Abstract
Osmotin-like proteins (OLPs) mediate defenses against abiotic and biotic stresses and fungal pathogens in plants. However, no OLPs have been functionally elucidated in poplar. Here, we report an osmotin-like protein designated PdOLP1 from Populus deltoides (Marsh.). Expression analysis showed that PdOLP1 transcripts were mainly present in immature xylem and immature phloem during vascular tissue development in P. deltoides. We conducted phenotypic, anatomical, and molecular analyses of PdOLP1-overexpressing lines and the PdOLP1-downregulated hybrid poplar 84K (Populus alba × Populus glandulosa) (Hybrid poplar 84K PagOLP1, PagOLP2, PagOLP3 and PagOLP4 are highly homologous to PdOLP1, and are downregulated in PdOLP1-downregulated hybrid poplar 84K). The overexpression of PdOLP1 led to a reduction in the radial width and cell layer number in the xylem and phloem zones, in expression of genes involved in lignin biosynthesis, and in the fibers and vessels of xylem cell walls in the overexpressing lines. Additionally, the xylem vessels and fibers of PdOLP1-downregulated poplar exhibited increased secondary cell wall thickness. Elevated expression of secondary wall biosynthetic genes was accompanied by increases in lignin content, dry weight biomass, and carbon storage in PdOLP1-downregulated lines. A PdOLP1 coexpression network was constructed and showed that PdOLP1 was coexpressed with a large number of genes involved in secondary cell wall biosynthesis and wood development in poplar. Moreover, based on transcriptional activation assays, PtobZIP5 and PtobHLH7 activated the PdOLP1 promoter, whereas PtoBLH8 and PtoWRKY40 repressed it. A yeast one-hybrid (Y1H) assay confirmed interaction of PtoBLH8, PtoMYB3, and PtoWRKY40 with the PdOLP1 promoter in vivo. Together, our results suggest that PdOLP1 is a negative regulator of secondary wall biosynthesis and may be valuable for manipulating secondary cell wall deposition to improve carbon fixation efficiency in tree species.
1. Introduction
Wood provides abundant biomass and important biomaterials for renewable sources and industrial products around the world. Wood formation includes multiple developmental stages, such as vascular cambium differentiation, vascular tissue division, cell expansion, cell wall thickening, and programmed cell death [1,2]. Cloning and transformation of genes related to wood development in tree species are important for cultivating new varieties of trees with fine quality and high yield. However, the functions of a large number of genes related to wood traits have not been discovered and the molecular mechanisms of secondary wall thickening and secondary xylem formation in forest trees remain largely unknown [3,4,5]. Hence, elucidation of the mechanisms underlying gene regulation in xylem formation and secondary cell wall synthesis during wood development would be instrumental in providing the molecular basis and technical approaches for genetic improvement of the wood properties of forest trees.
Osmotin-like proteins (OLPs) comprise a group of 24–26-kD proteins belonging to the pathogenesis-related protein 5 (PR-5) family that were originally discovered in tobacco cells (Nicotiana tabacum L. var Wisconsin 38) under osmotic stress conditions [6]. OLPs play important roles in growth and development. For example, higher accumulation of the Petunia (Petunia hybrida) osmotin-like protein PhOSM is found in the root, with lower levels detected in the leaf and flower, showing that PhOSM plays vital roles in normal development [7]. In addition, the rice osmotin protein gene OsOSM1 is mainly expressed in the leaf sheath at the booting stage and is associated with development [8]. Hybrid poplar (Populus deltoides × P. euramericana cv. ‘Nanlin895′) PeTLP with high homology to OLP genes has been isolated. Accumulation of PeTLP transcripts in stems indicates that PeTLP is associated with wood formation [9]. Involved in stem vascular tissue development, hybrid poplar (Populus trichocarpa × P. deltoides) TLP1, which is homologous to OLPs, is mainly expressed in vascular tissues of stems, petioles, and midveins [10]. Osmotins are osmoticum-induced proteins that can accumulate in large amounts in the cytoplasmic vesicles or vacuoles of plant cells. OLPs are produced in plants under different abiotic and biotic stresses [11,12]. For instance, OLP-transgenic tomato plants under saline stress exhibit increased secondary cell wall thicknesses of fibers and vessels, which act as an important mechanism for improving the resistance of plants to water stress [13]. Abdin et al. (2011) [14] showed that osmotin proteins do not contain a DNA-binding motif and may act as modulators of metabolic signaling or transcription factors in response to biotic and abiotic stresses. In addition, osmotin proteins may have a unique contribution to pathogenesis. Indeed, osmotins exhibit antifungal activity by inhibiting spore germination and lysis, hyphal growth, and spore viability [15].
Recent studies in woody plants (e.g., poplar and eucalyptus) as well as Arabidopsis have demonstrated that secondary wall formation is mediated by a complex transcriptional network. In this hierarchical network of transcription factors, a group of wood-associated NAC transcription factors (WNDs), such as PtrWND2B and Eucalyptus gunnii EgWND1, have been identified as functional orthologs of Arabidopsis NACs including SND1, NST1/NST2, and VND6/VND7 and have been identified as master switches that are able to activate the promoter activities of secondary wall biosynthetic genes [16,17,18]. Several lignin biosynthetic genes in transgenic plants have been assessed in investigating the function of wood formation-related genes involved in secondary cell wall formation. Reduced transcript levels for the PtoCCR2, PtoCAD1, and PtoC4H2 genes in Populus trichocarpa PtrWRKY19-overexpressing poplar plants were observed, suggesting that PtrWRKY19 negatively regulates the expression of secondary wall biosynthesis genes in poplar [19]. Furthermore, lignin biosynthetic genes, including COMT2, CCOAOMT1, C3H, 4CL5, CCR2, PAL4, HCT1, and CAD1 were upregulated in P. tomentosa (Carr.) PtoMYB92-overexpressing transgenic plants, suggesting that PtoMYB92 positively modulates secondary wall development in poplar [20]. Arabidopsis thaliana TALE homeodomain transcription factor KNOTTED ARABIDOPSIS THALIANA7 (KNAT7) interacts with a BELL1-LIKE HOMEODOMAIN protein BLH6, and this interaction may influence secondary cell wall development [21]. Sun et al., (2017) [22] showed that miR319 and its target TCP4 can activate secondary cell wall biosynthesis in Arabidopsis. Moreover, TCP4 possibly targets the promoter of VND7 to regulate its expression and is involved in xylem vessel differentiation. Overexpression of SbMyb60 has been shown to promote monolignol biosynthesis and leads to induced lignin deposition and changed cell wall composition in transgenic Sorghum bicolor plants [23]. Additionally, Li et al. (2019) [24] showed that Populus deltoides (Marsh.) proline-rich protein gene PdPRP might positively regulate secondary cell wall formation by promoting secondary wall thickening and expansion in both poplars and Arabidopsis. These studies have largely improved our knowledge of the complicated regulatory mechanisms of secondary cell wall formation in poplars and other woody plants.
P. deltoides (Marsh.), introduced from North America into China in 1972, is a black poplar species of the Aigeiros section in the genus Populus. Compared with other poplar tree species, P. deltoides is resistant to stress and disease; it is fast-growing and has fine quality and good environmental adaptability. Thus, P. deltoides is widely used worldwide as a major species for poplar gene function studies and transgenic breeding. However, compared with knowledge regarding the biological function of OLPs in Arabidopsis, Glycine max, and Nicotiana tabacum, the molecular mechanism of these proteins in poplar remains largely unknown.
In this study, we isolated and characterized the molecular functions of a P. deltoides osmotin-like protein gene, PdOLP1, in secondary cell wall formation. We altered PdOLP1 transcript levels by overexpression and suppression in the transgenic hybrid poplar 84K (Populus alba × Populus glandulosa) and analyzed its effect on growth and cell wall thickening. The PdOLP1 coexpression network indicated that genes related to secondary cell wall biosynthesis and wood development are affected by PdOLP1. Moreover, transcriptional activation assays showed that several transcription factors involved in wood formation activate or repress the promoter of PdOLP1. Our results reveal that modifying the PdOLP1 gene may enable cultivation of transgenic poplar lines with improved carbon sequestration and valuable properties for wood processing.
2. Results
2.1. Isolation and Characterization of PdOLP1
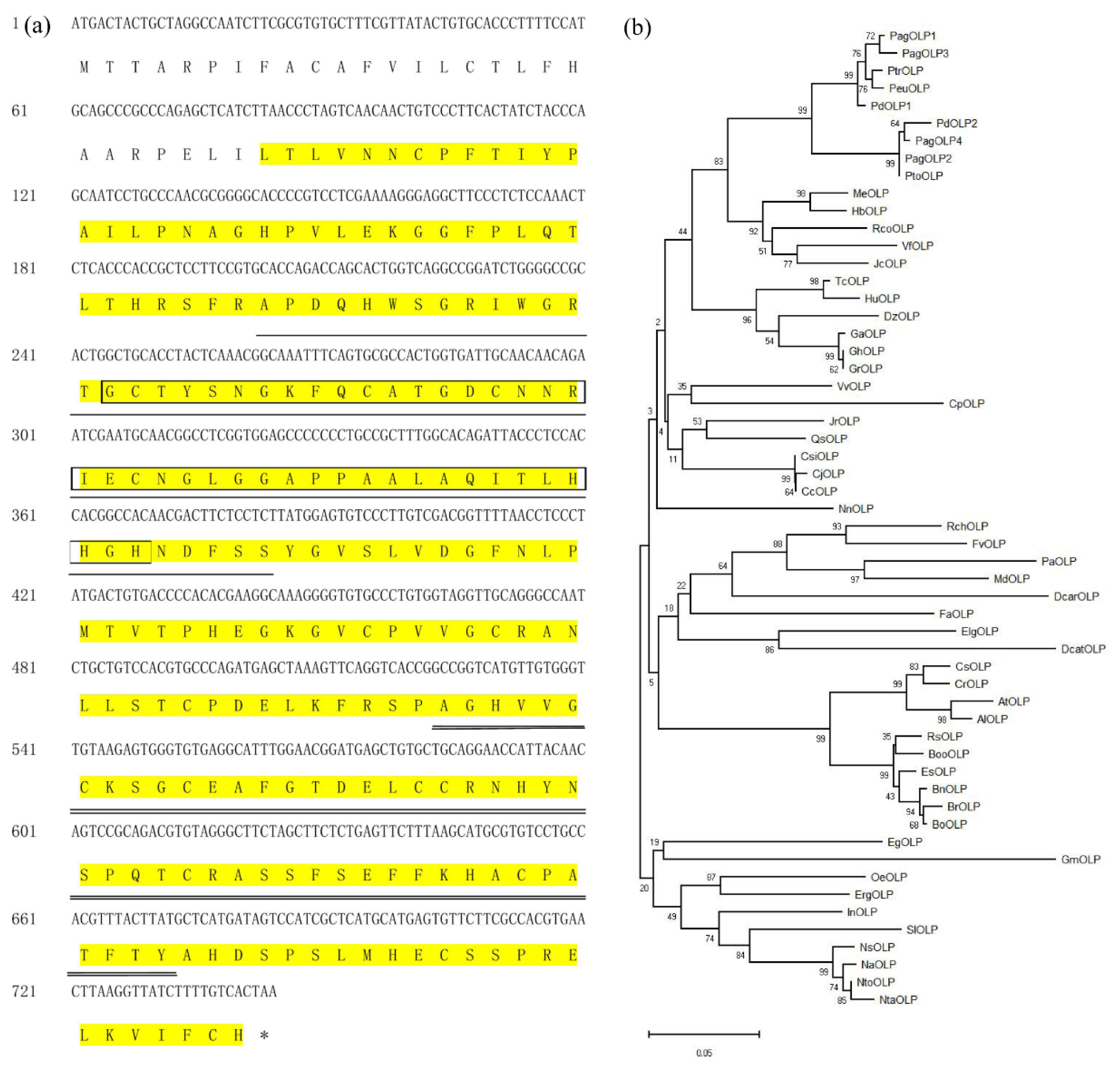
We isolated a 744 bp cDNA sequence from the immature xylem of 15-year-old P. deltoides (Figure 1a). The sequence has 97.6% homology with the open reading frame (ORF) of Populus trichocarpa PtOLP, and we named it PdOLP1. Another gene of P. deltoides (Podel.01G248000.1) has high protein similarity (93.2%) with PdOLP1, and this homologous gene was named PdOLP2. PagOLP1 (a gene of hybrid poplar 84K (Pop_G09G014129.T1)) exhibits high sequence similarity (97.2%) to PdOLP1. PagOLP2 (a gene of hybrid poplar 84K (Pop_A01G056866.T1)) has high sequence similarity (98.4%) to PdOLP2. PagOLP3 (a gene of hybrid poplar 84K (Pop_A09G059600.T1)) exhibits high sequence similarity (96.9%) to PdOLP1. PagOLP4 (a gene of hybrid poplar 84K (Pop_G01G020795.T1)) has high sequence similarity (89.0%) to PdOLP1 [25].

Figure 1.
Characterization of the PdOLP1 protein. (a) Amino acid sequence of the coding region of PdOPL. The sequence highlighted in yellow indicates the THN domain. The AWS domain is boxed. The VWC domain is underlined with a thin black line. FU domain is underlined with double black line. SMART (http://smart.embl-heidelberg.de/) was used to analyze the PdOPL1 protein sequence. (b) A phylogenetic tree of the OPL proteins from 52 plants was constructed based on the neighbor-joining method, including PdOPL1 (GenBank:MK052942), PdOLP2: (Podel.01G248000.1), PagOLP1: (Pop_G09G014129.T1), PagOLP2:.(Pop_A01G056866.T1), PagOLP3: (Pop_A09G059600.T1), PagOLP4: (Pop_G01G020795.T1), PtrOLP (GenBank:XP_024463852.1), PeuOLP (GenBank:XP_011042680.1), PtoOLP (GenBank:APA20308.1), MeOLP (GenBank:XP_021626825.1), HbOLP (GenBank:XP_021636655.1), RcOLP (GenBank:XP_002531364.1), DzOLP (GenBank:XP_022756598.1), CjOLP (GenBank:BAI63297.1), VfOLP (GenBank:ARV78462.1), TcOLP (GenBank:XP_007016468.1), CsiOLP (GenBank:XP_006488293.1), CcOLP (GenBank:XP_006424795.1), VvOLP (GenBank:XP_002281193.1), HuOLP (GenBank:XP_021278215.1), JrOLP (GenBank:XP_018823302.1), QsOLP (GenBank:XP_023907784.1), GhOLP (GenBank:XP_016689054.1), GrOLP (GenBank:XP_012446484.1), GaOLP (GenBank:XP_017650093.1), JcOLP (GenBank:XP_012065012.1), EsOLP (GenBank:XP_006409911.1), RsOLP (GenBank:XP_018463081.1), BnOLP (GenBank:XP_013676421.1), BrOLP (GenBank:XP_009140942.1), BooOLP (GenBank:XP_013632142.1), BoOLP (GenBank:AAO12209.1), RcOLP (GenBank:XP_024159050.1), OeOLP (GenBank:XP_022881590.1), FaOLP (GenBank:ABB86299.1), CsOLP (GenBank:XP_010470049.1), ErgOLP (GenBank:XP_012827362.1), FvOLP (GenBank:XP_004294572.1), InOLP (GenBank:XP_019199014.1), NtOLP (GenBank:XM_018769102.1), NnOLP (GenBank:XP_010259948.1), ElgOLP (GenBank:XP_010918389.1), EgOLP (GenBank:XP_010033142.1), NtOLP (GenBank: NP_001312698.1), PaOLP (GenBank:XP_021820385.1), AtOLP (GenBank:NP_001324474.1), AlOLP (GenBank:XP_020885214.1), DcarOLP (GenBank:XP_017251860.1), NsOLP (GenBank:XP_009800616.1), CpOLP (GenBank:XP_021887635.1), GmOLP (GenBank:XP_003538000.1), CrOLP (GenBank:XP_006294865.1), NaOLP (GenBank:XP_019237711.1), MdOLP (GenBank:XP_008357503.1). DcatOLP (GenBank:XP_020683326.1), SlOLP (GenBank:NP_001234714.1). The 51 OPL amino acid sequences are list in Table S5. Bar = 0.05 substitutions per site.
