Characterization of the UDP-glycosyltransferase UGT72 Family in Poplar and Identification of Genes Involved in the Glycosylation of Monolignols
Abstract
1. Introduction
2. Results
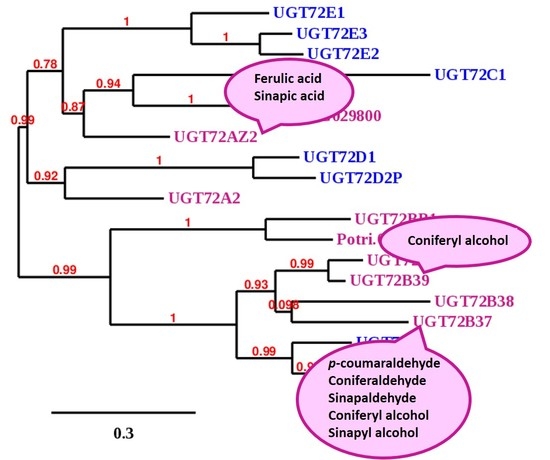
2.1. Cloning of the Full Length cDNA of Poplar UGT72 Genes
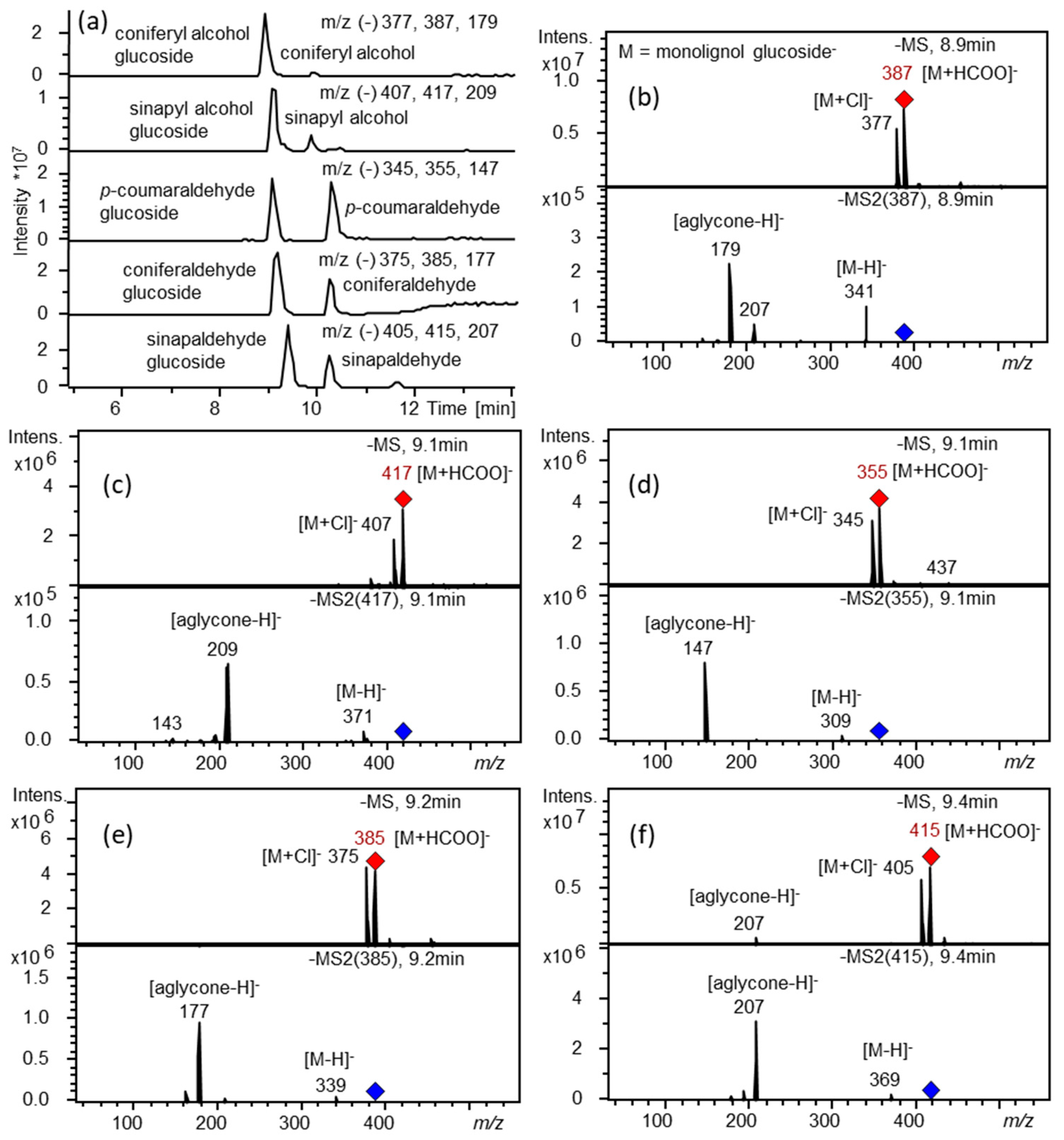
2.2. Recombinant UGT72B37 and UGT72B39 Proteins Glycosylate Monolignols
2.3. Accumulation of Monolignol Glycosides in Transgenic Poplar Overexpressing UGT72s
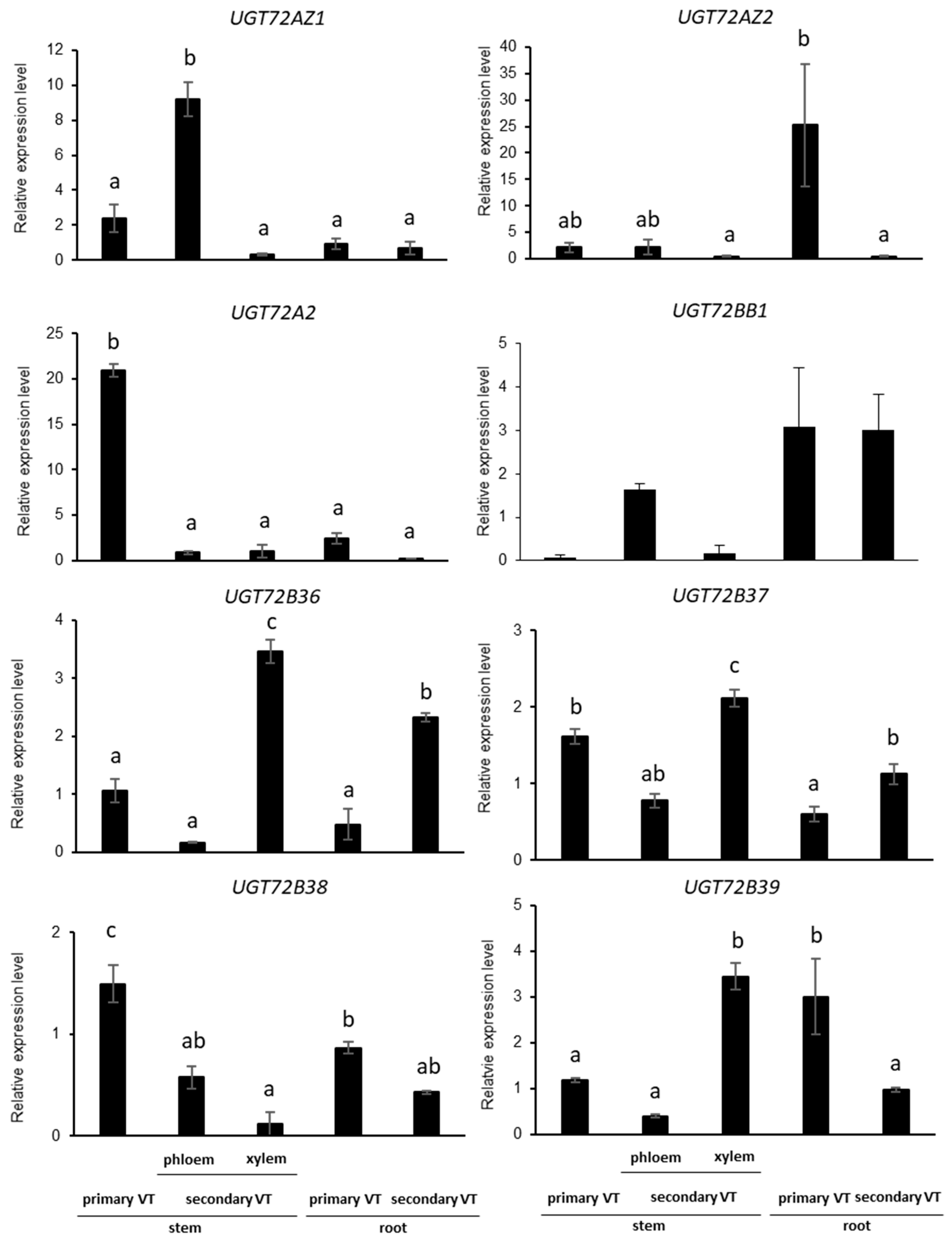
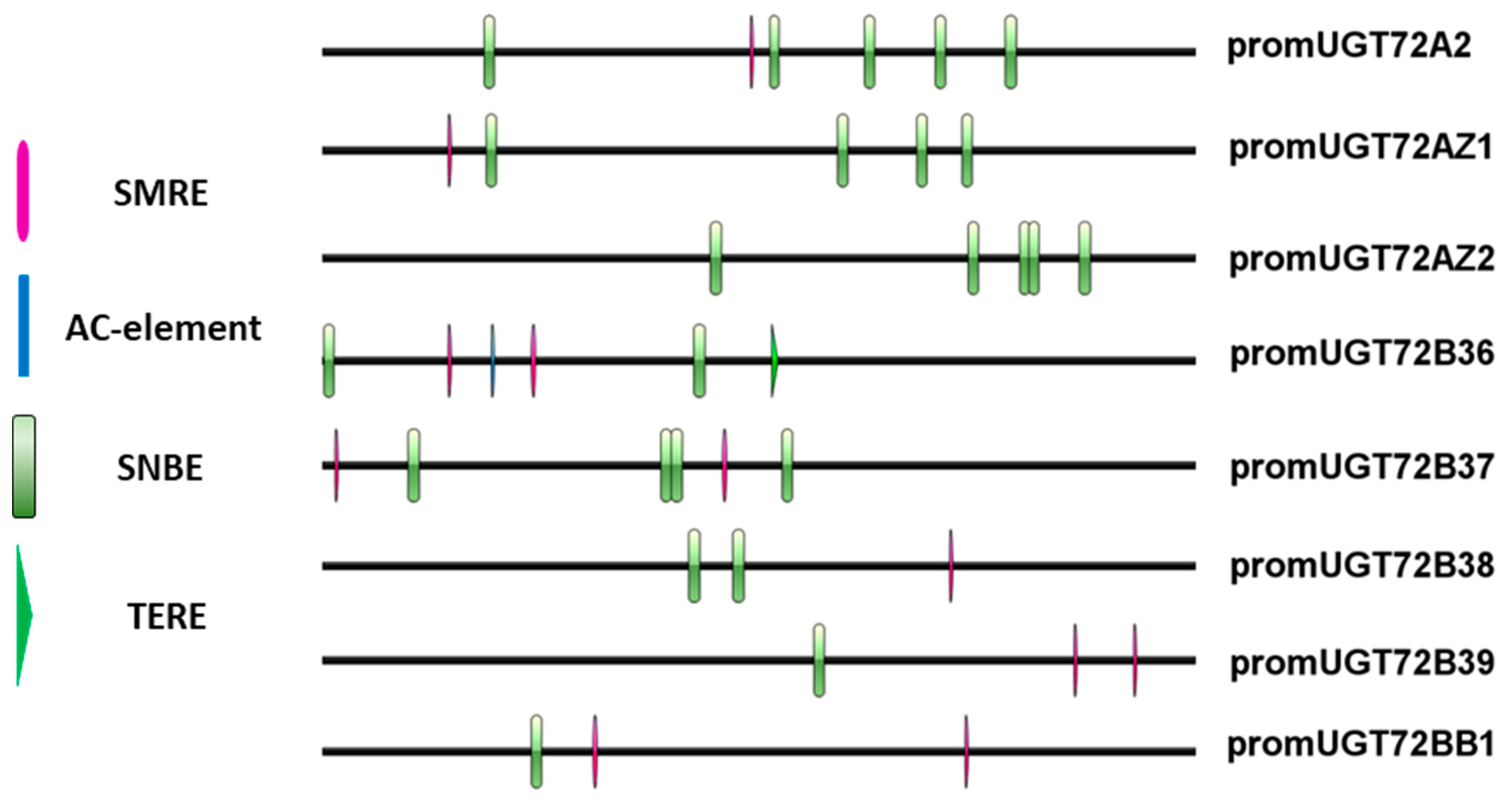
2.4. UGT72s Expression Profiles in Poplar
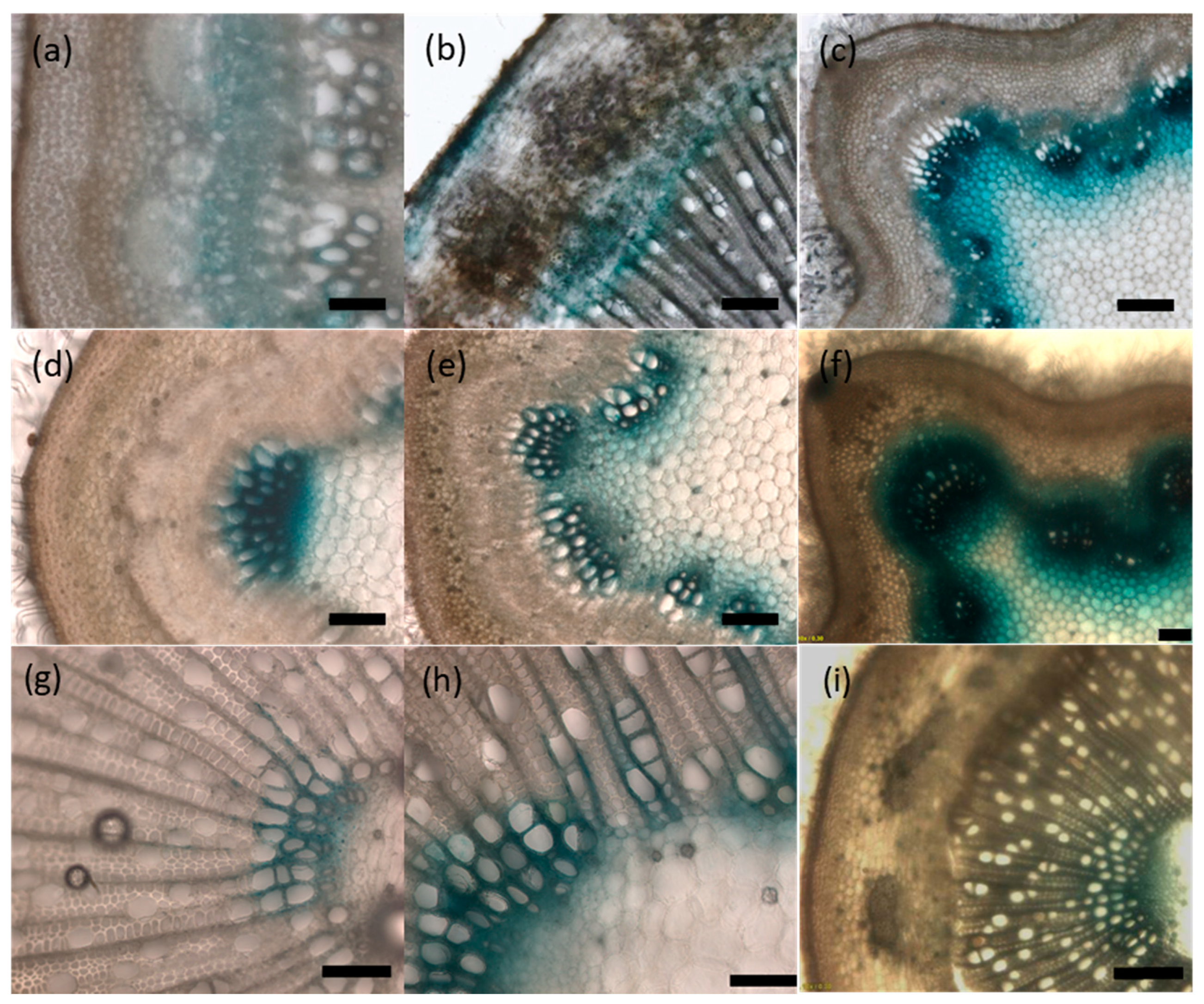
2.5. UGT72s Are Expressed within Vascular Tissues
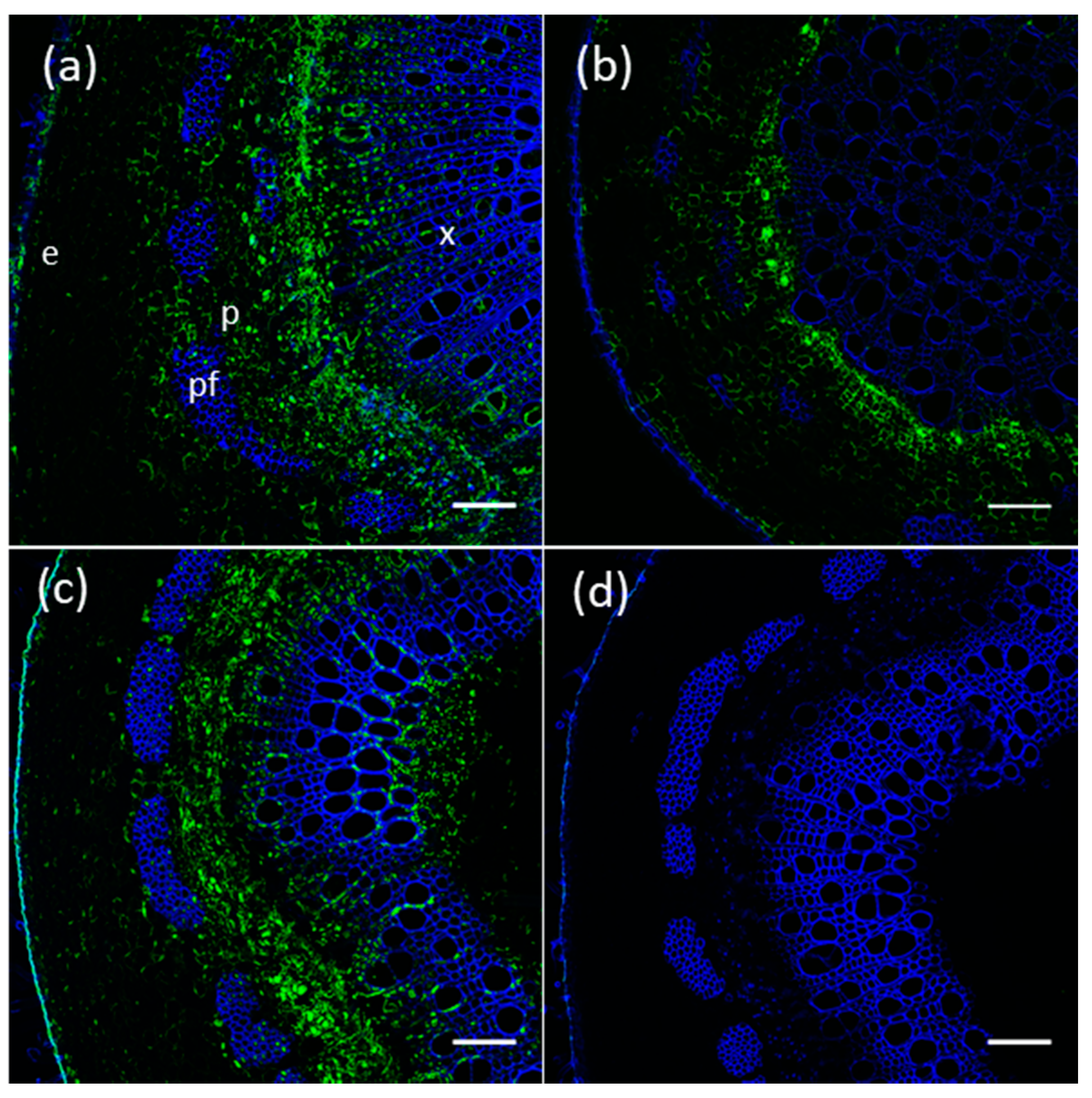
2.6. Proteins of Members of The Poplar UGT72 Family Preferentially Accumulate in Vascular Tissues
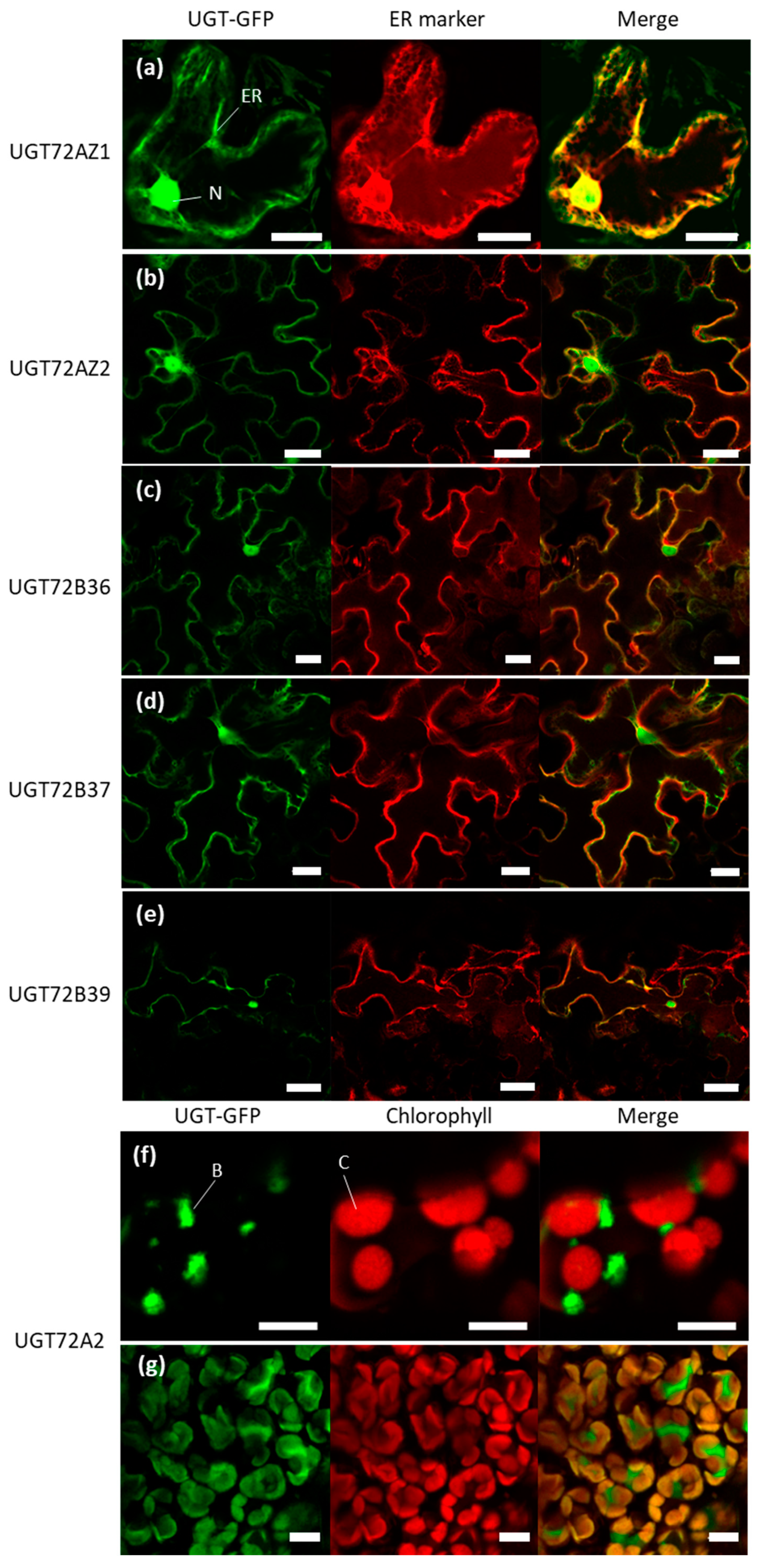
2.7. Subcellular Localization of the Poplar UGT72s
3. Discussion
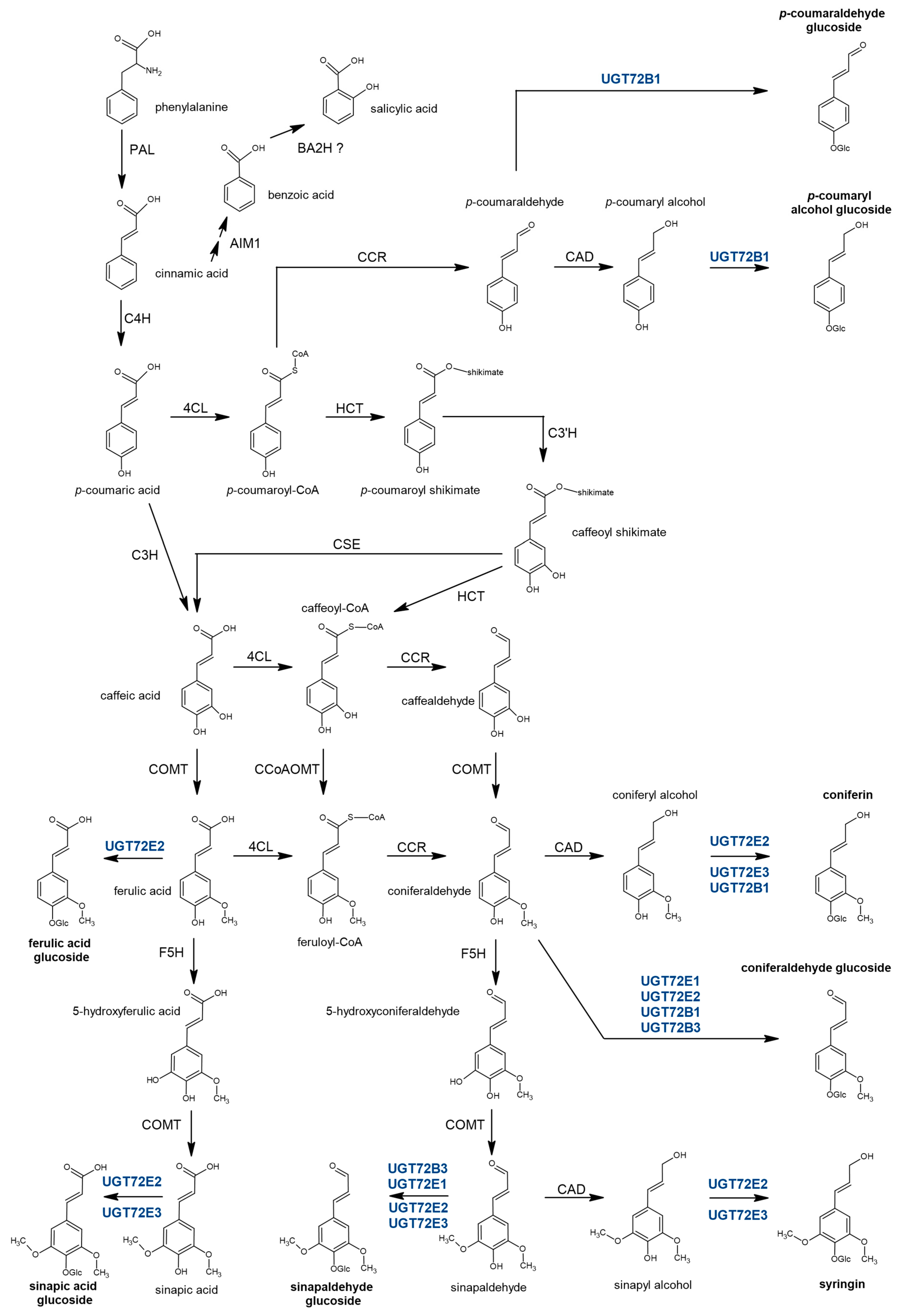
3.1. Some Members of the Poplar UGT72 Family Are Involved in the Glycosylation of Monolignols
3.2. UGT72s Are Expressed in the Vascular System
3.3. No Direct Impact of Poplar UGT72s Overexpression on Lignification Was Demonstrated
3.4. Differences in Subcellular Localization of Poplar UGT72s May Indicate Divergent Functions among the Different Members of the Same Family
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Cloning of UGT72s Coding and Promoter Sequences from P. Tremula × P. Alba
4.3. Phylogenetic Analysis
4.4. Expression Profile Analysis by RT-qPCR and Histochemical GUS Staining
4.5. Antibody Production, Immunoblotting and Immunolocalization
4.6. Subcellular Localization of the poplar UGT72s Fused with GFP
4.7. Enzymatic Assay on Recombinant Proteins Produced in E. Coli
4.8. Generation of Transgenic Poplars Upregulating UGT72s, Expressing Promoter-GUS or UGT72A2-GFP Constructs
4.9. Lignin Content
4.10. Metabolite Extraction, HPLC-UV-MS and LC-MS Analyses
4.11. Accession Uumbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, Functions, and Mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.; Lim, E.-K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, J.; Huss, B.; Creach, A.; Hawkins, S.; Neutelings, G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 2016, 7, 735. [Google Scholar] [CrossRef]
