Transcriptome Analysis Reveals the Neuro-Immune Interactions in Duck Tembusu Virus-Infected Brain
Abstract
1. Introduction
2. Results
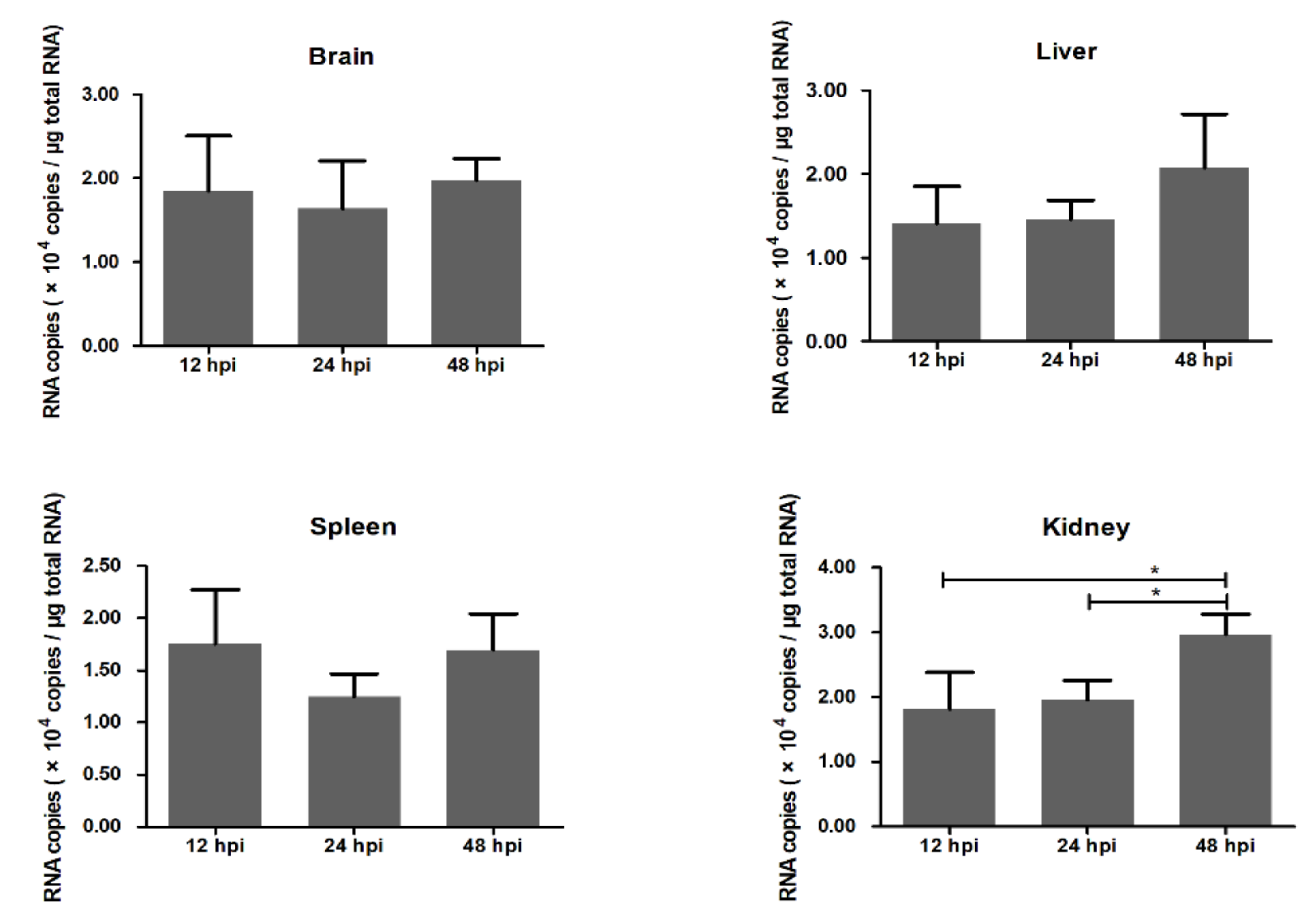
2.1. Detection of Viral Infections in the Brain and Other Organs
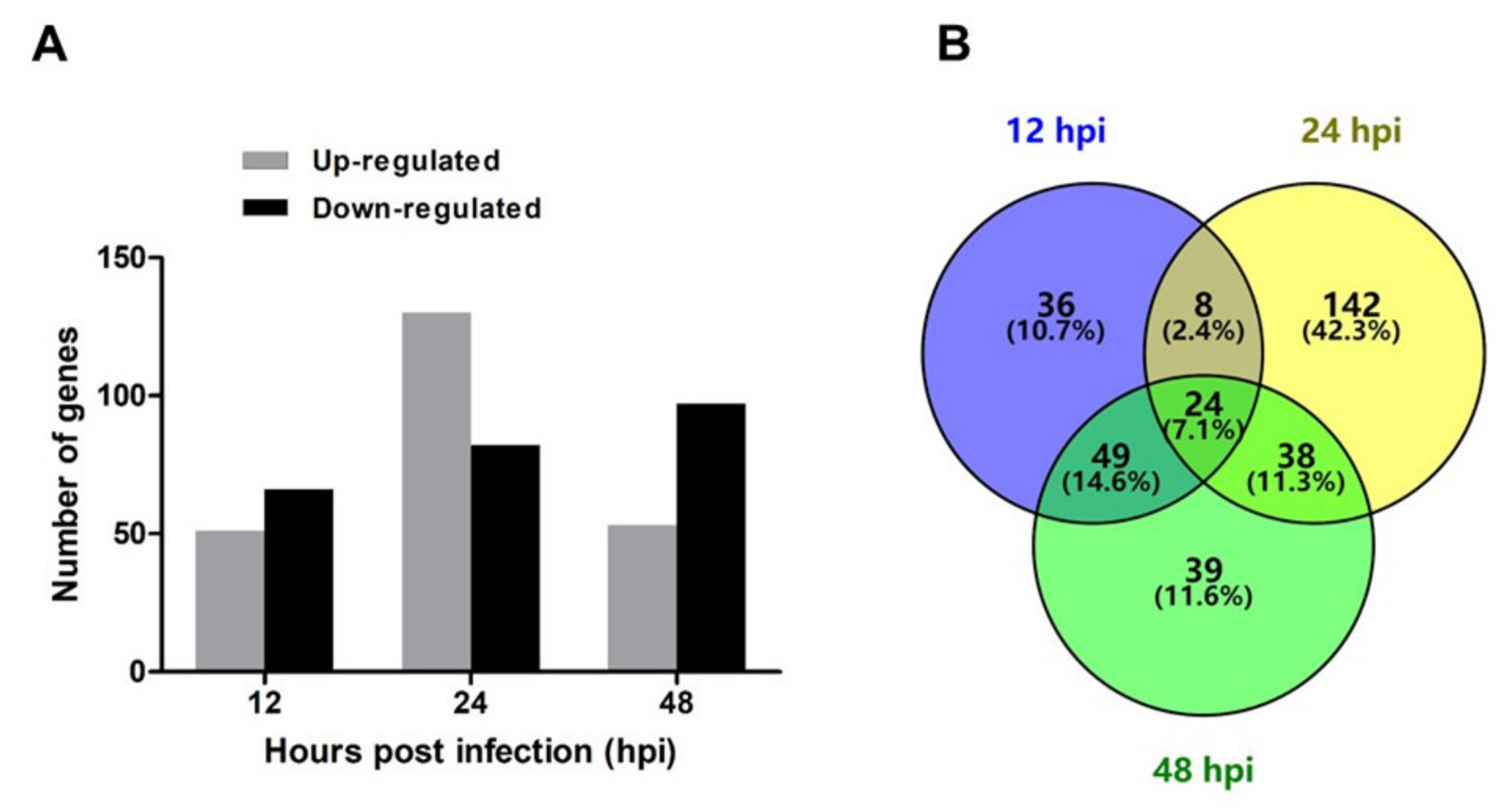
2.2. Identification of Differentially Expressed Genes (DEGs)
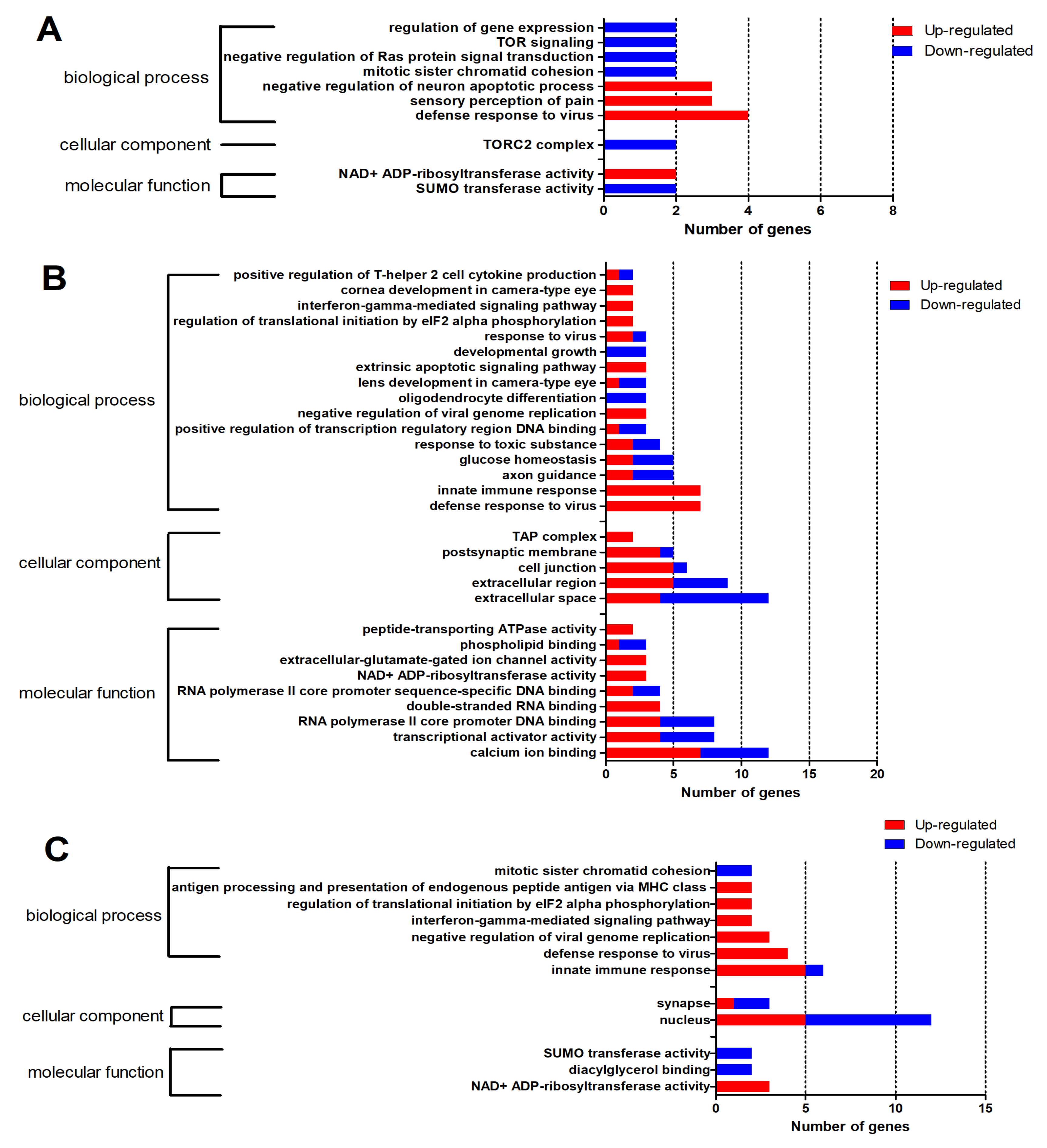
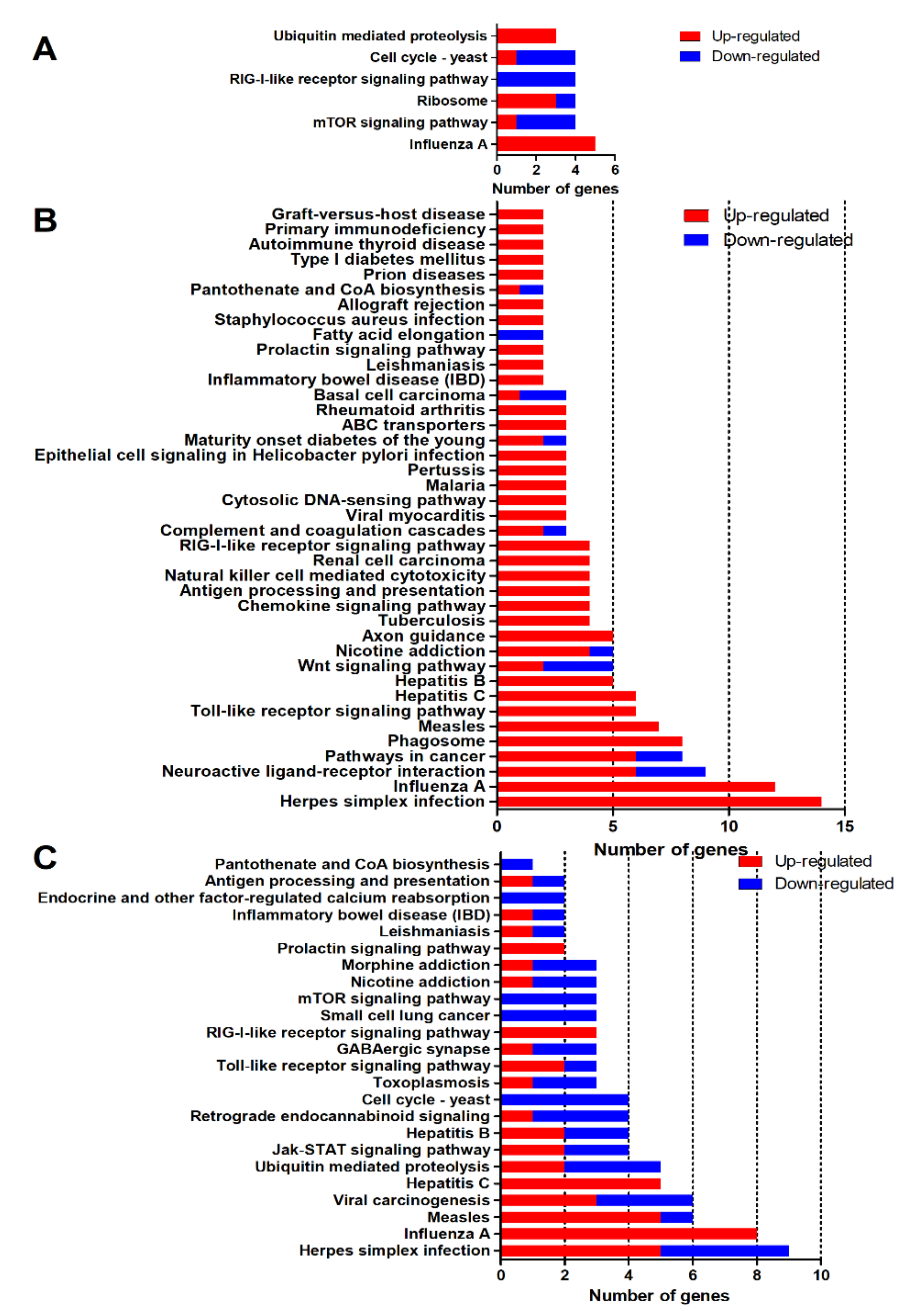
2.3. GO and KEGG Enrichment Analysis
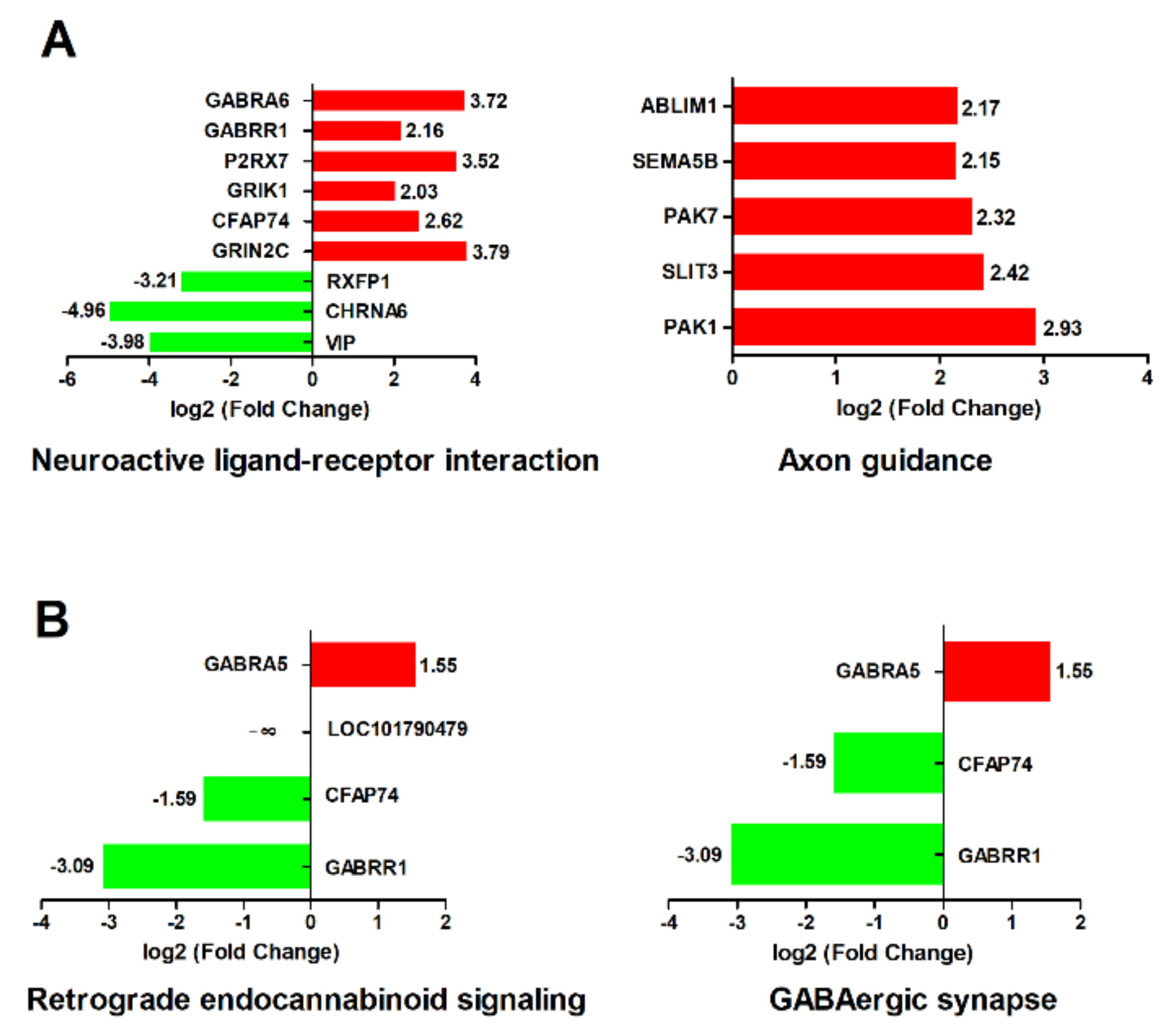
2.4. Pathways Related to Nervous System
2.5. Pathways Related to Immune Responses
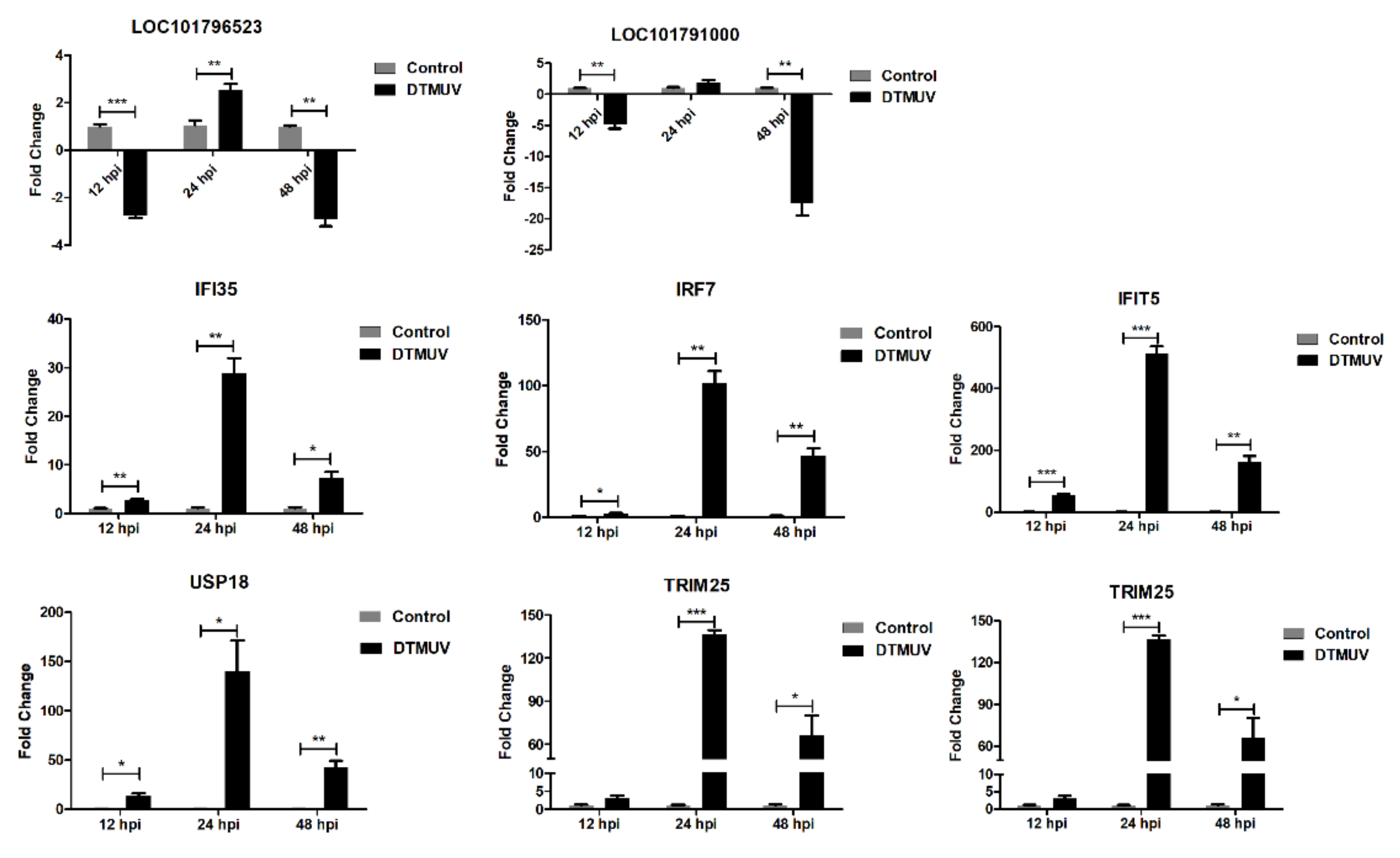
2.6. Validation of RNA-seq by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
3. Discussion
3.1. Genes Related to Nervous System
3.2. Immune Responses in Duck Brain
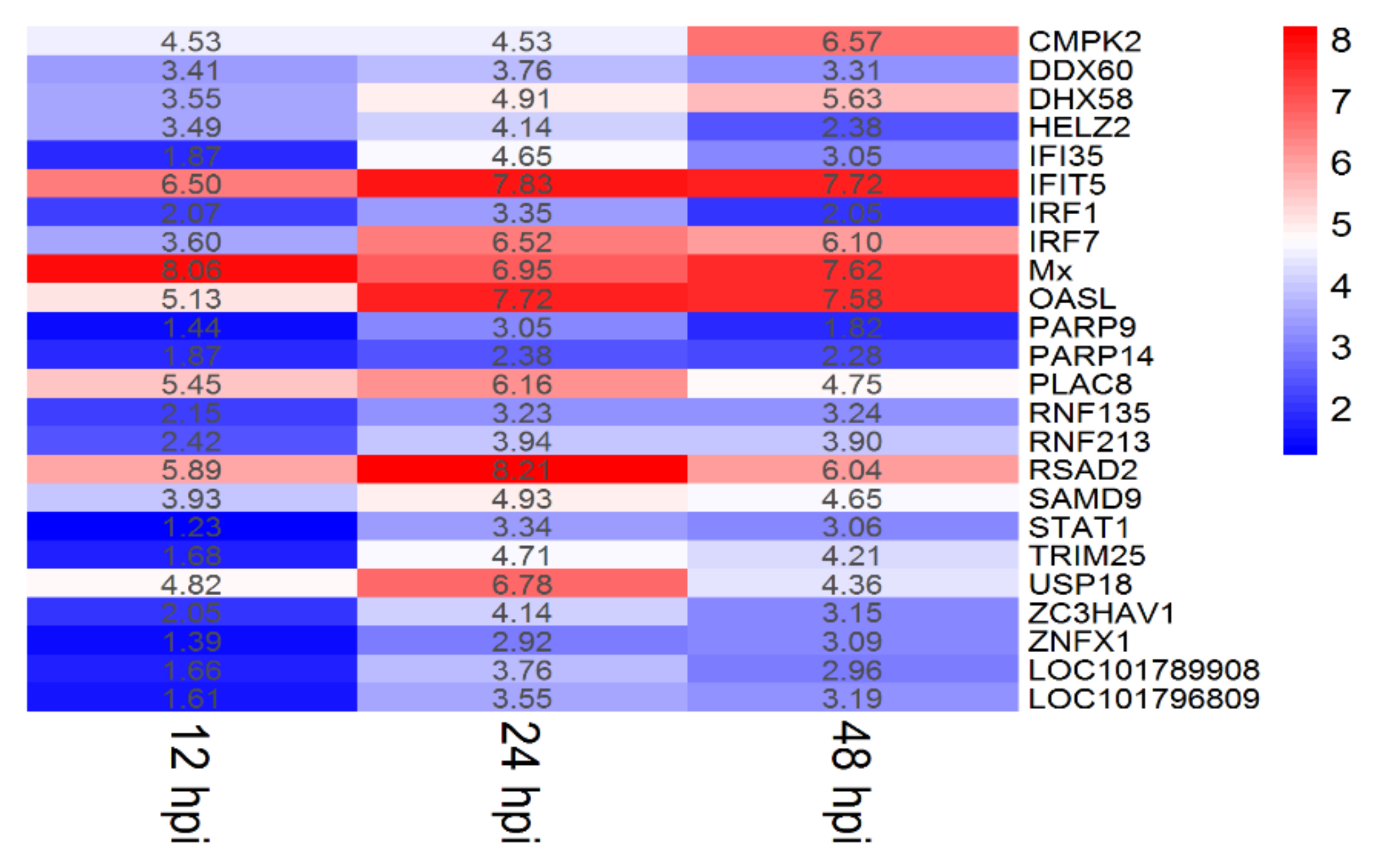
3.3. ISGs Response to DTMUV Infection in Duck Brain
4. Materials and Methods
4.1. Virus and Cells
4.2. Animal Experiment
4.3. Transcriptome cDNA Library Construction and Sequencing
4.4. Transcriptome Data Analysis
4.5. Differential Expression Analysis
