreme environments. Sci. Rep. 2019, 9, 4950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouda, A.; Eid, A.M.; Elsaied, A.; El-Belely, E.F.; Barghoth, M.G.; Azab, E.; Gobouri, A.A.; Hassan, S.E.-D. Plant growth-promoting endophytic bacterial community inhabiting the leaves of Pulicaria incisa (Lam.) DC inherent to arid regions. Plants 2021, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S. The soil microbiome influences grapevine-associated microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [Green Version]
- Wallace, J.G.; Kremling, K.A.; Kovar, L.L.; Buckler, E.S. Quantitative genetics of the maize leaf microbiome. Phytobiomes J. 2018, 2, 208–224. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; Tijana, G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef]
- Kecskeméti, E.; Berkelmann-Löhnertz, B.; Reineke, A. Are epiphytic microbial communities in the carposphere of ripening grape clusters (Vitis vinifera L.) different between conventional, organic, and biodynamic grapes? PLoS ONE 2016, 11, e0160852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steven, B.; Huntley, R.B.; Zeng, Q. The influence of flower anatomy and apple cultivar on the apple flower phytobiome. Phytobiomes 2018, 2, 171–179. [Google Scholar] [CrossRef] [Green Version]
- Aleklett, K.; Hart, M.; Shade, A. The microbial ecology of flowers: An emerging frontier in phyllosphere research. Botany 2014, 92, 253–266. [Google Scholar] [CrossRef]
- Griffiths, S.M.; Galambao, M.; Rowntree, J.; Goodhead, I.; Hall, J.; O’Brien, D.; Atkinson, N.; Antwis, R.E. Complex associations between cross-kingdom microbial endophytes and host genotype in ash dieback disease dynamics. J. Ecol. 2020, 108, 291–309. [Google Scholar] [CrossRef] [Green Version]
- Morelli, M.; Bahar, O.; Papadopoulou, K.K.; Hopkins, D.L.; Obradović, A. Role of endophytes in plant health and defense against pathogens. Front. Plant. Sci. 2020, 11, 1312. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, J.; Zhang, H.; Ji, G.; Zeng, L.; Li, Y.; Yu, C.; Fernando, W.D.; Chen, W. Bacterial blight induced shifts in endophytic microbiome of rice leaves and the enrichment of specific bacterial strains with pathogen antagonism. Front. Plant. Sci. 2020, 11, 963. [Google Scholar] [CrossRef] [PubMed]
- Cordovez, V.; Dini-Andreote, F.; Carrión, V.J.; Raaijmakers, J.M. Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef]
- Van der Heijden, M.G.; Hartmann, M. Networking in the plant microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar] [CrossRef]
- O'Neal, L.; Vo, L.; Alexandre, G. Specific root exudates compounds sensed by dedicated chemoreceptors shape Azospirillum brasilense chemotaxis in the rhizosphere. Appl. Environ. Microbiol. 2020, 86, e01026-20. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, J.; Riera, N.; Jin, T.; Li, J.; Wang, N. Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 2017, 5, 97. [Google Scholar] [CrossRef] [PubMed]
- Levy, A.; Gonzalez, I.S.; Mittelviefhaus, M.; Clingenpeel, S.; Paredes, S.H.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F. Genomic features of bacterial adaptation to plants. Nat. Genet. 2018, 50, 138–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Bacilio-Jiménez, M.; Aguilar-Flores, S.; Ventura-Zapata, E.; Pérez-Campos, E.; Bouquelet, S.; Zenteno, E. Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant. Soil 2003, 249, 271–277. [Google Scholar] [CrossRef]
- Kumar, R.; Bhatia, R.; Kukreja, K.; Behl, R.K.; Dudeja, S.S.; Narula, N. Establishment of Azotobacter on plant roots: Chemotactic response, development and analysis of root exudates of cotton (Gossypium hirsutum L.) and wheat (Triticum aestivum L.). J. Basic Microbiol. 2007, 47, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Zhang, N.; Du, W.; Zhang, H.; Liu, Y.; Fu, R.; Shao, J.; Zhang, G.; Shen, Q.; Zhang, R. Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. MPMI 2018, 31, 995–1005. [Google Scholar] [CrossRef] [Green Version]
- Mandal, S.M.; Chakraborty, D.; Dey, S. Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant. Signal. Behav. 2010, 5, 359–368. [Google Scholar] [CrossRef] [Green Version]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Saleh, D.; Charron, J.-B.; Jabaji, S. A crosstalk between Brachypodium root exudates, organic acids, and Bacillus velezensis B26, a growth promoting bacterium. Front. Microbiol. 2020, 11, 575578. [Google Scholar] [CrossRef]
- Walker, T.S.; Bais, H.P.; Grotewold, E.; Vivanco, J.M. Root exudation and rhizosphere biology. Plant. Physiol. 2003, 132, 44–51. [Google Scholar] [CrossRef] [Green Version]
- Sampedro, I.; Parales, R.E.; Krell, T.; Hill, J.E. Pseudomonas chemotaxis. FEMS Microbiol. Rev. 2015, 39, 17–46. [Google Scholar]
- Scharf, B.E.; Hynes, M.F.; Alexandre, G.M. Chemotaxis signaling systems in model beneficial plant–bacteria associations. Plant. Mol. Biol. Rep. 2016, 90, 549–559. [Google Scholar] [CrossRef]
- Ankati, S.; Podile, A.R. Metabolites in the root exudates of groundnut change during interaction with plant growth promoting rhizobacteria in a strain-specific manner. J. Plant. Physiol. 2019, 243, 153057. [Google Scholar] [CrossRef]
