Understanding Mechanisms Underlying Non-Alcoholic Fatty Liver Disease (NAFLD) in Mental Illness: Risperidone and Olanzapine Alter the Hepatic Proteomic Signature in Mice
Abstract
1. Introduction
2. Results
2.1. Animal Health
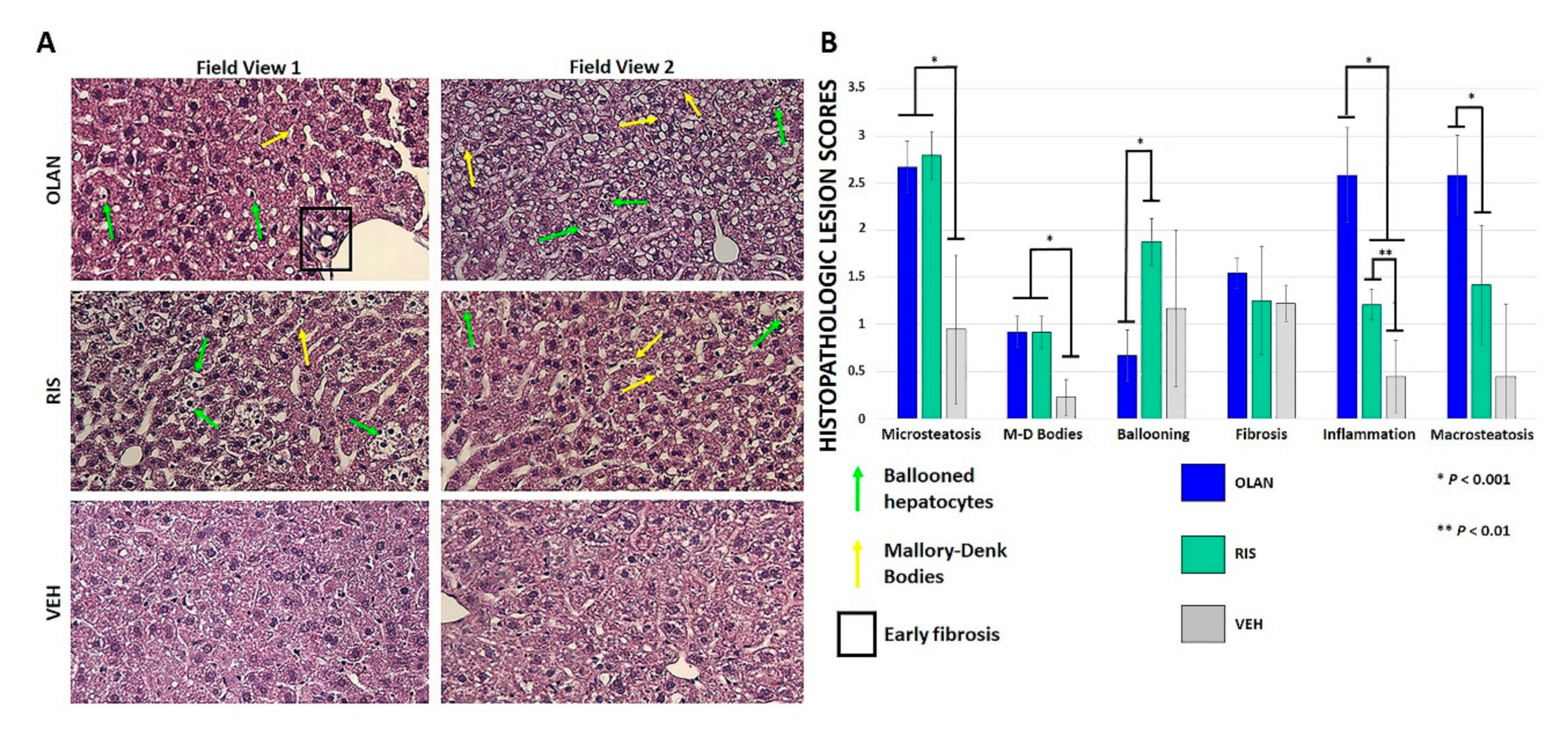
2.2. Atypical Antipsychotic Treatment Induced Histopathologic Changes Consistent with the Development of NAFLD
2.3. Atypical Antipsychotic Treatment Altered the Hepatic Proteome
2.4. Antipsychotic Treatment Altered Protein Expression in Hepatic Energy Metabolism Pathways
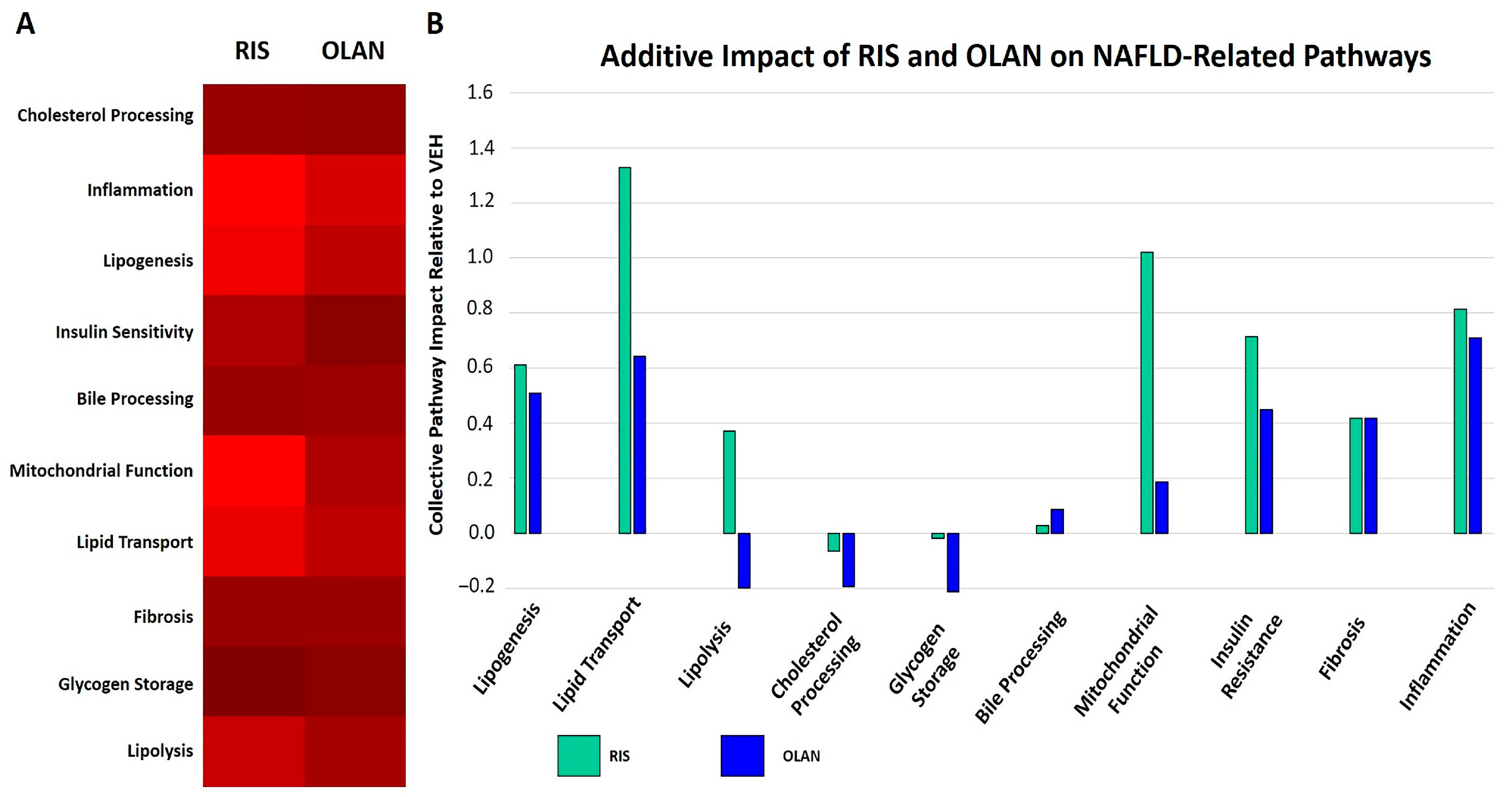
2.5. Antipsychotic Treatment Altered Expression of Proteins Associated with NAFLD
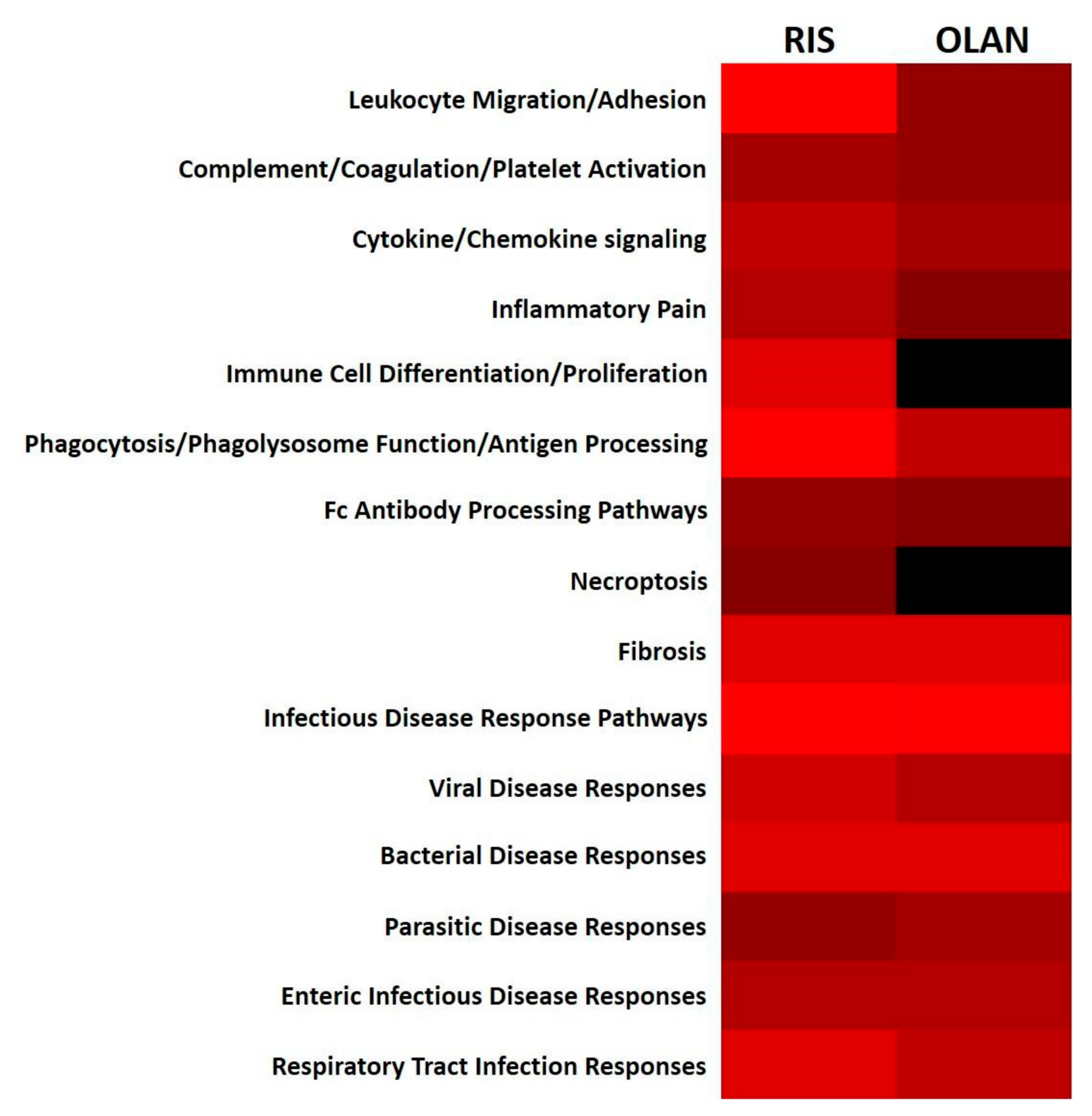
2.6. Antipsychotic Treatment Disrupted Hepatic Immune/Inflammatory Signaling Pathways
2.7. Antipsychotic Treatment Affected Pathways Regulated by the Autonomic Nervous System: A Unifying Mechanism
3. Discussion
4. Materials and Methods
4.1. Animals and Experimental Design
4.2. Drug Formulation and Dosing Strategy
4.3. Histopathology
4.4. Proteomic Methodology
4.5. Proteomic Analysis
4.6. RNA Analysis
4.7. Statistical Analysis
4.8. Data Statement
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chesney, E.; Goodwin, G.M.; Fazel, S. Risks of All-Cause and Suicide Mortality in Mental Disorders: A Meta-Review. World Psychiatry 2014, 13, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Vancampfort, D.; Stubbs, B.; Mitchell, A.J.; De Hert, M.; Wampers, M.; Ward, P.B.; Rosenbaum, S.; Correll, C.U. Risk of Metabolic Syndrome and Its Components in People with Schizophrenia and Related Psychotic Disorders, Bipolar Disorder and Major Depressive Disorder: A Systematic Review and Meta-Analysis. World Psychiatry 2015, 14, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Soto-Angona, Ó.; Anmella, G.; Valdés-Florido, M.J.; De Uribe-Viloria, N.; Carvalho, A.F.; Penninx, B.W.J.H.; Berk, M. Non-Alcoholic Fatty Liver Disease (NAFLD) as a Neglected Metabolic Companion of Psychiatric Disorders: Common Pathways and Future Approaches. BMC Med. 2020, 18, 261. [Google Scholar] [CrossRef] [PubMed]
- Mikolasevic, I.; Milic, S.; Turk Wensveen, T.; Grgic, I.; Jakopcic, I.; Stimac, D.; Wensveen, F.; Orlic, L. Nonalcoholic Fatty Liver Disease—A Multisystem Disease? World J. Gastroenterol. 2016, 22, 9488–9505. [Google Scholar] [CrossRef] [PubMed]
- Engin, A. Non-Alcoholic Fatty Liver Disease. Adv. Exp. Med. Biol. 2017, 960, 443–467. [Google Scholar] [CrossRef] [PubMed]
- Mouzaki, M.; Yodoshi, T.; Arce-Clachar, A.C.; Bramlage, K.; Fei, L.; Ley, S.L.; Xanthakos, S.A. Psychotropic Medications Are Associated with Increased Liver Disease Severity in Pediatric Nonalcoholic Fatty Liver Disease. J. Pediatric Gastroenterol. Nutr. 2019, 69, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Amir, M.; Yu, M.; He, P.; Srinivasan, S. Hepatic Autonomic Nervous System and Neurotrophic Factors Regulate the Pathogenesis and Progression of Non-Alcoholic Fatty Liver Disease. Front. Med. 2020, 7, 62. [Google Scholar] [CrossRef]
- Xu, H.; Zhuang, X. Atypical Antipsychotics-Induced Metabolic Syndrome and Nonalcoholic Fatty Liver Disease: A Critical Review. Neuropsychiatr. Dis. Treat 2019, 15, 2087–2099. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, C.; Xu, F.; Zhao, M.; Bergquist, J.; Yang, C.; Liu, X.; Tan, Y.; Wang, X.; Li, S.; et al. System Biology Analysis Reveals the Role of Voltage-Dependent Anion Channel in Mitochondrial Dysfunction during Non-Alcoholic Fatty Liver Disease Progression into Hepatocellular Carcinoma. Cancer Sci. 2020, 111, 4288–4302. [Google Scholar] [CrossRef]