Furthermore, sequence analysis showed that the cDNA sequence of PdOLP1 is 74–97.6% identical to the cDNA sequences of OLPs from 51 other plant species. PdOLP1 encodes a protein of 247 amino acids (Figure 1a) with a predicted molecular mass of 26.85 kD and a calculated isoelectric point (pI) of 7.86.
Comparison of PdOLP1 with 51 OLPs from other species revealed high homology between PdOLP1and PeuOLP (97% similarity) in P. euphratica, PtoOLP (88% similarity) in P. tomentosa, HbOLP (86% similarity) in Hevea brasiliensis, MeOLP (79% similarity) in Manihot esculenta, EgOLP (79% similarity) in Eucalyptus grandis, and AtOLP (72% similarity) in Arabidopsis thaliana (Figure 1b, Table S6).
The PdOLP1 protein sequence contains some typical protein motifs, including a THN domain, a VWC domain, and an AWS domain (Figure 1a, Figure S2). A THN domain that belongs to the thaumatin family and is also referred to as pathogenesis-related group 5 (PR5) exists between residues 28 and 274; it is related to plant pathogenesis and possesses antifungal activity. The VWC domain between residues 68 and 130 is involved in transcription, signal transduction, ribosomal and membrane transport and the proteasome. The AWS domain is situated between residues 82 and 123, with a role in gene regulation by methylation of lysine residues in histones and other proteins. Furin-like repeats of the FU domain, involved in the mechanism of signal transduction by receptor tyrosine kinases, may be present between residues 175 and 224. These analyses indicate that the PdOLP1 protein may play an important role in signal transduction, transcription, and methylation, among other processes.
2.2. Expression Pattern and Subcellular Localization of PdOLP1
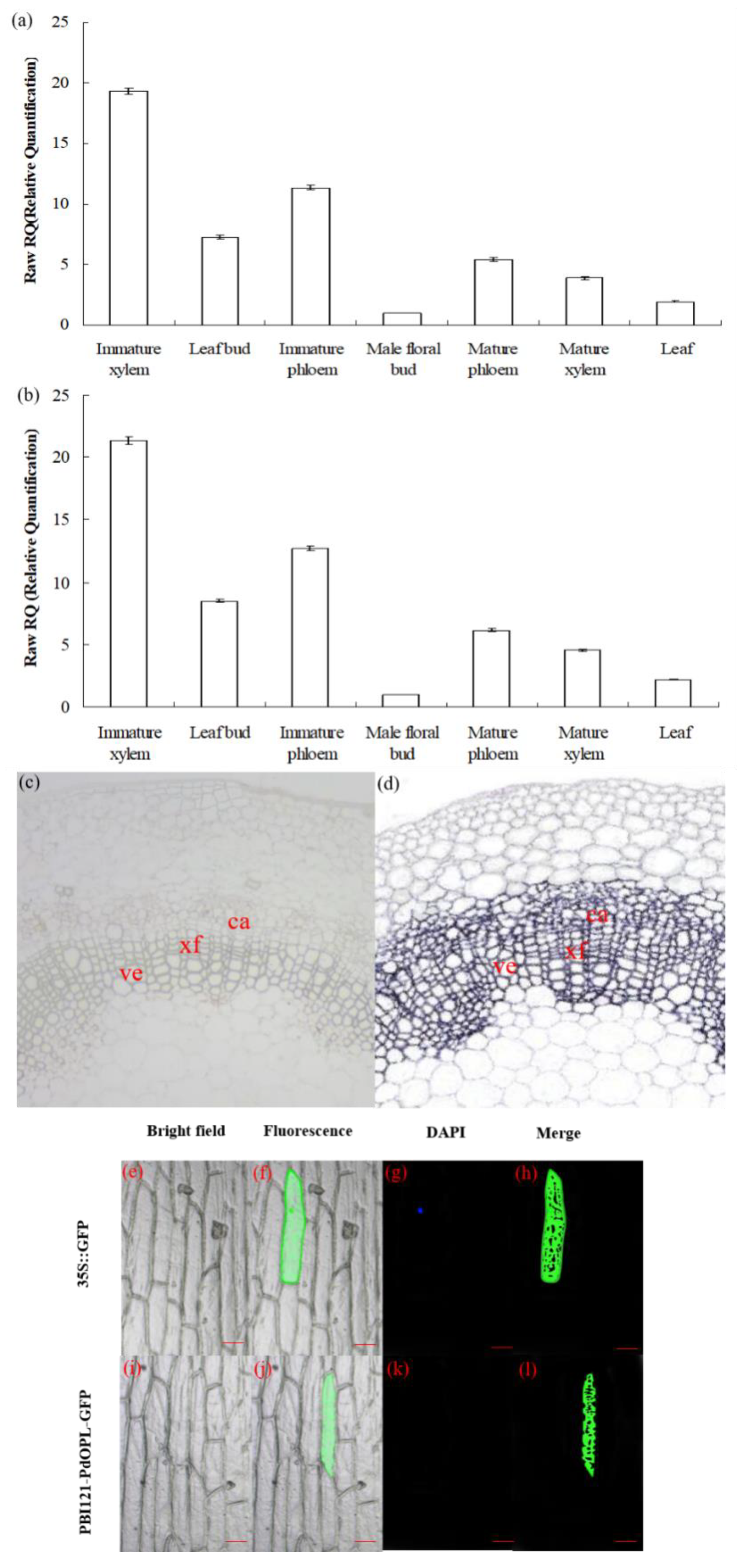
Quantitative real-time PCR (qRT-PCR) analysis revealed that PdOLP1 was differentially expressed at detectable levels in all tissues as tested. The relative expression levels of PdOLP1 in immature xylem and immature phloem of 15-year-old P. deltoides were higher than in any other examined tissues, whereas expression in the floral bud was the lowest (Figure 2a). The expression levels of PdOLP1 in different tissues were almost the same when using either TUA1 (Figure 2a) or UBQ1 (Figure 2b) as internal controls. In situ hybridization analysis of P. deltoides stem segments indicated the PdOLP1 gene to be specifically expressed in xylary fibers and vessels, with lower expression levels found in male floral buds (Figure 2c). There was no detectable hybridization signal in the control sections hybridized with PdOLP1 sense probes (Figure 2d). These results suggest that PdOLP1 is expressed predominantly in the immature xylem and immature phloem tissues undergoing secondary cell wall thickening. The PdOLP1 coding sequence was fused in-frame to the 5′ end of green fluorescent protein (GFP) (Figure S1); subcellular localization of PBI121-PdOLP1-GFP showed green fluorescence of the recombinant protein in the cytoplasm of onion epidermal cells (Figure 2j,l). In contrast, using the construct with GFP alone used as a control, green fluorescence was observed throughout the cells, including the cytoplasm, plasma membrane, and nucleus (4′,6-diamidino-2-phenylindole (DAPI) stained) (Figure 2f,h). These results suggest that PdOPL is located in the cytoplasmic compartment.
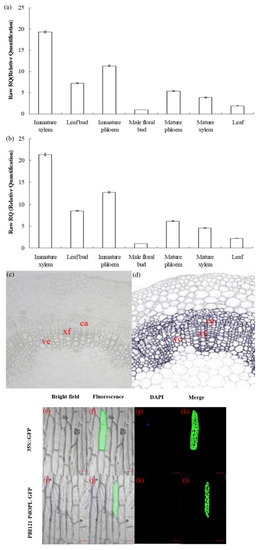
Figure 2.
Expression pattern and subcellular localization of the PdOLP1 protein. (a,b) PdOLP1 expression in various tissues was analyzed by quantitative real-time PCR (qRT-PCR). Expression was normalized to α-tubulin (TUA1) (a) and ubiquitin (UBQ1) reference genes (b). Error bars: ± standard error (SE) of three replicates. (c,d) In situ localization of PdOLP1 in P. deltoides stems. Sections of the stems were hybridized with digoxigenin-labeled antisense (c) or sense (d) probes. The hybridization signals are shown in purple. ve, vessel; xf, xylary fiber; ca, cambium. Scale bars = 25 µm. (e–l) Subcellular localization of PdOLP1. The green fluorescent signal of PBI121-PdOLP1-GFP was detected within the cytoplasm of onion epidermal cells (j,l), and green fluorescent protein (GFP) signal alone was localized in the cytoplasm, nucleus (4′,6-diamidino-2-phenylindole (DAPI) stained) and plasma membrane (f,h). Scale bars = 100 µm.
2.3. Verification of PdOLP1 Overexpression and Downregulation in Transgenic Poplar Plants
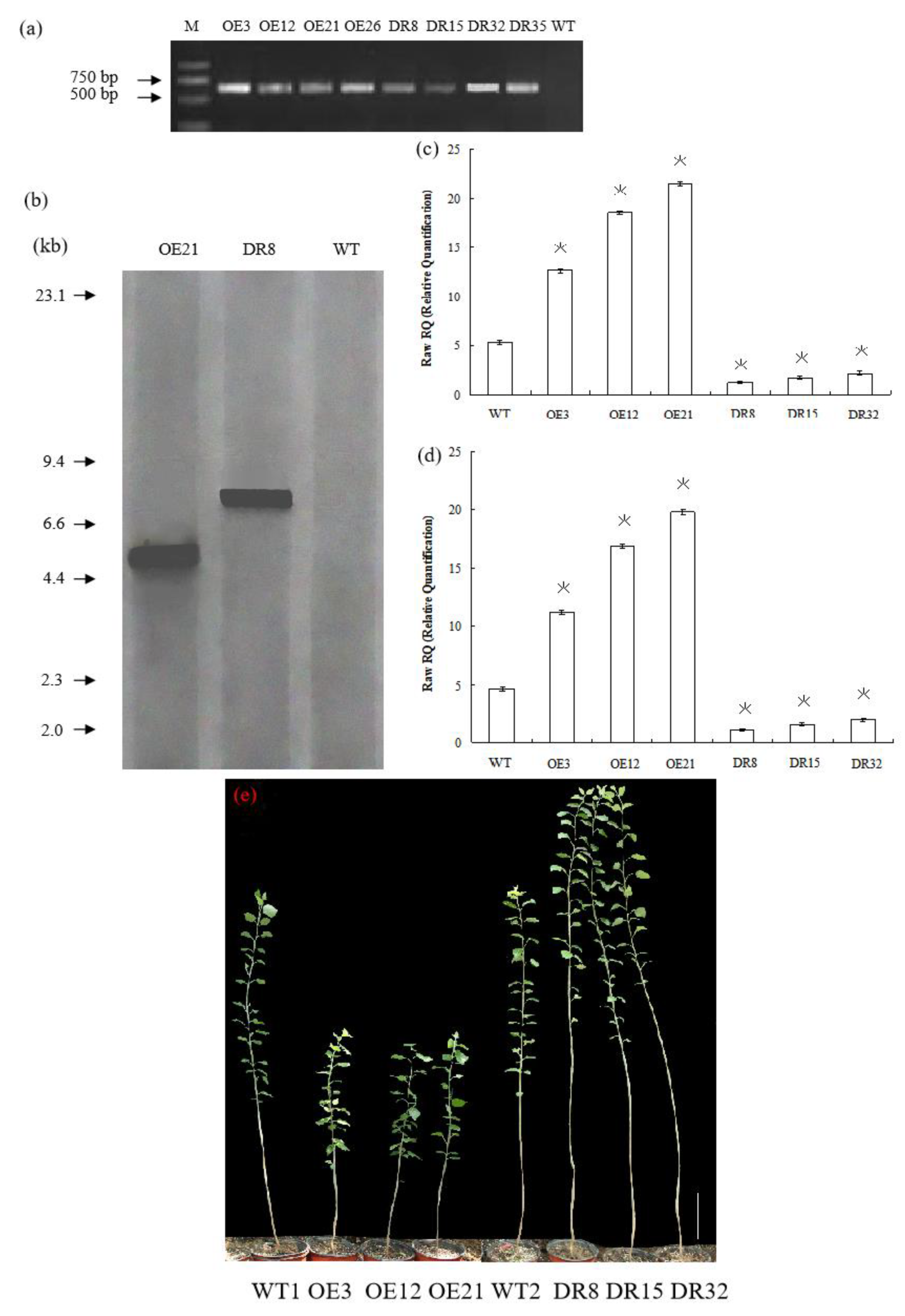
To investigate the biological function of PdOLP1, we overexpressed it under the control of the proNAC068 promoter in hybrid poplar (P. alba × P. glandulosa) of clone 84K. A total of 28 independent overexpression lines (pBI121-ProNAC068-sense PdOLP1) and 35 suppression lines (pBI121-ProNAC068-antisense PdOLP1) were obtained and examined by PCR (Figure 3a). At least one copy of the PdOLP1 gene was detected in three of the overexpression (OE3, OE12, OE21) and three of the knockdown (DR8, DR15, DR32) lines (Figure 3b) by southern hybridization, and no hybridization signals were detected in wildtype plants. This indicates that the constructs containing PdOLP1 had integrated into the genomes of the transformed poplar plants. qRT-PCR was used to further verify transgene expression in PdOLP1-overexpressing (OE) lines and PdOLP1-downregulated (DR) lines, revealing significant upregulation of PdOLP1 by 137.67%, 247.53%, and 303.65% in the PdOLP1-OE poplar lines OE3, OE12, and OE21, respectively, compared with non-transgenic (WT) poplar plants (p = 0.002, Figure 3c,d). PdOLP1 expression in the three independent PdOLP1-DR lines DR8, DR15 and DR32 was obviously reduced by 77.00%, 67.12%, and 59.64%, respectively, compared with the WT plants (p = 0.008, Figure 3c,d). The expression patterns found using TUA1 (Figure 3c) as an internal control were the same as those using UBQ1 (Figure 3d). These six transgenic poplar lines (OE3, OE12, OE21, DR8, DR15, and DR32) with higher or lower level expression of PdOLP1 were selected for further functional characterization.