- Cesari, A.; Paulucci, N.; López-Gómez, M.; Hidalgo-Castellanos, J.; Plá, C.L.; Dardanelli, M.S. Restrictive water condition modifies the root exudates composition during peanut-PGPR interaction and conditions early events, reversing the negative effects on plant growth. Plant. Physiol. Biochem. 2019, 142, 519–527. [Google Scholar] [CrossRef]
- Liu, C.-W.; Murray, J.D. The role of flavonoids in nodulation host-range specificity: An update. Plants 2016, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Oter, R.; Nakano, R.T.; Dombrowski, N.; Ma, K.-W.; Team, T.A.; McHardy, A.C.; Schulze-Lefert, P. Modular traits of the Rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell Host Microbe 2018, 24, 155–167. [Google Scholar] [CrossRef] [Green Version]
- Jiménez Bremont, J.F.; Marina, M.; Guerrero-Gonzalez, M.d.l.L.; Rossi, F.R.; Sánchez-Rangel, D.; Rodríguez-Kessler, M.; Ruiz, O.A.; Gárriz, A. Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front. Plant. Sci. 2014, 5, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Beskrovnaya, P.; Melnyk, R.A.; Hossain, S.S.; Khorasani, S.; O’Sullivan, L.R.; Wiesmann, C.L.; Bush, J.; Richard, J.D.; Haney, C.H. A genome-wide screen identifies genes in rhizosphere-associated Pseudomonas required to evade plant defenses. mBio 2018, 9, e00433-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleh, D.; Sharma, M.; Seguin, P.; Jabaji, S. Organic acids and root exudates of Brachypodium distachyon: Effects on chemotaxis and biofilm formation of endophytic bacteria. Can. J. Microbiol. 2020, 66, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Blair, P.M.; Land, M.L.; Piatek, M.J.; Jacobson, D.A.; Lu, T.-Y.S.; Doktycz, M.J.; Pelletier, D.A. Exploration of the biosynthetic potential of the Populus microbiome. mSystems 2018, 3, e00045-18. [Google Scholar] [CrossRef] [Green Version]
- Kwak, M.-J.; Kong, H.G.; Choi, K.; Kwon, S.-K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 36, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Raaijmakers, J.M.; de Hollander, M.; Mendes, R.; Tsai, S.M. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018, 12, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; De Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.; Gomez-Exposito, R.; Elsayed, S.S. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, E.J.; Vogel, C.M.; Ueoka, R.; Schäfer, M.; Ryffel, F.; Müller, D.B.; Probst, S.; Kreuzer, M.; Piel, J.; Vorholt, J.A. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nat. Microbiol. 2018, 3, 909–919. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S.; Stukenbrock, E.H.; Rose, L.E. Rapid evolution in plant-microbe interactions—An evolutionary genomics perspective. New Phytol. 2020, 226, 1256–1262. [Google Scholar] [CrossRef] [Green Version]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, V.K.; Jansson, C.; Baker, S.E.; Ahkami, A.H. Molecular mechanisms of plant-microbe interactions in the rhizosphere as targets for improving plant productivity. In Rhizosphere Biology: Interactions between Microbes and Plants; Gupta, V., Sharma, A., Eds.; Springer: Singapore, 2020; pp. 295–338. [Google Scholar]
- Mousa, W.K.; Shearer, C.; Limay-Rios, V.; Ettinger, C.L.; Eisen, J.A.; Raizada, M.N. Root-hair endophyte stacking in finger millet creates a physicochemical barrier to trap the fungal pathogen Fusarium graminearum. Nat. Microbiol. 2016, 1, 16167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chagas, F.O.; de Cassia Pessotti, R.; Caraballo-Rodriguez, A.M.; Pupo, M.T. Chemical signaling involved in plant–microbe interactions. Chem. Soc. Rev. 2018, 47, 1652–1704. [Google Scholar] [CrossRef]
- Khanna, K.; Jamwal, V.L.; Kohli, S.K.; Gandhi, S.G.; Ohri, P.; Bhardwaj, R.; Wijaya, L.; Alyemeni, M.N.; Ahmad, P. Role of plant growth promoting bacteria (PGPRs) as biocontrol agents of Meloidogyne incognita through improved plant defense of Lycopersicon esculentum. Plant. Soil 2019, 436, 325–345. [Google Scholar] [CrossRef]
- Schmidt, R.; de Jager, V.; Zühlke, D.; Wolff, C.; Bernhardt, J.; Cankar, K.; Beekwilder, J.; van Ijcken, W.; Sleutels, F.; De Boer, W. Fungal volatile compounds induce production of the secondary metabolite Sodorifen in Serratia plymuthica PRI-2C. Sci. Rep. 2017, 7, 862. [Google Scholar] [CrossRef]
- Korenblum, E.; Dong, Y.; Szymanski, J.; Panda, S.; Jozwiak, A.; Massalha, H.; Meir, S.; Rogachev, I.; Aharoni, A. Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3874–3883. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Kalia, V.C.; Gong, C.; Patel, S.K.; Lee, J.-K. Regulation of plant mineral nutrition by signal molecules. Microorganisms 2021, 9, 774. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.; Torres, M.; Blanco, L.; Béjar, V.; Sampedro, I.; Llamas, I. Plant growth-promoting activity and quorum quenching-mediated biocontrol of bacterial phytopathogens by Pseudomonas segetis strain P6. Sci. Rep. 2020, 10, 4121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grandclément, C.; Tannières, M.; Moréra, S.; Dessaux, Y.; Faure, D. Quorum quenching: Role in nature and applied developments. FEMS Microbiol. Rev. 2016, 40, 86–116. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Dessaux, Y.; Oger, P. Quorum sensing and quorum quenching: The yin and yang of bacterial communication. ChemBioChem 2009, 10, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C.; Patel, S.K.; Kang, Y.C.; Lee, J.-K. Quorum sensing inhibitors as antipathogens: Biotechnological applications. Biotechnol. Adv. 2019, 37, 68–90. [Google Scholar] [CrossRef]