- Cobbina, E.; Akhlaghi, F. Non-Alcoholic Fatty Liver Disease (NAFLD)—Pathogenesis, Classification, and Effect on Drug Metabolizing Enzymes and Transporters. Drug Metab. Rev. 2017, 49, 197–211. [Google Scholar] [CrossRef]
- Anty, R.; Gual, P. Pathogenesis of Non-Alcoholic Fatty Liver Disease. Presse Med. 2019, 48, 1468–1483. [Google Scholar] [CrossRef] [PubMed]
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the Epidemic of Nonalcoholic Fatty Liver Disease Demonstrates an Exponential Increase in Burden of Disease. Hepatology 2018, 67, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Benedict, M.; Zhang, X. Non-Alcoholic Fatty Liver Disease: An Expanded Review. World J. Hepatol. 2017, 9, 715–732. [Google Scholar] [CrossRef]
- Harris, L.W.; Guest, P.C.; Wayland, M.T.; Umrania, Y.; Krishnamurthy, D.; Rahmoune, H.; Bahn, S. Schizophrenia: Metabolic Aspects of Aetiology, Diagnosis and Future Treatment Strategies. Psychoneuroendocrinology 2013, 38, 752–766. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.J.; Vancampfort, D.; Sweers, K.; van Winkel, R.; Yu, W.; De Hert, M. Prevalence of Metabolic Syndrome and Metabolic Abnormalities in Schizophrenia and Related Disorders—A Systematic Review and Meta-Analysis. Schizophr. Bull. 2013, 39, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Papanastasiou, E. The Prevalence and Mechanisms of Metabolic Syndrome in Schizophrenia: A Review. Ther. Adv. Psychopharmacol. 2013, 3, 33–51. [Google Scholar] [CrossRef] [PubMed]
- Freyberg, Z.; Aslanoglou, D.; Shah, R.; Ballon, J.S. Intrinsic and Antipsychotic Drug-Induced Metabolic Dysfunction in Schizophrenia. Front. Neurosci. 2017, 11, 432. [Google Scholar] [CrossRef]
- Pillinger, T.; McCutcheon, R.A.; Vano, L.; Mizuno, Y.; Arumuham, A.; Hindley, G.; Beck, K.; Natesan, S.; Efthimiou, O.; Cipriani, A.; et al. Comparative Effects of 18 Antipsychotics on Metabolic Function in Patients with Schizophrenia, Predictors of Metabolic Dysregulation, and Association with Psychopathology: A Systematic Review and Network Meta-Analysis. Lancet Psychiatry 2020, 7, 64–77. [Google Scholar] [CrossRef]
- Siafis, S.; Tzachanis, D.; Samara, M.; Papazisis, G. Antipsychotic Drugs: From Receptor-Binding Profiles to Metabolic Side Effects. Curr. Neuropharmacol. 2018, 16, 1210–1223. [Google Scholar] [CrossRef]
- American Diabetes Association; American Psychiatric Association; American Association of Clinical Endocrinologists; North American Association for the Study of Obesity. Consensus Development Conference on Antipsychotic Drugs and Obesity and Diabetes. Diabetes Care 2004, 27, 596–601. [Google Scholar] [CrossRef]
- Beauchemin, M.; Geguchadze, R.; Guntur, A.R.; Nevola, K.; Le, P.T.; Barlow, D.; Rue, M.; Vary, C.P.H.; Lary, C.W.; Motyl, K.J.; et al. Exploring Mechanisms of Increased Cardiovascular Disease Risk with Antipsychotic Medications: Risperidone Alters the Cardiac Proteomic Signature in Mice. Pharmacol. Res. 2020, 152, 104589. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, K.T.; Enos, R.T.; Bader, J.E.; Sougiannis, A.T.; Carson, M.S.; Chatzistamou, I.; Carson, J.A.; Nagarkatti, P.S.; Nagarkatti, M.; Murphy, E.A. Prolonged High-Fat-Diet Feeding Promotes Non-Alcoholic Fatty Liver Disease and Alters Gut Microbiota in Mice. World J. Hepatol. 2019, 11, 619–637. [Google Scholar] [CrossRef] [PubMed]
- Recena Aydos, L.; Aparecida do Amaral, L.; Serafim de Souza, R.; Jacobowski, A.C.; Freitas dos Santos, E.; Rodrigues Macedo, M.L. Nonalcoholic Fatty Liver Disease Induced by High-Fat Diet in C57bl/6 Models. Nutrients 2019, 11, 3067. [Google Scholar] [CrossRef] [PubMed]
