A Machine Learning-Based Identification of Genes Affecting the Pharmacokinetics of Tacrolimus Using the DMETTM Plus Platform
Abstract
1. Introduction
2. Results
2.1. Subjects
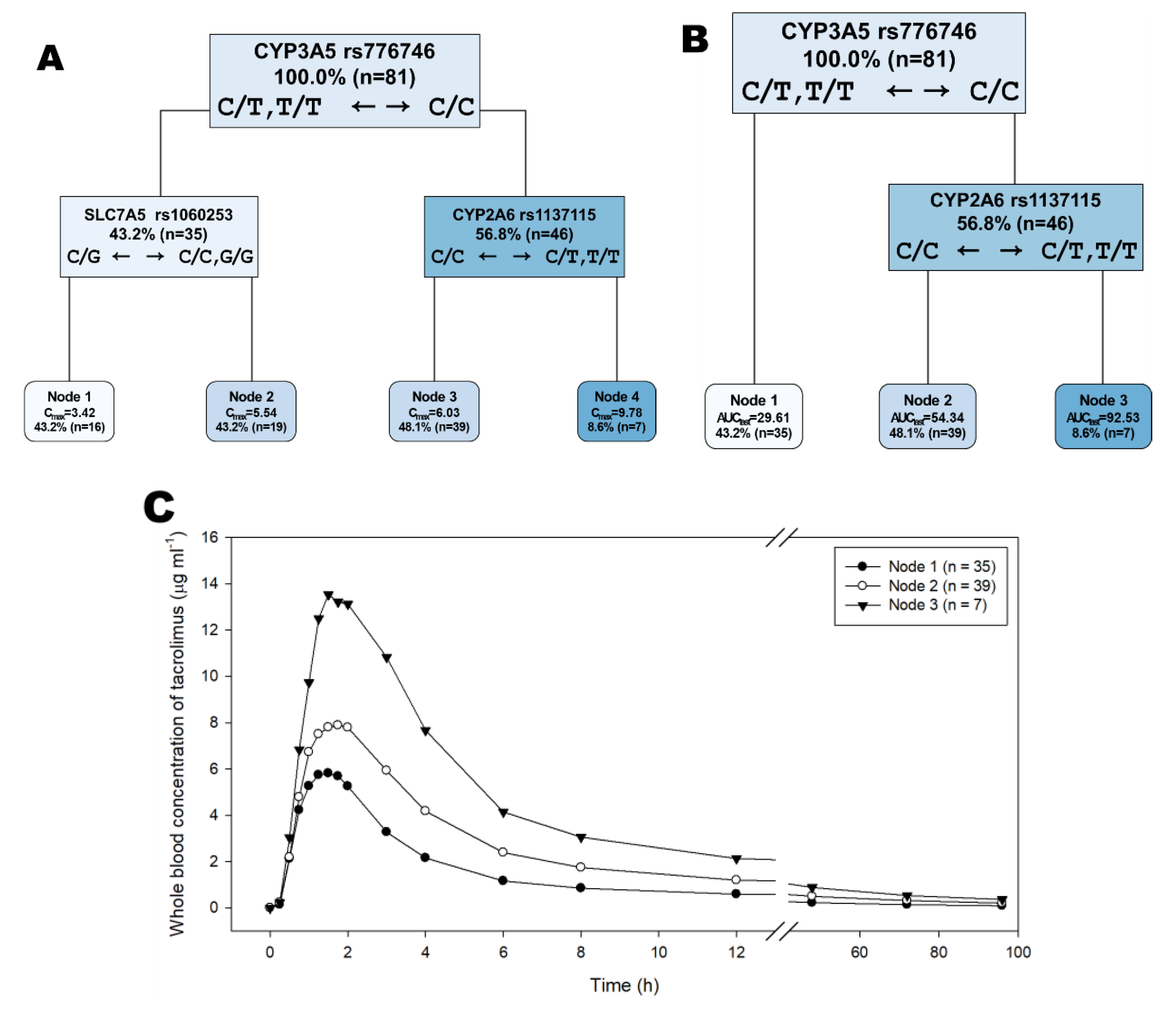
2.2. Genetic Associations with Tacrolimus Pharmacokinetics by Decision Tree, Random Forest, and LASSO Analyses
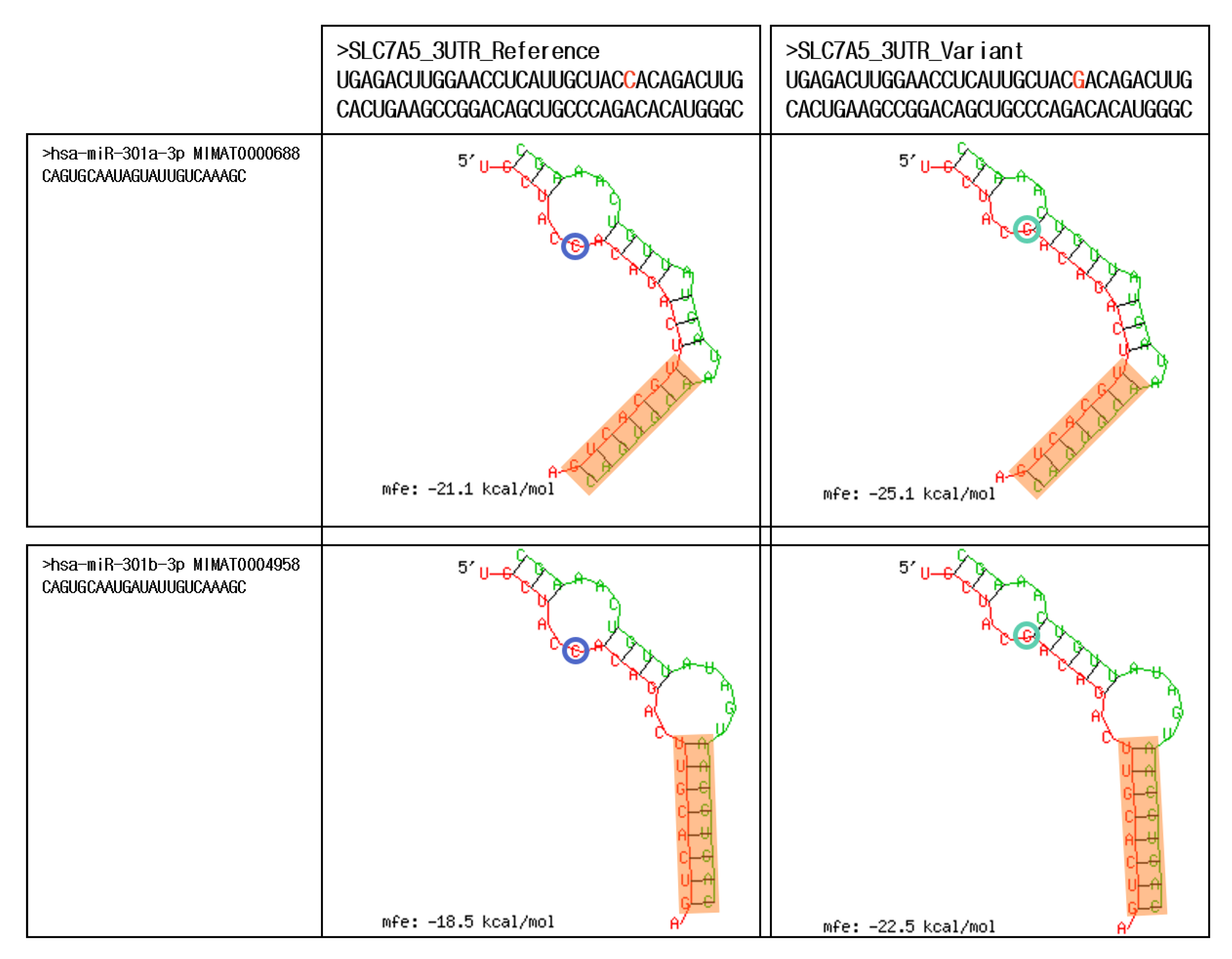
2.3. In silico Analysis of the SNPs in the 3′UTR
3. Discussion
4. Materials and Methods
4.1. Clinical Studies and Subjects
4.2. Determination of Tacrolimus Concentrations and Pharmacokinetic Analysis
4.3. DNA Extraction and Genotype Analysis
4.4. Statistical Analysis and Machine Learning Application
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tang, H.-L.; Xie, H.-G.; Yao, Y.; Hu, Y.-F. Lower tacrolimus daily dose requirements and acute rejection rates in the CYP3A5 nonexpressers than expressers. Pharm. Genom. 2011, 21, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Barratt, D.T.; Coller, J.K.; Sallustio, B.C.; Somogyi, A.A. CYP 3A5* 3 and ABCB 1 61A> G Significantly Influence Dose-adjusted Trough Blood Tacrolimus Concentrations in the First Three Months Post Kidney Transplantation. Basic Clin. Pharmacol. Toxicol. 2018, 123, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Dorr, C.R.; Wu, B.; Remmel, R.P.; Muthusamy, A.; Schladt, D.P.; Abrahante, J.E.; Guan, W.; Mannon, R.B.; Matas, A.J.; Oetting, W.S. Identification of genetic variants associated with tacrolimus metabolism in kidney transplant recipients by extreme phenotype sampling and next generation sequencing. Pharm. J. 2018, 19, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Haufroid, V.; Mourad, M.; Van Kerckhove, V.; Wawrzyniak, J.; De Meyer, M.; Eddour, D.C.; Malaise, J.; Lison, D.; Squifflet, J.-P.; Wallemacq, P. The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharm. Genom. 2004, 14, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.N.; Barama, A.; Poirier, C.; Vinet, B.; Roger, M. Cyp3A4, Cyp3A5, and MDR-1 genetic influences on tacrolimus pharmacokinetics in renal transplant recipients. Pharm. Genom. 2006, 16, 659–665. [Google Scholar] [CrossRef]
- Min, S.-I.; Kim, S.Y.; Ahn, S.H.; Min, S.-K.; Kim, S.H.; Kim, Y.S.; Moon, K.C.; Oh, J.M.; Kim, S.J.; Ha, J. CYP3A5* 1 allele: impacts on early acute rejection and graft function in tacrolimus-based renal transplant recipients. Transplantation 2010, 90, 1394–1400. [Google Scholar] [CrossRef]