- Kalia, V.C.; Purohit, H.J. Quenching the quorum sensing system: Potential antibacterial drug targets. Crit. Rev. Microbiol. 2011, 37, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef]
- Hida, A.; Oku, S.; Miura, M.; Matsuda, H.; Tajima, T.; Kato, J. Characterization of methyl-accepting chemotaxis proteins (MCPs) for amino acids in plant-growth-promoting rhizobacterium Pseudomonas protegens CHA0 and enhancement of amino acid chemotaxis by MCP genes overexpression. Biosci. Biotechnol. Biochem. 2020, 84, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Brettell, L.E.; Qiu, Z.; Singh, B.K. Microbiome-mediated stress resistance in plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, M.; Compton, K.K.; Mancl, J.M.; Webb, B.A.; Brown, A.M.; Scharf, B.E.; Schubot, F.D. Structure of the sensory domain of McpX from Sinorhizobium meliloti, the first known bacterial chemotactic sensor for quaternary ammonium compounds. Biochem. J. 2018, 475, 3949–3962. [Google Scholar] [CrossRef]
- Webb, B.A.; Compton, K.K.; del Campo, J.S.M.; Taylor, D.; Sobrado, P.; Scharf, B.E. Sinorhizobium meliloti chemotaxis to multiple amino acids is mediated by the chemoreceptor McpU. MPMI 2017, 30, 770–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudrappa, T.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rekha, K.; Baskar, B.; Srinath, S.; Usha, B. Plant-growth-promoting rhizobacteria Bacillus subtilis RR4 isolated from rice rhizosphere induces malic acid biosynthesis in rice roots. Can. J. Microbiol. 2018, 64, 20–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Weert, S.; Vermeiren, H.; Mulders, I.H.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; De Mot, R.; Lugtenberg, B.J. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. MPMI 2002, 15, 1173–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, K.J.; Vaknin, A.; Fuqua, C.; Kazmierczak, B.I. New twists and turns in bacterial locomotion and signal transduction. J. Bacteriol. 2019, 201, e00439-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Zheng, S.C.; Zhang, T.K.; Liu, Z.Y.; Wang, X.J.; Zhou, X.K.; Yang, C.G.; Duo, J.L.; Mo, M.H. Effect of nicotine from tobacco root exudates on chemotaxis, growth, biocontrol efficiency, and colonization by Pseudomonas aeruginosa NXHG29. Antonie Van Leeuwenhoek 2018, 111, 1237–1257. [Google Scholar] [CrossRef]
- Liu, H.; Khan, M.Y.; Carvalhais, L.C.; Delgado-Baquerizo, M.; Yan, L.; Crawford, M.; Dennis, P.G.; Singh, B.; Schenk, P.M. Soil amendments with ethylene precursor alleviate negative impacts of salinity on soil microbial properties and productivity. Sci. Rep. 2019, 9, 6892. [Google Scholar] [CrossRef]
- Li, P.; Ma, L.; Feng, Y.L.; Mo, M.H.; Yang, F.X.; Dai, H.F.; Zhao, Y.X. Diversity and chemotaxis of soil bacteria with antifungal activity against Fusarium wilt of banana. J. Ind. Microbiol. 2012, 39, 1495–1505. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhu, H.; Luo, S.; Yang, W.; Zhang, L.; Li, S.; Jin, Q.; Cao, Q.; Sun, S.; Xiao, M. Role of maize root exudates in promotion of colonization of Bacillus velezensis strain S3-1 in rhizosphere soil and root tissue. Curr. Microbiol. 2019, 76, 855–862. [Google Scholar] [CrossRef]
- Jiang, N.; Liu, W.; Li, Y.; Wu, H.; Zhang, Z.; Alexandre, G.; Elmerich, C.; Xie, Z. A chemotaxis receptor modulates nodulation during the Azorhizobium caulinodans-Sesbania rostrata symbiosis. Appl. Environ. Microbiol. 2016, 82, 3174–3184. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Hu, H.-J.; Li, J.-Y.; Wang, C.; Chen, S.-L.; Yan, S.-Z. Effects of the endophytic bacteria Bacillus cereus BCM2 on tomato root exudates and Meloidogyne incognita infection. Plant. Dis. 2019, 103, 1551–1558. [Google Scholar] [CrossRef]
- Russell, A.B.; Peterson, S.B.; Mougous, J.D. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Ofek-Lalzar, M.; Sela, N.; Goldman-Voronov, M.; Green, S.J.; Hadar, Y.; Minz, D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Carlström, C.I.; Field, C.M.; Bortfeld-Miller, M.; Müller, B.; Sunagawa, S.; Vorholt, J.A. Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat. Ecol. Evol. 2019, 3, 1445–1454. [Google Scholar] [CrossRef]
- Chhabra, R.; Kaur, S.; Vij, L.; Gaur, K. Exploring physiological and biochemical factors governing plant pathogen interaction: A review. Int. J. Curr. Microbiol. App. Sci. 2020, 9, 1650–1666. [Google Scholar] [CrossRef]
- Boyd, L.A.; Ridout, C.; O'Sullivan, D.M.; Leach, J.E.; Leung, H. Plant–pathogen interactions: Disease resistance in modern agriculture. Trends Genet. 2013, 29, 233–240. [Google Scholar] [CrossRef]
- Peyraud, R.; Dubiella, U.; Barbacci, A.; Genin, S.; Raffaele, S.; Roby, D. Advances on plant-pathogen interactions from molecular toward systems biology perspectives. Plant. J. 2017, 90, 720–737. [Google Scholar] [CrossRef]
- Seybold, H.; Demetrowitsch, T.J.; Hassani, M.A.; Szymczak, S.; Reim, E.; Haueisen, J.; Lübbers, L.; Rühlemann, M.; Franke, A.; Schwarz, K. A fungal pathogen induces systemic susceptibility and systemic shifts in wheat metabolome and microbiome composition. Nat. Commun. 2020, 11, 1910. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Singh, I.K. Molecular Aspects of Plant-Pathogen Interaction; Springer: Singapore, 2018; Volume 10. [Google Scholar]