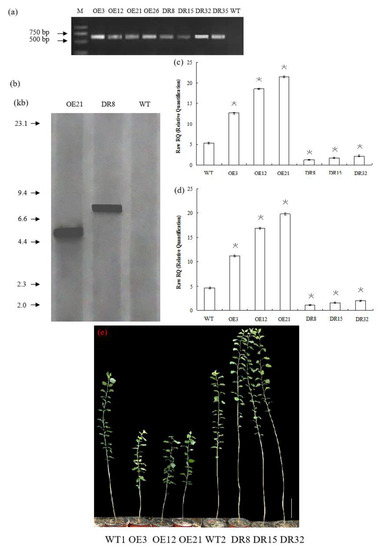
Figure 3.
PdOLP1 overexpression restricted development, and PdOLP1 downregulation promoted growth. (a) Transgenic poplar plants were identified by amplifying the NptII-sensitive marker (Table S3). M, DNA molecular weight marker; O3, O12, and O21, PdOLP1-overexpressing poplar plants; DR8, DR15 and DR32, PdOLP1-downregulated poplar plants. (b) Southern blot analysis of PCR identified transgenic poplar plants. Genomic DNA of the plants digested with XbaI was hybridized with the NptII probe. OE21, PdOLP1-OE hybrid transgenic poplar line; DR8, PdOLP1-DR poplar line; WT, wildtype. (c,d) Confirmation of PdOLP1 expression levels in the transgenic plants by qRT-PCR. TUA1 (c) and UBQ1 (d) were used as reference genes. Error bars represent the standard deviation from the mean. (e) Growth comparison of 150-day-old WT1 poplar and the PdOLP1-OE lines OE3, OE12 and OE21. Growth comparison of 150-day-old WT2 poplar and the PdOLP1-DR lines DR8, DR15 and DR32 (right). The scale bars correspond to 21.71 cm/in (e).
2.4. Overexpression of PdOLP1 Delayed Primary Growth and Repressed Secondary Wall Biosynthesis in Poplars
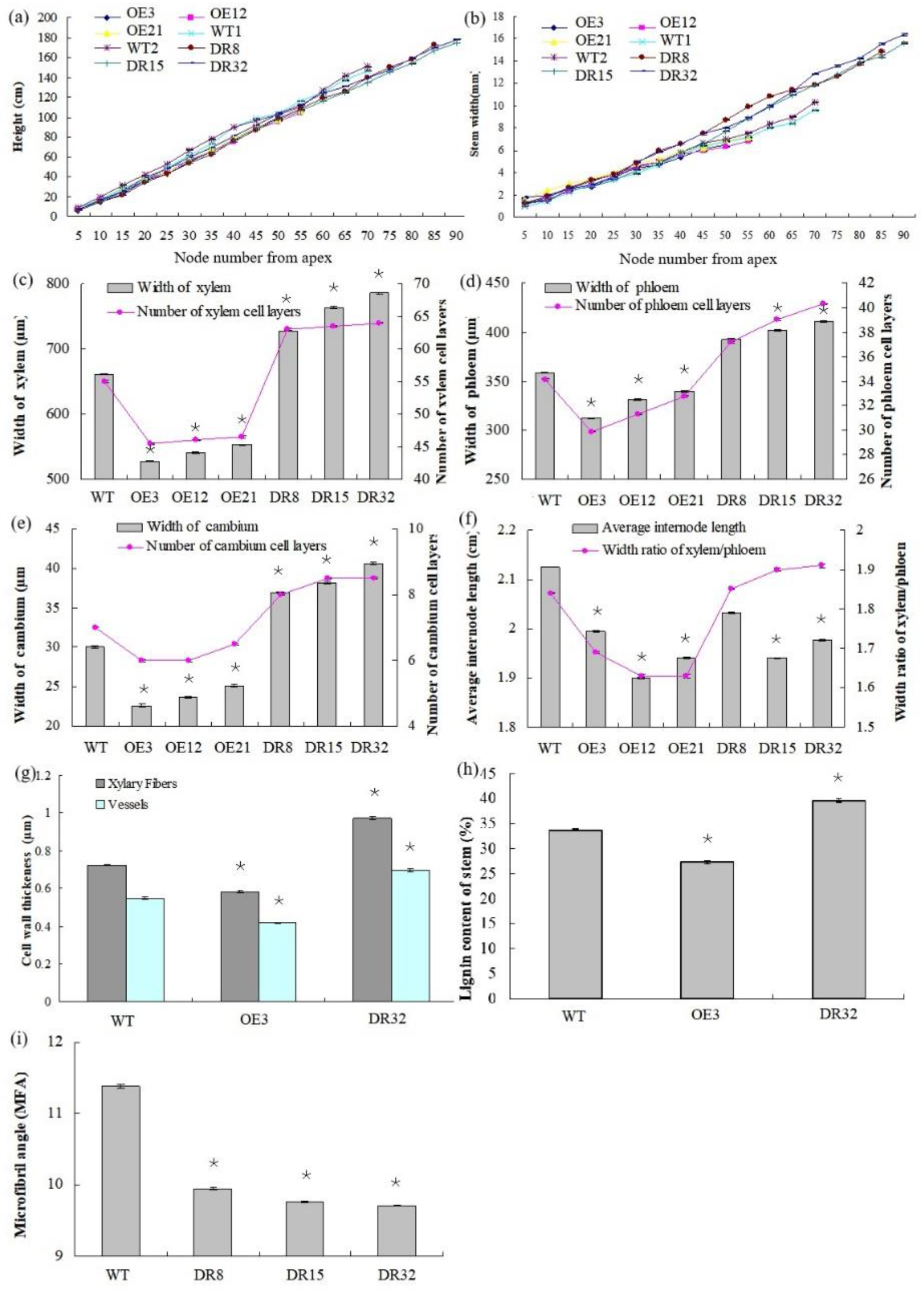
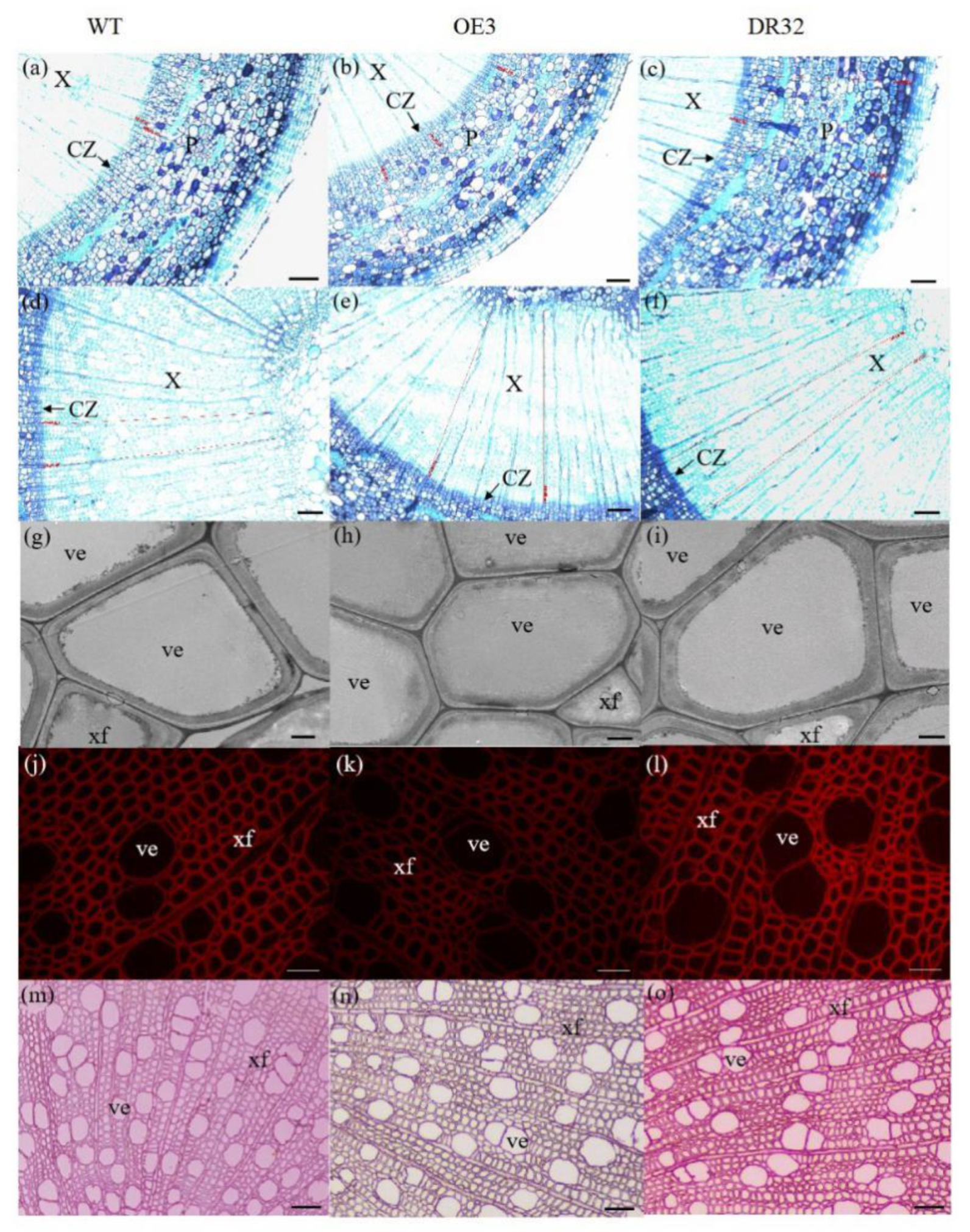
After growth for 1.5 years in a greenhouse with a 16/8 h light/dark cycle, PdOLP1-OE transgenic lines OE3, OE12 and OE21 showed 28.14%–34.47% reduction in plant height and a reduced diameter of 28.27–32.97% compared to WT plants (Figure 4a,b), respectively. Overexpression of PdOLP1 in hybrid poplar resulted in decreased radial width and number of cell layers of the xylem, cambium and phloem zones of the 15th internode compared with controls (Figure 4c–e, Figure 5a,b,d,e, Table S7). Reductions were also detected in the number of internodes, width ratio of xylem/phloem and average internode length in PdOLP1-OE plants (Figure 4f). Moreover, the cell walls of xylem fibers and vessels in the three PdOLP1-OE lines were significantly thinner than those in WT plants (p = 0.013, Figure 5g,h). To determine lignin modifications, we measured the Klason lignin content in the stems of WT and transgenic plants. The three tested PdOLP1-OE lines contained 18.99% lower lignin contents on average than WT plants (p = 0.003, Figure 4h). Weaker lignin autofluorescence was also observed in PdOLP1-OE OE3 plants compared to WT plants (Figure 5j,k). Phloroglucinol-HCl staining was employed to visualize lignin deposition and distribution in cell walls, and the 15th internodes of OE3 plant stem sections showed weaker staining in both the xylem vessels and interfascicular fibers (Figure 5m,n). These results demonstrate that upregulation of PdOLP1 weakened lignin deposition in transgenic plants and that PdOLP1 may negatively regulate secondary cell wall thickening of fibers and vessels in poplar stems.
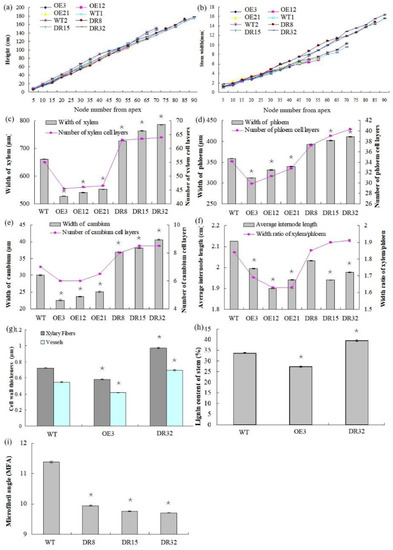
Figure 4.
Anatomical features of the stem cross-sections, growth, and wood property indexes of PdOLP1 transgenic plants. (a,b) PdOLP1-OE plants showed smaller shoots and a decreased stem diameter compared to WT plants. Plant height (a) and stem width (b) were measured at every fifth node from the apex. (c) Xylem widths (left panel) and numbers of xylem cell layers (right panel) in transgenic or WT plants. (d) Phloem widths (left panel) and numbers of phloem cell layers (right panel) in transgenic or WT plants. (e) Cambium widths (left panel) and numbers of cambium cell layers (right panel) in transgenic or WT plants. (f) The average internode lengths (left panel) and the xylem: phloem width ratios (right panel) in transgenic or WT lines. (g) Thicknesses of vessels and fibers in the stems of transgenic or WT plants. The values are the mean ± standard error (SE) of 20 cells. (h) Lignin content of transgenic or WT plants. (i) Analysis of the microfibril angle (MFA) in PdOLP1 transgenic poplar plants. The PdOLP1-DR poplar lines had lower MFAs than the control line. Significant differences between the transgenic plants and WT plants are marked with asterisks (* p < 0.05). WT: wildtype poplar; WT1: control for PdOLP1-OE plant; WT2: control for the PdOLP1-DR plant; OE3, OE12 and OE21: PdOLP1-OE poplar lines; DR8, DR15 and DR32: PdOLP1-DR plants.
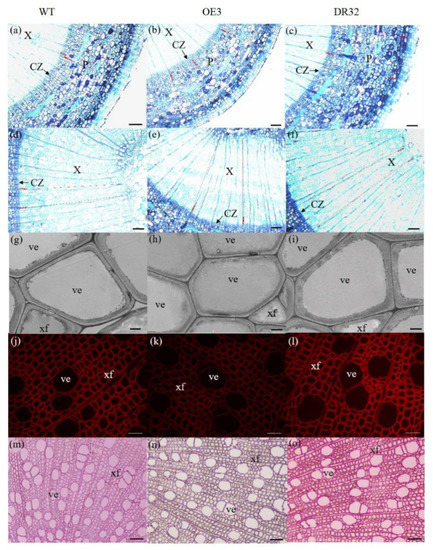
Figure 5.