- Kumar, V.; Hainaut, M.; Delhomme, N.; Mannapperuma, C.; Immerzeel, P.; Street, N.R.; Henrissat, B.; Mellerowicz, E.J. Poplar carbohydrate-active enzymes: Whole-genome annotation and functional analyses based on RNA expression data. Plant J. 2019, 99, 589–609. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef]
- Hughes, J.; Hughes, M.A. Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq. 1994, 5, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Osmani, S.A.; Bak, S.; Møller, B.L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 2009, 70, 325–347. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Sangwan, R.S.; Sangwan, N.S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 714–739. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Tsuyama, T.; Takabe, K. Distribution of lignin and lignin precursors in differentiating xylem of Japanese cypress and poplar. J. Wood Sci. 2014, 60, 353–361. [Google Scholar] [CrossRef]
- Freudenberg, K.; Harkin, J.M. The glucosides of cambial sap of spruce. Phytochemistry 1963, 2, 189–193. [Google Scholar] [CrossRef]
- Rolando, C.; Daubresse, N.; Pollet, B.; Jouanin, L.; Lapierre, C. Lignification in poplar plantlets fed with deuterium-labelled lignin precursors. C. R. Biol. 2004, 327, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Terazawa, M.; Okuyama, H.; Miyake, M. Isolation of coniferin and syringin from the cambial tissue and inner-bark of some angiospermous woods. J. Jpn. Wood Res. Soc. 1984, 30, 409–412. [Google Scholar]
- Fukushima, K.; Taguchi, S.; Matsui, N.; Yasuda, S. Heterogeneous distribution of monolignol glucosides in the stems of Magnolia kobus. Mokuzai Gakkaishi 1996, 42, 1029–1031. [Google Scholar]
- Aoki, D.; Okumura, W.; Akita, T.; Matsushita, Y.; Yoshida, M.; Sano, Y.; Fukushima, K. Microscopic distribution of syringin in freeze-fixed Syringa vulgaris stems. Plant Direct 2019, 3, e00155. [Google Scholar] [CrossRef]
- Aoki, D.; Nomura, K.; Hashiura, M.; Imamura, Y.; Miyata, S.; Terashima, N.; Matsushita, Y.; Nishimura, H.; Watanabe, T.; Katahira, M.; et al. Evaluation of ring-5 structures of guaiacyl lignin in Ginkgo biloba L. using solid- and liquid-state 13C NMR difference spectroscopy. Holzforschung 2019, 73, 1083–1092. [Google Scholar] [CrossRef]