- Pathma, J.; Raman, G.; Kennedy, R.K.; Bhushan, L.S. Recent advances in plant-microbe interaction. In Microbial Diversity, Interventions and Scope; Sharma, S., Sharma, N., Sharma, M., Eds.; Springer: Singapore, 2020; pp. 23–49. [Google Scholar]
- Zipfel, C.; Oldroyd, G.E. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef]
- Gupta, R.; Lee, S.E.; Agrawal, G.K.; Rakwal, R.; Park, S.; Wang, Y.; Kim, S.T. Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front. Plant. Sci. 2015, 6, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, P.J.P.; Colaianni, N.R.; Fitzpatrick, C.R.; Dangl, J.L. Beyond pathogens: Microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 2019, 49, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, M.; Xiong, C.; Gao, C.; Tsui, C.K.; Zhou, X.; Wang, M.-M.; Zhang, A.-M.; Cai, L. Disease-induced changes in plant microbiome assembly and functional adaptation. Res. Seq. 2020. [Google Scholar] [CrossRef]
- Musonerimana, S.; Bez, C.; Licastro, D.; Habarugira, G.; Bigirimana, J.; Venturi, V. Pathobiomes revealed that Pseudomonas fuscovaginae and Sarocladium oryzae are independently associated with rice sheath rot. Microb. Ecol. 2020, 80, 627–642. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Grube, M.; Köberl, M. The plant microbiome explored: Implications for experimental botany. J. Exp. Bot. 2016, 67, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Prashar, P.; Kapoor, N.; Sachdeva, S. Rhizosphere: Its structure, bacterial diversity and significance. Rev. Environ. Sci. Biotechnol. 2014, 13, 63–77. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babalola, O.O.; Fadiji, A.E.; Enagbonma, B.J.; Alori, E.T.; Ayilara, M.S.; Ayangbenro, A.S. The nexus between plant and plant microbiome: Revelation of the networking strategies. Front. Microbiol. 2020, 11, 2128. [Google Scholar] [CrossRef] [PubMed]
- Sergaki, C.; Lagunas, B.; Lidbury, I.; Gifford, M.L.; Schäfer, P. Challenges and approaches in microbiome research: From fundamental to applied. Front. Plant. Sci. 2018, 9, 1205. [Google Scholar] [CrossRef]
- Berg, M.; Koskella, B. Nutrient-and dose-dependent microbiome-mediated protection against a plant pathogen. Curr. Biol. 2018, 28, 2487–2492. [Google Scholar] [CrossRef] [Green Version]
- Ravanbakhsh, M.H.; Kowalchuk, G.A.; Jousset, A. Root-associated microorganisms reprogram plant life history along the growth-stress resistance tradeoff. ISME J. 2019, 13, 3093–3101. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Jain, J.; Singh, J. The rhizosphere microbiome: Microbial communities and plant health. In Plant Microbiome Paradigm; Varma, A., Tripathi, S., Prasad, R., Eds.; Springer: Cham, Germany, 2020; pp. 175–190. [Google Scholar]
- Ritpitakphong, U.; Falquet, L.; Vimoltust, A.; Berger, A.; Métraux, J.P.; L’Haridon, F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016, 210, 1033–1043. [Google Scholar] [CrossRef] [Green Version]
- Okubara, P.A.; Paulitz, T.C. Root defense responses to fungal pathogens: A molecular perspective. In Root Physiology: From Gene to Function; Lambers, H., Colmer, T., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 215–226. [Google Scholar]
- De Coninck, B.; Timmermans, P.; Vos, C.; Cammue, B.P.; Kazan, K. What lies beneath: Belowground defense strategies in plants. Trends Plant. Sci. 2015, 20, 91–101. [Google Scholar] [CrossRef]
- Qu, Q.; Zhang, Z.; Peijnenburg, W.; Liu, W.; Lu, T.; Hu, B.; Chen, J.; Chen, J.; Lin, Z.; Qian, H. Rhizosphere microbiome assembly and its impact on plant growth. J. Agric. Food Chem. 2020, 68, 5024–5038. [Google Scholar] [CrossRef]
- Gonzalez, M.; Pujol, M.; Metraux, J.P.; Gonzalez-Garcia, V.; Bolton, M.D.; Borrás-Hidalgo, O. Tobacco leaf spot and root rot caused by Rhizoctonia solani Kühn. Mol. Plant Pathol. 2011, 12, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant. Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, W.-Y.; Ren, L.-X.; Ran, W.; Shen, Q.-R. Allelopathic effects of root exudates from watermelon and rice plants on Fusarium oxysporum f. sp. niveum. Plant Soil 2010, 336, 485–497. [Google Scholar] [CrossRef]
- Turrà, D.; el Ghalid, M.; Rossi, F.; di Pietro, A. Fungal pathogen uses sex pheromone receptor for chemotropic sensing of host plant signals. Nature 2015, 527, 521–524. [Google Scholar] [CrossRef]
- Katan, J. Diseases caused by soilborne pathogens: Biology, management and challenges. J. Plant. Pathol. 2017, 99, 305–315. [Google Scholar]
- Andrighetti, T.; Bohar, B.; Lemke, N.; Sudhakar, P.; Korcsmaros, T. MicrobioLink: An integrated computational pipeline to infer functional effects of microbiome-host interactions. Cells 2020, 9, 1278. [Google Scholar] [CrossRef]
- Antoniou, A.; Tsolakidou, M.-D.; Stringlis, I.A.; Pantelides, I.S. Rhizosphere microbiome recruited from a suppressive compost improves plant fitness and increases protection against vascular wilt pathogens of tomato. Front. Plant. Sci. 2017, 8, 2022. [Google Scholar] [CrossRef] [Green Version]