4.6. qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Su, J.; Li, S.; Hu, X.; Yu, X.; Wang, Y.; Liu, P.; Lu, X.; Zhang, G.; Hu, X.; Liu, D. Duck egg-drop syndrome caused by byd virus, a new tembusu-related flavivirus. PLoS ONE 2011, 6, e18106. [Google Scholar] [CrossRef]
- Liu, P.; Lu, H.; Li, S.; Wu, Y.; Gao, G.F.; Su, J. Duck egg drop syndrome virus: An emerging tembusu-related flavivirus in china. Sci. China Life Sci. 2013, 56, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Zhang, C.; Liu, Y.; Liu, Y.; Ye, W.; Han, J.; Ma, G.; Zhang, D.; Xu, F.; Gao, X.; et al. Tembusu virus in ducks, China. Emerg Infect. Dis. 2011, 17, 1873–1875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, S.; Mahalingam, S.; Wang, M.; Cheng, A. An updated review of avian-origin tembusu virus: A newly emerging avian flavivirus. J. Gen. Virol. 2017, 98. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Li, X.F.; Liu, L.; Xu, Y.P.; Qin, C.-F. The emerging duck flavivirus is not pathogenic for primates and is highly sensitive to mammal interferon signaling. J. Virol. 2016, 90, 6538–6548. [Google Scholar] [CrossRef]
- Tang, Y.; Gao, X.; Diao, Y.; Feng, Q.; Yu, C. Tembusu virus in human, china. Transbound. Emerg. Dis. 2013, 60, 193–196. [Google Scholar] [CrossRef]