- Aoki, D.; Matsushita, Y.; Fukushima, K. Cryo-TOF-SIMS visualization of water-soluble compounds in plants. In ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2018; Volume 1286, pp. 137–150. ISBN 9780841232969. [Google Scholar]
- Aoki, D.; Hanaya, Y.; Akita, T.; Matsushita, Y.; Yoshida, M.; Kuroda, K.; Yagami, S.; Takama, R.; Fukushima, K. Distribution of coniferin in freeze-fixed stem of Ginkgo biloba L. by cryo-TOF-SIMS/SEM. Sci. Rep. 2016, 6, 31525. [Google Scholar] [CrossRef] [PubMed]
- Terashima, N. Non-destructive approaches to identify the ultrastructure of lignified ginkgo cell walls. Int. J. Dev. Biol. 2007, 1, 170–177. [Google Scholar]
- Tsuji, Y.; Fukushima, K. Behavior of monolignol glucosides in angiosperms. J. Agric. Food Chem. 2004, 52, 7651–7659. [Google Scholar] [CrossRef] [PubMed]
- Väisänen, E.E.; Smeds, A.I.; Fagerstedt, K.V.; Teeri, T.H.; Willför, S.M.; Kärkönen, A. Coniferyl alcohol hinders the growth of tobacco BY-2 cells and Nicotiana benthamiana seedlings. Planta 2015, 242, 747–760. [Google Scholar] [CrossRef]
- Liu, C.J. Deciphering the enigma of lignification: Precursor transport, oxidation, and the topochemistry of lignin assembly. Mol. Plant 2012, 5, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chantreau, M.; Sibout, R.; Hawkins, S. Plant cell wall lignification and monolignol metabolism. Front. Plant Sci. 2013, 4, 220. [Google Scholar] [CrossRef] [PubMed]
- Dima, O.; Morreel, K.; Vanholme, B.; Kim, H.; Ralph, J.; Boerjan, W. Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 2015, 27, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.C.; Liu, C.J. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 22728–22733. [Google Scholar] [CrossRef]
- Alejandro, S.; Lee, Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Lee, Y.; Geldner, N.; Fernie, A.R.; et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2012, 22, 1207–1212. [Google Scholar] [CrossRef]
- Vermaas, J.V.; Dixon, R.A.; Chen, F.; Mansfield, S.D.; Boerjan, W.; Ralph, J.; Crowley, M.F.; Beckham, G.T. Passive membrane transport of lignin-related compounds. Proc. Natl. Acad. Sci. USA 2019, 116, 23117–23123. [Google Scholar] [CrossRef]
- Lim, E.K.; Li, Y.; Parr, A.; Jackson, R.; Ashford, D.A.; Bowles, D.J. Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J. Biol. Chem. 2001, 276, 4344–4349. [Google Scholar] [CrossRef]