- Anand, A.; Uppalapati, S.R.; Ryu, C.M.; Allen, S.N.; Kang, L.; Tang, Y.; Mysore, K.S. Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant. Physiol. 2008, 146, 703–715. [Google Scholar] [CrossRef] [Green Version]
- Peeters, N.; Guidot, A.; Vailleau, F.; Valls, M. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 2013, 14, 651–662. [Google Scholar] [CrossRef] [Green Version]
- Barelli, L.; Waller, A.S.; Behie, S.W.; Bidochka, M.J. Plant microbiome analysis after Metarhizium amendment reveals increases in abundance of plant growth-promoting organisms and maintenance of disease-suppressive soil. PLoS ONE 2020, 15, e0231150. [Google Scholar] [CrossRef]
- Snelders, N.C.; Kettles, G.J.; Rudd, J.J.; Thomma, B.P. Plant pathogen effector proteins as manipulators of host microbiomes? Mol. Plant Pathol. 2018, 19, 257. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [Green Version]
- Durán, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 2018, 175, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Penton, C.R.; Gupta, V.; Tiedje, J.M.; Neate, S.M.; Ophel-Keller, K.; Gillings, M.; Harvey, P.; Pham, A.; Roget, D.K. Fungal community structure in disease suppressive soils assessed by 28S LSU gene sequencing. PLoS ONE 2014, 9, e93893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, J.-Y.; Han, S.; Hong, H.-J.; Cho, H.; Kim, D.; Kwon, Y.; Kwon, S.-K.; Crüsemann, M.; Lee, Y.B.; Kim, J.F. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016, 10, 119–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hol, W.G.; Garbeva, P.; Hordijk, C.; Hundscheid, M.P.; Gunnewiek, P.J.K.; Van Agtmaal, M.; Kuramae, E.E.; De Boer, W. Non-random species loss in bacterial communities reduces antifungal volatile production. Ecology 2015, 96, 2042–2048. [Google Scholar] [CrossRef] [Green Version]
- Carrión, V.J.; Cordovez, V.; Tyc, O.; Etalo, D.W.; de Bruijn, I.; de Jager, V.C.; Medema, M.H.; Eberl, L.; Raaijmakers, J.M. Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 2018, 12, 2307–2321. [Google Scholar] [CrossRef] [Green Version]
- Gómez Expósito, R.; de Bruijn, I.; Postma, J.; Raaijmakers, J.M. Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front. Microbiol. 2017, 8, 2529. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef]
- Mazurier, S.; Corberand, T.; Lemanceau, P.; Raaijmakers, J.M. Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J. 2009, 3, 977–991. [Google Scholar] [CrossRef]
- Latz, E.; Eisenhauer, N.; Rall, B.C.; Allan, E.; Roscher, C.; Scheu, S.; Jousset, A. Plant diversity improves protection against soil-borne pathogens by fostering antagonistic bacterial communities. J. Ecol. 2012, 100, 597–604. [Google Scholar] [CrossRef]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Fan, X.; Wang, Y.; Kusstatscher, P.; Duan, J.; Wu, S.; Chen, S.; Qiao, K.; Wang, Y.; Ma, B. Bacterial seed endophyte shapes disease resistance in rice. Nat. Plants 2021, 7, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Kwak, Y.-S.; Weller, D.M. Take-all of wheat and natural disease suppression: A review. Plant. Pathol. J. 2013, 29, 125. [Google Scholar] [CrossRef] [PubMed]
- Bonanomi, G.; Antignani, V.; Capodilupo, M.; Scala, F. Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases. Soil Biol. Biochem. 2010, 42, 136–144. [Google Scholar] [CrossRef]
- Santhanam, R.; Weinhold, A.; Goldberg, J.; Oh, Y.; Baldwin, I.T. Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc. Natl. Acad. Sci. USA 2015, 112, E5013–E5020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunziker, L.; Bönisch, D.; Groenhagen, U.; Bailly, A.; Schulz, S.; Weisskopf, L. Pseudomonas strains naturally associated with potato plants produce volatiles with high potential for inhibition of Phytophthora infestans. Appl. Environ. Microbiol. 2015, 81, 821–830. [Google Scholar] [CrossRef] [Green Version]
- Peralta, A.L.; Sun, Y.; McDaniel, M.D.; Lennon, J.T. Crop rotational diversity increases disease suppressive capacity of soil microbiomes. Ecosphere 2018, 9, e02235. [Google Scholar] [CrossRef]
- Ab Rahman, S.F.S.; Singh, E.; Pieterse, C.M.; Schenk, P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef] [Green Version]
- Finkel, O.M.; Castrillo, G.; Paredes, S.H.; González, I.S.; Dangl, J.L. Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant. Biol. 2017, 38, 155–163. [Google Scholar] [CrossRef]
- Castrillo, G.; Teixeira, P.J.P.L.; Paredes, S.H.; Law, T.F.; de Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Mieczkowski, P.; Jones, C.D. Root microbiota drive direct integration of phosphate stress and immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 2011, 77, 3202–3210. [Google Scholar] [CrossRef] [Green Version]
- Vogel, C.; Bodenhausen, N.; Gruissem, W.; Vorholt, J.A. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 2016, 212, 192–207. [Google Scholar] [CrossRef] [Green Version]
- Haney, C.H.; Samuel, B.S.; Bush, J.; Ausubel, F.M. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat. Plants 2015, 1, 15051. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wang, J.; Yang, N.; Wen, Z.; Sun, X.; Chai, Y.; Ma, Z. Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat. Commun. 2018, 9, 3429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahu, K.P.; Kumar, A.; Patel, A.; Kumar, M.; Gopalakrishnan, S.; Prakash, G.; Rathour, R.; Gogoi, R. Rice blast lesions: An Unexplored phyllosphere microhabitat for novel antagonistic bacterial species against Magnaporthe oryzae. Microb. Ecol. 2020, 81, 731–745. [Google Scholar] [CrossRef]