- Platt, G.S.; Way, H.J.; Bowen, E.T.W.; Simpson, D.I.H.; Hill, M.N.; Kamath, S.; Bendell, P.J.E.; Heathcote, O.H.U. Arbovirus infections in sarawak, October 1968–February 1970 tembusu and sindbis virus isolations from mosquitoes. Trans. R. Soc. Trop. Med. Hyg. 1975, 69, 65–71. [Google Scholar] [CrossRef]
- Tang, Y.; Diao, Y.; Chen, H.; Ou, Q.; Liu, X.; Gao, X.; Yu, C.; Wang, L. Isolation and genetic characterization of a tembusu virus strain isolated from mosquitoes in shandong, china. Transbound Emerg. Dis. 2015, 62, 209–216. [Google Scholar] [CrossRef]
- Sanisuriwong, J.; Yurayart, N.; Thontiravong, A.; Tiawsirisup, S. Duck tembusu virus detection and characterization from mosquitoes in duck farms, thailand. Transbound Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Benzarti, E.; Linden, A.; Desmecht, D.; Garigliany, M. Mosquito-borne epornitic flaviviruses: An update and review. J. Gen. Virol. 2019, 100, 119–132. [Google Scholar] [CrossRef]
- Petersen, L.R.; Brault, A.C.; Nasci, R.S. West nile virus: Review of the literature. JAMA 2013, 310, 308–315. [Google Scholar] [CrossRef]
