Early Modulation of Circulating MicroRNAs Levels in HER2-Positive Breast Cancer Patients Treated with Trastuzumab-Based Neoadjuvant Therapy
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics and Response to Therapy
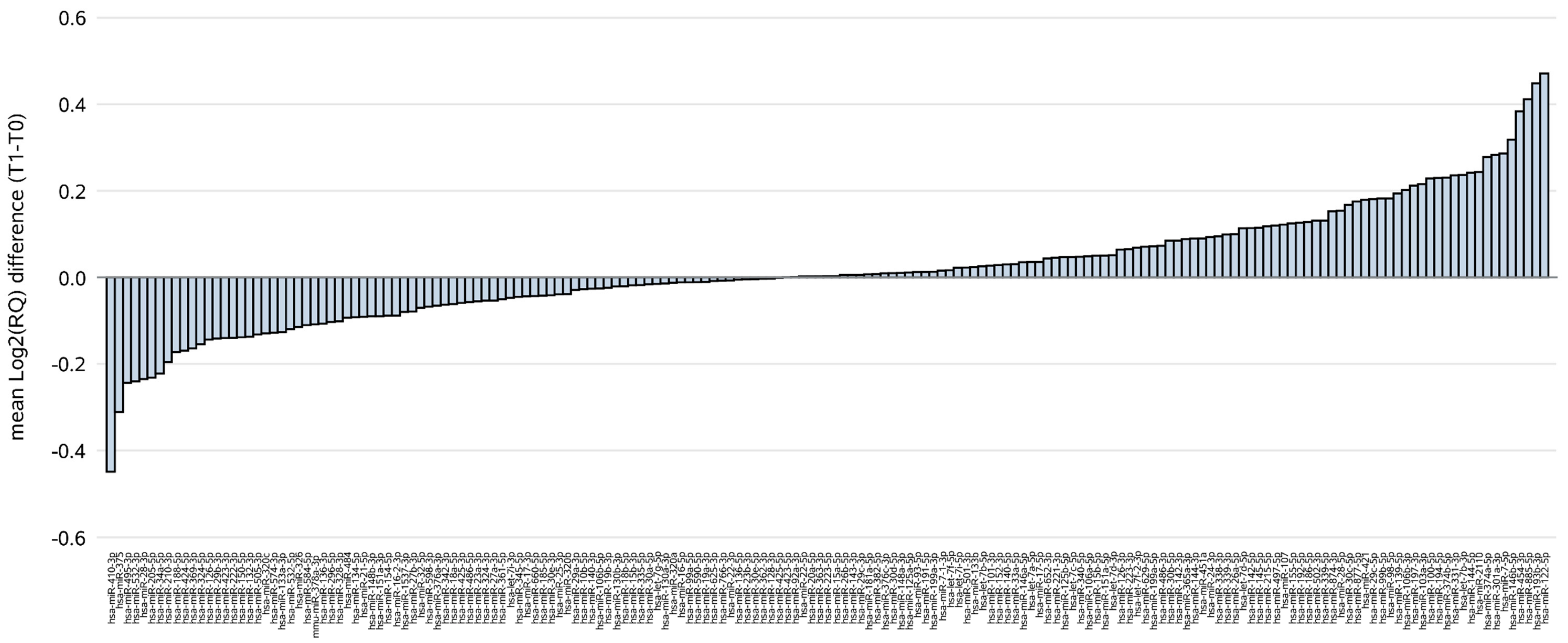
2.2. ct-miRNA Level Changes during Treatment with Trastuzumab
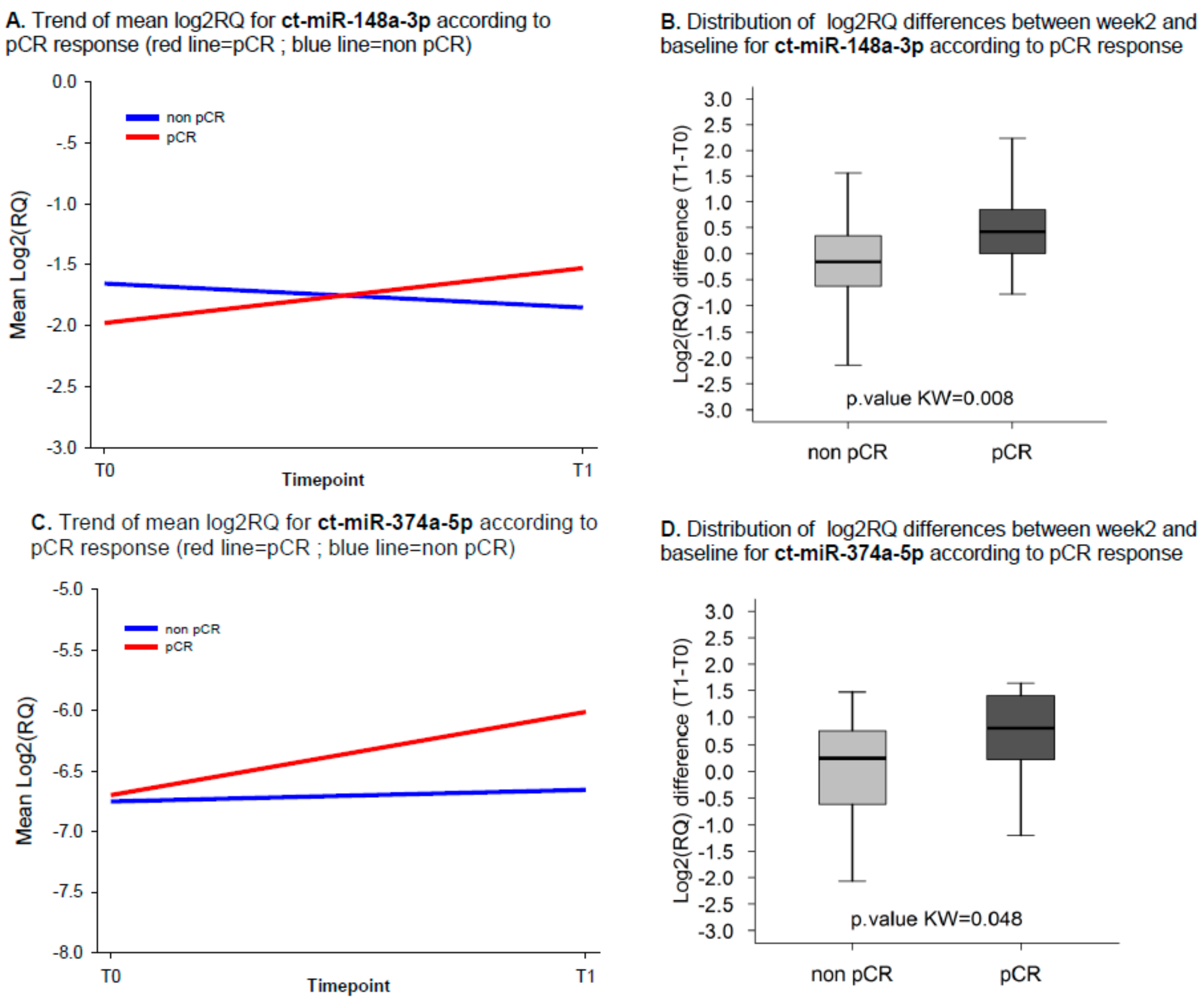
2.3. ct-miRNA Changes Are Associated with pCR
2.4. Univariate and Multivariate Logistic Models
2.5. Changes of ct-miR148a-3p Jointly Considered with the Evaluation of ct-miRNA140-5p at Week 2
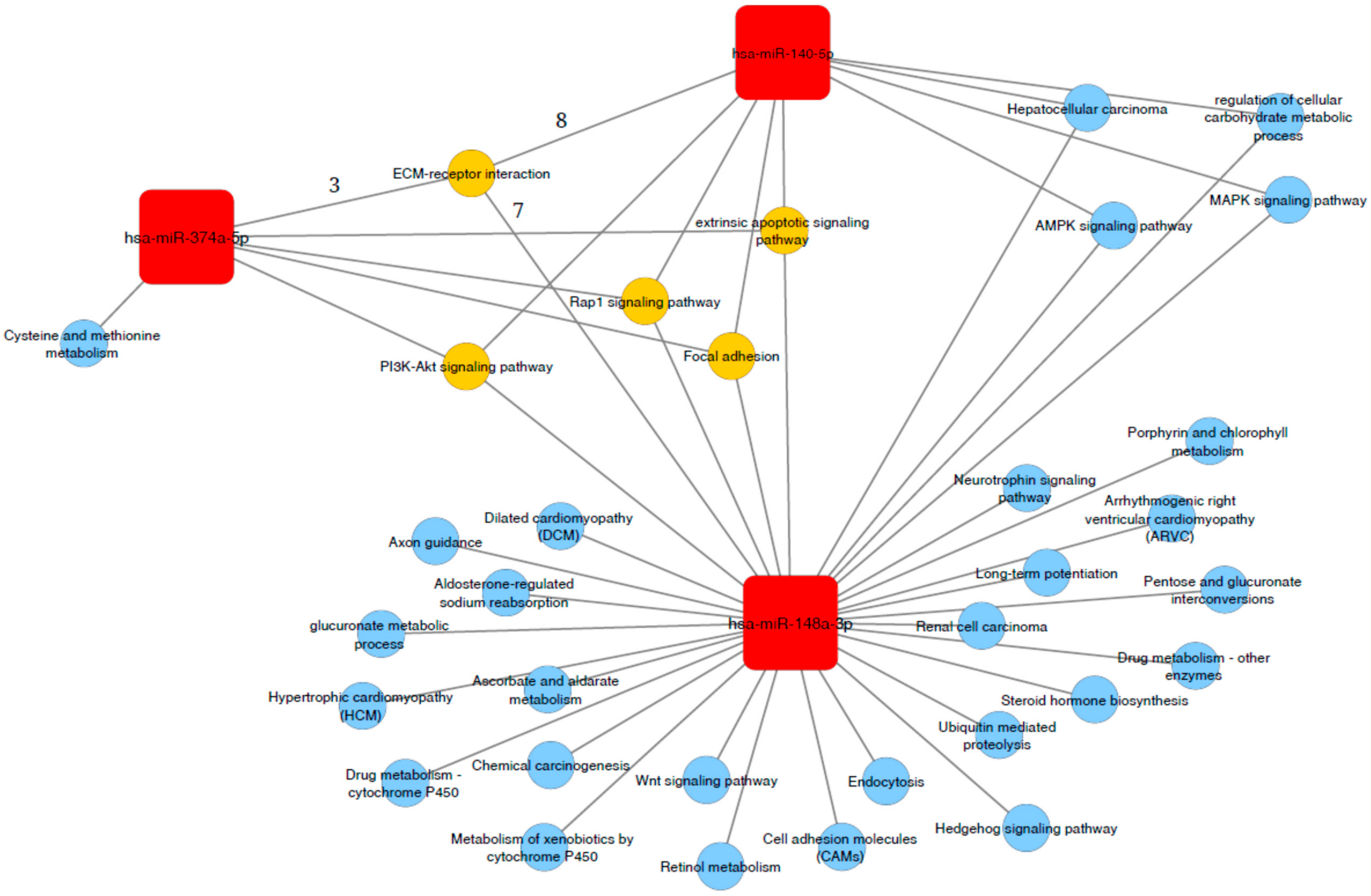
2.6. Association of miR-140-5p, miR 148a-3p and mir-374a-5p with Functional Related Pathways
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Processing
4.2. ct-miRNA Profiling Data Processing
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef]
- Guiu, S.; Mouret Reynier, M.A.; Toure, M.; Coudert, B. Predictive factors of response in HER2-positive breast cancer treated by neoadjuvant therapy. J. Oncol. 2013, 2013, 854121. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Von Minckwitz, G.; Schneeweiss, A.; Paepke, S.; Lehmann, A.; Rezai, M.; Zahm, D.M.; Sinn, P.; Khandan, F.; Eidtmann, H.; et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (HER2) therapy in primary HER2-overexpressing breast cancer. J. Clin. Oncol. 2014, 32, 3212–3220. [Google Scholar] [CrossRef] [PubMed]
- Scaltriti, M.; Nuciforo, P.; Bradbury, I.; Sperinde, J.; Agbor-Tarh, D.; Campbell, C.; Chenna, A.; Winslow, J.; Serra, V.; Parra, J.L.; et al. High HER2 expression correlates with response to the combination of lapatinib and trastuzumab. Clin. Cancer Res. 2015, 21, 569–576. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Campbell, C.; Savas, P.; Nuciforo, P.; Aura, C.; De Azambuja, E.; Eidtmann, H.; Ellis, C.E.; Baselga, J.; et al. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: A secondary analysis of the NeoALTTO trial. JAMA Oncol. 2015, 1, 448–454. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease -latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Berindan-Neagoe, I.; Calin, G.A. Molecular pathways: MicroRNAs, cancer cells, and microenvironment. Clin. Cancer Res. 2014, 20, 6247–6253. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Müller, V.; Gade, S.; Steinbach, B.; Loibl, S.; von Minckwitz, G.; Untch, M.; Schwedler, K.; Lübbe, K.; Schem, C.; Fasching, P.A.; et al. Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: A translational research project within the Geparquinto trial. Breast Cancer Res. Treat. 2014, 147, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Yuan, P.; Zhao, Z.T.; Yang, Z.; Wang, T.; Zhao, J.D.; Luo, Y.; Ma, F.; Wang, J.Y.; Fan, Y.; et al. A miRNA-based signature predicts development of disease recurrence in HER2 positive breast cancer after adjuvant trastuzumab-based treatment. Sci. Rep. 2016, 6, 33825. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.J.; Santarpia, L.; Kim, J.; Esteva, F.J.; Moretti, E.; Buzdar, A.U.; Di Leo, A.; Le, X.F.; Bast, R.C., Jr.; Park, S.T.; et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer 2012, 118, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Di Cosimo, S.; Appierto, V.; Pizzamiglio, S.; Tiberio, P.; Iorio, M.V.; Hilbers, F.; De Azambuja, E.; De La Peña, L.; Izquierdo, M.; Huober, J.; et al. Plasma miRNA levels for predicting therapeutic response to neoadjuvant treatment in HER2-positive breast cancer: Results from the NeoALTTO trial. Clin. Cancer Res. 2019, 25, 3887–3895. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Bradbury, I.; Eidtmann, H.; Di Cosimo, S.; De Azambuja, E.; Aura, C.; Gómez, H.; Dinh, P.; Fauria, K.; Van Dooren, V.; et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): A randomised, open-label, multicentre, phase 3 trial. Lancet 2012, 379, 633–640. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenet. 2018, 10, 59. [Google Scholar] [CrossRef]