Anatomical transverse sections of PdOLP1 transgenic poplar. The 15th internodes of 1.5-year-old plants were used to observe the anatomical characteristics of the stem cross-sections. (a–f) Toluidine blue staining of stem sections of a WT plant (a,d), PdOLP1-OE-transgenic plant (b,e) and PdOLP1-DR-transgenic plant (c,f). Scale bars = 100 µm. (g–i) Scanning electron microscopy of sections analyzed in WT (g), OE (h) and DR (i) plants. Scale bars = 5 µm. (j–l) Lignin autofluorescence was observed in WT (j), OE (k) and DR (l), Scale bars = 400 µm. (m–o) Phloroglucinol-HCl staining of WT (m), OE (n) and DR (o) plants. Scale bars = 400 µm. C, cortex cells; CZ, cambial zone; Pi, pith; P, secondary phloem; ve, vessel; X, secondary xylem; xf, xylem fiber.
2.5. Downregulation of PdOLP1 Promotes Vigorous Growth and Causes Ectopic Thickening of the Secondary Wall in Poplar
1.5-year-old PdOLP1-downregulated poplar lines displayed evident alterations in primary growth; plant heights were increased at least 16.10%, and stem widths were 56.88% thicker than those of WT plants (Figure 4a,b). The PdOLP1-downregulated lines individually produced more secondary xylem and secondary phloem than did the WT plants, with a mean increase in xylem radial width of 14.75% and an increase of 15.45% in xylem cell layers (Figure 4c, Figure 5a,c,d,f, Table S7).
Based on quantitative analysis, the cell wall thicknesses of the xylem fibers and vessels of the three PdOLP1-DR plants were significantly thicker than those of WT plants (p = 0.009, Figure 5g,i). Thus, lignin accumulation was significantly increased by 17.51% in stems of PdOLP1-DR poplars compared to WT plants (p = 0.005, Figure 4h). Confocal microscopy of lignin autofluorescence revealed lignified secondary wall thickening in WT stem cross-sections, but with higher signals in PdOLP1-downregulated DR8 plants (Figure 5j,l). Phloroglucinol-HCl staining of lignin in stem sections showed an intense red stain of the secondary cell walls in DR8 plants (Figure 5m,o), but less intense staining was detected in WT plants (Figure 5m). Therefore, downregulation of PdOLP1 in poplar causes high levels of lignification in stem xylem cells.
2.6. Wood Index, Total Biomass, and Carbon Storage Is Obviously Changed in PdOLP1-DR Poplars
The microfibril angle (MFA) values of 1.5-year-old PdOLP1-transgenic plants were measured, and a reduction of 12.69–14.86% was found compared to WT plants, with statistically significant differences (p = 0.005, Figure 4i). The PdOLP1-DR transgenic plants also had a significantly higher wood density, with up to a 20.39% increase compared to WT plants. Therefore, it is possible that PdOLP1 may play an important role in improving wood mechanical properties in poplar. Obvious differences in shoot and root biomass between transgenic and nontransgenic plants were also found when dry weight was analyzed. PdOLP1-DR plants had a 30.05% higher dry weight biomass than WT plants (p = 0.008), whereas the dry weight biomass in PdOLP1-OE plants was decreased by 22.56% compared to WT (p = 0.006, Table S8). Biotic sequestration is an effective way to weaken the influence of continuously rising atmospheric CO2 concentrations through increased carbon sequestration by forest trees [26]. Indeed, the carbon stock was significantly enhanced by 31.63% in the three tested PdOLP1-DR poplar lines (p = 0.005, Table S8), suggesting improved carbon storage.
2.7. Overexpression or Downregulation of PdOLP1 Affects Expression of Cell Wall Biosynthetic Genes in Transgenic Poplars
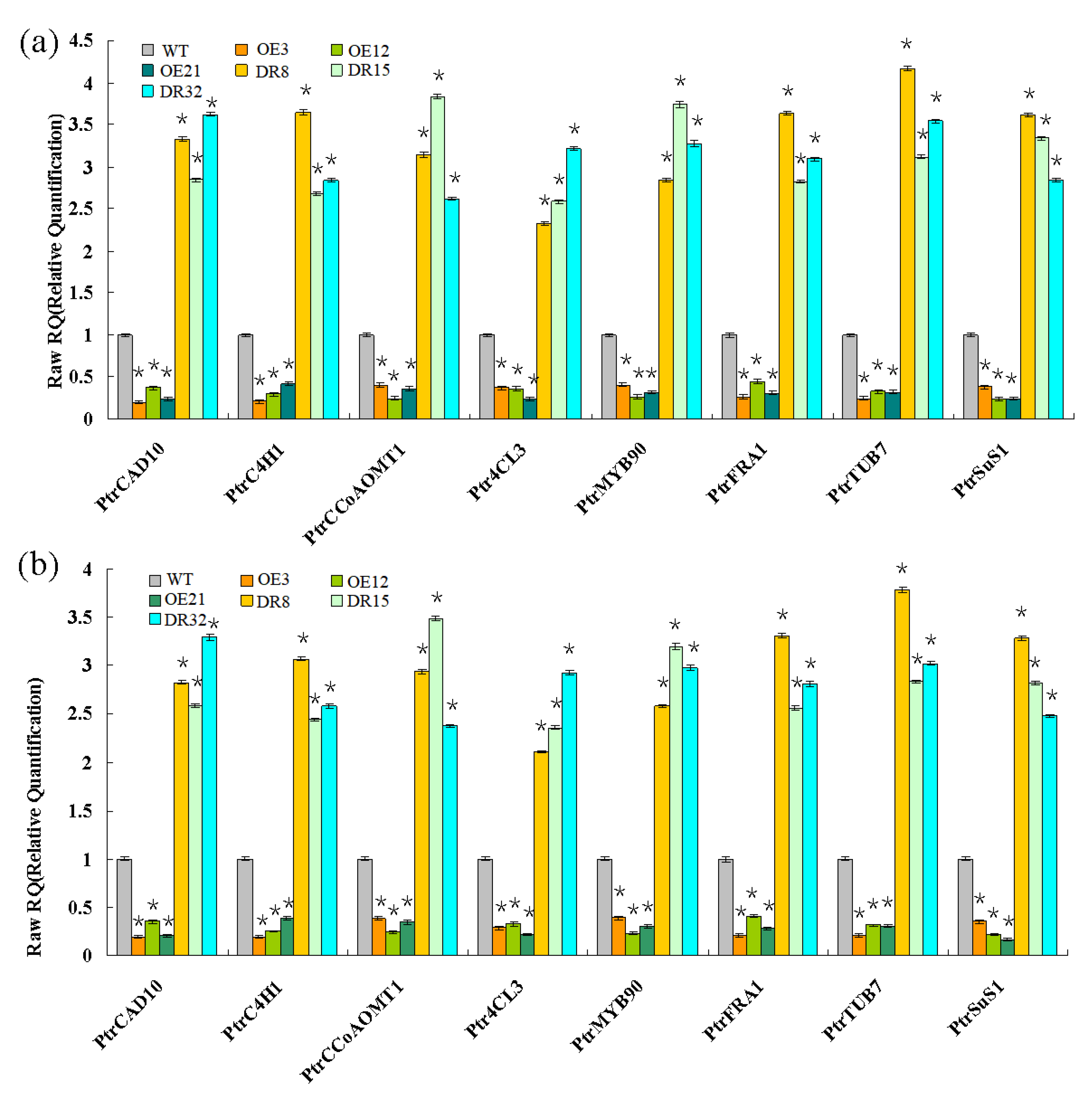
Expression of secondary wall biosynthetic genes was investigated in 1.5-year-old PdOLP1 transgenic poplar. Two internal control genes and eight genes associated with the phenylpropanoid pathway and microfibril angle formation were selected to confirm primer amplification efficiency and specificity (Table S4, Figures S3 and S4). The results from qRT-PCR analysis revealed that the transcript levels of Ptr4CL3, PtrCAD10, PtrC4H1, PtrCCoAOMT1, and PtrMYB90 were significantly repressed in the PdOLP1-OE poplar plants (Figure 6). In addition, the expression of microfibril angle-related genes PtrSuS1, PtrTUB7, and PtrFRA1 was obviously downregulated in PdOLP1-OE poplar compared with WT plants (Figure 6). However, downregulation of PdOLP1 in poplar resulted in upregulation of the eight genes examined. Compared with WT plants, in the PdOLP1-DR lines, the expression level of Ptr4CL3 increased by 132.30%–221.80% (p = 0.004), and that of PtrCCoAOMT1 increased by 152.30–274.10% (p = 0.006); in the PdOLP1-OE lines, PtrSuS1 expression decreased by 61.2–75.10% (p = 0.003) and PtrCAD10 decreased by 62.6–79.80% (p = 0.002). These results of these experiments indicate that PdOLP1 may function as a negative regulator of secondary wall biosynthesis in poplar.
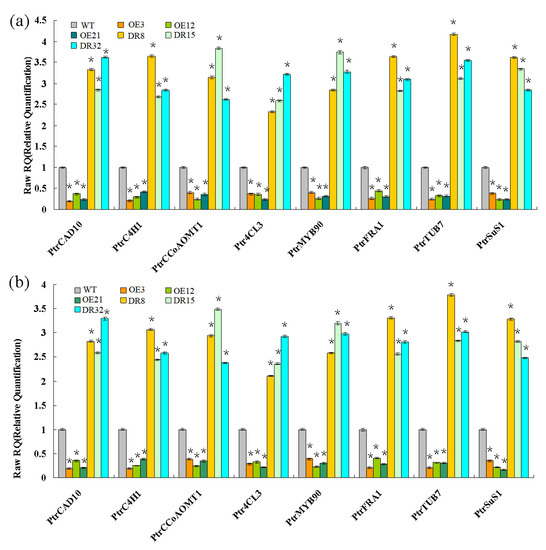
Figure 6.
Wood formation-related genes analyzed by qRT-PCR. Eight genes involved in secondary wall biosynthesis were evaluated: Ptr4CL3, PtrCAD10, PtrC4H1, and PtrCCoAOMT1, PtrMYB90, PtrFRA1, PtrSuS1, and PtrTUB7. TUA1 (a) and UBQ1 (b) were used as internal control genes, and transcript levels of genes in WT plants were set to 1. The data are the mean ± SE from three biological replicates. Asterisks indicate significant differences between transgenic and WT plants (* p < 0.05).
2.8. Coexpression Network of PdOLP1 in Poplars
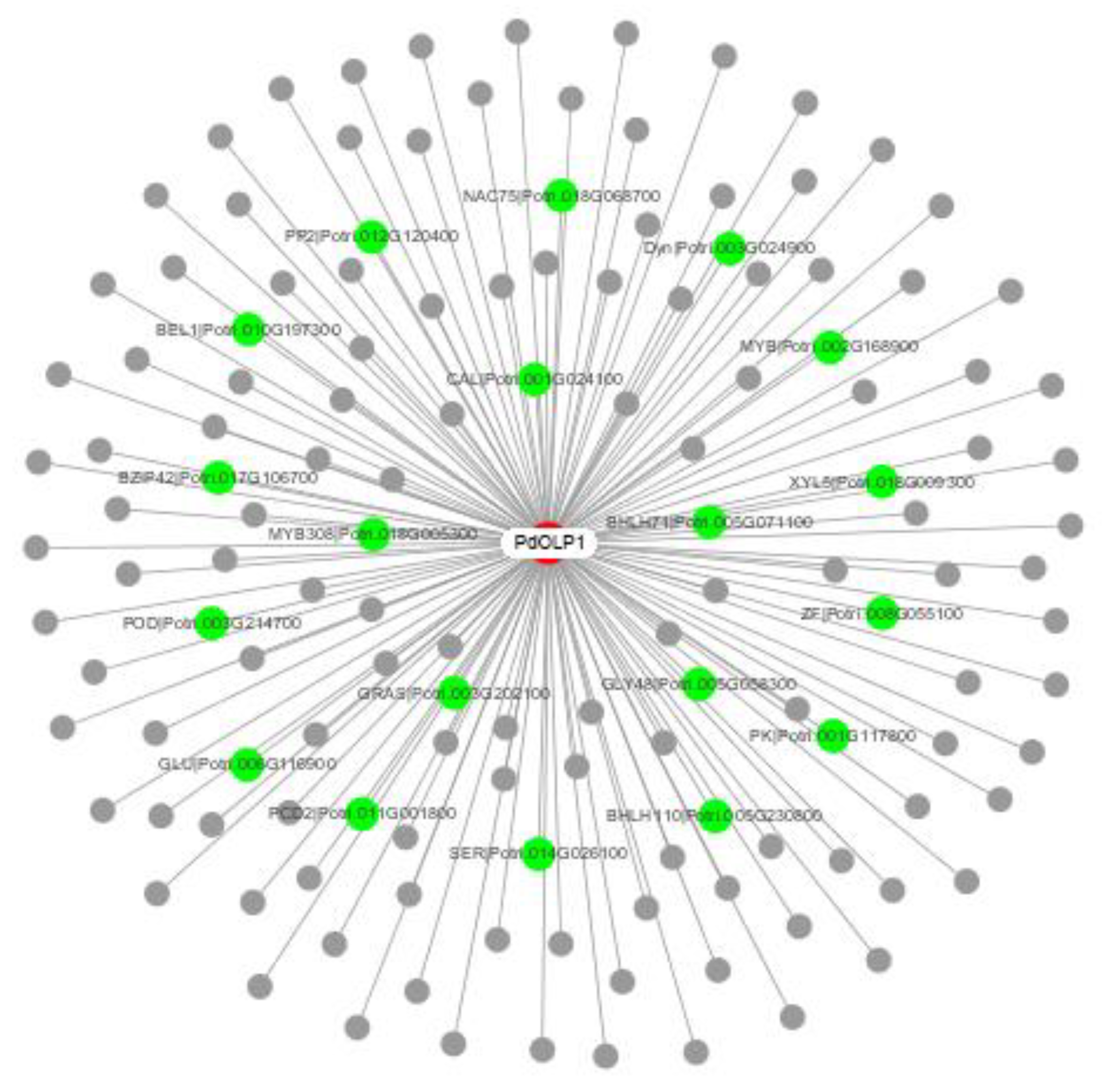
To elucidate the molecular mechanism of PdOLP1 in poplar growth and development, a PdOLP1 coexpression network was constructed (Figure 7 and Table 1). A total of 140 genes were identified in the coexpression network. Among these, 19 genes (15.70%) are associated with wood formation. For example, NACs are involved in the regulation of secondary cell wall biosynthesis and zinc finger protein (ZFs)-induced secondary cell wall thickening in the stem, bZIP can bind to the regulatory element that controls xylem-specific gene expression, glycosyl transferases participate in cell wall biosynthesis, and glucomannan-synthases are believed to have a role in hemicellulose biosynthesis. bHLH transcription factor regulates early xylem development, and MYBs play a vital role in regulating secondary wall formation; several bHLH and MYB transcription factor members including bHLH71, bHLH110, and MYB308 were coexpressed with PdOLP1. Furthermore, some genes related to protein kinase family proteins such as leucine-rich-repeat receptor-like kinase, which is associated with fiber development and secondary cell wall formation, were coexpressed with PdOLP1.
