Abstract
Plants adjust their architecture to a constantly changing environment, requiring adaptation of differential growth. Despite their importance, molecular switches, which define growth transitions, are largely unknown. Apical hook development in dark grown Arabidopsis thaliana (A. thaliana) seedlings serves as a suitable model for differential growth transition in plants. Here, we show that the phytohormone auxin counteracts the light-induced growth transition during apical hook opening. We, subsequently, identified genes which are inversely regulated by light and auxin. We used in silico analysis of the regulatory elements in this set of genes and subsequently used natural variation in gene expression to uncover correlations between underlying transcription factors and the in silico predicted target genes. This approach uncovered that MADS box transcription factor AGAMOUS-LIKE 8 (AGL8)/FRUITFULL (FUL) modulates apical hook opening. Our data shows that transient FUL expression represses the expression of growth stimulating genes during early phases of apical hook development and therewith guards the transition to growth promotion for apical hook opening. Here, we propose a role for FUL in setting tissue identity, thereby regulating differential growth during apical hook development.
1. Introduction
Dicotyledonous plants form an apical hook when seeds are germinating in the soil. It protects the shoot apical meristem and the cotyledons when seedlings are growing through the soil seeking for light [1]. Apical hook development is a very suitable model to study differential growth regulation, due to its non-vital nature during in vitro plant cultivation [1,2,3,4]. During the apical hook formation phase, the PIN- and AUX1/LAX-dependent intercellular auxin transport machinery establishes an auxin signaling maximum at the inner side of the hook [5,6,7,8]. At this site, auxin asymmetrically represses growth, leading to the curvature of the hypocotyl and apical hook formation [5,7,9]. Afterwards, a light sensitive growth promotion machinery induces cell expansion at the inner side of the apical hook enabling its opening (Figure 1A) [2]. Accordingly, the apical hook is a unique model system to address conditional transitions of differential growth repression to promotion [2,10]. Here, we address the molecular pathways that define apical hook opening and aimed to identify transcription factors that steer this differential growth transition.
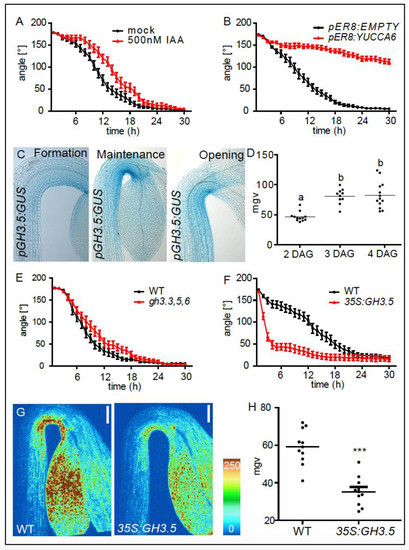
Figure 1.
Auxin represses light induced growth transition in apical hooks. (A) Kinetics of light triggered apical hook opening in dark grown wild type (WT) seedlings in the maintenance phase (3 DAG) in the presence or absence of 500 nM indole-3-acetic-acid (IAA). (B) Kinetics of light triggered apical hook opening in dark grown pER8:EMPTY and pER8:YUCCA6 seedlings in the presence of 10 µM estradiol. (C) Expression patterns of pGH3.5:GUS in the formation (2 DAG), maintenance (3 DAG), and the opening (4 DAG) phase. (D) Graph depicts mean grey values (mgv) of the pGH3.5:GUS intensity at the inner side of the hook (statistical significance was tested using a 1-way-ANOVA and a Bonferroni post-hoc test, p < 0.05, n ≥ 10 apical hooks per developmental stage. Letters indicate different significance classes). (E,F). Kinetics of light triggered apical hook opening in dark grown WT, gh3.3,5,6 and p35S:GH3.5 seedlings in the maintenance phase (3DAG). (G) DR5 promoter activity as shown by pDR5:GFP signal intensity in 3-day-old (maintenance phase), dark grown WT and p35S:GH3.5 seedlings as visualized using confocal microscopy. Color code (blue to red) depicts (low to high) pDR5:GFP signal intensity. (H) Graph depicts mean grey values of pDR5:GFP intensity at the inner side of the hook. Statistical significance was tested using an unpaired Student’s t-test, *** p < 0.0001, n ≥ 10 seedlings per line.