- Spence, C.; Alff, E.; Johnson, C.; Ramos, C.; Donofrio, N.; Sundaresan, V.; Bais, H. Natural rice rhizospheric microbes suppress rice blast infections. BMC Plant. Biol. 2014, 14, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Kesten, C.; Gámez-Arjona, F.M.; Menna, A.; Scholl, S.; Dora, S.; Huerta, A.I.; Huang, H.Y.; Tintor, N.; Kinoshita, T.; Rep, M. Pathogen-induced pH changes regulate the growth-defense balance in plants. EMBO J. 2019, 38, e101822. [Google Scholar] [CrossRef]
- Lazcano, C.; Boyd, E.; Holmes, G.; Hewavitharana, S.; Pasulka, A.; Ivors, K. The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions. Sci. Rep. 2021, 11, 3188. [Google Scholar] [CrossRef] [PubMed]
- Garbeva, P.; Postma, J.; Van Veen, J.; Van Elsas, J. Effect of above-ground plant species on soil microbial community structure and its impact on suppression of Rhizoctonia solani AG3. Environ. Microbiol. 2006, 8, 233–246. [Google Scholar] [CrossRef]
- Vannier, N.; Agler, M.; Hacquard, S. Microbiota-mediated disease resistance in plants. PLoS Pathog. 2019, 15, e1007740. [Google Scholar] [CrossRef] [Green Version]
- Ryu, C.-M.; Farag, M.A.; Hu, C.-H.; Reddy, M.S.; Kloepper, J.W.; Paré, P.W. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004, 134, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Pršić, J.; Ongena, M. Elicitors of plant immunity triggered by beneficial bacteria. Front. Plant Sci. 2020, 11, 594530. [Google Scholar] [CrossRef]
- Iavicoli, A.; Boutet, E.; Buchala, A.; Métraux, J.-P. Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. MPMI 2003, 16, 851–858. [Google Scholar] [CrossRef] [Green Version]
- Ongena, M.; Jourdan, E.; Adam, A.; Paquot, M.; Brans, A.; Joris, B.; Arpigny, J.L.; Thonart, P. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 2007, 9, 1084–1090. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; van Bentum, S.; van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.; Feussner, I.; Pieterse, C.M. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef] [Green Version]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef] [Green Version]
- Pieterse, C.M.; van Pelt, J.A.; Verhagen, B.W.; Ton, J.; van Wees, S.C.; Léon-Kloosterziel, K.M.; Van Loon, L. Induced systemic resistance by plant growth-promoting rhizobacteria. Symbiosis 2003, 35, 39–54. [Google Scholar]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Schornack, S.; Marsh, J.F.; Gobbato, E.; Schwessinger, B.; Eastmond, P.; Schultze, M.; Kamoun, S.; Oldroyd, G.E. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr. Biol. 2012, 22, 2242–2246. [Google Scholar] [CrossRef] [Green Version]
- Zamioudis, C.; Pieterse, C.M. Modulation of host immunity by beneficial microbes. MPMI 2012, 25, 139–150. [Google Scholar] [CrossRef] [Green Version]
- Kloppholz, S.; Kuhn, H.; Requena, N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 2011, 21, 1204–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plett, J.M.; Kemppainen, M.; Kale, S.D.; Kohler, A.; Legué, V.; Brun, A.; Tyler, B.M.; Pardo, A.G.; Martin, F. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr. Biol. 2011, 21, 1197–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brotman, Y.; Landau, U.; Cuadros-Inostroza, Á.; Takayuki, T.; Fernie, A.R.; Chet, I.; Viterbo, A.; Willmitzer, L. Trichoderma-plant root colonization: Escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 2013, 9, e1003221. [Google Scholar] [CrossRef]
- Lakshmanan, V.; Castaneda, R.; Rudrappa, T.; Bais, H.P. Root transcriptome analysis of Arabidopsis thaliana exposed to beneficial Bacillus subtilis FB17 rhizobacteria revealed genes for bacterial recruitment and plant defense independent of malate efflux. Planta 2013, 238, 657–668. [Google Scholar] [CrossRef]
- Millet, Y.A.; Danna, C.H.; Clay, N.K.; Songnuan, W.; Simon, M.D.; Werck-Reichhart, D.; Ausubel, F.M. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant. Cell 2010, 22, 973–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubey, A.; Malla, M.A.; Khan, F.; Chowdhary, K.; Yadav, S.; Kumar, A.; Sharma, S.; Khare, P.K.; Khan, M.L. Soil microbiome: A key player for conservation of soil health under changing climate. Biodivers. Conserv. 2019, 28, 2405–2429. [Google Scholar] [CrossRef]
- Bonanomi, G.; Lorito, M.; Vinale, F.; Woo, S.L. Organic amendments, beneficial microbes, and soil microbiota: Toward a unified framework for disease suppression. Annu. Rev. Phytopathol. 2018, 56, 1–20. [Google Scholar] [CrossRef]
- Ling, N.; Zhu, C.; Xue, C.; Chen, H.; Duan, Y.; Peng, C.; Guo, S.; Shen, Q. Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol. Biochem. 2016, 99, 137–149. [Google Scholar] [CrossRef]
- Porter, S.S.; Bantay, R.; Friel, C.A.; Garoutte, A.; Gdanetz, K.; Ibarreta, K.; Moore, B.M.; Shetty, P.; Siler, E.; Friesen, M.L. Beneficial microbes ameliorate abiotic and biotic sources of stress on plants. Funct. Ecol. 2020, 34, 2075–2086. [Google Scholar] [CrossRef] [Green Version]
- Stringlis, I.A.; de Jonge, R.; Pieterse, C.M. The age of coumarins in plant-microbe interactions. Plant Cell Physiol. 2019, 60, 1405–1419. [Google Scholar] [CrossRef] [Green Version]