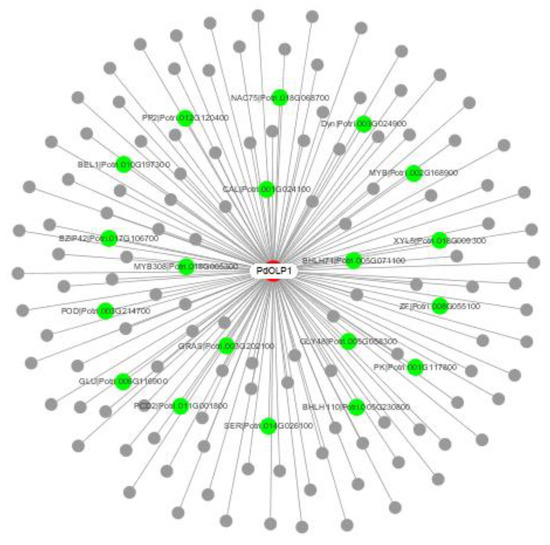
Figure 7.
PdOLP1 coexpression network. Among the 140 genes coexpressed with PdOLP1, 19 (green nodes) are involved in wood formation based on previous research. The white node is PdOLP1; gray nodes are other coexpressed genes.

Table 1.
Genes related to wood formation in the PdOLP1 coexpression network.
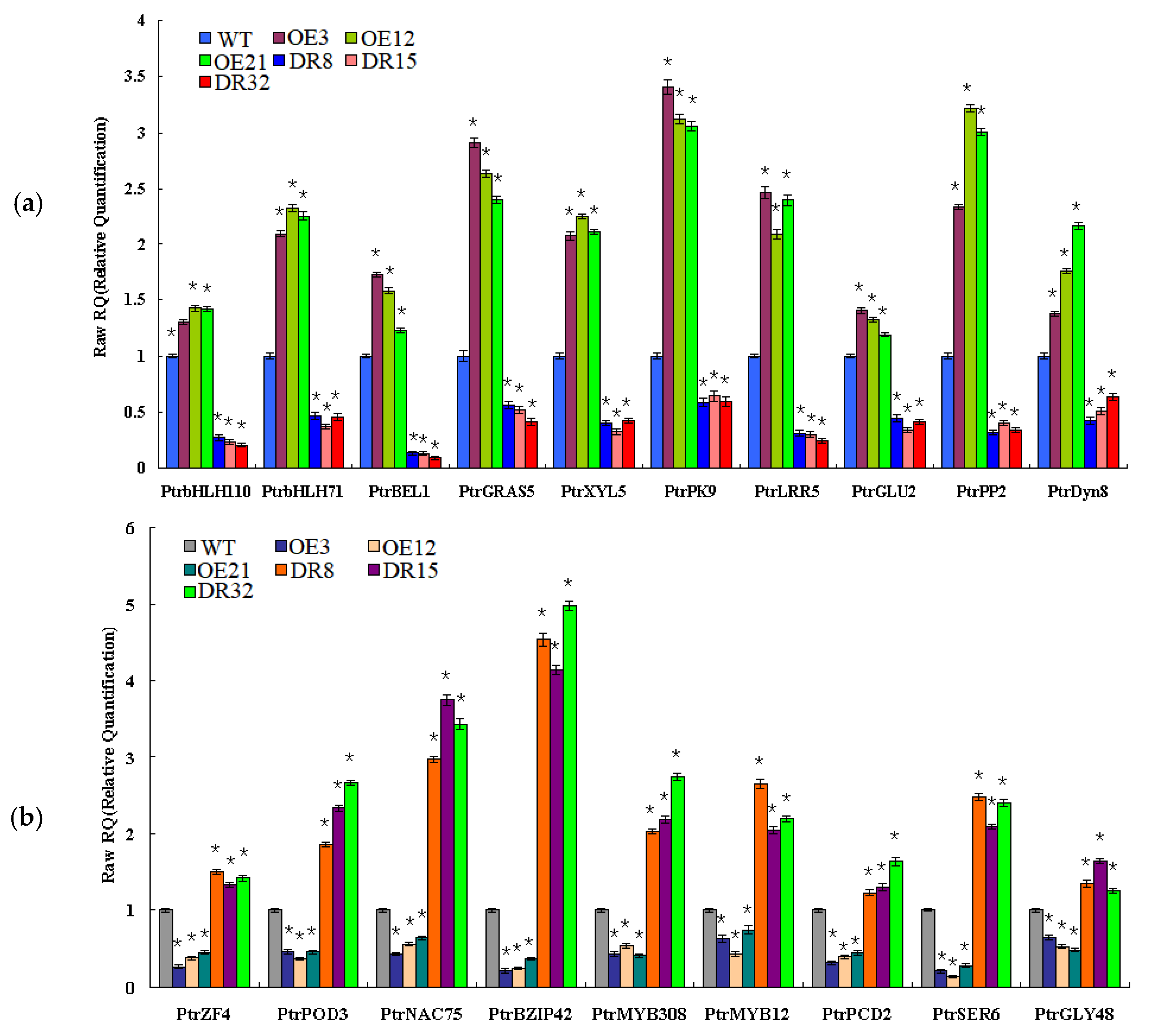
To validate the network constructed by the core of PdOLP1, changes in the expression of the 19 genes related to wood formation in the PdOLP1 coexpression network were compared in 3-month-old PdOLP1-OE, PdOLP1-DR and WT plants. Among the 19 genes, 10 were upregulated in PdOLP1-OE plants, and 9 were upregulated in PdOLP1-DR plants (Figure 8). Of the genes upregulated in PdOLP1-OE plants, 2 were upregulated by >2-fold in PdOLP1-OE lines, such as PtrPK9 and PtrPP2 (2.0- to 2.5-fold) (Figure 8). Of the upregulated genes in PdOLP1-DR plants, 3 were upregulated by >2-fold (p value <0.5), such as PtrNAC75 and PtrbZIP42 (2.0- to 4.0-fold) (Figure 8). The expression patterns of coexpressed genes in transgenic poplar plants suggest that PdOLP1 cooperates with a large number of wood-formation-related genes to regulate secondary growth and development.
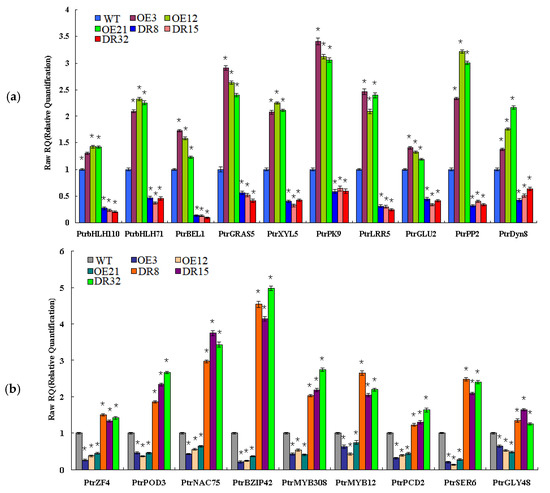
Figure 8.
Expression patterns of secondary cell wall biosynthesis genes from the coexpression network in PdOLP1-OE and PdOLP1-DR plants and poplar protoplasts. (a) Expression patterns of wood formation-related genes upregulated in PdOLP1-OE plants and downregulated in PdOLP1-DR plants. (b) Expression patterns of secondary wall biosynthesis genes upregulated in PdOLP1-DR plants and downregulated in PdOLP1-OE plants. UBQ1 was used as a reference gene, and the expression level of each gene in the WT background was set to 1. Asterisks indicate a significant difference between the WT and the transgenic lines (* p < 0.05).
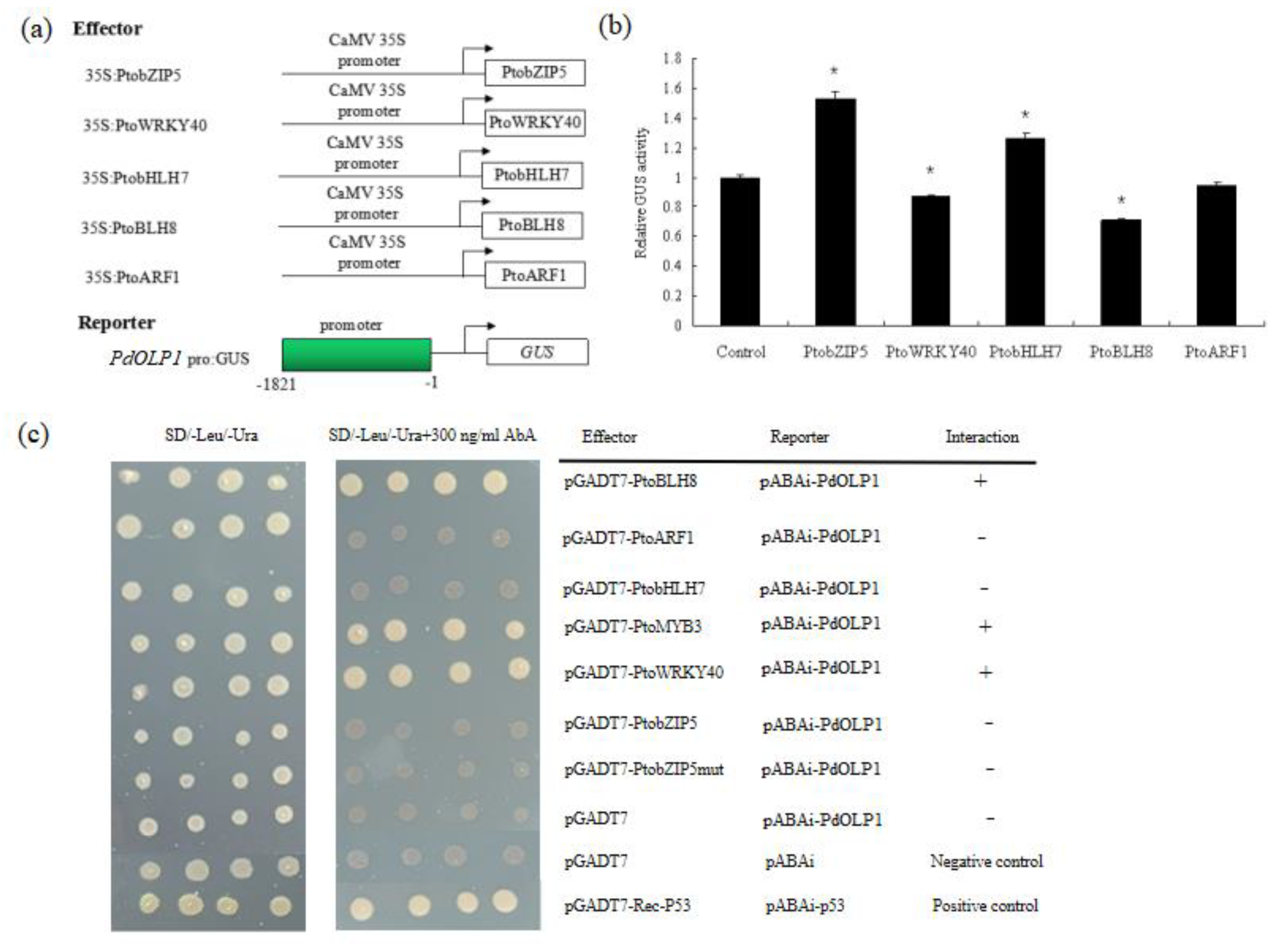
2.9. The PdOLP1 Promoter Is Activated by PtobZIP5 and PtobHLH7, and Inhibited by PtoBLH8 and PtoWRKY40
Several binding motifs such as “ACACNNG”, “TGAC”, and “CANNTG” were detected in the promoter of PdOLP1 (Table S9). This suggests that wood-associated transcription factors such as bZIP, WRKY, bHLH, BELL1-LIKE HOMEODOMAIN protein (BLH) and auxin response factor (ARF) may regulate PdOLP1 expression through promoter binding. In this study, we employed a transient expression assay in tobacco to investigate whether the PdOLP1 promoter can be activated or inhibited by PtobZIP5, PtoWRKY40, PtobHLH7, PtoBLH8 and PtoARF1. Subsequent assays of GUS activity in transiently transfected tobaccos demonstrated that PtobZIP5 and PtobHLH7 were able to activate expression of PdOLP1 and that PtoBLH8 and PtoWRKY40 were able to inhibit its expression. However, there were some differences in the activation or inhibition levels. PtobZIP5 and PtobHLH7 increased PdOLP1 activation by 53.36% and 26.63% compared to the control, respectively. PtoBLH8 and PtoWRKY40 decreased PdOLP1 expression by 29.27% and 13.42% compared to the control, respectively (Figure 9b).
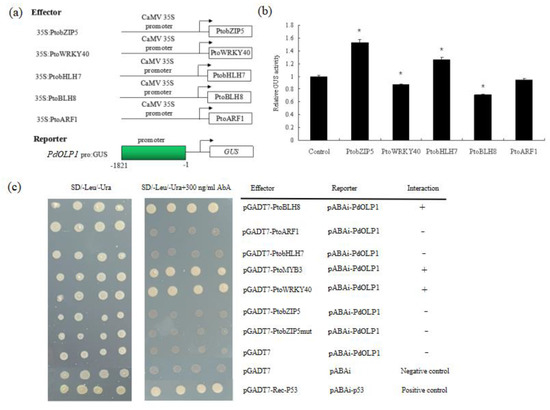
Figure 9.
Activation of the PdOLP1 promoter by PtobZIP5 and PtobHLH7 and inhibition by PtoBLH8 and PtoWRKY40. (a) Diagrams of the reporter and effector constructs used for transactivation analysis. (b) Transcriptional activity assays in tobacco leaves showed that PtoBLH8 and PtoWRKY40 repress PdOLP1 promoter-driven GUS expression and that PtobZIP5 and PtobHLH7 activate GUS reporter gene expression. GUS expression in tobacco leaves not transfected with any effector was used as a control and was set to 1. Values are the mean ± standard error (SE) of three biological replicates. (c) Yeast one-hybrid (Y1H) assay showing the interaction between PtoBLH8, PtoMYB3, and PtoWRKY40 and the PdOLP1 promoter. The yeast strains were grown on SD/-Leu/-Ura and SD/-Leu/-Ura +300 ng/mL Aureobasidin A (AbA) for 3 d.