2. Results and Discussion
Besides its role in apical hook formation, asymmetric auxin signaling has also been proposed to maintain the apical hook [11]. However, a direct inhibiting role for auxin in apical hook opening has not been experimentally shown so far. Therefore, we tested whether auxin indeed inhibits the light induced growth transition between the apical hook maintenance and opening phase. Wild type seedlings were grown in the dark until the maintenance phase (3-days-old). We then transferred the seedlings to growth medium supplemented with 500 nM of the natural auxin, indole-3-acetic-acid (IAA) or solvent control (mock) solution (DMSO), exposed them to a light stimulus, and subsequently performed time lapse imaging (Figure 1A,B). Seedlings exposed to exogenous IAA displayed a slower light-induced apical hook opening compared to mock treated seedlings (Figure 1A). Next, we endogenously induced auxin biosynthesis during apical hook opening, using the transgenic estradiol inducible pER8:YUCCA6 line. Three-day-old dark grown seedlings (apical hooks in maintenance phase) were transferred on estradiol and the light-induced apical hook opening kinetics were monitored. While the empty vector control seedlings (pER8:EMPTY) displayed normal opening kinetics, YUCCA6 induction largely prohibited the light induced apical hook opening, suggesting that the increased auxin biosynthesis prohibits apical hook opening (Figure 1B). This set of data confirms that auxin signaling indeed represses light triggered apical hook opening.
Since auxin and light play a profound role in apical hook opening, it is to be expected that molecular factors that are inversely regulated by both auxin and light may play a crucial role in this growth transition. Previous work described an auxin and light dependent role for GH3.5 in hypocotyl growth [12]. We therefore hypothesized that GH3.5 may also steer the growth transition towards apical hook opening. GH3.5 encodes an IAA-amido synthase and is known to conjugate the natural auxin IAA to amino acids, thereby decreasing the active pool of free IAA in the cell [13,14]. In agreement, GUS staining of dark grown transgenic pGH3.5:GUS seedlings revealed GH3.5 expression during apical hook development (Figure 1C,D). Apart from the asymmetric GH3.5 expression, we observed increased expression towards the maintenance and opening phase, suggesting a role in growth transition (Figure 1C,D). The asymmetry and developmentally defined (stage dependent) expression of GH3.5 is remarkable, because auxin conjugation is rather known to contribute to cellular auxin homeostasis [15]. We tested whether GH3.5, besides its homeostatic role in safeguarding cellular auxin levels, could play an additional role in the transition of differential growth during apical hook opening. We performed time-lapse imaging of dark grown apical hooks in the maintenance phase that were exposed to a light stimulus (Figure 1E,F). GH3 genes are highly redundant [14] and only gh3.3,5,6 triple loss-of-function mutants displayed slower apical hook opening, when compared to wild type (Figure 1E). On the contrary, GH3.5 overexpressing seedlings showed faster apical hook opening compared to WT seedlings (Figure 1F). Since GH3.5 activity is known to adenylate IAA to amino acids and reduces free IAA levels in the cell [13], we assessed the impact of GH3.5 activity on nuclear auxin signaling during apical hook development by crossing a p35S:GH3.5 line with the auxin signaling output reporter pDR5:GFP. Analysis of apical hooks in the maintenance phase (3-day-old dark grown seedlings), revealed that GH3.5 overexpression substantially decreases pDR5:GFP signaling, confirming a negative impact of GH3.5 on nuclear auxin signaling in the apical hook (Figure 1G,H). These data suggest that GH3.5 stimulates apical hook opening, presumably by decreasing nuclear auxin signaling at the inner side of the hook structure.
In the absence of a light stimulus, apical hooks still open eventually at a slower pace due to a steady decrease in auxin accumulation at the inner side of the hook [2,7]. In accordance, we also observed a faster apical hook opening in dark grown GH3.5 overexpressing seedlings compared to WT seedlings in the absence of light (Figure S1).
This set of data confirms the negative role of auxin in light induced apical hook opening. Moreover, this approach shows that auxin and light dependency of molecular factors could hint at a potential role in growth transition during apical hook development. Accordingly, we aimed to identify novel molecular factors for growth transition in apical hooks, depicting genes that are oppositely regulated by auxin and light. Therefore, dark grown WT A. thaliana seedlings were transferred (three days after germination (3 DAG) under green light (mimicking dark growth conditions) to growth medium supplemented with DMSO (solvent control) or with 500 nM IAA for 1 h. We then dissected the shoots for subsequent RNA extraction and RNA sequencing (RNA-seq). Analysis of RNA-seq transcriptomics data revealed 241 genes that were significantly (p < 0.001) and at least two-fold up- or down-regulated upon auxin treatment (Table S1). We used the genevestigator software to compare our dataset with a previously reported RNA-seq dataset (AT-00706) in which shoot tissue was dissected from 4-day-old dark grown seedlings that were conditionally exposed to light for four hours (www.genevestigator.com). We identified in total 39 genes, displaying opposite regulation by auxin and light (Figure 2A; Table S2). Notably, GH3.5 is also significantly upregulated by IAA (log2FC = 0.94) and downregulated by light (log2FC = 1.76). However, GH3.5 has not been included in the candidate gene list, because of the commonly used threshold cut off (log2FC > 1).