- Tavira, B.; Coto, E.; Diaz-Corte, C.; Alvarez, V.; López-Larrea, C.; Ortega, F. A search for new CYP3A4 variants as determinants of tacrolimus dose requirements in renal-transplanted patients. Pharm. Genom. 2013, 23, 445–448. [Google Scholar] [CrossRef]
- Hesselink, D.A.; Bouamar, R.; Elens, L.; Van Schaik, R.H.; Van Gelder, T. The role of pharmacogenetics in the disposition of and response to tacrolimus in solid organ transplantation. Clin. Pharm. 2014, 53, 123–139. [Google Scholar] [CrossRef]
- Kuehl, P.; Zhang, J.; Lin, Y.; Lamba, J.; Assem, M.; Schuetz, J.; Watkins, P.B.; Daly, A.; Wrighton, S.A.; Hall, S.D. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 2001, 27, 383. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Liu, R.; Zhang, Y.-L.; Liu, M.-Z.; Hu, Y.-F.; Shao, M.-J.; Zhu, L.-J.; Xin, H.-W.; Feng, G.-W.; Shang, W.-J. Application of machine-learning models to predict tacrolimus stable dose in renal transplant recipients. Sci. Rep. 2017, 7, 42192. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.N.; McLoughlin, K.; Nicholas, A.B.; Allen, J.; Weaver, S.C.; Forrester, N.; Guerbois, M.; Jaing, C. Characterization of genetic variability of Venezuelan equine encephalitis viruses. PLoS ONE 2016, 11, e0152604. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, J.S.; Bonham, L.W.; Sears, R.L.; Klein, E.; Karydas, A.; Kramer, J.H.; Miller, B.L.; Coppola, G. Decision tree analysis of genetic risk for clinically heterogeneous Alzheimer’s disease. BMC Neurol. 2015, 15, 47. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.A.; Polley, E.C.; Briggs, F.B. Random forests for genetic association studies. Stat. Appl. Genet. Mol. Biol. 2011, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Stephan, J.; Stegle, O.; Beyer, A. A random forest approach to capture genetic effects in the presence of population structure. Nat. Commun. 2015, 6, 7432. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Jiang, F.; An, H.; Park, H.; Choi, J.; Lee, H. A pharmacogenomic study on the pharmacokinetics of tacrolimus in healthy subjects using the DMET TM Plus platform. Pharm. J. 2017, 17, 174–179. [Google Scholar]