- Voges, M.J.; Bai, Y.; Schulze-Lefert, P.; Sattely, E.S. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. USA 2019, 116, 12558–12565. [Google Scholar] [CrossRef] [Green Version]
- Cotton, T.A.; Pétriacq, P.; Cameron, D.D.; Al Meselmani, M.; Schwarzenbacher, R.; Rolfe, S.A.; Ton, J. Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 2019, 13, 1647–1658. [Google Scholar] [CrossRef] [Green Version]
- Hart, M.M.; Antunes, P.M.; Chaudhary, V.B.; Abbott, L.K. Fungal inoculants in the field: Is the reward greater than the risk? Funct. Ecol. 2018, 32, 126–135. [Google Scholar] [CrossRef] [Green Version]
- Kong, Z.; Hart, M.; Liu, H. Paving the way from the lab to the field: Using synthetic microbial consortia to produce high-quality crops. Front. Plant Sci. 2018, 9, 1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vyas, P.; Kumar, D.; Dubey, A.; Kumar, A. Screening and characterization of Achromobacter xylosoxidans isolated from rhizosphere of Jatropha curcas L. (energy crop) for plant-growth-promoting traits. J. Adv. Res. Biotech. 2018, 3, 1–8. [Google Scholar] [CrossRef]
- Del Carmen Orozco-Mosqueda, M.; del Carmen Rocha-Granados, M.; Glick, B.R.; Santoyo, G. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 2018, 208, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Molina-Romero, D.; Baez, A.; Quintero-Hernández, V.; Castañeda-Lucio, M.; Fuentes-Ramírez, L.E.; Bustillos-Cristales, M.d.R.; Rodríguez-Andrade, O.; Morales-García, Y.E.; Munive, A.; Muñoz-Rojas, J. Compatible bacterial mixture, tolerant to desiccation, improves maize plant growth. PLoS ONE 2017, 12, e0187913. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Rajkumar, M.; Oliveira, R.S.; Zhang, C.; Freitas, H. Potential of plant beneficial bacteria and arbuscular mycorrhizal fungi in phytoremediation of metal-contaminated saline soils. J. Hazard. Mater. 2019, 379, 120813. [Google Scholar] [CrossRef] [PubMed]
- Chihaoui, S.-A.; Trabelsi, D.; Jdey, A.; Mhadhbi, H.; Mhamdi, R. Inoculation of Phaseolus vulgaris with the nodule-endophyte Agrobacterium sp. 10C2 affects richness and structure of rhizosphere bacterial communities and enhances nodulation and growth. Arch. Microbiol. 2015, 197, 805–813. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Koprivova, A.; Kopriva, S. Pinpointing secondary metabolites that shape the composition and function of the plant microbiome. J. Exp. Bot. 2020, 72, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Solís, D.; Zetter-Salmón, E.; Contreras-Pérez, M.; del Carmen Rocha-Granados, M.; Macías-Rodríguez, L.; Santoyo, G. Pseudomonas stutzeri E25 and Stenotrophomonas maltophilia CR71 endophytes produce antifungal volatile organic compounds and exhibit additive plant growth-promoting effects. Biocatal. Agric. Biotechnol. 2018, 13, 46–52. [Google Scholar] [CrossRef]
- Tsolakidou, M.-D.; Stringlis, I.A.; Fanega-Sleziak, N.; Papageorgiou, S.; Tsalakou, A.; Pantelides, I.S. Rhizosphere-enriched microbes as a pool to design synthetic communities for reproducible beneficial outputs. FEMS Microbiol. Ecol. 2019, 95, fiz138. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Singh, S.; Kumar Sarma, B.; Bahadur Singh, H. Microbial consortium-mediated reprogramming of defence network in pea to enhance tolerance against Sclerotinia sclerotiorum. J. Appl. Microbiol. 2012, 112, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, S.; Ramamoorthy, V.; Samiyappan, R.; Subbian, P. Intercropping system with combined application of Azospirillum and Pseudomonas fluorescens reduces root rot incidence caused by Rhizoctonia bataticola and increases seed cotton yield. J. Phytopathol. 2013, 161, 405–411. [Google Scholar] [CrossRef]
- George, C.; Kohler, J.; Rillig, M.C. Biochars reduce infection rates of the root-lesion nematode Pratylenchus penetrans and associated biomass loss in carrot. Soil Biol. Biochem. 2016, 95, 11–18. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, J.; Wang, B.; Yoshikuni, Y. Microbiome engineering: Synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 2020, 39, 244–261. [Google Scholar] [CrossRef]
- Paungfoo-Lonhienne, C.; Rentsch, D.; Robatzek, S.; Webb, R.I.; Sagulenko, E.; Näsholm, T.; Schmidt, S.; Lonhienne, T.G. Turning the table: Plants consume microbes as a source of nutrients. PLoS ONE 2010, 5, e11915. [Google Scholar] [CrossRef] [Green Version]
- Wintermans, P.C.; Bakker, P.A.; Pieterse, C.M. Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol. Biol. Rep. 2016, 90, 623–634. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Nomura, K.; Wang, X.; Sohrabi, R.; Xu, J.; Yao, L.; Paasch, B.C.; Ma, L.; Kremer, J.; Cheng, Y. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 2020, 580, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Horton, M.W.; Bodenhausen, N.; Beilsmith, K.; Meng, D.; Muegge, B.D.; Subramanian, S.; Vetter, M.M.; Vilhjálmsson, B.J.; Nordborg, M.; Gordon, J.I. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 2014, 5, 5320. [Google Scholar] [CrossRef]