- Gaibani, P.; Rossini, G. An overview of usutu virus. Microbes Infect. 2017, 19, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Pardigon, N. Pathophysiological mechanisms of flavivirus infection of the central nervous system. Transfus Clin. Biol 2017, 24, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Barichello, T.; Stubbs, B.; Köhler, C.A.; Carvalho, A.F.; Maes, M. Zika virus as an emerging neuropathogen: Mechanisms of neurovirulence and neuro-immune interactions. Mol. Neurobiol. 2018, 55, 4160–4184. [Google Scholar] [CrossRef] [PubMed]
- Yun, T.; Ye, W.; Ni, Z.; Zhang, D.; Zhang, C. Identification and molecular characterization of a novel flavivirus isolated from pekin ducklings in china. Vet. Microbiol. 2012, 157, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Homonnay, Z.G.; Kovács, E.W.; Bányai, K.; Albert, M.; Fehér, E.; Mató, T.; Tatár-Kis, T.; Palya, V. Tembusu-like flavivirus (perak virus) as the cause of neurological disease outbreaks in young pekin ducks. Avian Pathol. 2014, 43, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.Y.; Diao, Y.X.; Wang, J.; Liu, X.; Lu, A.L.; Zhang, L.; Ge, P.P.; Hao, D.M. Tembusu virus infection in cherry valley ducks: The effect of age at infection. Vet. Microbiol. 2014, 168, 16–24. [Google Scholar] [CrossRef]