- Kim, Y.K.; Kim, A.; Park, S.J.; Lee, H. new tablet formulation of tacrolimus with smaller interindividual variability may become a better treatment option than the conventional capsule formulation in organ transplant patients. Drug. Des. Devel. Ther. 2017, 11, 2861. [Google Scholar] [CrossRef]
- Bosó, V.; Herrero, M.J.; Bea, S.; Galiana, M.; Marrero, P.; Marqués, M.R.; Hernández, J.; Sánchez-Plumed, J.; Poveda, J.L.; Aliño, S.F. Increased hospital stay and allograft disfunction in renal transplant recipients with Cyp2c19 AA variant in SNP rs4244285. Drug Metab. Dispos. 2013, 41, 480–487. [Google Scholar] [CrossRef]
- Brooks, E.; Tett, S.E.; Isbel, N.M.; Staatz, C.E. Population pharmacokinetic modelling and Bayesian estimation of tacrolimus exposure: is this clinically useful for dosage prediction yet? Clin. Pharmacokinet. 2016, 55, 1295–1335. [Google Scholar] [CrossRef]
- Jacobson, P.A.; Oetting, W.S.; Brearley, A.M.; Leduc, R.; Guan, W.; Schladt, D.; Matas, A.J.; Lamba, V.; Julian, B.A.; Mannon, R.B. Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation 2011, 91, 300. [Google Scholar] [CrossRef] [PubMed]
- Kamdem, L.K.; Streit, F.; Zanger, U.M.; Brockmöller, J.; Oellerich, M.; Armstrong, V.W.; Wojnowski, L. Contribution of CYP3A5 to the in vitro hepatic clearance of tacrolimus. Clin. Chem. 2005, 51, 1374–1381. [Google Scholar] [CrossRef] [PubMed]
- Lamba, J.; Hebert, J.M.; Schuetz, E.G.; Klein, T.E.; Altman, R.B. PharmGKB summary: very important pharmacogene information for CYP3A5. Pharm. Genom. 2012, 22, 555. [Google Scholar] [CrossRef] [PubMed]
- Niioka, T.; Satoh, S.; Kagaya, H.; Numakura, K.; Inoue, T.; Saito, M.; Narita, S.; Tsuchiya, N.; Habuchi, T.; Miura, M. Comparison of pharmacokinetics and pharmacogenetics of once-and twice-daily tacrolimus in the early stage after renal transplantation. Transplantation 2012, 94, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rubio, G.; López-Flores, L.A.; Ramírez-Venegas, A.; Noé-Díaz, V.; García-Gómez, L.; Ambrocio-Ortiz, E.; Sánchez-Romero, C.; Hernández-Zenteno, R.D.J.; Sansores, R.H.; Falfán-Valencia, R. Genetic polymorphisms in CYP2A6 are associated with a risk of cigarette smoking and predispose to smoking at younger ages. Gene 2017, 628, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Bloom, A.J.; Harari, O.; Martinez, M.; Zhang, X.; McDonald, S.A.; Murphy, S.E.; Goate, A. A compensatory effect upon splicing results in normal function of the CYP2A6* 14 allele. Pharm. Genom. 2013, 23, 107. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Grimson, A.; Farh, K.K.-H.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef]