- Lim, E.-K.; Jackson, R.G.; Bowles, D.J. Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett. 2005, 579, 2802–2806. [Google Scholar] [CrossRef]
- Lin, J.S.; Huang, X.X.; Li, Q.; Cao, Y.; Bao, Y.; Meng, X.F.; Li, Y.J.; Fu, C.; Hou, B.K. UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 2016, 88, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Lanot, A.; Hodge, D.; Lim, E.K.; Vaistij, F.E.; Bowles, D.J. Redirection of flux through the phenylpropanoid pathway by increased glucosylation of soluble intermediates. Planta 2008, 228, 609–616. [Google Scholar] [CrossRef]
- Huang, Y.; Li, C.Y.; Qi, Y.; Park, S.; Gibson, S.I. SIS8, a putative mitogen-activated protein kinase kinase kinase, regulates sugar-resistant seedling development in Arabidopsis. Plant J. 2014, 77, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic acid biosynthesis in plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef]
- Shi, R.; Wang, J.P.; Lin, Y.-C.; Li, Q.; Sun, Y.-H.; Chen, H.; Sederoff, R.R.; Chiang, V.L. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 2017, 245, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Sundell, D.; Street, N.R.; Kumar, M.; Mellerowicz, E.J.; Kucukoglu, M.; Johnsson, C.; Kumar, V.; Mannapperuma, C.; Delhomme, N.; Nilsson, O.; et al. Aspwood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 2017, 29, 1585–1604. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.H. Secondary cell walls: Biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 2015, 56, 195–214. [Google Scholar] [CrossRef]
- Pyo, H.; Demura, T.; Fukuda, H. TERE; a novel cis-element responsible for a coordinated expression of genes related to programmed cell death and secondary wall formation during differentiation of tracheary elements. Plant J. 2007, 51, 955–965. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Mitsuda, N.; Ohtani, M.; Ohme-Takagi, M.; Kato, K.; Demura, T. VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011, 66, 579–590. [Google Scholar] [CrossRef]
- Yang, M.; Fehl, C.; Lees, K.V.; Lim, E.K.; Offen, W.A.; Davies, G.J.; Bowles, D.J.; Davidson, M.G.; Roberts, S.J.; Davis, B.G. Functional and informatics analysis enables glycosyltransferase activity prediction. Nat. Chem. Biol. 2018, 14, 1109–1117. [Google Scholar] [CrossRef]