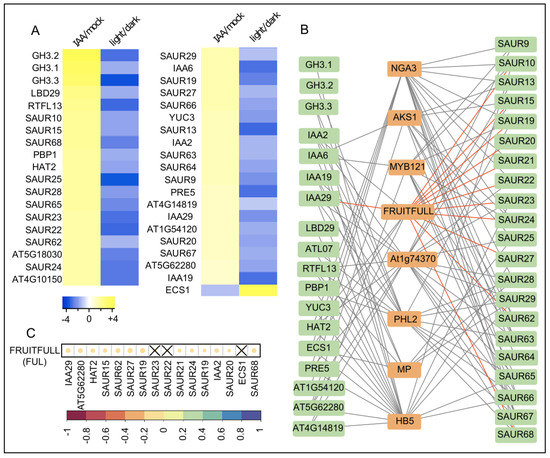
Figure 2.
Putative molecular components in differential growth control in apical hook hooks. (A) Heat map represents genes that are oppositely regulated by the auxin IAA and light in dark grown seedlings. Cut off values were 2-fold expression difference (−1 ≥ log2 ≥ 1) and p < 0.001. Color code blue to yellow depicts the log2 values of strongly decreased to strongly increased expression compared to the control condition. (B) Gene regulatory network depicts the transcription factors (orange boxes) which putatively regulate the auxin- and light-regulated candidate genes (green boxes) according to SeqEnrich [16]. Gray lines connect the transcription factors with their putative target genes. The connecting lines with FUL are highlighted in red. (C) Diagram represents the correlation between the expression of FRUITFULL and its target genes in 727 Arabidopsis thaliana natural variations [17] Color code (red to blue) depicts highly negative to highly positive Pearson correlation coefficients. The size of the circles also represents the correlation strength between the expression of FUL and its putative target genes. Non-significant (p ≥ 0.05) correlations are indicated with a cross.
We hypothesize that some of these differentially controlled genes may be relevant for growth control and hence could be controlled by transcription factors that define differential growth. In order to identify molecular factors that may define auxin and light dependent growth transitions, we screened in silico for common transcriptional regulators of these auxin- and light-dependent genes, using the previously established SeqEnrich script [16]. Thereby, we identified eight transcription factors, which presumably bind to regulatory elements in upstream regions of the identified genes (statistically significant level of p < 0.05) (Figure 2B; Table S3).
Subsequently, we assessed which transcription factors are likely to affect the predicted target genes, using natural variation of gene expression [17]. We assumed that, in case of a simple transcription factor-target gene scenario, the expression of a transcription factor correlates (whether positively or negatively) with the expression of its individual target genes in natural accessions of A. thaliana. We therefore calculated the Pearson correlation coefficient for the expression of the eight identified transcription factors and their individual target gene expression in 727 A thaliana accessions (Figure 2C; Figure S2). Three out of eight transcription factors, such as AKS1, FRUITFULL and HB5, correlated significantly (p < 0.05) with the expression of more than half of their predicted target genes, suggesting a potential role in differential growth control. In agreement, HB5 binds the promoters of the auxin responsive BODENLOS (BDL/IAA12) and EXPANSIN3 and thereby regulates their expression [18,19]. Moreover, HB5 and its target gene EXPANSIN3 are important for cell elongation in hypocotyls [19]. Accordingly, we conclude that our approach is suitable to identify molecular players affecting growth processes.
Interestingly, we also identified AGAMOUS LIKE8(AGL8)/FRUITFULL(FUL) as a transcription factor with a putative role in differential growth. AGL8/FUL expression significantly negatively correlated with a high proportion of its putative target genes in 727 Arabidopsis thaliana accessions (Figure 2C; Figure S2). AGL8/FUL is a member of the MADS box transcription factor family which are widely conserved throughout the domain of eukaryotes. Compared to animals, MADS box genes highly expanded during plant evolution. The subfamily of AGLs, which contains 42 members in Arabidopsis, has been shown to primarily define flower as well as fruit development and, hence, are considered as potential targets for crop improvements [20,21,22,23]. AGL8/FUL has also been reported to be important for the regulation of meristem activity during fruit development, but also for light dependent shoot branching [24,25,26]. However, a potential role of AGL8/FUL in differential growth processes, such as during apical hook development, remains to be assessed. This triggered our interest to further investigate the role of AGL8/FUL in light triggered apical hook opening.
Using the pFUL:GUS transcriptional reporter lines, we observed that AGL8 is expressed during apical hook formation with a decrease towards the opening phase (Figure 3A,B). Time-lapse analysis of light triggered apical hook opening revealed that knock-out mutants ful-2 and ful-7 loss-of-function mutants displayed an accelerated apical hook opening (Figure 3C,D), while AGL8/FUL overexpressing seedlings displayed a severe delay in apical hook opening when compared to WT seedlings (Figure 3E). This set of data suggests that AGL8/FUL represses growth promoting genes and thereby plays a regulatory role in defining growth transition during apical hook development.
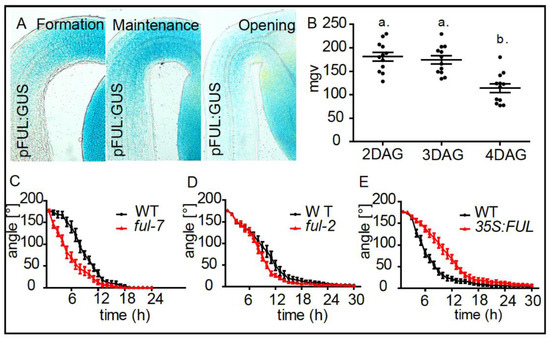
Figure 3.