- Chen, X.; Marszałkowska, M.; Reinhold-Hurek, B. Jasmonic acid, not salicyclic acid restricts endophytic root colonization of rice. Front. Plant Sci. 2020, 10, 1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitter, B.; Pfaffenbichler, N.; Flavell, R.; Compant, S.; Antonielli, L.; Petric, A.; Berninger, T.; Naveed, M.; Sheibani-Tezerji, R.; von Maltzahn, G. A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 2017, 8, 11. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Jaramillo, J.E.; de Hollander, M.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M.; Carrión, V.J. Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome 2019, 7, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balint-Kurti, P.; Simmons, S.J.; Blum, J.E.; Ballaré, C.L.; Stapleton, A.E. Maize leaf epiphytic bacteria diversity patterns are genetically correlated with resistance to fungal pathogen infection. MPMI 2010, 23, 473–484. [Google Scholar] [CrossRef] [Green Version]
- Wagner, M.R.; Roberts, J.H.; Balint-Kurti, P.; Holland, J.B. Heterosis of leaf and rhizosphere microbiomes in field-grown maize. New Phytol. 2020, 228, 1055–1069. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; de Hollander, M.; Raaijmakers, J.M. The wild side of plant microbiomes. Microbiome 2018, 6, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morella, N.M.; Weng, F.C.-H.; Joubert, P.M.; Metcalf, C.J.E.; Lindow, S.; Koskella, B. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc. Natl. Acad. Sci. USA 2020, 117, 1148–1159. [Google Scholar] [CrossRef]
- Latz, M.A.; Kerrn, M.H.; Sørensen, H.; Collinge, D.B.; Jensen, B.; Brown, J.K.; Madsen, A.M.; Jørgensen, H.J.L. Succession of the fungal endophytic microbiome of wheat is dependent on tissue-specific interactions between host genotype and environment. Sci. Total Environ. 2021, 759, 143804. [Google Scholar] [CrossRef] [PubMed]
| Root Exudates | Microbial Receptors 1 | Microbes | Plant Species | Effects | References |
|---|---|---|---|---|---|
| Alanine | CtaA, CtaB, CtaC and CtaD | Pseudomonas protegens CHA0 | Tobacco | Suppresses root disease | [58] |
| Arginine | McpA | Azospirillum caulinodans | Sesbania rostrata | Regulates root colonization and flagella synthesis | [59] |
| Chitinase | McpU | Pseudomonas sp. RP2 | Arachis hypogaea | Increases immunity | [55] |
| Choline | McpX | Sinorhizobium meliloti | Alfalfa | Facilitates nitrogen-fixation in root nodules | [60] |
| Citric acid | McpU | Sinorhizobium meliloti | Alfalfa | Regulates root colonization | [61] |
| Citric acid | McpU | Bacillus subtilis | Arabidopsis thaliana | Enhances root binding of Bacillus subtilis | [62] |
| Ethylene | ETR1 | Azospirillum brasilense | Wheat | Modulates plant morphology | [63] |
| Malic acid | McpU | Bacillus subtilis | Rice | Improves nutrient assimilation and pH regulation | [64] |
| Malic acid | McpA | Pseudomonas fluorescens | Tomato | Improves plant growth and nutrient acquisition | [65] |
| Methyl-glucoside | McpA | Bacillus amyloliquefaciens | Cucumber | Modulates chemotaxis mobility and enhances immunity | [48] |
| Nicotine | McpU | Pseudomonas aeruginosa | Tobacco | Increases biocontrol efficiency against bacterial wilt | [66] |
| Oxalic acid | TlpA1 | Azospirillum caulinodan | Sesbania rostrata | Increases plant growth | [67] |
| Proline | IcpB | Pseudomonas aeruginosa | Cucumber | Shows antifungal activity against Fusarium oxysporum | [68] |
| Proline | McpB | Bacillus velezensis | Maize | Regulates swarming motility and biofilm formation | [69] |
| Proline | McpU | Sinorhizobium meliloti | Alfalfa | Directs flagellar motor rotation and root colonization | [54] |
| Succinic acid | TlpA1 | Azospirillum brasilense | Wheat | Increases plant growth, root volume, and crop yield | [41] |
| Succinic acid | TlpA1 | Bacillus velezensis | Brachypodium distachyon | Regulates biofilm formation | [51] |
| Tryptophan | IcpB | Azorhizobium caulinodans | Sesbania rostrata | Modulates nodulation and nitrogen fixation | [70] |
| Tryptophan | IcpB | Bacillus cereus | Tomato | Reduces the damage of Medoidogyne incognita | [71] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noman, M.; Ahmed, T.; Ijaz, U.; Shahid, M.; Azizullah; Li, D.; Manzoor, I.; Song, F. Plant–Microbiome Crosstalk: Dawning from Composition and Assembly of Microbial Community to Improvement of Disease Resilience in Plants. Int. J. Mol. Sci. 2021, 22, 6852. https://doi.org/10.3390/ijms22136852
Noman M, Ahmed T, Ijaz U, Shahid M, Azizullah, Li D, Manzoor I, Song F. Plant–Microbiome Crosstalk: Dawning from Composition and Assembly of Microbial Community to Improvement of Disease Resilience in Plants. International Journal of Molecular Sciences. 2021; 22(13):6852. https://doi.org/10.3390/ijms22136852
Chicago/Turabian StyleNoman, Muhammad, Temoor Ahmed, Usman Ijaz, Muhammad Shahid, Azizullah, Dayong Li, Irfan Manzoor, and Fengming Song. 2021. "Plant–Microbiome Crosstalk: Dawning from Composition and Assembly of Microbial Community to Improvement of Disease Resilience in Plants" International Journal of Molecular Sciences 22, no. 13: 6852. https://doi.org/10.3390/ijms22136852
APA StyleNoman, M., Ahmed, T., Ijaz, U., Shahid, M., Azizullah, Li, D., Manzoor, I., & Song, F. (2021). Plant–Microbiome Crosstalk: Dawning from Composition and Assembly of Microbial Community to Improvement of Disease Resilience in Plants. International Journal of Molecular Sciences, 22(13), 6852. https://doi.org/10.3390/ijms22136852