- Li, N.; Lv, C.; Yue, R.; Shi, Y.; Wei, L.; Chai, T.; Liu, S. Effect of age on the pathogenesis of duck tembusu virus in cherry valley ducks. Front. Microbiol. 2015, 6, 581. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Zhang, L.; Wang, Y.; Su, J. Duck tembusu virus exhibits neurovirulence in balb/c mice. Virol. J. 2013, 10, 260. [Google Scholar] [CrossRef] [PubMed]
- Ti, J.; Zhang, L.; Li, Z.; Zhao, D.; Zhang, Y.; Li, F.; Diao, Y. Effect of age and inoculation route on the infection of duck tembusu virus in goslings. Vet. Microbiol 2015, 181, 190–197. [Google Scholar] [CrossRef]
- Pashirzad, M.; Fiuji, H.; Khazei, M.; Moradi-Binabaj, M.; Ryzhikov, M.; Shabani, M.; Avan, A.; Hassanian, S.M. Role of wnt3a in the pathogenesis of cancer, current status and prospective. Mol. Biol. Rep. 2019, 46, 5609–5616. [Google Scholar] [CrossRef] [PubMed]
- Arrázola, M.S.; Silva-Alvarez, C.; Inestrosa, N.C. How the wnt signaling pathway protects from neurodegeneration: The mitochondrial scenario. Front. Cell. Neurosci. 2015, 9, 166. [Google Scholar]
- Cisternas, P.; Salazar, P.; Silva-Alvarez, C.; Barros, L.F.; Inestrosa, N.C. Activation of wnt signaling in cortical neurons enhances glucose utilization through glycolysis. J. Biol. Chem. 2016, 291, 25950–25964. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.C.; Lakshmanan, G.; Crawford, S.E.; Gu, Y.; Engel, J.D. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat. Genet. 2000, 25, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Tsarovina, K.; Pattyn, A.; Stubbusch, J.; Muller, F.; van der Wees, J.; Schneider, C.; Brunet, J.F.; Rohrer, H. Essential role of gata transcription factors in sympathetic neuron development. Development 2004, 131, 4775–4786. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, T.; Takako, N.; Hamada, M.; Maeda, A.; Fujioka, Y.; Kuroha, T.; Huber, R.E.; Hasegawa, S.L.; Rao, A.; Yamamoto, M.; et al. Gata3 participates in a complex transcriptional feedback network to regulate sympathoadrenal differentiation. Development 2006, 133, 3871–3881. [Google Scholar] [CrossRef]
- Koopman, F.A.; Stoof, S.P.; Straub, R.H.; Maanen, M.A.V.; Tak, P.P. Restoring the balance of the autonomic nervous system as an innovative approach to the treatment of rheumatoid arthritis. Mol. Med. 2011, 17, 937–948. [Google Scholar] [CrossRef]
- Waxenbaum, J.A.; Varacallo, M. Anatomy, Autonomic Nervous System. In Statpearls, Treasure Island (FL). 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK539845/ (accessed on 10 March 2020).