FRUITFULL defines apical hook opening. (A) Expression patterns of pFUL:GUS in the formation (2 DAG), maintenance (3 DAG) and the opening (4 DAG) phase. (B) Graph depicts mean grey values (mgv) of the pFUL:GUS intensity at the inner side of the hook (statistical significance was tested using a 1-way-ANOVA and a Bonferroni post-hoc test, p < 0.05, n ≥ 10 apical hooks per developmental stage. Letters indicate different significance classes). (C–E) Kinetics of light triggered apical hook opening in dark grown WT, ful-7, ful-2, and p35S:FUL seedlings.
It is unknown whether all 16 predicted target genes are indeed direct targets of AGL8/FUL. However, based on ChIPseq, AGL8/FUL presumably binds to the promoters of GH3.5 and SAUR10 [25]. Moreover, AGL8/FUL was shown to directly bind and consequently represses SAUR10 [25], which was among our putative target genes. Similar to GH3.5, the transcriptional SAUR10 reporter displayed enhanced expression during maintenance and opening phases of apical hook development (Figure 4A,B). Accordingly, we used these GH3.5 and SAUR10 marker lines to further assess the potential role of AGL8/FUL during growth transitions in apical hooks.
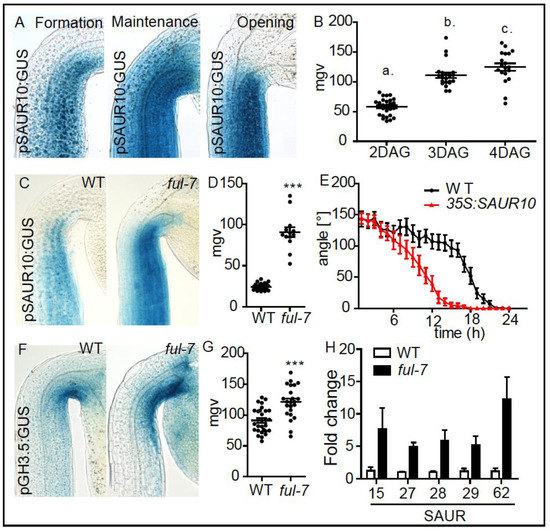
Figure 4.
FUL-dependent control of SAUR10 in apical hooks. (A) Expression patterns of pSAUR10:GUS in the formation (2 DAG), maintenance (3 DAG) and the opening (4 DAG) phase. (B) Graph depicts mean grey values of the pSAUR10:GUS intensity at the inner side of the hook (statistical significance was tested using a 1-way-ANOVA and a Bonferroni post-hoc test, p < 0.05, n ≥ 10 apical hooks per developmental stage. Letters indicate different significance classes). (C) pSAUR10 promoter activity in 3-day-old (maintenance phase), dark grown WT and ful-7 seedlings. (D) Graph depicts mean grey values of pSAUR10:GUS intensity in the apical hook area. Statistical significance was tested using an unpaired Student’s t-test, *** p < 0.0001, n ≥ 10 seedlings per line. (E) Kinetics of light-triggered apical hook opening in dark grown WT and p35S:SAUR10 seedlings in the early maintenance phase (2-day-old). (F) pGH3.5 promoter activity in 3-day-old (maintenance phase), dark grown WT and ful-7 seedlings. (G) Graph depicts mean grey values of pGH3.5:GUS intensity in the apical hook area. Statistical significance was tested using an unpaired Student’s t-test, *** p < 0.0001, n ≥ 10 seedlings per line. (H) Graph depicts Q-PCR measured relative expression levels of putative FUL target genes in dark grown shoots in the maintenance phase (3 DAG) of ful-7 compared to WT seedlings. The statistical significant difference between WT and ful-7 was demonstrated using a 2-way-ANOVA test including a Bonferroni post-hoc test. p < 0.0001.
In order to test whether AGL8/FUL affects SAUR10 and/or GH3.5 expression during apical hook development, we investigated pSAUR10:GUS and pGH3.5:GUS reporter lines crossed into a ful-7 knock-out mutant. During the maintenance phase, we detected an increased SAUR10 and GH3.5 expression in the apical hook of ful-7 mutants when compared to wild type control lines (Figure 4C,D,F,G). Using qPCR, we also detected higher expression of other SAUR genes in dissected apical hook tissues, which were among our list of 16 potential target genes, in the agl8 mutant background (Figure 4H). This data suggests that AGL8/FUL negatively affects the expression of its target genes, such as SAUR10, in apical hooks (Figure 4C,D). Ectopic expression of SAUR10 caused a faster light triggered apical hook opening compared to wild type seedlings (Figure 4E), which is reminiscent of the overexpression of GH3.5 (Figure 1F). Our results suggest that de-repression of FUL target genes, such as SAUR10, induce faster growth transition during apical hook development.