- Hamam, R.; Hamam, D.; Alsaleh, K.A.; Kassem, M.; Zaher, W.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017, 8, e3045. [Google Scholar] [CrossRef]
- Li, Y.; Deng, X.; Zeng, X.; Peng, X. The role of Mir-148a in cancer. J. Cancer 2016, 7, 1233–1241. [Google Scholar] [CrossRef]
- Bian, H.; Zhou, Y.; Zhou, D.; Zhang, Y.; Shang, D.; Qi, J. The latest progress on miR-374 and its functional implications in physiological and pathological processes. J. Cell Mol. Med. 2019, 23, 3063–3076. [Google Scholar] [CrossRef]
- Rothe, F.; Ignatiadis, M.; Chaboteaux, C.; Haibe-Kains, B.; Kheddoumi, N.; Majjaj, S.; Badran, B.; Fayyad-Kazan, H.; Desmedt, C.; Harris, A.L.; et al. Global microRNA expression profiling identifies miR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE 2011, 6, e20980. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, Y.; Jasper, J.; Lykken, E.; Alexander, P.B.; Markowitz, G.J.; McDonnell, D.P.; Li, Q.J.; Wang, X.F. miR-148a functions to suppress metastasis and serves as a prognostic indicator in triple-negative breast cancer. Oncotarget 2016, 7, 20381–20394. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, Q.; Xu, Q.; Liu, L.; Jiang, B. MiR-148a inhibits angiogenesis by targeting ERBB3. J. Biomed. Res. 2011, 25, 170–177. [Google Scholar] [CrossRef]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013, 5, 3–13. [Google Scholar] [CrossRef]
- Cosentino, C.; Di Domenico, M.; Porcellini, A.; Cuozzo, C.; De Gregorio, G.; Santillo, M.R.; Agnese, S.; Di Stasio, R.; Feliciello, A.; Migliaccio, A.; et al. The p85 regulatory subunit of PI3K mediates cAMP-PKA and insulin biological effects on MCF-7 cell growth and motility. Sci. World J. 2014, 2014, 565839. [Google Scholar]