- Mauri, M.C.; Paletta, S.; Maffini, M.; Colasanti, A.; Dragogna, F.; Di Pace, C.; Altamura, A.C. Clinical Pharmacology of Atypical Antipsychotics: An Update. EXCLI J. 2014, 13, 1163–1191. [Google Scholar]
- Ader, M.; Kim, S.P.; Catalano, K.J.; Ionut, V.; Hucking, K.; Richey, J.M.; Kabir, M.; Bergman, R.N. Metabolic Dysregulation with Atypical Antipsychotics Occurs in the Absence of Underlying Disease A Placebo-Controlled Study of Olanzapine and Risperidone in Dogs. Diabetes 2005, 54, 862–871. [Google Scholar] [CrossRef]
- Houseknecht, K.L.; Robertson, A.S.; Zavadoski, W.; Gibbs, E.M.; Johnson, D.E.; Rollema, H. Acute Effects of Atypical Antipsychotics on Whole-Body Insulin Resistance in Rats: Implications for Adverse Metabolic Effects. Neuropsychopharmacology 2007, 32, 289–297. [Google Scholar] [CrossRef]
- Martins, P.J.F.; Haas, M.; Obici, S. Central Nervous System Delivery of the Antipsychotic Olanzapine Induces Hepatic Insulin Resistance. Diabetes 2010, 59, 2418–2425. [Google Scholar] [CrossRef]
- DeFronzo, R.A. Insulin Resistance, Lipotoxicity, Type 2 Diabetes and Atherosclerosis: The Missing Links. The Claude Bernard Lecture 2009. Diabetologia 2010, 53, 1270–1287. [Google Scholar] [CrossRef]
- Schaffer, J.E. Lipotoxicity: Many Roads to Cell Dysfunction and Cell Death: Introduction to a Thematic Review Series. J. Lipid Res. 2016, 57, 1327–1328. [Google Scholar] [CrossRef]
- Czech, M.P. Insulin Action and Resistance in Obesity and Type 2 Diabetes. Nat. Med. 2017, 23, 804–814. [Google Scholar] [CrossRef]
- Hong, F.; Pan, S.; Guo, Y.; Xu, P.; Zhai, Y. PPARs as Nuclear Receptors for Nutrient and Energy Metabolism. Molecules 2019, 24, 2545. [Google Scholar] [CrossRef] [PubMed]
- Lamas Bervejillo, M.; Ferreira, A.M. Understanding Peroxisome Proliferator-Activated Receptors: From the Structure to the Regulatory Actions on Metabolism. Adv. Exp. Med. Biol. 2019, 1127, 39–57. [Google Scholar] [CrossRef] [PubMed]
- Astapova, O.; Leff, T. Adiponectin and PPARγ: Cooperative and Interdependent Actions of Two Key Regulators of Metabolism. Vitam. Horm. 2012, 90, 143–162. [Google Scholar] [CrossRef] [PubMed]
- Iwabu, M.; Okada-Iwabu, M.; Yamauchi, T.; Kadowaki, T. Adiponectin/AdipoR Research and Its Implications for Lifestyle-Related Diseases. Front. Cardiovasc. Med. 2019, 6, 116. [Google Scholar] [CrossRef]
- Straub, L.G.; Scherer, P.E. Metabolic Messengers: Adiponectin. Nat. Metab. 2019, 1, 334–339. [Google Scholar] [CrossRef]
- Albaugh, V.L.; Vary, T.C.; Ilkayeva, O.; Wenner, B.R.; Maresca, K.P.; Joyal, J.L.; Breazeale, S.; Elich, T.D.; Lang, C.H.; Lynch, C.J. Atypical Antipsychotics Rapidly and Inappropriately Switch Peripheral Fuel Utilization to Lipids, Impairing Metabolic Flexibility in Rodents. Schizophr. Bull. 2012, 38, 153–166. [Google Scholar] [CrossRef]
- Qi, Y.; Xu, R. Roles of PLODs in Collagen Synthesis and Cancer Progression. Front. Cell. Dev. Biol. 2018, 6, 66. [Google Scholar] [CrossRef]
- Shoelson, S.E.; Lee, J.; Goldfine, A.B. Inflammation and Insulin Resistance. J. Clin. Invest. 2006, 116, 1793–1801. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, R.; Xiong, Y.; Du, F.; Zhu, S. A Vicious Circle between Insulin Resistance and Inflammation in Nonalcoholic Fatty Liver Disease. Lipids Health Dis. 2017, 16, 203. [Google Scholar] [CrossRef]
- Zatterale, F.; Longo, M.; Naderi, J.; Raciti, G.A.; Desiderio, A.; Miele, C.; Beguinot, F. Chronic Adipose Tissue Inflammation Linking Obesity to Insulin Resistance and Type 2 Diabetes. Front. Physiol. 2020, 10, 1607. [Google Scholar] [CrossRef]