In this work, we experimentally confirmed the negative impact of exogenous as well as endogenous auxin on apical hook opening which counteracts the effect of light. We used these features to unveil differentially controlled genes. Using in silico predictions of overlapping regulatory elements, we identified novel transcription factors with a presumable role in defining differential growth transitions. We subsequently used natural variation in gene expression as a means to further assess if the expression of the identified transcription factors may positively or negatively correlate with the predicted target genes. These approaches led to the identification of AGL8/FUL as a negative regulator of apical hook opening by repressing growth promoting genes, such as SAUR10.
We propose that the decrease in FUL/AGL8 expression towards the opening phase allows the de-repression of growth promoting genes, such as GH3.5 and SAUR10, which will initiate growth transition and subsequently apical hook opening. Intriguingly, GH3.5 and SAUR10 get up- and down-regulated upon IAA and light treatment in dissected hypocotyls, respectively. On the other hand, auxin signaling decreases during light-induced apical hook opening, but GH3.5 and SAUR10 both increase during this condition. This observation pinpoints at tissue specific regulation, which warrants further investigation. In this regards, it remains to be seen how precisely FUL/AGL8 integrates developmental and environmental stimuli in order to accurately execute its function during apical hook development.
Interestingly, another MADS box family member, AGL9, also has been described to bind the GH3.5 promoter [27] and AGL15 interferes with auxin signaling by repressing auxin signaling genes, such as TIR1, ARF3, ARF6, and ARF8 [28,29]. Moreover, light perception impacts on transcription and alternative splicing of various AGLs [30]. Accordingly, AGLs could function possibly in part redundantly as a signaling nexus for context specific growth responses by integrating auxin and light signals. AGLs have been studied extensively before, with a strong focus on their role in flower and fruit development. Our findings propose a regulatory role for AGAMOUS LIKE genes in controlling context specific growth transitions.
3. Material and Methods
3.1. Plant Material
A. thaliana ecotype Columbia-0 was used as the wild type. p35S:GH3.5 (WES1D) [12], gh3.3,5,6 [31] pGH3.5:GUS (pWES1:GUS) [12] pER8:EMPTY, pER8:YUC6 [32], ful-2 [33], ful-7 [25], p35S:FUL [25], p35S:SAUR10 [25], pSAUR10:GUS [25], pFUL:GUS [25], and pSAUR10:GUS/ful-7 [25] have been described previously. We crossed the pDR5:GFP reporter line with the 35S:GH3.5 line in order to obtain the pDR5:GFP/p35S:GH3.5 line. We crossed the pGH3.5:GUS and ful-7 line in order to obtain the pGH3.5:GUS/ful-7 line.
3.2. Growth Conditions
Seeds were sterilized with 70% ethanol followed by drying in a sterile hood. Plants were grown under long-day (16 h light/8 h dark) conditions at 20 °C. Seedlings were grown on ½ Murashige and Skoog (MS) in vitro plates supplemented with 0.8% agar. For the depicted treatments, the MS medium was supplemented with 500 nM indole-3-acetic acid (IAA) or 10 µM estradiol. As a mock treatment, an equivalent volume of DMSO was added to the medium. The seeds were stratified at 4 °C for two days in the dark and then were exposed to 8 h of light (22 °C) and subsequently grown vertically in the dark (20 °C) until the corresponding time points. For light-induced apical hook-opening experiments, three days old dark-grown seedlings (apical hooks in maintenance phase) were exposed to constant low light (4 µmol m–2 s–1) for the duration of time indicated in figure legends. Notably, dark grown 35:SAUR10 seedlings display a premature apical hook opening in the dark. We therefore assessed the apical hook opening in 35:SAUR10 as well as their Col-0-WT controls in the early maintenance phase (2-days-old), when apical hooks were still maintained.
3.3. Expression Analysis via GUS Staining
Dark-grown seedlings were collected during the formation, maintenance and/or opening phases of apical hook development as indicated in the text. GUS staining was performed and quantified as described previously [34]. Before staining, seedlings were fixed in 80% cold acetone for 20 min on ice, and subsequently washed three times using MonoQ water before GUS staining. The seedlings were stained for 3 h (pGH3.5:GUS and pSAUR10:GUS) and 2 h (pFUL:GUS). The stained seedlings were mounted in chloralhydrate, incubated overnight at 4 °C and subsequently imaged using a ZEISS upright light microscope equipped with a camera. Images have been analyzed as described previously [34].
3.4. Time-Lapse Imaging of Apical Hook Kinetics
Dark-grown seedlings in the maintenance phase were transferred on in vitro growth plates supplemented with the indicated chemicals and subsequently imaged in a customized light-sealed imaging box equipped with a white light source and a Canon mirror reflex camera (EOS035 Canon Rebel T3i) operated by the Canon EOS utility software. Images were taken and analyzed every hour for a duration of 30 h. Angles between the cotyledons and the hypocotyl axis were measured using the Fiji software (http://rsb.info.nih.gov/ij/). The complementary angle is given in the graphs. At least three independent experiments were performed and the sample size is indicated in the figure legends. To display potential conditional variability among the experimental replicates, all raw measurements have been included in Table S4.