- Gonzalez-Covarrubias, V.; Martínez-Magaña, J.J.; Coronado-Sosa, R.; Villegas-Torres, B.; Genis-Mendoza, A.D.; Canales-Herrerias, P.; Nicolini, H.; Soberón, X. Exploring variation in known pharmacogenetic variants and its association with drug response in different Mexican populations. Pharm. Res. 2016, 33, 2644–2652. [Google Scholar] [CrossRef]
- Medhasi, S.; Pinthong, D.; Pasomsub, E.; Vanwong, N.; Ngamsamut, N.; Puangpetch, A.; Chamnanphon, M.; Hongkaew, Y.; Pratoomwun, J.; Limsila, P. Pharmacogenomic study reveals new variants of drug metabolizing enzyme and transporter genes associated with steady-state plasma concentrations of risperidone and 9-hydroxyrisperidone in Thai autism spectrum disorder patients. Front. Pharmacol. 2016, 7, 475. [Google Scholar] [CrossRef]
- Van Looy, S.; Verplancke, T.; Benoit, D.; Hoste, E.; Van Maele, G.; De Turck, F.; Decruyenaere, J. A novel approach for prediction of tacrolimus blood concentration in liver transplantation patients in the intensive care unit through support vector regression. Crit. Care 2007, 11, R83. [Google Scholar] [CrossRef] [PubMed]
- Thishya, K.; Vattam, K.K.; Naushad, S.M.; Raju, S.B.; Kutala, V.K. Artificial neural network model for predicting the bioavailability of tacrolimus in patients with renal transplantation. PLoS ONE 2018, 13, e0191921. [Google Scholar] [CrossRef] [PubMed]
- Franconi, F.; Campesi, I. Pharmacogenomics, pharmacokinetics and pharmacodynamics: interaction with biological differences between men and women. Br. J. Pharmacol. 2014, 171, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Undre, N.; Baccarani, U.; Britz, R.; Popescu, I. Pharmacokinetic Profile of Prolonged-Release Tacrolimus When Administered via Nasogastric Tube in De Novo Liver Transplantation: A Sub-Study of the DIAMOND Trial. Ann. Transplant. 2019, 24, 268. [Google Scholar] [CrossRef]