- Katsarou, A.; Moustakas, I.I.; Pyrina, I.; Lembessis, P.; Koutsilieris, M.; Chatzigeorgiou, A. Metabolic Inflammation as an Instigator of Fibrosis during Non-Alcoholic Fatty Liver Disease. World J. Gastroenterol. 2020, 26, 1993–2011. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, S.; Rhodes, F.; Srivastava, A.; Trembling, P.M.; Rosenberg, W.M. Inflammation and Fibrosis in Chronic Liver Diseases Including Non-Alcoholic Fatty Liver Disease and Hepatitis C. World J. Gastroenterol. 2020, 26, 109–133. [Google Scholar] [CrossRef] [PubMed]
- Arrese, M.; Cabrera, D.; Kalergis, A.M.; Feldstein, A.E. Innate Immunity and Inflammation in NAFLD/NASH. Dig Dis Sci 2016, 61, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Dawood, R.M.; El-Meguid, M.A.; Salum, G.M.; El Awady, M.K. Key Players of Hepatic Fibrosis. J Interferon Cytokine Res 2020, 40, 472–489. [Google Scholar] [CrossRef] [PubMed]
- Franceschetti, L.; Bonomini, F.; Rodella, L.F.; Rezzani, R. Critical Role of NFκB in the Pathogenesis of Non-Alcoholic Fatty Liver Disease: A Widespread Key Regulator. Curr. Mol. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sutti, S.; Albano, E. Adaptive Immunity: An Emerging Player in the Progression of NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Stine, J.G.; Intagliata, N.; Northup, P.G.; Caldwell, S.H. Nonalcoholic Fatty Liver Disease, Portal Vein Thrombosis and Coagulation: More Questions Than Answers? Transplantation 2017, 101, e281–e282. [Google Scholar] [CrossRef]
- Mizuno, K.; Ueno, Y. Autonomic Nervous System and the Liver. Hepatol. Res. 2017, 47, 160–165. [Google Scholar] [CrossRef]
- Leung, J.Y.T.; Barr, A.M.; Procyshyn, R.M.; Honer, W.G.; Pang, C.C.Y. Cardiovascular Side-Effects of Antipsychotic Drugs: The Role of the Autonomic Nervous System. Pharmacol. Ther. 2012, 135, 113–122. [Google Scholar] [CrossRef]
- Scigliano, G.; Ronchetti, G. Antipsychotic-Induced Metabolic and Cardiovascular Side Effects in Schizophrenia: A Novel Mechanistic Hypothesis. CNS Drugs 2013, 27, 249–257. [Google Scholar] [CrossRef]
- Motyl, K.J.; DeMambro, V.E.; Barlow, D.; Olshan, D.; Nagano, K.; Baron, R.; Rosen, C.J.; Houseknecht, K.L. Propranolol Attenuates Risperidone-Induced Trabecular Bone Loss in Female Mice. Endocrinology 2015, 156, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, L.; Scrushy, M.; Meng, F.; Lairmore, T.C.; Alpini, G.; Glaser, S. Biliary Epithelium: A Neuroendocrine Compartment in Cholestatic Liver Disease. Clin. Res. Hepatol. Gastroenterol. 2018, 42, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, M.; Karunanayake, T.; Wier, J.; Hsu, N.; Yang, X. Network Modeling Approaches and Applications to Unravelling Non-Alcoholic Fatty Liver Disease. Genes 2019, 10, 966. [Google Scholar] [CrossRef] [PubMed]
- May, M.; Beauchemin, M.; Vary, C.; Barlow, D.; Houseknecht, K.L. The Antipsychotic Medication, Risperidone, Causes Global Immunosuppression in Healthy Mice. PLoS ONE 2019, 14, e0218937. [Google Scholar] [CrossRef] [PubMed]
- May, M.; Slitzky, M.; Rostama, B.; Barlow, D.; Houseknecht, K.L. Antipsychotic-Induced Immune Dysfunction: A Consideration for COVID-19 risk. Brain Behav. Immun. Health 2020, 6, 100097. [Google Scholar] [CrossRef] [PubMed]
- Drzyzga, L.; Obuchowicz, E.; Marcinowska, A.; Herman, Z.S. Cytokines in Schizophrenia and the Effects of Antipsychotic Drugs. Brain Behav. Immun. 2006, 20, 532–545. [Google Scholar] [CrossRef]
- Correll, C.U. Cardiometabolic Risk of Second-Generation Antipsychotic Medications during First-Time Use in Children and Adolescents. JAMA 2009, 302, 1765–1773. [Google Scholar] [CrossRef]