- Lim, E.-K.; Baldauf, S.; Li, Y.; Elias, L.; Worrall, D.; Spencer, S.P.; Jackson, R.G.; Taguchi, G.; Ross, J.; Bowles, D.J. Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 2003, 13, 139–145. [Google Scholar] [CrossRef]
- Han, D.Y.; Lee, H.R.; Kim, B.G.; Ahn, J.H. Biosynthesis of ferulic acid 4-O-glucoside and feruloyl glucoside using Escherichia coli harboring regioselective glucosyltransferases. Appl. Biol. Chem. 2016, 59, 481–484. [Google Scholar] [CrossRef]
- Lanot, A.; Hodge, D.; Jackson, R.G.; George, G.L.; Elias, L.; Lim, E.-K.; Vaistij, F.E.; Bowles, D.J. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 2006, 48, 286–295. [Google Scholar] [CrossRef]
- Achnine, L.; Huhman, D.V.; Farag, M.A.; Sumner, L.W.; Blount, J.W.; Dixon, R.A. Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 2005, 41, 875–887. [Google Scholar] [CrossRef]
- Chapelle, A. Caractérisation de gènes de β-glucosidase et d’UDP-glycosyltransférase potentiellement impliqués dans la lignification chez Arabidopsis thaliana. Ph.D. Thesis, Université Paris-Sud, Orsay, France, 2009. [Google Scholar]
- McCarthy, R.L.; Zhong, R.; Ye, Z.H. Secondary wall NAC binding element (SNBE), a key Cis-acting element required for target gene activation by secondary wall NAC master switches. Plant Signal. Behav. 2011, 6, 1282–1285. [Google Scholar] [CrossRef]
- Ohashi-Ito, K.; Oda, Y.; Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 2010, 22, 3461–3473. [Google Scholar] [CrossRef]
- Kim, W.C.; Ko, J.H.; Han, K.H. Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol. Biol. 2012, 78, 489–501. [Google Scholar] [CrossRef]
- Zhong, R.; McCarthy, R.L.; Haghighat, M.; Ye, Z.-H. The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS ONE 2013, 8, e69219. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.L.; Zhong, R.; Fowler, S.; Lyskowski, D.; Piyasena, H.; Carleton, K.; Spicer, C.; Ye, Z.H. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 2010, 51, 1084–1090. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.H. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 2012, 53, 368–380. [Google Scholar] [CrossRef]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef]
- Ko, J.H.; Jeon, H.W.; Kim, W.C.; Kim, J.Y.; Han, K.H. The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 2014, 114, 1099–1107. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef] [PubMed]