3.5. Confocal Microscopy
Confocal microscopy was done with a Leica SP5 or Leica SP2 upright confocal scanning microscope (Leica). Fluorescence signals for GFP (excitation 488 nm, emission peak 509 nm) were detected with a 40× (dry) objective. Fluorescence signal intensity was analyzed using the Fiji software (http://rsb.info.nih.gov/ij/) software and data were statistically evaluated with Microsoft Excel 2010 and Graphpad Prism 5.
3.6. RNA Sequencing
Seedlings were grown on sterile nylon meshes for 3 days in the dark until the apical hook maintenance phase. The seedlings on the meshes were then transferred under green light to in vitro growth plates supplemented with 500 nM IAA. After 1 h incubation in the dark, 15–20 shoots per sample were dissected and transferred to a 2 mL Eppendorf tube with 2 glass beads for flash freezing in liquid nitrogen. RNA has been isolated using the innuPREP Plant RNA kit (Analytic Jena, Jena, Germany). Libraries were generated with the NEBNext Ultra II RNA Library Prep Kit for Illumina with poly (A) enrichment, after which the RNA was degraded using Ribozero (New England Biolabs, Ipswich, MA, USA). The sequencing was performed on an Illumina HiSeq2500 with 250 bp paired ended fragments.
A total of nine single-end 50 bp long read data-sets, three biological replicates per sample, were obtained from a single lane of Illumina sequencing. Reads were mapped to the Arabidopsis thaliana reference genome TAIR10 (The Arabidopsis Genome Initiative 2000) using TopHat v2.0.13 [35] allowing for a maximum of two mismatches. Minimum and maximum intron lengths and minimum anchor length were fixed at 60, 6000, and 12 nt, respectively, and the --b2-very-sensitive and --microexon-search options were enabled. Pearson correlation coefficients between the replicates were calculated using the transcript expression values obtained by StringTie v1.0.4 [36]. Transcript quantification was limited to the TAIR10 annotated transcripts, excluding the chloroplast and mitochondrial chromosomes. Good correlation was obtained for the replicates of all three samples (R2 > 0.95 or higher).
Differentially expressed genes (DEG) were identified using Cuffdiff 2 [37], with default parameters, for the following comparison: WT + mock vs. WT + IAA (Table 1).

Table 1.
Differentially expressed genes (DEG) between mock treated and IAA treated shoots.
The raw sequencing data has been deposited at the Sequence Read Archive (SRA) of NCBI (SAMN14353622 and SAMN14353623).
3.7. qRT-PCR
We dissected shoots from dark grown seedlings in the maintenance phase (3DAG) and transferred them to a 2 mL Eppendorf tube with 2 glass beads for flash freezing in liquid nitrogen. For each sample, 15–20 shoots were pooled. For each line, 3 biological replicates were grown, extracted and analyzed. The tissue was ground using a milling machine (Retsch) after which the RNA was extracted using the InnuPREP Plant RNA Kit (Analytic Jena, Jena, Germany). We synthesized cDNA using the iSCRIPT cDNA Synthesis Kit from Bio-Rad and according to the manufacturer’s recommendation. Q-PCR was carried out in a C1000 Touch Thermal Cycler equipped with the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA), using a Takyon qPCR Kit for SYBER assay (Eurogentec, Liège, Belgium) and according to the manufacturer’s recommendation. We normalized the expression of the target genes with the expression of the Eif4a and UBL5 household genes.
3.8. In Silico Analysis
For the performance of the in silico analysis mentioned in the manuscript, we made use of the Genevestigator (Experiment AT-00706 [38]) (https://genevestigator.com), Cytoscape (https://cytoscape.org) and the Seqenrichtools as described before [16,39,40].
3.9. Data Analysis
We used Excel to organize data and MVapp (https://mvapp.kaust.edu.sa) [41] and the GraphPad Prism 5 software for statistical analysis and graphing. For statistical analysis of the raw data, we used Student’s t-tests or one/two-way ANOVAs and Bonferroni post-hoc tests as indicated in the figure legends. The most representative images are shown throughout the article. The experiments have been performed in triplicates or more.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/17/6438/s1. Figure S1: Kinetics of apical hook development in Col-0-WT and p35S:GH3.5 dark grown seedlings in the absence of a light stimulus. Figure S2: Correlation between the expression of the transcription factors and their putative target genes in 727 Arabidopsis thaliana natural variations. Table S1: Differential gene expression of auxin and mock-treated dark grown hypocotyls. Table S2: Genes displaying inverse regulation by light and auxin as well as the enrichment of regulatory elements in candidate gene list. Table S3: Expression of the transcription factor candidates and their putative target genes in 727 A. thaliana natural variations. Table S4: Raw data measurements of the individual apical hook kinetics replicates.