- Jänig, W. Sympathetic nervous system and inflammation: A conceptual view. Autonomic Neurosci. Basic Clin. 2014, 182, 4–14. [Google Scholar]
- Ebihara, M.; Ohba, H.; Ohno, S.I.; Yoshikawa, T. Genomic organization and promoter analysis of the human nicotinic acetylcholine receptor α6 subunit (chnra6) gene: Alu and other elements direct transcriptional repression. Gene 2002, 298, 101–108. [Google Scholar] [CrossRef]
- Exley, R.; Clements, M.A.; Hartung, H.; McIntosh, J.M.; Cragg, S.J. Alpha6-containing nicotinic acetylcholine receptors dominate the nicotine control of dopamine neurotransmission in nucleus accumbens. Neuropsychopharmacology 2008, 33, 2158–2166. [Google Scholar] [CrossRef]
- Streicher, F.; Jouvenet, N. Stimulation of innate immunity by host and viral rnas. Trends Immunol. 2019, 40, 1134–1148. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, Y.; Li, R.; Liu, J.; Zhang, J.; Cai, Y.; Liu, S.; Chai, T.; Wei, L. Immune responses of ducks infected with duck tembusu virus. Front. Microbiol. 2015, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, K.R.; Bruns, A.M.; Horvath, C.M. Mda5 and lgp2: Accomplices and antagonists of antiviral signal transduction. J. Virol. 2014, 88, 8194–8200. [Google Scholar] [CrossRef]
- Bruns, A.M.; Leser, G.P.; Lamb, R.A.; Horvath, C.M. The innate immune sensor lgp2 activates antiviral signaling by regulating mda5-RNA interaction and filament assembly. Mol. Cell 2014, 55, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.M.; Horvath, C.M. Lgp2 synergy with mda5 in rlr-mediated rna recognition and antiviral signaling. Cytokine 2015, 74, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Huo, H.; Zhao, L.; Wang, D.; Chen, X.; Chen, H. Lgp2 plays a critical role in mda5-mediated antiviral activity against duck enteritis virus. Mol. Immunol. 2019, 116, 160–166. [Google Scholar] [CrossRef]
- Verma, R.; Bharti, K. Toll like receptor 3 and viral infections of nervous system. J. Neurol. Sci. 2017, 372, 40–48. [Google Scholar] [CrossRef]
- Wang, T.; Town, T.; Alexopoulou, L.; Anderson, J.F.; Fikrig, E.; Flavell, R.A. Toll-like receptor 3 mediates west nile virus entry into the brain causing lethal encephalitis. Nat. Med. 2004, 10, 1366–1373. [Google Scholar] [CrossRef]
- Daffis, S.; Samuel, M.A.; Suthar, M.S.; Gale, M.; Diamond, M.S. Toll-like receptor 3 has a protective role against west nile virus infection. J. Virol. 2008, 82, 10349–10358. [Google Scholar] [CrossRef]
- Han, Y.W.; Choi, J.Y.; Uyangaa, E.; Kim, S.B.; Kim, J.H.; Kim, B.S.; Kim, K.; Eo, S.K. Distinct dictation of japanese encephalitis virus-induced neuroinflammation and lethality via triggering tlr3 and tlr4 signal pathways. PLoS Pathog. 2014, 10, e1004319. [Google Scholar] [CrossRef]
- Momburg, F.; Mullbacher, A.; Lobigs, M. Modulation of transporter associated with antigen processing (tap)-mediated peptide import into the endoplasmic reticulum by flavivirus infection. J. Virol. 2001, 75, 5663–5671. [Google Scholar] [CrossRef] [PubMed]
- Hershkovitz, O.; Zilka, A.; Bar-Ilan, A.; Abutbul, S.; Davidson, A.; Mazzon, M.; Kummerer, B.M.; Monsoengo, A.; Jacobs, M.; Porgador, A. Dengue virus replicon expressing the nonstructural proteins suffices to enhance membrane expression of hla class i and inhibit lysis by human nk cells. J. Virol 2008, 82, 7666–7676. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dong, M.; Wang, X.G. The implication and significance of beta 2 microglobulin: A conservative multifunctional regulator. Chin. Med. J. 2016, 129, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.K.; He, Y.; Park, J.-S.; Bieri, G.; Snethlage, C.E.; Lin, K.; Gontier, G.; Wabl, R.; Plambeck, K.E.; Udeochu, J. B2-microglobulin is a systemic pro-aging factor that impairs cognitive function and neurogenesis. Nat. Med. 2015, 21, 932. [Google Scholar] [CrossRef]
- Schoggins, J.W. Interferon-stimulated genes: What do they all do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Shaw, A.E.; Hughes, J.; Gu, Q.; Behdenna, A.; Singer, J.B.; Dennis, T.; Orton, R.J.; Varela, M.; Gifford, R.J.; Wilson, S.J.; et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type i interferon responses. PLoS Biol. 2017, 15, e2004086. [Google Scholar] [CrossRef]
- Zhou, X.; Michal, J.J.; Zhang, L.; Ding, B.; Lunney, J.K.; Liu, B.; Jiang, Z. Interferon induced ifit family genes in host antiviral defense. Int J. Biol. Sci. 2013, 9, 200–208. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, W.; Liu, Y.; Qi, H.; Hao, C.; Zhang, W.; Gao, M.; Wang, J.; Ma, B. Molecular cloning and mrna expression of ifit5 in tissues of ducklings infected with virulent duck hepatitis a virus type 3. Res. Vet. Sci. 2019, 124, 256–262. [Google Scholar] [CrossRef]