- Kowalchuk, C.; Castellani, L.N.; Chintoh, A.; Remington, G.; Giacca, A.; Hahn, M.K. Antipsychotics and Glucose Metabolism: How Brain and Body Collide. Am. J. Physiol. Endocrinol. Metab. 2018, 316, E1–E15. [Google Scholar] [CrossRef]
- Kim, H.J.; Wilson, C.; Deusen, T.V.; Millard, H.; Qayyum, Z.; Parke, S. Metabolic Syndrome in Child and Adolescent Psychiatry. Psychiatr. Ann. 2020, 50, 326–333. [Google Scholar] [CrossRef]
- Müller, N.; Weidinger, E.; Leitner, B.; Schwarz, M.J. The Role of Inflammation in Schizophrenia. Front. Neurosci. 2015, 9, 372. [Google Scholar] [CrossRef]
- Gaughran, F. Immunity and Schizophrenia: Autoimmunity, Cytokines, and Immune Responses. Int. Rev. Neurobiol. 2002, 52, 275–302. [Google Scholar] [CrossRef] [PubMed]
- Schuld, A.; Hinze-Selch, D.; Pollmächer, T. Cytokine Network in Patients with Schizophrenia and Its Significance for the Pathophysiology of the Illness. Nervenarzt 2004, 75, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Calarge, C.A.; Ivins, S.D.; Motyl, K.J.; Shibli-Rahhal, A.A.; Bliziotes, M.M.; Schlechte, J.A. Possible Mechanisms for the Skeletal Effects of Antipsychotics in Children and Adolescents. Ther. Adv. Psychopharmacol. 2013, 3, 278–293. [Google Scholar] [CrossRef] [PubMed]
- Motyl, K.J.; Beauchemin, M.; Barlow, D.; Le, P.T.; Nagano, K.; Treyball, A.; Contractor, A.; Baron, R.; Rosen, C.J.; Houseknecht, K.L. A Novel Role for Dopamine Signaling in the Pathogenesis of Bone Loss from the Atypical Antipsychotic Drug Risperidone in Female Mice. Bone 2017, 103, 168–176. [Google Scholar] [CrossRef]
- Caldez, M.J.; Bjorklund, M.; Kaldis, P. Cell Cycle Regulation in NAFLD: When Imbalanced Metabolism Limits Cell Division. Hepatol. Int. 2020, 14, 463–474. [Google Scholar] [CrossRef]
- Wojcikowski, J.; Anna Daniel, W. The Role of the Nervous System in the Regulation of Liver Cytochrome P450. Curr. Drug Metab. 2011, 12, 124–138. [Google Scholar] [CrossRef]
- Jensen, K.J.; Alpini, G.; Glaser, S. Hepatic Nervous System and Neurobiology of the Liver. Compr. Physiol. 2013, 3, 655–665. [Google Scholar] [CrossRef]
- Bär, K.-J. Cardiac Autonomic Dysfunction in Patients with Schizophrenia and Their Healthy Relatives—A Small Review. Front. Neurol. 2015, 6, 139. [Google Scholar] [CrossRef]
- Esler, M.; Rumantir, M.; Wiesner, G.; Kaye, D.; Hastings, J.; Lambert, G. Sympathetic Nervous System and Insulin Resistance: From Obesity to Diabetes. Am. J. Hypertens. 2001, 14, 304S–309S. [Google Scholar] [CrossRef]
- Oliveira, C.; Silveira, E.A.; Rosa, L.; Santos, A.; Rodrigues, A.P.; Mendonça, C.; Silva, L.; Gentil, P.; Rebelo, A.C. Risk Factors Associated with Cardiac Autonomic Modulation in Obese Individuals. J. Obes. 2020, 2020. [Google Scholar] [CrossRef]
- Grassi, G.; Biffi, A.; Dell’Oro, R.; Quarti Trevano, F.; Seravalle, G.; Corrao, G.; Perseghin, G.; Mancia, G. Sympathetic Neural Abnormalities in Type 1 and Type 2 Diabetes: A Systematic Review and Meta-Analysis. J. Hypertens. 2020, 38, 1436–1442. [Google Scholar] [CrossRef] [PubMed]
- Savoy, Y.E.; Ashton, M.A.; Miller, M.W.; Nedza, F.M.; Spracklin, D.K.; Hawthorn, M.H.; Rollema, H.; Matos, F.F.; Hajos-Korcsok, E. Differential Effects of Various Typical and Atypical Antipsychotics on Plasma Glucose and Insulin Levels in the Mouse: Evidence for the Involvement of Sympathetic Regulation. Schizophr. Bull. 2010, 36, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Hattori, S.; Kishida, I.; Suda, A.; Miyauchi, M.; Shiraishi, Y.; Fujibayashi, M.; Tsujita, N.; Ishii, C.; Ishii, N.; Moritani, T.; et al. Effects of Four Atypical Antipsychotics on Autonomic Nervous System Activity in Schizophrenia. Schizophr. Res. 2018, 193, 134–138. [Google Scholar] [CrossRef] [PubMed]