- Bollhöner, B.; Jokipii-Lukkari, S.; Bygdell, J.; Stael, S.; Adriasola, M.; Muñiz, L.; Van Breusegem, F.; Ezcurra, I.; Wingsle, G.; Tuominen, H. The function of two type II metacaspases in woody tissues of Populus trees. New Phytol. 2018, 217, 1551–1565. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Yamaguchi, M.; Tamura, T.; Nakano, Y.; Nishikubo, N.; Yoneda, A.; Kato, K.; Kubo, M.; Kajita, S.; Katayama, Y.; et al. Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 2015, 56, 242–254. [Google Scholar] [CrossRef]
- Wang, Y.W.; Wang, W.C.; Jin, S.H.; Wang, J.; Wang, B.; Hou, B.K. Over-expression of a putative poplar glycosyltransferase gene, PtGT1, in tobacco increases lignin content and causes early flowering. J. Exp. Bot. 2012, 63, 2799–2808. [Google Scholar] [CrossRef]
- König, S.; Feussner, K.; Kaever, A.; Landesfeind, M.; Thurow, C.; Karlovsky, P.; Gatz, C.; Polle, A.; Feussner, I. Soluble phenylpropanoids are involved in the defense response of Arabidopsis against Verticillium longisporum. New Phytol. 2014, 202, 823–837. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef]
- Wang, J.P.; Liu, B.; Sun, Y.; Chiang, V.L.; Sederoff, R.R. Enzyme-enzyme interactions in monolignol biosynthesis. Front. Plant Sci. 2019, 9, 1942. [Google Scholar] [CrossRef]
- Bassard, J.E.; Richert, L.; Geerinck, J.; Renault, H.; Duval, F.; Ullmann, P.; Schmitt, M.; Meyer, E.; Mutterer, J.; Boerjan, W.; et al. Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 2012, 24, 4465–4482. [Google Scholar] [CrossRef]
- Ono, N.N.; Qin, X.; Wilson, A.E.; Li, G.; Tian, L. Two UGT84 family glycosyltransferases catalyze a critical reaction of hydrolyzable tannin biosynthesis in pomegranate (Punica granatum). PloS ONE 2016, 11, e0156319. [Google Scholar] [CrossRef]
- Gallage, N.J.; Jørgensen, K.; Janfelt, C.; Nielsen, A.J.Z.; Naake, T.; Duński, E.; Dalsten, L.; Grisoni, M.; Møller, B.L. The intracellular localization of the vanillin biosynthetic machinery in pods of Vanilla planifolia. Plant Cell Physiol. 2018, 59, 304–318. [Google Scholar] [CrossRef]
- Knudsen, C.; Gallage, N.J.; Hansen, C.C.; Møller, B.L.; Laursen, T. Dynamic metabolic solutions to the sessile life style of plants. Nat. Prod. Rep. 2018, 35, 1140–1155. [Google Scholar] [CrossRef]