- Ashizawa, M.; Okayama, H.; Ishigame, T.; Thar Min, A.K.; Saito, K.; Ujiie, D. miRNA-148a-3p regulates immunosuppression in DNA mismatch repair-deficient colorectal cancer by targeting PD-L1. Mol. Cancer Res. 2019, 17, 1403–1413. [Google Scholar]
- Tao, S.; He, H.; Chen, Q.; Yue, W. GPER mediated estradiol reduces miR-148a to promote HLA-G expression in breast cancer. Biochem. Biophys. Res. Commun. 2014, 451, 74–78. [Google Scholar] [CrossRef]
- Genua, M.; Pandini, G.; Sisci, D.; Castoria, G.; Maggiolini, M.; Vigneri, R.; Belfiore, A. Role of cyclic AMP response element–binding protein in insulin-like growth factor-I receptor up-regulation by sex steroids in prostate cancer cells. Cancer Res. 2009, 69, 7270–7277. [Google Scholar] [CrossRef]
- Migliaccio, A.; Di Domenico, M.; Castoria, G.; Nanayakkara, M.; Lombardi, M.; de Falco, A.; Bilancio, A.; Varricchio, L.; Ciociola, A.; Auricchio, F. Steroid receptor regulation of epidermal growth factor signaling through Src in breast and prostate cancer cells: Steroid antagonist action. Cancer Res. 2005, 65, 10585–10593. [Google Scholar] [CrossRef]
- Curley, M.D.; Sabnis, G.J.; Wille, L.; Adiwijaya, B.S.; Garcia, G.; Moyo, V.; Kazi, A.A.; Brodie, A.; MacBeath, G. Seribantumab, an anti-ERBB3 antibody, delays the onset of eesistance and restores sensitivity to letrozole in an estrogen receptor-positive breast cancer model. Mol. Cancer Ther. 2015, 14, 2642–2652. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, L.Z.; Yin, Y.; He, J.; Li, Q.; Qian, X.; You, Y.; Lu, Z.; Peiper, S.C.; Shu, Y.; et al. Regulatory circuit of PKM2/NF-κB/miR-148a/152-modulated tumor angiogenesis and cancer progression. Oncogene 2015, 34, 5482–5493. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Xue, P.; Zhang, L.; Pan, R.; Cai, Z.; He, Z.; Sun, J.; Zheng, M. Prediction of key genes and pathways involved in trastuzumab-resistant gastric cancer. World J. Surg. Oncol. 2018, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Thiel, G.; Cibelli, G. Regulation of life and death by the zinc finger transcription factor Egr-1. J. Cell Physiol. 2002, 193, 287–292. [Google Scholar] [CrossRef] [PubMed]
- De Melo Gagliato, D.; Jardim, D.L.; Marchesi, M.S.; Hortobagyi, G.N. Mechanisms of resistance and sensitivity to anti-HER2 therapies in HER2+ breast cancer. Oncotarget 2016, 7, 64431–64446. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Verderio, P.; Bottelli, S.; Ciniselli, C.; Pierotti, M.A.; Gariboldi, M.; Pizzamiglio, S. NqA: An R-based algorithm for the normalization and analysis of microRNA quantitative real-time polymerase chain reaction data. Anal. Biochem. 2014, 461, 7–9. [Google Scholar] [CrossRef]
- Ciniselli, C.M.; Lecchi, M.; Gariboldi, M.; Verderio, P.; Daidone, M.G. Workflow for circulating miRNA identification and development in cancer research: Methodological considerations. In Precision Medicine Tools and Quantitative Approaches; Elsevier: London, UK, 2017. [Google Scholar]
- Sticht, C.; de la Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
| Variable | Characteristics | N | % |
|---|---|---|---|
| Age (years) | median (range) | 48.5 (25;70) | |
| ER status | Positive | 26 | 50 |
| Negative | 26 | 50 | |
| Nodal status | N0/1 | 43 | 83 |
| N2 | 9 | 17 | |
| Tumor size | ≤5 cm | 33 | 63 |
| >5 cm | 19 | 36 | |
| pCR | No | 36 | 69 |
| Yes | 16 | 31 | |
| Planned surgery | Conservative | 12 | 23 |
| Not Conservative | 40 | 77 | |
| Difference Log2RQ (Week2-Baseline) | Univariate | Multivariate * | ||
|---|---|---|---|---|
| OR (95%CI) | p-Value | OR (95%CI) | p-Value | |
| ct-miR-148a-3p | 3.42 (1.24;9.46) | 0.018 | 3.42 (1.23;9.46) | 0.018 |