Author Contributions
Conceptualization, E.B. and J.K.-V.; Investigation, M.F., A.G., J.D., C.B. and E.B.; Formal analysis, M.F., A.G., P.V., J.D., C.B. and E.B.; Resources, M.I.F. and A.G.; Supervision, E.B., M.K. and J.K.-V.; Writing—original draft, E.B.; Writing—review and editing, M.F., P.V., J.K.-V., M.K. and E.B.; Funding acquisition, J.K.-V. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Vienna Research Group (VRG) program of the Vienna Science and Technology Fund (WWTF) and the European Research Council (ERC) (Starting Grant 639478-AuxinER) (to J.K.-V.).
Acknowledgments
We are grateful to Marian Bemer, Gerco Angenent, and Paul Staswick for sharing published material, the BOKU-VIBT Imaging Centre for access and expertise, and the VBCBF for RNA sequencing.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| AGL | AGAMOUS-LIKE |
| FUL | FRUITFULL |
| IAA | indole-3-acetic-acid |
| DAG | days after germination |
| mgv | mean gray value |
| FC | fold change |
| DEG | differentially expressed genes |
| 3 DAG | 3 days after germination |
References
- Abbas, M.; Alabadi, D.; Blázquez, M.A. Differential growth at the apical hook: All roads lead to auxin. Front. Plant Sci. 2013, 4, 441. [Google Scholar] [CrossRef] [PubMed]
- Béziat, C.; Barbez, E.; Feraru, M.I.; Lucyshyn, D.; Kleine-Vehn, J. Light triggers PILS-dependent reduction in nuclear auxin signaling for growth transition. Nat. Plants 2017, 3, 17105. [Google Scholar] [CrossRef] [PubMed]
- Mazzella, M.A.; Casal, J.J.; Muschietti, J.; Fox, A.R. Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 2014, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Raz, V.; Ecker, J.R. Regulation of differential growth in the apical hook of Arabidopsis. Development 1999, 126, 3661–3668. [Google Scholar] [PubMed]
- Boutté, Y.; Crosnier, M.-T.; Carraro, N.; Traas, J.; Satiat-Jeunemaitre, B. The plasma membrane recycling pathway and cell polarity in plants: Studies on PIN proteins. J. Cell Sci. 2006, 119, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Vandenbussche, F.; Petrasek, J.; Žádníková, P.; Hoyerová, K.; Pešek, B.; Raz, V.; Swarup, R.; Bennett, M.; Zažímalová, E.; Benkova, E.; et al. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 2010, 137, 597–606. [Google Scholar] [CrossRef]
- Zadnikova, P.; Petrasek, J.; Marhavý, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerová, K.; Morita, M.T.; Tasaka, M.; Hejatko, J.; et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef]
- Zhu, Q.; Gallemi, M.; Pospisil, J.; Zadnikova, P.; Strnad, M.; Benkova, E. Root gravity response module guides differential growth determining both root bending and apical hook formation in Arabidopsis. Development 2019, 146, dev175919. [Google Scholar] [CrossRef]
- Jonsson, K.; Boutté, Y.; Singh, R.K.; Gendre, D.; Bhalerao, R.P. Ethylene Regulates Differential Growth via BIG ARF-GEF-Dependent Post-Golgi Secretory Trafficking in Arabidopsis. Plant Cell 2017, 29, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Béziat, C.; Kleine-Vehn, J. The Road to Auxin-Dependent Growth Repression and Promotion in Apical Hooks. Curr. Biol. 2018, 28, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Chen, R.; Li, P.; Yu, Y.; Zheng, R.; Ge, D.; Zheng, W.; Wang, X.; Gu, Y.; Gelová, Z.; et al. TMK1-mediated auxin signaling regulates differential growth of the apical hook. Nature 2019, 568, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Seo, P.J.; Lee, A.-K.; Jung, J.-H.; Kim, Y.-S.; Park, C.-M. An Arabidopsis GH3 Gene, Encoding an Auxin-Conjugating Enzyme, Mediates Phytochrome B-Regulated Light Signals in Hypocotyl Growth. Plant Cell Physiol. 2007, 48, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Park, J.-Y.; Kim, Y.-S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.-Y.; Kim, J.; Lee, Y.-H.; Park, C.-M. GH3-mediated Auxin Homeostasis Links Growth Regulation with Stress Adaptation Response in Arabidopsis. J. Boil. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis Enzyme Family That Conjugates Amino Acids to Indole-3-Acetic Acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef]