- National Research Council of the National Academies; Institute for Laboratory Animals Research; Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals: Eighth Edition; National Academies Press: Washington, DC, USA, 2010; ISBN 978-0-309-18663-6. [Google Scholar]
- Aman, M.G.; Vinks, A.A.; Remmerie, B.; Mannaert, E.; Ramadan, Y.; Masty, J.; Lindsay, R.L.; Malone, K. Plasma Pharmacokinetic Characteristics of Risperidone and Their Relationship to Saliva Concentrations in Children with Psychiatric or Neurodevelopmental Disorders. Clin. Ther. 2007, 29, 1476–1486. [Google Scholar] [CrossRef]
- Jensen, V.S.; Tveden-Nyborg, P.; Zacho-Rasmussen, C.; Quaade, M.L.; Ipsen, D.H.; Hvid, H.; Fledelius, C.; Wulff, E.M.; Lykkesfeldt, J. Variation in Diagnostic NAFLD/NASH Read-Outs in Paired Liver Samples from Rodent Models. J. Pharmacol. Toxicol. Methods 2020, 101, 106651. [Google Scholar] [CrossRef]
- Gillet, L.; Navarro, P.; Tate, S.; Rost, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted Data Extraction of the MS/MS Spectra Generated by Data-Independent Acquisition: A New Concept for Consistent and Accurate Proteome Analysis. Mol. Cell. Proteomics 2012, 11, O111.016717. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New Approach for Understanding Genome Variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]


| Predicted Dysregulated Functions with RIS | Up | Down | Associated Proteins | |
|---|---|---|---|---|
| Predicted metabolic effects | Energy production (all ATP-generating pathways combined) | 41 | 62 | BCAT2, CS, AGXT, ABAT, NDUFA8, NDUFB8, NDUFS1, COX6B1, CYB5R3, ALDH7A1, TALDO1, ACAA2, ACLY, PRKCA, PDGFRB, GNAS, PGAM2, ENO3, LRP1, ALR1, AOX3, ADIPOQ, HIBADH, COMT, MTHFD1, ECI2, NUDT7, DGKZ, EPHA2, ITGA3, SLC27A1, BLVRB, SEC22B, SMARCC1, ASGR1, BTD |
| Amino acid production | 32 | 37 | BCAT2, CS, ADIPOQ, LAP3, PGAM2, TALDO1, ENO3, ABAT, ACLY, PCBD2, ALR1, DGKZ, GNMT, AGXT, ALDH7A1, PRKCA, GNAS, FTCD, AMDHD1, ATIC, MTHFD1, EPHA2, PDGFRB, ITGA3, AOX3, COMT, ACAA2, HIBADH, TCN2 | |
| Glycolysis | 36 | 7 | ABAT, ACLY, ALDH7A1, AOX3, COX6B1, CS, EPHA2, GNAS, ITGA3, LAP3, NDUFS1, PDGFRB, PGAM2, PRKCA, TALDO1, ALR1, ENO3, | |
| Lipolysis | 27 | 9 | ABAT, ACAA2, ACLY, ADIPOQ, ALDH7A1, BLVRB, COX6B1, CS, ECI2, GNAS, MECR, PDGFRB, PRKCA, SLC27A1, AOX3, NUDT7 | |
| Lipogenesis | 24 | 2 | ABAT, ACAA2, ADIPOQ, ALDH7A1, ALR1, AOX3, DGKZ, GNAS, MECR, PRKCA, SLC27A1, TCN2, SEC22B | |
| TCA/OxPhos/ETC | 16 | 9 | ADIPOQ, BCAT2, NDUFA8, NDUFB8, PGAM2, ALDH7A1, ALR1, COX6B1, ENO3, NDUFS1 | |
| Nucleic acid synthesis | 7 | 13 | EPHA2, GNAS, ITGA3, PDGFRB, PRKCA, SMARCC1, TCN2, ALR1, AMDHD1, ATIC, FTCD, MTHFD1, NUDT7, TALDO1 | |
| Lysosomal function | 10 | 3 | ADIPOQ, CTSS, GSTA3, HGS, LAMP1, RAB11FIP5, SUMF1, RPN2, SEC22B, VAPA | |
| Mitochondrial biogenesis | 11 | 0 | ADIPOQ, GNAS, PDGFRB, PRKCA, SLC27A1, SMARCC1 | |
| Predicted mitogenic effects | Migration/motility | 55 | 21 | DGKZ, ENAH, EPHA2, GNAS, HSPB1, IL1RAP, ITGA3, MSN, NDUFA8, PDGFRB, PRKCA, ROCK2, ADIPOQ, AFDN, ASGR1, ELAVL1, INPP5D, MYH9, NDUFS1 |
| Cell survival | 49 | 6 | CRYAB, DGKZ, EPHA2, GNAS, GSTA3, HSPB1, IL1RAP, ITGA3, NDUFA8, PDGFRB, PRKCA, RNF31, ADIPOQ, ASGR1, ELAVL1, INPP5D, NDUFS1 | |
| Cell proliferation | 46 | 6 | DGKZ, EPHA2, GNAS, HSPB1, IL1RAP, ITGA3, NDUFA8, NDUFB8, PDGFRB, PRKCA, ADIPOQ, ASGR1, ELAVL1, INPP5D, NDUFS1 | |