- Ramakrishna, N.; Vishwottam, K.; Puran, S.; Manoj, S.; Santosh, M.; Wishu, S.; Koteshwara, M.; Chidambara, J.; Gopinadh, B.; Sumatha, B. Liquid chromatography–negative ion electrospray tandem mass spectrometry method for the quantification of tacrolimus in human plasma and its bioanalytical applications. J. Chromatogr. B Biomed. Appl. 2004, 805, 13–20. [Google Scholar] [CrossRef]
- Consortium, G.P. A global reference for human genetic variation. Nature 2015, 526, 68. [Google Scholar] [CrossRef]
- Loh, W.Y. Classification and regression trees. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011, 1, 14–23. [Google Scholar] [CrossRef]
- Deconinck, E.; Hancock, T.; Coomans, D.; Massart, D.; Vander Heyden, Y. Classification of drugs in absorption classes using the classification and regression trees (CART) methodology. J. Pharm. Biomed. Anal. 2005, 39, 91–103. [Google Scholar] [CrossRef]
- Kim, Y.; Wojciechowski, R.; Sung, H.; Mathias, R.A.; Wang, L.; Klein, A.P.; Lenroot, R.K.; Malley, J.; Bailey-Wilson, J.E. Evaluation of Random Forests Performance for Genome-Wide Association Studies in the Presence of Interaction Effects. BMC Proc. 2009, 3, S64. [Google Scholar] [CrossRef]
- Wu, T.T.; Chen, Y.F.; Hastie, T.; Sobel, E.; Lange, K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics 2009, 25, 714–721. [Google Scholar] [CrossRef]
| Gene | SNP | Location | Reference Allele | Variant Allele | Reference Allele Frequency | Variant Allele Frequency | ||
|---|---|---|---|---|---|---|---|---|
| 1000 Genomes * | Our Data ** | 1000 Genomes * | Our Data ** | |||||
| CYP3A5 | rs776746 | Splice acceptor | T | C | 0.379 | 0.253 | 0.621 | 0.747 |
| CYP2A6 | rs1137115 | Exon | T | C | 0.239 | 0.136 | 0.761 | 0.864 |
| SLC7A5 *** | rs1060253 | 3′UTR | G | C | 0.698 | 0.370 | 0.302 | 0.630 |
| Gene | SNP and Genotype | Location | Reference Allele | Variant Allele | Reference Allele Frequency | Variant Allele Frequency | Importance | ||
|---|---|---|---|---|---|---|---|---|---|
| 1000 Genomes * | Our Data ** | 1000 Genomes * | Our data ** | ||||||
| Cmax | |||||||||
| CYP3A5 | rs776746 | Splice acceptor | T | C | 0.379 | 0.253 | 0.621 | 0.747 | 0.28524489 |
| SLCO3A1 | rs2190748 | Intron | G | A | 0.517 | 0.525 | 0.483 | 0.475 | 0.14800742 |
| ADC1 | rs1049793 | Exon | C | G | 0.627 | 0.358 | 0.373 | 0.642 | 0.13512953 |
| SLC7A5 | rs1060253 | 3′UTR | G | C | 0.698 | 0.370 | 0.302 | 0.630 | 0.11857793 |