Regulation of PdOLP1 gene expression by these transcription factors was confirmed by a Y1H assay, which detected interaction of the PtoBLH8, PtoMYB3, and PtoWRKY40 protein with the PdOLP1 promoter in vivo (Figure 9c). The full-length PtobZIP5, PtoWRKY40, PtoMYB3, PtobHLH7, PtoBLH8, and PtoARF1 gene and the PdOLP1 promoter fragments were cloned into pGADT7 and ABAi vectors, respectively. Although yeast strains carrying the pGADT7-PtoBLH8, pGADT7-PtoMYB3, and pGADT7-PtoWRKY40 constructs grew normally on selective medium, the empty vector control did not, indicating a direct interaction between PtoBLH8, PtoMYB3, and PtoWRKY40 protein with the PdOLP1 promoter, respectively (Figure 9c).
3. Discussion
Secondary walls of woody plants display various changes in their properties and composition [27,28]. These differences are reflected in the cell wall components and thickness, cell morphology and size, cell layers and width of the xylem or phloem region. Isolating and transforming genes that regulate secondary wall thickening and development in plants can be useful in analyzing the molecular mechanism of wood-formation-associated proteins. OLPs were previously found to play vital roles in plant organ or tissue development. Pluskota et al. (2019) reported that tomato (Solanum lycopersicum L.) SlNP24, encoding an osmotin protein, was mainly localized in the micropylar region of tomato seed endosperm, and plays an important role in germinating tomato seeds [29]. Previous studies have also identified that osmotin or OLPs accumulate in response to biotic and abiotic stresses, such as drought, salt, or cold stress, and possible mechanisms underlying the defense function of OLPs against biotic and abiotic stresses were proposed [11,12]. In the present study, we demonstrate a new mechanism by which the poplar OLP gene PdOLP1 is associated with secondary cell wall development, and the effect of PdOLP1 overexpression or downregulation on secondary cell wall formation was investigated in poplar.
According to the study of Raghothama et al. (1997), expression of the fusion vector consisting of an osmotin promoter fused with a GUS reporter gene was increased in the stem, leaves, and flowers of transgenic tobacco, with the greatest increases in the epidermis and vascular parenchyma of the stem [30]. Chowdhury et al. (2017) detected tissue-specific expression of sesame SindOLP in the stems, leaves, roots, and fruit of SindOLP-transgenic line-7 [31]. Wang et al. (2013) isolated hybrid poplar (Populus deltoides × P. euramericana cv. ‘Nanlin895′) PeTLP, which show high homology to OLP genes. Increased accumulation of PeTLP transcripts in stems but low transcription levels detected in roots and leaves have also been reported [9]. Additionally, Dafoe et al. (2010) [32] cloned hybrid poplar (Populus trichocarpa × P. deltoides) TLP1, which is homologous to OLPs, expressed predominantly in the vascular tissues of stems, midveins and petioles and localizes in phloem parenchyma cells [10]. In our research, the highest level of PdOLP1 expression was found in immature xylem and immature phloem (Figure 2a,b). This tissue-specific expression pattern of PdOLP1 in P. deltoides is in accordance with the expression of PeTLP in hybrid poplar (Populus deltoides × P. euramericana cv. ‘Nanlin895′) [9] and TLP1 in hybrid poplar (Populus trichocarpa × P. deltoides) [10]. Such preferential expression in immature xylem and phloem tissues indicates that PdOLP1 is involved in secondary wall development and wood formation in poplar.
Green fluorescent protein (GFP) is used as a marker of protein localization and provides a method to study gene function [33,34]. In recent years, stable or transient GFP expression has been carried out in the model plant Arabidopisis thaliana, rice, poplars and other plants [35,36]. DNA particle bombardment can be used to obtain transgenic poplar [37], rice cowpea [38], peanut [39], bean [40], and soybean [41]. Kim et al. (2010) [42] found that lemon (Citrus jambhiri Lush.) can also be used for this. RlemTLP encodes a thaumatin-like protein that is homologous to osmotin. Subcellular localization analysis showed that GFP-tagged RlemTLP predominantly localized to both the periphery of the cytoplasm and the plasma membrane [42]. In our study, the recombination protein PBI121-PdOLP1-GFP localized to the cytoplasm in onion epidermal cells (Figure 2e–l). This cytoplasm localization of PBI121-PdOLP1-GFP protein coincides with the subcellular localization of the OLP gene in other plants, such as lemon [42].
Expression effects of target genes under the control of the CaMV 35S promoter is stable and continuous, with no clear differences among various organs/tissues or between different developmental stages. However, constitutive and stable transgene expression is not beneficial to the long-term growth and development of transgenic plants [43,44]. Tissue-specific or inducible promoters can be used for driving target gene expression in specific tissues or locations. NAC transcription factors act as upstream switches associated with secondary cell wall development and vascular tissue differentiation [16,17,18]. In previous studies of ProNAC068::GUS-transgenic poplar plants (the promoter of P. deltoides PtoNAC068 replaced the CaMV 35S promoter), GUS expression was mainly detected in vascular tissues, especially in xylem regions of stem cross-sections compared to the untransformed plants [45]. Therefore, the ProNAC068 promoter is tissue specific, and can be used to study the function of target genes by regulating genes specifically expressed in vascular tissues. To further analyze the role of PdOLP1 in regulating secondary wall biosynthesis, we replaced the CaMV 35S promoter with the ProNAC068 promoter to drive PdOLP1 expression specifically in vascular tissues. Compared with untransformed poplar plants, expression of PdOLP1 was increased by 137.67–303.65% in PdOLP1-OE (PdOLP1 driven by the ProNAC068 promoter) poplar, and secondary wall deposition and expansion were detected in PdOLP1-OE poplar lines.
The sequence similarity of the four genes (PagOLP1, PagOLP2, PagOLP3, and PagOLP4) in hybrid poplar 84K is very high, more than 88%. Although we designed multiple pairs of primers, we unable to distinguish the expression of each gene. Therefore, we believe that the phenotype of PdOLP1-downregulated poplar is caused by inhibition of multiple endogenous genes of hybrid poplar 84K and does not merely involve suppression of the target gene (PdOLP1). Because of the high similarity of these endogenous genes, if we only inhibit the expression of one of the most homologous endogenous genes of hybrid poplar 84K, such as PagOLP1, the expression of other highly homologous endogenous genes can reverse the repressive effect of this endogenous gene (PagOLP1), and we may not observe a phenotype caused by gene inhibition. Antisense suppression can effectively inhibit the expression of several endogenous genes; thus, the function of the homologous gene cannot be complemented by other highly homologous genes. We generated PdOLP1-overexpressing and PdOLP1-downregulated hybrid lines. The phenotype of antisense inhibition is opposite to that of overexpression, which showed that the phenotype obtained by antisense inhibition was correct. Therefore, antisense inhibition was used to suppress multiple endogenous genes, resulting in a phenotype opposite to that of gene overexpression. Accordingly, the function of this gene can be confirmed from two aspects: gene overexpression and gene inhibition.
Plant cell walls have heterogeneous structures that affect xylem or phloem cell morphology and size and provide a supporting structure to promote plant growth and development [19,24]. Dafoe et al. (2010) detected expression of TLP (OLPs homologous gene) in sieve elements and phloem parenchyma cells by using a TLP antiserum [10]. Another study found that tomato OLP-transgenic plants exhibit increases in root biomass and total biomass both in the normal state and under stress conditions [43]. In our research, phloroglucinol–HCl staining and lignin autofluorescence revealed more fluorescent signals in PdOLP1-DR DR8 plants (Figure 5j,l,m,o), indicating that PdOLP1 downregulation induced lignin deposition. Moreover, PdOLP1 downregulation increased primary growth (such as stem elongation and enlargement) and dry weight biomass and facilitated secondary wall thickening and expansion in xylem zones. The radial widths and number of cell layers in the cambium, xylem and phloem tissues and the secondary cell wall thickness of vessels and fibers were significantly increased in PdOLP1-DR poplars (Figure 4c–e,g). This result is similar to the findings in Betula platyphylla BplMYB46-overexpressing birch plants, whereby the fresh weight, lignin deposition, and secondary cell wall thickness were increased in BplMYB46-overexpressing birch plants [46]. Together, these studies suggest that PdOLP1 may function as a negative regulator that promotes ligninification and secondary wall deposition and therefore plays a vital role in poplar development under normal or stress conditions.
Microfibril angle (MFA) is a wood chemical parameter characteristic of strength and stiffness [47]. A lower MFA indicates higher lumber strength and rigidity [48], which is valuable for improving wood mechanical properties in forest tree breeding. The MFA in Pinus patula was measured and it influences the modulus of elasticity of wood [49]. Derba-Maceluch et al. (2015) downregulated hybrid poplar (Populus tremula × tremuloides) PtxtXyn10A and altered the cellulose MFA in lumber fibers [50]. A previous study revealed that the MFA values in transgenic hybrid poplar (P. davidiana × P. bolleana) expressing antisense PdREM, PdRanBP, or PdCYTOB constructs were significantly lower those in untransformed plants [28,51,52]. In this research, the stem MFAs of PdOLP1-DR poplars were 12.69%–14.86% lower than in non-transgenic lines (Figure 4i). Therefore, PdOLP1 can contribute to physical mechanical strength improvement in poplar wood. Indeed, expression of several MFA-associated genes, such as PtrSuS1 [53], PtrFRA1 [54], PtrTUB7 [55], and PtrCCR7 [56], was increased in PdOLP1-DR plants, suggesting the functional mechanism by which the MFA was reduced in the transgenics plants.
To further elucidate the molecular mechanism of PdOLP1 in poplar, a coexpression network mediated by PdOLP1 was constructed (Figure 7 and Table 1). Consistent with the potential role of PdOLP1 as a negative regulator, most positive transcriptional regulators in the previously constructed network were repressed in PdOLP1-OE plants. For example, Zhong et al. (2011) [18] found that P. trichocarpa PtrNAC, PtrMYB, and PtrZF transcription factors can activate expression of lignin, xylan and cellulose biosynthesis genes in poplar. In our study, four (PtrNAC75, PtrZF4, PtrMYB308, and PtrMYB12) of these positive transcription factors were repressed in PdOLP1-OE plants, which may indirectly suggest that lignin synthesis genes were downregulated in PdOLP1-OE poplar plants. bHLH transcription factors are important regulators of development in plant. Yan et al. (2013) [57] overexpressed Sorghum bicolor SbbHLH1 in Arabidopsis thaliana, markedly downregulating the lignin biosynthesis genes COMT, HCT, PAL1, CCR1, and 4CL1, and reducing the lignin content of transgenic plants. In this study, two bHLH genes, PtrbHLH110 and PtrbHLH71, were upregulated in PdOLP1-OE plants and may downregulate the lignin synthesis genes and decrease the lignin content. Sasaki et al. (2006) found that P. alba cell wall peroxidase-cationic (CWPO-C) is essential for the lignification of the secondary xylem [58]. We found that PtrPOD3 was upregulated in PdOLP1-DR lines, indicating that peroxidase is responsible for promoting lignification of the cell wall in PdOLP1-DR poplar lines. Additionally, Liu et al. (2014) overexpressed PtrBEL1 in Arabidopsis and revealed a decreased cell wall thickness of interficular fibers (IF) compared to the wildtype plants and the IF secondary cell wall thickness was increased in bel1 relative to wildtype plants [21]. In our study, PtrBEL1 was overexpressed in PdOLP1-OE poplar plants, which may suggest that the thinner xylem fibers and vessels of the cell walls of PdOLP1-OE plants may be caused by PdOLP1 through upregulation of PtrBEL1. This result supports our hypothesis that overexpression of PdOLP1 influences the expression of genes involved in secondary wall biosynthesis, which are coexpressed with PdOLP1.
Because secondary wall biosynthetic genes were repressed by PdOLP1 in transgenic poplar and also regulated by transcription factors such as bHLH, BLH, and WRKY [21,57], we sought to determine whether secondary wall biosynthetic genes are regulated in through the transcription factor-PdOLP1 interaction. Transient expression assays in tobacco leaves were used, and we found that the PdOLP1 promoter could be activated by PtobHLH7 and PtobZIP5 and repressed by PtoWRKY40 and PtoBLH8 (Figure 9b). Yan et al. [57] (2013) found that S. bicolor SbbHLH1 overexpression in Arabidopsis caused downregulation of the lignin biosynthesis genes, which suggests that the bHLH transcription factor is a negative regulator of lignin biosynthesis in plants. In our research, PtobHLH7 was able to activate the promoter of PdOLP1 by using the transcriptional activity assays. Therefore, we deduce that bHLH may regulate expression of OLP to influence the lignin biosynthetic genes in poplars.
Furthermore, direct evidence for the binding of these transcription factors to the promoter of the PdOLP1 gene was obtained using a Y1H assay, which showed that PtoBLH8, PtoMYB3, and PtoWRKY40 could bind directly to the PdOLP1 promoter (Figure 9c). Accordingly, there is reason to believe that secondary wall biosynthetic genes are regulated through the transcription factor-PdOLP1 interaction. Important transcription factors in wood formation such as PtoBLH8, PtoMYB3, and PtoWRKY40 can interact with PdOLP1 and this interaction can further influence the expression of secondary wall biosynthetic genes in PdOLP1-overexpression or -knockdown lines, affecting secondary cell wall biosynthesis and wood development in poplar.
Taken together, this study provides new data to help understand the function of PdOLP1 as a negative regulator of the lignin biosynthesis pathway and secondary wall formation in poplar. PdOLP1 is an OLP that displays cytoplasmic localization and preferential expression in immature xylem and phloem tissues. Functional identification revealed that decreased expression of PdOLP1 increases the number of cell layers and radial width in the xylem and phloem regions, increases the expression of genes involved in secondary cell wall biosynthesis, and thickens the fibers and vessels of the xylem cell walls in downregulated lines. Moreover, overexpression of PdOLP1 resulted in thinner fibers and vessels of the cell walls in xylem cells. The coexpression network showed that PdOLP1 cooperates with secondary wall biosynthesis genes, providing a molecular basis to reveal the role of PdOLP1 participation in wood development. Furthermore, the PdOLP1 promoter is activated or inhibited by transcription factors related to wood formation, such as PtobHLH7, PtoBLH8 and PtoWRKY40; PtoBLH8, PtoMYB3 and PtoWRKY40 can directly regulate transcription of the PdOLP1 gene. This research may be valuable for cultivating new clones of tree species with an elevated wood yield and strength and improved bioenergy traits.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Fifteen-year-old Populus deltoides plants were used to isolate the PdOLP1 gene and analyze the gene expression pattern. Leaves, immature phloem, mature phloem, immature xylem, mature xylem, male flower buds and leaf buds were harvested three times from different regions of three P. deltoides plants for determination (Table S1). Hybrid poplar trees (Populus alba × Populus glandulosa) of clone 84K were used for genetic transformation experiments to characterize PdOLP1 function. Branches from the current year of a 15-year-old P. deltoides tree were chosen for in situ hybridization.