| Protein production | 13 | 37 | EIF5, EPHA2, FAU, GNAS, HNRNPC, HSPB1, ITGA3, PDGFRB, PRKCA, TRA2B, AARS, ABAT, AGXT, ALDH7A1, ALR1, AMDHD1, ATIC, CRYAB, DDOST, EIF5B, FTCD, GANAB, HGS, HYOU1, NOP56, PABPC1, PUF60, RAB11FIP5, REX02, RPN2, RPS3A1, SEC22B, SNRPB, SNRPG, TALDO1, TARS | |
| Cancer | 25 | 7 | DGKZ, GNAS, GSTA3, ITGA3, KLK3, PDGFRB, PRKCA, ROCK2, EPHA2, HPGD, SMARCC1 | |
| Predicted tissue-damaging effects | Oxidation/ROS generation | 34 | 10 | ABAT, AGXT, ALDH7A1, ALR1, AOX3, BLVRB, COMT, COX6B1, DDOST, EPHA2, GANAB, GNAS, HGS, YOU1, ITGA3, NDUFA8, NDUFB8, PDGFRB, PRKCA, RAB11FIP5, ROCK2, RPN2, ADIPOQ, COX6B1, GSTA3, LAP3, NDUFS1, SMARCC1, |
| Disrupted elimination of toxins/xenobiotics | 8 | 7 | GSTA3, LAP3, ALR1, AOX3, COX6B1, NUDT7, TALDO1 | |
| Protein misfolding | 9 | 1 | DDOST, GANAB, LAP3, RPN2, VCP |
| Predicted Dysregulated Functions of OLAN | Up | Down | Associated Proteins | |
|---|---|---|---|---|
| Predicted metabolic functions | Energy production (all ATP-generating pathways combined) | 7 | 27 | G6PD, GLB1, NPC2, RPL34, AK2, AMY2B, ATP2B4, COX6B1, ENO3, GNAI, PNP, TNNC2, VAPA |
| Nucleic acid synthesis | 7 | 15 | G6PD, MTHFD1L, AK2, ATIC, ENO3, MTAP, PNP | |
| Amino acid production | 7 | 10 | G6PD, GLB1, MTHFD1L, ATIC, ENO3, MTAP, PNP | |
| Glycolysis | 6 | 10 | ATP2B4, G6PD, GLB1, P2RX4, AK2, AMY2B, ENO3 | |
| Lipogenesis | 12 | 5 | AGPAT3, G6PD, GNAI, HMGA2, HNRNPD, NPC2, ATP2B4, ENO3, VAPA | |
| Lipolysis | 5 | 10 | AGPAT3, GNAI1, NPC2, AMY2B, COX6B1, CYCS, DECR2, ENO3, VAPA | |
| TCA/OxPhos/ETC | 4 | 9 | G6PD, GLB1, RPL34, COX6B1, CYCS, ENO3, PNP | |
| Glycogen production | 2 | 9 | ENO3, GNAI, ATP2B4, P2RX4 | |
| Predicted mitogenic effects | Cell survival | 21 | 14 | AGPAT3, ATP2B4, CYCS, FMOD, GNAI1, P2RX4, SGPL1, GLB1, GNAI1, INPP5D, LUM, PIP4P2, PNP, TOP2B |
| Cell proliferation | 8 | 15 | AGPAT3, FMOD, GLB1, GNAI1, SGPL1, ATP2B4, CTC1, ELAVL1, INPP5D, P2RX4, PIP4P2, SGPL1, TOP2B | |
| Cancer | 13 | 5 | CYCS, G6PD, GNAI, HMGA2, PLOD3, LUM, TOP2B, TRIM32 | |
| Migration/motility | 6 | 9 | AGPAT3, CFL1, GLB1, SGPL1, ATP2B4, GNAI1, INPP5D, PIP4P2 | |
| Protein production | 7 | 6 | FAU, HNRNPC, NUP98, RPL34, RPL8, RPS15, RPS4Y1, ATP2B4, GNAI, RNPS1, TOP2B | |
| Predicted immune effects | Inflammation/cytokines | 23 | 9 | ACE, AMY2B, ATIC, CD68, CFL1, CNTN1, CYCS, G6PD, GLB1, GNAI1, MBL1, SGPL1, ELAVL1, INPP5D, MTHFD1L |
| Immune activation | 10 | 13 | CD68, CFL1, DYNCH1H1, FMOD, G6PD, GLB1, GNAI, MBL1, AK2, CNTN1, CYCS, ELAVL1, INPP5D, PIP4P2, PNP | |
| Cell microbe response | 14 | 7 | ACE, CFL1, CYCS, GNAI1, NCL, NUP98, SEPT2 | |
| Predicted GI/nutrient handling | GI function/nutrient absorption | 5 | 17 | GLB1, GNAI1, NPC2, AMY2B, AQP1, ATP2B4, TNNC2, VAPA |
| Bile processing | 5 | 10 | GLB1, GNAI1, NPC2, AQP1, ATP2B4, P2RX4, TNNC2, VAPA | |
| Cholesterol processing | 5 | 5 | AMY2B, HNRNPD, NPC2, AQP1, VAPA | |
| Predicted tissue damaging effects | Oxidation/ROS generation | 17 | 3 | AMY2B, CNTN1, COX6B1, CYCS, GLB1, GNAi1, PLOD3, PNP, G6PD |
| Fibrosis | 9 | 1 | COX6B1, FMOD, GLB1, GNAI, PLOD3, NPC2 | |
| Predicted intracellular functions | Cytoskeletal dynamics/intracellular transport | 13 | 5 | CFL1, CMT4F, DYNCH1H1, GNAI1, RAB31, SRP19, VPS13C, PIP4P2, SEPT2, VAPA |
| Export of intracellular calcium ions | 8 | 2 | ATP2B4, P2RX4, GNAI1 | |
| Predicted Blood physiology | Blood pressure | 7 | 7 | ACE, DYNCH1H1, GNAI1, AQP1, ATP2B4 |
| Risperidone | |||||
| Upregulated | Name | # KEGG Pathways | Downregulated | Name | # KEGG Pathways |