- Bi, K.R.; Han, K.K.; Liu, Q.T.; Zhao, D.M.; Huang, X.M.; Liu, Y.Z.; Yang, J.; Li, Y. Molecular cloning, characterization, and expression of duck 2′-5′-oligoadenylate synthetase-like gene. Gene 2017, 629, 43–51. [Google Scholar] [CrossRef]
- Dumbrepatil, A.B.; Ghosh, S.; Zegalia, K.A.; Malec, P.A.; Hoff, J.D.; Kennedy, R.T.; Marsh, E.N.G. Viperin interacts with the kinase irak1 and the e3 ubiquitin ligase traf6, coupling innate immune signaling to antiviral ribonucleotide synthesis. J. Biol Chem. 2019, 294, 6888–6898. [Google Scholar] [CrossRef]
- Lindqvist, R.; Overby, A.K. The role of viperin in antiflavivirus responses. DNA Cell Biol. 2018, 37, 725–730. [Google Scholar] [CrossRef]
- Zhou, Q.; Gu, X.; Xu, Z.; Huang, R.; Yin, D.; Ayaz, S.; Lyu, X.; Wang, G. Duck viperin can express in transfected bhk-21 cells and inhibit germination of duck tembusu virus. Chin. J. Cell. Mol. Immunol. 2019, 35, 412–418. [Google Scholar]
- Li, L.; An, H.; Sun, M.; Dong, J.; Yuan, J.; Hu, Q. Identification and genomic analysis of two duck-origin tembusu virus strains in southern china. Virus Genes 2012, 45, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. Hisat: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by rna-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Liu, Z.; Fu, Y.; Ji, Y.; Wei, J.; Zhu, Q. Development and validation of one-step sybr green real-time rt-pcr for the rapid detection of newly emerged duck tembusu virus. Avian Dis. 2013, 57, 595–601. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]

| Samples | Raw Reads | Clean Reads 1 | Clean Reads Ratio 2 | Total Mapped Reads3 | Total Mapped Ratio 4 |
|---|---|---|---|---|---|
| 12 h-Control | 52,151,546 | 51,155,090 | 98.09% | 39,377,403 | 76.98% |
| 24 h-Control | 50,060,388 | 49,233,950 | 98.35% | 38,039,893 | 77.26% |
| 48 h-Control | 52,859,536 | 51,873,096 | 98.13% | 39,886,284 | 76.89% |
| 12 h-Infected | 58,372,706 | 57,108,946 | 97.84% | 43,294,720 | 75.81% |
| 24 h-Infected | 40,835,734 | 40,030,410 | 98.03% | 30,830,799 | 77.02% |
| 48 h-Infected | 50,284,812 | 49,197,138 | 97.84% | 37,552,725 | 76.33% |
| Time Point | Pathway | p-Value | Up-Regulated | Down-Regulated |
|---|---|---|---|---|
| 12 h | RIG-I-like receptor signaling pathway | 3.43 × 10−4 | DHX58, IRF7, TRIM25 | — |
| 24 h | RIG-I-like receptor signaling pathway | 3.64 × 10−4 | DHX58, IRF7, TRIM25, IFIH1 | — |
| Toll-like receptor signaling pathway | 2.57 × 10−5 | TLR2, TLR3, STAT1, IRF7, LOC101791385, LOC101791569 | — | |
| Chemokine signaling pathway | 1.38 × 10−2 | STAT1, PAK1, LOC101791385, LOC101791569 | — | |
| Antigen processing and presentation | 2.93 × 10−4 | TAP1, TAP2, LOC101797502, LOC101790497 | — | |
| Natural killer cell mediated cytotoxicity | 3.31 × 10−3 | PAK1, BID, LOC101797502, LOC101790497 | — | |
| Complement and coagulation cascades | 1.18 × 10−2 | C1S, C1R | LOC101801552 | |
| 48 h | RIG-I-like receptor signaling pathway | 1.03 × 10−3 | DHX58, IRF7, TRIM25 | — |
| Toll-like receptor signaling pathway | 1.84 × 10−3 | STAT1, IRF7 | TLR2 | |
| JAK-STAT signaling pathway | 9.03 × 10−4 | STAT1, LOC101793812 | LOC101803793, LOC101789923 | |
| Antigen processing and presentation | 9.13 × 10−3 | B2M | LOC101797502 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Huang, Y.; Li, L.; Dong, J.; Liao, M.; Sun, M. Transcriptome Analysis Reveals the Neuro-Immune Interactions in Duck Tembusu Virus-Infected Brain. Int. J. Mol. Sci. 2020, 21, 2402. https://doi.org/10.3390/ijms21072402
Zhang J, Huang Y, Li L, Dong J, Liao M, Sun M. Transcriptome Analysis Reveals the Neuro-Immune Interactions in Duck Tembusu Virus-Infected Brain. International Journal of Molecular Sciences. 2020; 21(7):2402. https://doi.org/10.3390/ijms21072402
Chicago/Turabian StyleZhang, Junqin, Yunzhen Huang, Linlin Li, Jiawen Dong, Ming Liao, and Minhua Sun. 2020. "Transcriptome Analysis Reveals the Neuro-Immune Interactions in Duck Tembusu Virus-Infected Brain" International Journal of Molecular Sciences 21, no. 7: 2402. https://doi.org/10.3390/ijms21072402
APA StyleZhang, J., Huang, Y., Li, L., Dong, J., Liao, M., & Sun, M. (2020). Transcriptome Analysis Reveals the Neuro-Immune Interactions in Duck Tembusu Virus-Infected Brain. International Journal of Molecular Sciences, 21(7), 2402. https://doi.org/10.3390/ijms21072402