- Machettira, A.B.; Groß, L.E.; Tillmann, B.; Weis, B.L.; Englich, G.; Sommer, M.S.; Königer, M.; Schleiff, E. Protein-induced modulation of chloroplast membrane morphology. Front. Plant. Sci. 2012, 2. [Google Scholar] [CrossRef]
- Perello, C.; Llamas, E.; Burlat, V.; Ortiz-Alcaide, M.; Phillips, M.A.; Pulido, P.; Rodriguez-Concepcion, M. Differential subplastidial localization and turnover of enzymes involved in isoprenoid biosynthesis in chloroplasts. PLoS ONE 2016, 11, e0150539. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; Von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef]
- Baldacci-Cresp, F.; Moussawi, J.; Leplé, J.-C.; Van Acker, R.; Kohler, A.; Candiracci, J.; Twyffels, L.; Spokevicius, A.V.; Bossinger, G.; Laurans, F.; et al. PtaRHE1, a Populus tremula × Populus alba RING-H2 protein of the ATL family, has a regulatory role in secondary phloem fibre development. Plant. J. 2015, 82, 978–990. [Google Scholar] [CrossRef]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Baldacci-Cresp, F.; Sacré, P.-Y.; Twyffels, L.; Mol, A.; Vermeersch, M.; Ziemons, E.; Hubert, P.; Pérez-Morga, D.; El Jaziri, M.; de Almeida Engler, J.; et al. Poplar–root knot nematode interaction: A model for perennial woody species. Mol. Plant.-Microbe Interact. 2016, 29, 560–572. [Google Scholar] [CrossRef]
- Hemerly, A.S.; Ferreira, P.; de Almeida Engler, J.; Van Montagu, M.; Engler, G.; Inze, D. cdc2a expression in Arabidopsis is linked with competence for cell division. Plant. Cell 1993, 5, 1711–1723. [Google Scholar] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.C.; Giri, A.; Daniilidis, M.; Sun, G.; Härtl, K.; Hoffmann, T.; Schwab, W. Structural and functional analysis of UGT92G6 suggests an evolutionary link between mono- and disaccharide glycoside-forming transferases. Plant Cell Physiol. 2018, 59, 857–870. [Google Scholar] [CrossRef]
- Leple, J.C.; Brasileiro, A.C.M.; Michel, M.F.; Delmotte, F.; Jouanin, L. Transgenic poplars: Expression of chimeric genes using four different constructs. Plant Cell Rep. 1992, 11, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, R.; Vanholme, R.; Storme, V.; Mortimer, J.; Dupree, P.; Boerjan, W. Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol. Biofuels 2013, 6, 46. [Google Scholar] [CrossRef]