| PRKCA | Protein kinase C alpha | 53 | ALDH7A1 | Aldehyde dehydrogenase 7, member a1 | 14 |
| GNAS | Guanine nucleotide-binding protein, alpha-stimulating | 40 | COX6B1 | Cytochrome C oxidase subunit 6B1 | 11 |
| PDGFRB | Platelet-derived growth factor receptor beta | 18 | AOX3 | Aldehyde oxidase 3 | 9 |
| ROCK2 | Rho-associated coiled-coil-containing protein kinase 2 | 15 | ENO3 | Enolase 3 | 8 |
| ITGA3 | Integrin alpha 3 | 11 | AGXT | Alanine-glyoxylate and serine-pyruvate aminotransferase | 7 |
| GSTA3 | Glutathione-S-transferase A3 | 10 | CS | Citrate synthase oxidoreductase core subunit S1 | 7 |
| NDUFA8/ NDUFB8 | Inner mitochondrial membrane complex 1 NADH:ubiquinone oxidoreductase subunit A8/B8 | 7/7 | NDUFS1 | NADH ubiquinone | 7 |
| PGAM2 | Phosphoglycerate mutase 2 | 7 | ABAT | 4-Aminobutyrate aminotransferase | 7 |
| BCAT2 | Branched-chain amino acid transaminase 2 | 7 | ALR1 | Aldo-keto reductase family 1 member B | 6 |
| AFDN | Afadin | 6 | |||
| INPP5D | Inositol polyphosphate 5-phosphatase D | 6 | |||
| ACAA2 | Acetyl CoA acyltransferase 2 | 6 | |||
| ADIPOQ | Adiponectin, C1Q, and collagen domain-containing | 6 | |||
| DGKZ | Diacylglycerol kinase-zeta | 6 | |||
| EPHA2 | EPH-ephrin receptor A2 | 5 | |||
| Olanzapine | |||||
| Upregulated | Name | # KEGG Pathways | Downregulated | Name | # KEGG Pathways |
| GLB1 | Beta-galactosidase | 12 | GNAI1 | G-protein subunit alpha inhibitory-1 | 37 |
| G6PD | Glucose 6 phosphate dehydrogenase | 10 | CYCS | Somatic cytochrome C | 26 |
| ATP2B4 | ATPase plasma membrane Ca+2 transporting 1 | 9 | COX6B1 | Cytochrome C oxidase subunit 6B1 | 11 |
| INPP5D | Inositol polyphosphate 5-phosphatase D | 6 | ENO3 | Enolase 3 | 8 |
| AGPAT3 | 1-Acylglycerol-3-phosphate O-acyltransferase-3 | 5 | ACE | Angiotensin I-converting enzyme | 7 |
| CFL1 | Cofilin 1 | 5 | PNP | Purine nucleoside phosphorylase | 6 |
| AK2 | Adenylate kinase 2 | 5 | |||
| ATIC | 5-Aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 5 | |||
| AMY2B | Amylase alpha 2B | 5 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rostama, B.; Beauchemin, M.; Bouchard, C.; Bernier, E.; Vary, C.P.H.; May, M.; Houseknecht, K.L. Understanding Mechanisms Underlying Non-Alcoholic Fatty Liver Disease (NAFLD) in Mental Illness: Risperidone and Olanzapine Alter the Hepatic Proteomic Signature in Mice. Int. J. Mol. Sci. 2020, 21, 9362. https://doi.org/10.3390/ijms21249362
Rostama B, Beauchemin M, Bouchard C, Bernier E, Vary CPH, May M, Houseknecht KL. Understanding Mechanisms Underlying Non-Alcoholic Fatty Liver Disease (NAFLD) in Mental Illness: Risperidone and Olanzapine Alter the Hepatic Proteomic Signature in Mice. International Journal of Molecular Sciences. 2020; 21(24):9362. https://doi.org/10.3390/ijms21249362
Chicago/Turabian StyleRostama, Bahman, Megan Beauchemin, Celeste Bouchard, Elizabeth Bernier, Calvin P. H. Vary, Meghan May, and Karen L. Houseknecht. 2020. "Understanding Mechanisms Underlying Non-Alcoholic Fatty Liver Disease (NAFLD) in Mental Illness: Risperidone and Olanzapine Alter the Hepatic Proteomic Signature in Mice" International Journal of Molecular Sciences 21, no. 24: 9362. https://doi.org/10.3390/ijms21249362
APA StyleRostama, B., Beauchemin, M., Bouchard, C., Bernier, E., Vary, C. P. H., May, M., & Houseknecht, K. L. (2020). Understanding Mechanisms Underlying Non-Alcoholic Fatty Liver Disease (NAFLD) in Mental Illness: Risperidone and Olanzapine Alter the Hepatic Proteomic Signature in Mice. International Journal of Molecular Sciences, 21(24), 9362. https://doi.org/10.3390/ijms21249362