| ct-miR-374a-5p | 2.24 (0.92;5.48) | 0.077 | 2.31 (0.93;5.78) | 0.073 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Cosimo, S.; Appierto, V.; Pizzamiglio, S.; Silvestri, M.; Baselga, J.; Piccart, M.; Huober, J.; Izquierdo, M.; de la Pena, L.; Hilbers, F.S.; et al. Early Modulation of Circulating MicroRNAs Levels in HER2-Positive Breast Cancer Patients Treated with Trastuzumab-Based Neoadjuvant Therapy. Int. J. Mol. Sci. 2020, 21, 1386. https://doi.org/10.3390/ijms21041386
Di Cosimo S, Appierto V, Pizzamiglio S, Silvestri M, Baselga J, Piccart M, Huober J, Izquierdo M, de la Pena L, Hilbers FS, et al. Early Modulation of Circulating MicroRNAs Levels in HER2-Positive Breast Cancer Patients Treated with Trastuzumab-Based Neoadjuvant Therapy. International Journal of Molecular Sciences. 2020; 21(4):1386. https://doi.org/10.3390/ijms21041386
Chicago/Turabian StyleDi Cosimo, Serena, Valentina Appierto, Sara Pizzamiglio, Marco Silvestri, José Baselga, Martine Piccart, Jens Huober, Miguel Izquierdo, Lorena de la Pena, Florentine S. Hilbers, and et al. 2020. "Early Modulation of Circulating MicroRNAs Levels in HER2-Positive Breast Cancer Patients Treated with Trastuzumab-Based Neoadjuvant Therapy" International Journal of Molecular Sciences 21, no. 4: 1386. https://doi.org/10.3390/ijms21041386
APA StyleDi Cosimo, S., Appierto, V., Pizzamiglio, S., Silvestri, M., Baselga, J., Piccart, M., Huober, J., Izquierdo, M., de la Pena, L., Hilbers, F. S., de Azambuja, E., Untch, M., Pusztai, L., Pritchard, K., Nuciforo, P., Vincent-Salomon, A., Symmans, F., Apolone, G., de Braud, F. G., ... Daidone, M. G. (2020). Early Modulation of Circulating MicroRNAs Levels in HER2-Positive Breast Cancer Patients Treated with Trastuzumab-Based Neoadjuvant Therapy. International Journal of Molecular Sciences, 21(4), 1386. https://doi.org/10.3390/ijms21041386