- Foster, C.E.; Martin, T.M.; Pauly, M. Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) Part I: Lignin. J. Vis. Exp. 2010, 37, e1745. [Google Scholar] [CrossRef] [PubMed]


| Substrates | UGT72AZ2 (Group 1) | UGT72B37 (Group 4) | UGT72B39 (Group 4) |
|---|---|---|---|
| p-coumaraldehyde | − | + | − |
| coniferaldehyde | − | + | − |
| sinapaldehyde | − | + | − |
| coniferyl alcohol | − | + | + |
| sinapyl alcohol | − | + | − |
| trans-cinnamic acid | − | − | − |
| p-coumaric acid | − | − | − |
| caffeic acid | − | − | − |
| ferulic acid | + | − | − |
| sinapic acid | + | − | − |
| salicylic acid | − | − | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Speeckaert, N.; Adamou, N.M.; Hassane, H.A.; Baldacci-Cresp, F.; Mol, A.; Goeminne, G.; Boerjan, W.; Duez, P.; Hawkins, S.; Neutelings, G.; et al. Characterization of the UDP-glycosyltransferase UGT72 Family in Poplar and Identification of Genes Involved in the Glycosylation of Monolignols. Int. J. Mol. Sci. 2020, 21, 5018. https://doi.org/10.3390/ijms21145018
Speeckaert N, Adamou NM, Hassane HA, Baldacci-Cresp F, Mol A, Goeminne G, Boerjan W, Duez P, Hawkins S, Neutelings G, et al. Characterization of the UDP-glycosyltransferase UGT72 Family in Poplar and Identification of Genes Involved in the Glycosylation of Monolignols. International Journal of Molecular Sciences. 2020; 21(14):5018. https://doi.org/10.3390/ijms21145018
Chicago/Turabian StyleSpeeckaert, Nathanael, Nassirou Mahamadou Adamou, Hadjara Amadou Hassane, Fabien Baldacci-Cresp, Adeline Mol, Geert Goeminne, Wout Boerjan, Pierre Duez, Simon Hawkins, Godfrey Neutelings, and et al. 2020. "Characterization of the UDP-glycosyltransferase UGT72 Family in Poplar and Identification of Genes Involved in the Glycosylation of Monolignols" International Journal of Molecular Sciences 21, no. 14: 5018. https://doi.org/10.3390/ijms21145018
APA StyleSpeeckaert, N., Adamou, N. M., Hassane, H. A., Baldacci-Cresp, F., Mol, A., Goeminne, G., Boerjan, W., Duez, P., Hawkins, S., Neutelings, G., Hoffmann, T., Schwab, W., El Jaziri, M., Behr, M., & Baucher, M. (2020). Characterization of the UDP-glycosyltransferase UGT72 Family in Poplar and Identification of Genes Involved in the Glycosylation of Monolignols. International Journal of Molecular Sciences, 21(14), 5018. https://doi.org/10.3390/ijms21145018