4.2. In Situ Hybridization
The 30-bp PdOLP1 nucleotide sequence was separately isolated and used for the synthesis of sense and antisense probes labeled with digoxigenin (DIG) following the DIG RNA Labeling mix instructions (Roche Diagnostics, Indianapolis, IN, USA). The tenth internodes of newly emerged branches of P. deltoides trees were sampled and fixed in FAA (formalin-acetic acid-alcohol) solution overnight. Stem specimens were embedded in paraffin wax and cut into 10-μm sections using a rotary Leica RM 6025 microtome (Leica Microsystems, Wetzlar, Germany) and hybridized with digoxigenin-labeled PdOLP1 sense or antisense probes (Table S2). Hybridization signals were immunohistochemically detected using alkaline phosphatase-conjugated antibodies against digoxigenin. Images were obtained under an inverted fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
4.3. Cloning of PdOLP1, Vector Construction and Genetic Transformation
Total RNA was extracted from the immature xylem (IX) of P. deltoides using RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) as described in the manufacturer’s instructions. The full-length cDNA fragment encoding PdOLP1 was amplified by PCR with gene-specific primers (Table S3) based on PtOLP (XM_024608084.1) from P. trichocarpa by PCR. The PCR products were subcloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and sequenced. The coding sequence (CDS) of PdOLP1 with the stop codon was inserted into the expression vector pBI121-ProNAC068-GUS (GUS expression driven by the ProNAC068 promoter was mainly detected in wood-forming tissues of poplar trees) in forward and reverse directions using the NEBuilder HiFi DNA Assembly master mix (New England BioLabs, E2621L) (the primers are listed in Table S3, Figure S1). These plant binary vectors (Figure S1) were transformed into Agrobacterium tumefaciens (strain GV3101) and subsequently transformed into hybrid poplar (P. alba × P. glandulosa) of clone 84K using the method described by Zhang et al. (2008) [79]. Rooted plantlets were acclimatized in a greenhouse at 25 °C under a 16-/8- h light/dark cycle.
4.4. Detection of Transgenic Plants
Transformed poplar plants were detected by PCR with gene-specific primers for the NptII gene (Table S3). Positive plants were further confirmed by Southern hybridization. Twenty-five micrograms of purified genomic DNA from transgenic and nontransformed poplar plants was digested with XbaI (New England Biolabs), followed by agarose gel electrophoresis and transfer to a nylon-based membrane (Hybond-N +, Amersham) using Vacuum Blotting Model 785 (BioRad, Hercules, CA, USA). The DNA fragment amplified from the NptII gene was labeled with biotin (BIO)-dUTP and used as a probe for hybridization [52]. Specific experimental methods followed the instructions of North2South Biotin Random Prime Kit and the chemiluminescent nucleic acid hybridization and detection kit (Pierce, Rockford, IL, USA).
4.5. Gene Expression Analyzed by Quantitative Real-Time PCR (qRT-PCR)
The level of PdOLP1 expression in different 15-year-old P. deltoides tissues and in the stems of 1.5-year-old PdOLP1 transgenic lines was detected by qRT-PCR using the primers listed in Table S3. qRT-PCR was performed using an ABI Prism 7500 sequence detector (Applied Biosystems, Foster City, CA, USA) with SYBR ® Premix Ex TaqTM Kit (TaKaRa, Dalian, China); α-tubulin (TUA1) and ubiquitin (UBQ1) were used as control genes [80] for internal standardization of qRT-PCR data. Each PCR (final volume 20 μL) contained 1× SYBR Green PCR Master Mix, 200 nM of primers, and 1 μL of first-strand cDNA. Three replicates were conducted in parallel, and data analysis was performed following ABI Prism 7500 Sequence Detection System Users Guide. PCR primers for wood formation genes and genes coexpressed with PdOLP1 were designed using Primer Premier 5.0 software (Premier Biosoft Int., Palo Alto, CA, USA), as listed in Table S4 and Table S5. Each gene was measured with three biological and three technical replicates, and error bars represent the standard error (SE) of the three replicates. The relative expression levels of target genes were analyzed using the 2−ΔΔCT method (the comparative Ct method) [81]. Standard curves were analyzed to compare the gene-specific PCR efficiency from 10-fold series dilutions of the mixed cDNA templates for each primer pair. Slope values and correlation coefficients (R2) were calculated from the standard curve, and the PCR amplification efficiencies (E) were analyzed by the following formula: E = (10−1/slope-1) × 100 [82].
4.6. Subcellular Localization
The full-length coding sequence (CDS) of PdOLP1 without the termination codon was amplified and ligated to the N-terminus of GFP driven by the cauliflower mosaic virus (CaMV) 35S promoter. The vector 35S-GFP was transformed simultaneously in parallel as a control. All of these constructs were transformed into onion epidermal cells via particle bombardment [83]. The onion epidermis was mounted onto glass slides. For the DNA construct, 2 µg of DNA was mixed with 50 µL of 2.5 M CaCl2, 25 µL of 0.1 M spermidine, and 50 µL of gold microcarrier, vortexed vigorously for 2.5 min, and centrifuged at 10,000 rpm (5810R, Eppendorf, Germany) in a microcentrifuge for 15 s. The pellet was washed with 180 µL of ethanol and then resuspended in 30 µL of ethanol. The DNA-gold particles were bombarded into cells at a pressure of 1,100 lb/in2 using the Biolistic Particle Delivery System 1000/He (Bio-Rad). The nuclei of of the transfected onion cells were stained with 4′,6-diamidino-2-phenylindole (DAPI), and GFP fluorescent signals were detected using an inverted fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
4.7. Histological Analysis and Microscopy
The 15th internodes on the stems of 1.5-year-old poplar plants were fixed in FAA buffer and then embedded in paraffin. The stem segments were cut into 8-μm-thick sections with a microtome (Leica Microsystems, Wetzlar, Germany) and stained with 0.05% (w/v) toluidine blue O or stained for lignin with phloroglucinol-HCl, followed cby observation under an inverted fluorescence microscope (Carl Zeiss, Oberkochen, Germany). Lignin autofluorescence was also assessed under a confocal laser microscope (Zeiss, Jena, Germany). We used 0.2-cm-thick stem sections of PdOLP1 transgenic and nontransgenic lines for scanning electron microscopy (S-4800, HITACHI, Tokyo, Japan). Secondary wall thickness of vessels and fibers was measured, and at least three transformed lines were measured.
4.8. Microfibril Angle Measurement
We measured the angles of stem blocks of 1.5-year-old transformed and non-transformed poplar lines at 5 cm above the pot. The microfibril angle (MFA) was measured as described previously [84]. Glacial acetic acid/hydrogen peroxide solution (1:1, v/v) was used to dissociate the fibers of different samples at 60 °C overnight, and polarized microscopy was used to measure individual fibers of each sample (Olympus BX51; Olympus, Melville, NY, USA).
4.9. Determination of Lignin Content, Wood Density, Total Biomass, and Carbon Storage
Lignin content of 1.5-year-old transformed and non-transformed poplar plants was evaluated using dry extract-free cell wall residue (CWR), which was ground to powder and passed through a 0.5-mm sieve before solvent extraction (2:1 [v/v] toluene:ethanol, ethanol and then water). The lignin contents of basal stem segments from transgenic and nontransgenic plants were measured according to the Klason procedure [85]. Three biological replicates were performed. Drainage method is used to measure volume, drying method is used to measure wood quality and total biomass, and basic density is determined by wood quality/wood volume. The specific steps refer to the methods of national standards of GB/T 1933–2009 Determination of wood density. We crushed, mixed, and dried the sample, and 2mg was used for testing of carbon storage by using Vario EL cube element analyzer. For detailed methods as described previously [86].
4.10. Phylogenetic Analysis and Statistical Analyses
The CDSs of OLPs were identified from the website (available online: http://www.phytozome.com, accessed on: 6 June 2019) and the Swedish Center for Tree Functional Genomics project database (Populus DB). Multiple sequence alignments were generated with the Clustal W program (available online: www.ebi.ac.uk/clustalw/, accessed on: 10 June 2019). The phylogenetic relationships of OLPs were performed using MEGA version 7.0 [87] by the neighbor-joining method. Measurement data for plant height, cell wall thickness, and lignin content, etc, were subjected to statistical analysis using one-way analysis of variance. Quantitative differences between two groups of data in all comparisons were statistically significant (p < 0.05; ANOVA, Fisher test).
4.11. Transient Expression Assay
Promoter fragments of the PdOLP1 gene were amplified by PCR using the gene-specific primers listed in Table S3. The amplified fragments were fused to the GUS reporter gene in the modified pCAMBIA 1305 vector to generate reporter constructs. To create the effector constructs, the PtoWRKY40, PtoARF1, PtobZIP5, PtoBLH8, and PtobHLH7 coding regions were amplified from poplar (Populus tomentosa) xylem cDNA (primers in Table S3) and ligated between the 35S promoter and the NOS terminator after removing GUS from the pBI121 vector. Tobacco (Nicotiana benthamiana) leaves were injected with Argrobacterium cells containing the reporter and effector plasmids [88]. Briefly, A. tumefaciens GV3101 carrying the constructed plasmids was cultured to OD600 = 0.5; the samples were combined at equal volumes, incubated at room temperature without shaking for 3 h and infiltrated into N. benthamiana leaves. The GUS expression level was measured by spectrophotometry [89]. In each experiment, the expression level of the GUS reporter gene in N. benthamiana leaves transfected with the reporter construct alone was used as the control. Data are presented as the mean ± SE of three independent experiments.
4.12. Yeast One-Hybrid Assay
The yeast one-hybrid assay was performed by using Matchmaker Gold Yeast One-Hybrid System (Clontech, http://www.clontech.com/, accessed on: 3 January 2020) following the manufacturer’s instructions. The promoter sequence of the PdOLP1 gene was ligated to the vector pAbAi (Y1H Gold), and the plasmid was linearized and integrated into the yeast genome. Yeast cells were then transformed with the pGADT7-AD vector (Y1H Gold) carrying the PtoWRKY40, PtoARF1, PtobZIP5, PtoMYB3, PtoBLH8, or PtobHLH7 coding sequence. The protein–DNA interaction was determined based on the ability of the transformed yeast to grow on SD/-Leu/-Ura medium with Aureobasidin A (AbA) according to the manufacturer’s protocol.
4.13. Coexpression Network Analysis
The coexpression data for PdOLP1 were obtained from Phytozome (available online: https://phytozome.jgi.doe.gov/pz/portal.html, accessed on: 15 July 2019). For Populus genome-wide coexpression network construction, transcriptome data from 24 various P. trichocarpa tissues [90] were used. Genes with Pearson correlation coefficients >0.85 were selected to build the coexpression network of PdOLP1. The network diagram was generated according to Cytoscape software [91] (The Cytoscape v1.1 Core was downloaded from http://www.cytoscape.org/, accessed on: 27 July 2019).
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/11/3993/s1.
Author Contributions
Conceptualization, S.L. and J.Z.; investigation, S.L., F.L., W.M. and X.G.; resources, S.W. and J.C.; writing—original draft preparation, S.L.; writing—review and editing, S.L. and J.Z.; validation, L.S., M.D., C.D. (Chenxi Dong), and X.D.; project administration, C.D. (Changjun Ding), J.S., Y.Z., X.X. and D.D.; funding acquisition, Y.X. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key Program on Transgenic Research (Grant No. 2018ZX08020002), the National Natural Science Foundation of China (Grant No. 31700531, 31770705 and 31400570), and the Open Fund of State Key Laboratory of Tree Genetics and Breeding (Chinese Academy of Forestry) (Grant No. TGB2013002).