| AUClast | |||||||||
| CYP3A5 | rs776746 | Splice acceptor | T | C | 0.379 | 0.253 | 0.621 | 0.747 | 1.5377314 |
| SLCO3A1 | rs2190748 | Intron | G | A | 0.517 | 0.525 | 0.483 | 0.475 | 0.3333521 |
| CYP2A6 | rs1137115 | Exon | T | C | 0.239 | 0.136 | 0.761 | 0.864 | 0.1921316 |
| NR1I2 | rs3814055 | Exon | C | T | 0.678 | 0.710 | 0.322 | 0.290 | 0.1419874 |
| Gene | SNP | Location | Reference Allele | Variant Allele | Reference Allele Frequency | Variant Allele Frequency | Coefficient | ||
|---|---|---|---|---|---|---|---|---|---|
| 1000 Genomes * | Our Data ** | 1000 Genomes * | Our Data ** | ||||||
| Cmax | |||||||||
| CYP3A5 | rs776746 | Splice acceptor | T | C | 0.379 | 0.253 | 0.621 | 0.747 | 0.13331 |
| CBR1 | rs3787728 | Intron | T | C | 0.270 | 0.519 | 0.730 | 0.481 | 0.07863 |
| NAT2 | rs1208 | Exon | G | A, T | 0.323 | 0.025 | 0.677 | 0.975 | 0.07224 |
| AUClast | |||||||||
| CYP3A5 | rs776746 | Splice acceptor | T | C | 0.379 | 0.253 | 0.621 | 0.747 | 0.36133 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gim, J.-A.; Kwon, Y.; Lee, H.A.; Lee, K.-R.; Kim, S.; Choi, Y.; Kim, Y.K.; Lee, H. A Machine Learning-Based Identification of Genes Affecting the Pharmacokinetics of Tacrolimus Using the DMETTM Plus Platform. Int. J. Mol. Sci. 2020, 21, 2517. https://doi.org/10.3390/ijms21072517
Gim J-A, Kwon Y, Lee HA, Lee K-R, Kim S, Choi Y, Kim YK, Lee H. A Machine Learning-Based Identification of Genes Affecting the Pharmacokinetics of Tacrolimus Using the DMETTM Plus Platform. International Journal of Molecular Sciences. 2020; 21(7):2517. https://doi.org/10.3390/ijms21072517
Chicago/Turabian StyleGim, Jeong-An, Yonghan Kwon, Hyun A Lee, Kyeong-Ryoon Lee, Soohyun Kim, Yoonjung Choi, Yu Kyong Kim, and Howard Lee. 2020. "A Machine Learning-Based Identification of Genes Affecting the Pharmacokinetics of Tacrolimus Using the DMETTM Plus Platform" International Journal of Molecular Sciences 21, no. 7: 2517. https://doi.org/10.3390/ijms21072517
APA StyleGim, J.-A., Kwon, Y., Lee, H. A., Lee, K.-R., Kim, S., Choi, Y., Kim, Y. K., & Lee, H. (2020). A Machine Learning-Based Identification of Genes Affecting the Pharmacokinetics of Tacrolimus Using the DMETTM Plus Platform. International Journal of Molecular Sciences, 21(7), 2517. https://doi.org/10.3390/ijms21072517