- Rosquete, M.R.; Barbez, E.; Kleine-Vehn, J. Cellular Auxin Homeostasis: Gatekeeping Is Housekeeping. Mol. Plant 2012, 5, 772–786. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.G.; Walker, P.L.; Pulgar-Vidal, N.C.; Belmonte, M.F. SeqEnrich: A tool to predict transcription factor networks from co-expressed Arabidopsis and Brassica napus gene sets. PLoS ONE 2017, 12, e0178256. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Huang, S.-S.C.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Castanon, R.; Nery, J.R.; Barragan, C.; He, Y.; et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef]
- De Smet, I.; Lau, S.; Ehrismann, J.S.; Axiotis, I.; Kolb, M.; Kientz, M.; Weijers, D.; Jürgens, G. Transcriptional repression of BODENLOS by HD-ZIP transcription factor HB5 in Arabidopsis thaliana. J. Exp. Bot. 2013, 64, 3009–3019. [Google Scholar] [CrossRef]
- Stamm, P.; Topham, A.T.; Mukhtar, N.K.; Jackson, M.D.; Tomé, D.F.; Beynon, J.; Bassel, G.W. The Transcription Factor ATHB5 Affects GA-Mediated Plasticity in Hypocotyl Cell Growth during Seed Germination. Plant Physiol. 2016, 173, 907–917. [Google Scholar] [CrossRef]
- Garceau, D.; Batson, M.K.; Pan, I.L. Variations on a theme in fruit development: The PLE lineage of MADS-box genes in tomato (TAGL1) and other species. Planta 2017, 246, 313–321. [Google Scholar] [CrossRef]
- Theissen, G.; Saedler, H. Plant biology. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Li, H.; Wen, J.; Mysore, K.S.; Wang, X.-B.; Pei, Y.; Niu, L.; Lin, H. Functional Specialization of Duplicated AGAMOUS Homologs in Regulating Floral Organ Development of Medicago truncatula. Front. Plant Sci. 2018, 9, 854. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, D.D.; Batista, R.A.; Roszak, P.; Köhler, C. Auxin production couples endosperm development to fertilization. Nat. Plants 2015, 1, 15184. [Google Scholar] [CrossRef] [PubMed]
- Balanza, V.; Martínez-Fernández, I.; Sato, S.; Yanofsky, M.F.; Kaufmann, K.; Angenent, G.C.; Bemer, M.; Ferrandiz, C. Genetic control of meristem arrest and life span in Arabidopsis by a FRUITFULL-APETALA2 pathway. Nat. Commun. 2018, 9, 565. [Google Scholar] [CrossRef]
- Bemer, M.; Van Mourik, H.; Muiño, J.M.; Ferrandiz, C.; Kaufmann, K.; Angenent, G.C. FRUITFULL controls SAUR10 expression and regulates Arabidopsis growth and architecture. J. Exp. Bot. 2017, 68, 3391–3403. [Google Scholar] [CrossRef]
- Gu, Q.; Ferrándiz, C.; Yanofsky, M.F.; Martienssen, R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 1998, 125, 1509–1517. [Google Scholar]
- Kaufmann, K.; Muiño, J.M.; Jáuregui, R.; Airoldi, C.A.; Smaczniak, C.; Krajewski, P.; Angenent, G.C. Target Genes of the MADS Transcription Factor SEPALLATA3: Integration of Developmental and Hormonal Pathways in the Arabidopsis Flower. PLoS Boil. 2009, 7, e1000090. [Google Scholar] [CrossRef]
- Zheng, Q.; Zheng, Y.; Ji, H.; Burnie, W.; Perry, S.E. Gene Regulation by the AGL15 Transcription Factor Reveals Hormone Interactions in Somatic Embryogenesis. Plant Physiol. 2016, 172, 2374–2387. [Google Scholar] [CrossRef]
- Zheng, Y.; Ren, N.; Wang, H.; Stromberg, A.J.; Perry, S.E. Global Identification of Targets of the Arabidopsis MADS Domain Protein AGAMOUS-Like15. Plant Cell 2009, 21, 2563–2577. [Google Scholar] [CrossRef]
- Shikata, H.; Hanada, K.; Ushijima, T.; Nakashima, M.; Suzuki, Y.; Matsushita, T. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 18781–18786. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mongélard, G.; Floková, K.; Pacurar, D.I.; Novak, O.; Staswick, P.; Kowalczyk, M.; Păcurar, M.; Demailly, H.; Geiss, G.; et al. Auxin Controls Arabidopsis Adventitious Root Initiation by Regulating Jasmonic Acid Homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [PubMed]
- Béziat, C.; Kleine-Vehn, J.; Feraru, E. Histochemical staining of β-glucuronidase and its spatial quantification. Methods Mol. Biol 2017, 1497, 73–80. [Google Scholar]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Boil. 2013, 14, R36. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2012, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-J.; Wu, S.-H.; Wu, J.-F.; Lin, W.-D.; Wu, Y.-C.; Tsai, T.-Y.; Tsai, H.-L.; Wu, S.-H. Translational Landscape of Photomorphogenic Arabidopsis. Plant Cell 2013, 25, 3699–3710. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Saade, S.; Agarwal, G.; Gao, G.; Pailles, Y.; Morton, M.; Awlia, M.; Tester, M. MV app-multivariate analysis application for streamlined data analysis and curation. Plant Physiol. 2019, 180, 1261–1276. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).