Acknowledgments
We are grateful to Xiaohua Su (Research Institute of Forestry, Chinese Academy of Forestry), Mengzhu Lu and Junhui Wang (Research Institute of Forestry, Chinese Academy of Forestry) for kind help and valuable suggestions.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Gorshkova, T.; Brutch, N.; Chabbert, B.; Deyholos, M.; Hayashi, T.; Lev-Yadun, S.; Mellerowicz, E.J.; Morvan, C.; Neutelings, G.; Pilate, G. Plant Fiber Formation: State of the Art, Recent and Expected Progress, and Open Questions. Crit. Rev. Plant Sci. 2012, 31, 201–228. [Google Scholar] [CrossRef]
- Schuetz, M.; Smith, R.; Ellis, B. Xylem tissue specification, patterning, and differentiation mechanisms. J. Exp. Bot. 2012, 64, 11–31. [Google Scholar] [CrossRef] [PubMed]
- Hertzberg, M.; Aspeborg, H.; Schrader, J.; Andersson, A.; Erlandsson, R.; Blomqvist, K.; Bhalerao, R.; Uhlén, M.; Teeri, T.T.; Lundeberg, J.; et al. A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. USA 2001, 98, 14732–14737. [Google Scholar] [CrossRef]
- Yang, J.; Park, S.; Kamdem, D.P.; Keathley, D.E.; Retzel, E.; Paule, C.; Kapur, V.; Han, K.-H. Novel gene expression profiles define the metabolic and physiological processes characteristic of wood and its extractive formation in a hardwood tree species, Robinia pseudoacacia. Plant Mol. Biol. 2003, 52, 935–956. [Google Scholar] [CrossRef]
- Dungey, H.S.; Matheson, A.C.; Kain, D.; Evans, R. Genetics of wood stiffness and its component traits inPinus radiata. Can. J. For. Res. 2006, 36, 1165–1178. [Google Scholar] [CrossRef]
- Choi, D.S.; Hong, J.K.; Hwang, B.K. Pepper osmotin-like protein 1 (CaOSM1) is an essential component for defense response, cell death, and oxidative burst in plants. Planta 2013, 238, 1113–1124. [Google Scholar] [CrossRef]
- Byoun, B.H.; Kim, S.G. Isolation and Molecular Characterization of the Osmotin gene in Petunia hybrida. Genes Genom. 1997, 19, 340. [Google Scholar]
- Xue, X.; Cao, Z.X.; Zhang, X.T.; Wang, Y.; Zhang, Y.F.; Chen, Z.X.; Pan, X.B.; Zuo, S.M. Overexpression of OsOSM1 Enhances Resistance to Rice Sheath Blight. Plant Dis. 2016, 100, 1634–1642. [Google Scholar] [CrossRef]
- Wang, L.; Yang, L.; Zhang, J.; Dong, J.; Yu, J.; Zhou, J.; Zhuge, Q. Cloning and characterization of a thaumatin-like protein gene PeTLP in Populus deltoides × P. euramericana cv. ‘Nanlin895’. Acta Physiol. Plant. 2013, 35, 2985–2998. [Google Scholar] [CrossRef]
- Dafoe, N.J.; Gowen, B.E.; Constabel, C.P. Thaumatin-like proteins are differentially expressed and localized in phloem tissues of hybrid poplar. BMC Plant Biol. 2010, 10, 191. [Google Scholar] [CrossRef]
- Gimeno, J.; Gadea, J.; Forment, J.; Valle, J.P.; Santiago, J.; Martínez-Godoy, M.A.; Yenush, L.; Bellés, J.M.; Brumos, J.; Colmenero-Flores, J.; et al. Shared and novel molecular responses of mandarin to drought. Plant Mol. Biol. 2009, 70, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Vasavirama, K.; Kirti, P.B. Increased resistance to late leaf spot disease in transgenic peanut using a combination of PR genes. Funct. Integr. Genom. 2012, 12, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.A.; Kumari, P.H.; Jawahar, G.; Prashanth, S.; Suravajhala, P.; Katam, R.; Sivan, P.; Rao, K.S.; Kirtie, P.B.; Kishora, P.B.K. Beyond just being foot soldiers-osmotin like protein (OLP) and chitinase (Chi11) genes act as sentinels to confront salt, drought, and fungal stress tolerance in tomato. Environ. Exp. Bot. 2016, 132, 53–65. [Google Scholar] [CrossRef]
- Abdin, M.Z.; Kiran, U.; Alam, A. Analysis of osmotin, a PR protein as metabolic modulator in plants. Bioinformation 2011, 5, 336–340. [Google Scholar] [CrossRef]
- Campos, M.D.; Silva, M.S.; Magalhaes, C.P.; Ribeiro, S.G.; Sarto, R.P.D.; Vieira, E.A.; Grossi, M.F.S. Expression in Escherichia coli, purification, refolding and anti-fungal activity of an osmotin from Solanum nigrum. Microb. Cell Factories 2008, 7, 7. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z. The poplar PtrWNDs are transcriptional activators of secondary cell wall biosynthesis. Plant Signal. Behav. 2010, 5, 469–472. [Google Scholar] [CrossRef]
- Zhong, R.; McCarthy, R.L.; Lee, C.; Ye, Z. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 2011, 157, 1452–1468. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, X.; Yang, F.; Fan, D.; Jiang, Y.; Luo, K. PtrWRKY19, a novel WRKY transcription factor, contributes to the regulation of pith secondary wall formation in Populus trichocarpa. Sci. Rep. 2016, 6, 18643. [Google Scholar] [CrossRef]
- Li, C.; Wang, X.; Ran, L.; Tian, Q.; Fan, D.; Luo, K. PtoMYB92 is a Transcriptional Activator of the Lignin Biosynthetic Pathway During Secondary Cell Wall Formation in Populus tomentosa. Plant Cell Physiol. 2015, 56, 2436–2446. [Google Scholar] [CrossRef]
- Liu, Y.; You, S.; Taylor-Teeples, M.; Li, W.L.; Schuetz, M.; Brady, S.M.; Douglas, C.J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 Interact and Regulate Secondary Cell Wall Formation via Repression of REVOLUTA. Plant Cell 2014, 26, 4843–4861. [Google Scholar] [CrossRef]
- Sun, X.; Wang, C.; Xiang, N.; Li, X.; Yang, S.; Du, J.; Yang, Y.; Yang, Y. Activation of secondary cell wall biosynthesis by miR319-targeted TCP4 transcription factor. Plant Biotechnol. J. 2017, 15, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Scully, E.; Gries, T.; Palmer, N.A.; Sarath, G.; Funnell-Harris, D.L.; Baird, L.M.; Twigg, P.; Seravalli, J.; Clemente, T.E.; Sattler, S.E. Overexpression of SbMyb60 in Sorghum bicolor impacts both primary and secondary metabolism. New Phytol. 2017, 217, 82–104. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, Y.; Ding, C.; Gao, X.; Wang, R.; Mo, W.; Lv, F.; Wang, S.; Liu, L.; Tang, Z.; et al. Proline-rich protein gene PdPRP regulates secondary wall formation in poplar. J. Plant Physiol. 2019, 233, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Bai, S.; Ma, J.; Zhang, L.; Shao, F.; Zhang, K.; Yang, Y.; Sun, T.; Huang, J.; Zhou, Y.; et al. The genome of Populus alba × Populus tremula var. glandulosa clone 84K. DNA Res. 2019, 26, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Lemus, R.; Lal, R. Bioenergy Crops and Carbon Sequestration. Crit. Rev. Plant Sci. 2005, 24, 1–21. [Google Scholar] [CrossRef]
- Showalter, A.M. Structure and function of plant cell wall proteins. Plant Cell 1993, 5, 9–23. [Google Scholar]
- Li, S.-F.; Huang, Q.; Zhang, B.; Zhang, J.; Liu, X.; Lu, M.; Hu, Z.; Ding, C.; Su, X. Small GTP-binding protein PdRanBP regulates vascular tissue development in poplar. BMC Genet. 2016, 17, 96. [Google Scholar] [CrossRef]
- Pluskota, W.; Pupel, P.; Głowacka, K.; Okorska, S.B.; Jerzmanowski, A.; Nonogaki, H.; Górecki, R.J. Jasmonic acid and ethylene are involved in the accumulation of osmotin in germinating tomato seeds. J. Plant Physiol. 2019, 232, 74–81. [Google Scholar] [CrossRef]
- Raghothama, K.G.; Maggio, A.; Narasimhan, M.L.; Kononowicz, A.K.; Wang, G.; D’Urzo, M.P.; Hasegawa, P.M.; Bressan, R.A. Tissue-specific activation of the osmotin gene by ABA, C2H4 and NaCl involves the same promoter region. Plant Mol. Biol. 1997, 34, 393–402. [Google Scholar] [CrossRef]
- Chowdhury, S.; Basu, A.; Kundu, S. Overexpression of a New Osmotin-Like Protein Gene (SindOLP) Confers Tolerance against Biotic and Abiotic Stresses in Sesame. Front. Plant Sci. 2017, 8, 410. [Google Scholar] [CrossRef] [PubMed]
- Ibragimova, N.N.; Mokshina, N.E.; Gorshkova, T. Cell wall proteins of flax phloem fibers. Russ. J. Bioorg. Chem. 2012, 38, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [PubMed]
- Reichel, C.; Mathur, J.; Eckes, P.; Langenkemper, K.; Koncz, C.; Schell, J.; Reiss, B.; Maas, C. Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc. Natl. Acad. Sci. USA 1996, 93, 5888–5893. [Google Scholar] [CrossRef]
- Zhang, D.; Xu, Z.; Cao, S.; Chen, K.; Li, S.; Liu, X.; Gao, C.; Zhang, B.; Zhou, Y. An uncanonical CCCH tandem Zinc-Finger protein represses secondary wall synthesis and controls mechanical strength in rice. Mol. Plant 2018, 11, 163–174. [Google Scholar] [CrossRef]
- DeVantier, Y.A.; Moffatt, B.; Jones, C.; Charest, P.J. Microprojectile-mediated DNA delivery to the Salicaceae family. Can. J. Bot. 1993, 71, 1458–1466. [Google Scholar] [CrossRef]
- Su, X.; Chu, Y.; Li, H.; Hou, Y.; Zhang, B.; Huang, Q.; Hu, Z.; Huang, R.; Tian, Y. Expression of multiple resistance genes enhances tolerance to environmental stressors in transgenic poplar (Populus × euramericana ‘Guariento’). PLoS ONE 2011, 6, e24614. [Google Scholar] [CrossRef]
- Ikea, J.; Ingelbrecht, I.; Uwaifo, A.; Thottappilly, G. Stable gene transformation in cowpea (Vigna unguiculata L. Walp.) using particle gun method. Afr. J. Biotechnol. 2003, 2, 211–218. [Google Scholar]
- Brar, G.S.; Cohen, B.A.; Vick, C.L.; Johnson, G.W. Recovery of transgenic peanut (Arachis hypogaea L.) plants from elite cultivars utilizing ACCELLR technology. Plant J. 1994, 5, 745–753. [Google Scholar] [CrossRef]
- Russell, D.; Wallace, K.; Bathe, J.; Martinell, B.; McCabe, D. Stable transformation of Phaseolus vulgaris via electric-discharge mediated particle acceleration. Plant Cell Rep. 1993, 12, 165–169. [Google Scholar] [CrossRef]
- Christou, P.; McCabe, D.E.; Swain, W.F. Stable Transformation of Soybean Callus by DNA-Coated Gold Particles. Plant Physiol. 1988, 87, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-G.; Fukumoto, T.; Tatano, S.; Gomi, K.; Ohtani, K.; Tada, Y.; Akimitsu, K. Molecular cloning and characterization of a thaumatin-like protein-encoding cDNA from rough lemon. Physiol. Mol. Plant Pathol. 2009, 74, 3–10. [Google Scholar] [CrossRef]
- He, G.; Wang, M.; Chen, H.; Zhao, S.; Lu, M. Cloning and functional analysis of a vascular tissue-specific promoter from Populus tomentosa. For. Res. 2010, 23, 77–183. [Google Scholar]
- Li, S.F.; Fan, C.M.; Li, Y.; Zhang, J.H.; Sun, J.S.; Chen, Y.H.; Tian, C.Y.; Su, X.H.; Lu, M.Z.; Liang, C.Z.; et al. Effects of drought and salt-stresses on gene expression in Caragana korshinskii seedlings revealed by RNA-seq. BMC Genom. 2016, 17, 200. [Google Scholar] [CrossRef]
- Han, X.; He, G.; Zhao, S.; Guo, C.; Lu, M. Expression Analysis of Two NAC Transcription Factors PtNAC068 and PtNAC154 from Poplar. Plant Mol. Biol. Rep. 2011, 30, 370–378. [Google Scholar] [CrossRef]
- Guo, H.; Wang, Y.; Wang, L.; Hu, P.; Wang, Y.; Jia, Y.; Zhang, C.; Zhang, Y.; Zhang, Y.; Wang, C.; et al. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2016, 15, 107–121. [Google Scholar] [CrossRef]
- Cave, I.D.; Walker, J.C.F. Stiffness of wood in fast-grown plantation softwoods, the influence of microfibril angle. For. Prod. J. 1994, 44, 43–48. [Google Scholar]
- Evans, R.; Ilic, J. Rapid prediction of wood stiffness from microfibril angle and density. For. Prod. J. 2001, 51, 53–57. [Google Scholar]
- Wessels, B.; Malan, F.S.; Kidd, M.; Rypstra, T. The variation of microfibril angle in South African grown Pinus patula and its influence on the stiffness of structural lumber. South. For. J. For. Sci. 2015, 77, 213–219. [Google Scholar] [CrossRef]
- Derba-Maceluch, M.; Awano, T.; Takahashi, J.; Lucenius, J.; Ratke, C.; Kontro, I.; Busse-Wicher, M.; Kosík, O.; Tanaka, R.; Winzell, A.; et al. Suppression of xylan endotransglycosylasePtxtXyn10Aaffects cellulose microfibril angle in secondary wall in aspen wood. New Phytol. 2014, 205, 666–681. [Google Scholar] [CrossRef]
- Li, S.F.; Su, X.H.; Zhang, B.Y.; Huang, Q.J.; Chu, Y.G.; Ding, C.J. Functional identification of wood-property candidate gene PdCYTOB in Populus deltoides. Chin. Bull. Bot. 2011, 46, 642–651. [Google Scholar]
- Li, S.; Su, X.; Zhang, B.; Huang, Q.; Hu, Z.; Lu, M. Molecular cloning and functional analysis of the Populus deltoides remorin gene PdREM. Tree Physiol. 2013, 33, 1111–1121. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coleman, H.D.; Yan, J.; Mansfield, S.D. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. USA 2009, 106, 13118–13123. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Burk, D.H.; Morrison, W.H.; Ye, Z.-H. A Kinesin-Like Protein Is Essential for Oriented Deposition of Cellulose Microfibrils and Cell Wall Strength. Plant Cell 2002, 14, 3101–3117. [Google Scholar] [CrossRef] [PubMed]
- Spokevicius, A.V.; Southerton, S.G.; Macmillan, C.; Qiu, D.; Gan, S.; Tibbits, J.F.; Moran, G.F.; Bossinger, G. β-tubulin affects cellulose microfibril orientation in plant secondary fibre cell walls. Plant J. 2007, 51, 717–726. [Google Scholar] [CrossRef]
- Thumma, B.; Nolan, M.F.; Evans, R.; Moran, G.F. Polymorphisms in Cinnamoyl CoA Reductase (CCR) Are Associated With Variation in Microfibril Angle in Eucalyptus spp. Genetics 2005, 171, 1257–1265. [Google Scholar] [CrossRef]
- Zhou, G.K.; Zhong, R.; Richardson, E.; Morrison, W.; Nairn, C.; Wood-Jones, A.; Ye, Z.H. The Poplar Glycosyltransferase GT47C is Functionally Conserved with Arabidopsis Fragile Fiber8. Plant Cell Physiol. 2006, 47, 1229–1240. [Google Scholar] [CrossRef]
- Sasaki, S.; Baba, K.; Nishida, T.; Tsutsumi, Y.; Kondo, R. The cationic cell-wall-peroxidase having oxidation ability for polymeric substrate participates in the late stage of lignification of Populus alba L. Plant Mol. Biol. 2006, 62, 797–807. [Google Scholar] [CrossRef]
- Yan, L.; Xu, C.; Kang, Y.; Gu, T.; Wang, D.; Zhao, S.; Xia, G. The heterologous expression in Arabidopsis thaliana of sorghum transcription factor SbbHLH1 downregulates lignin synthesis. J. Exp. Bot. 2013, 64, 3021–3032. [Google Scholar] [CrossRef]
- He, Z.-H.; Fujiki, M.; Kohorn, B.D. A Cell Wall-associated, Receptor-like Protein Kinase. J. Biol. Chem. 1996, 271, 19789–19793. [Google Scholar] [CrossRef]
- Hematy, K.; Sado, P.-E.; Van Tuinen, A.; Rochange, S.; Desnos, T.; Balzergue, S.; Pelletier, S.; Renou, J.-P.; Höfte, H. A Receptor-like Kinase Mediates the Response of Arabidopsis Cells to the Inhibition of Cellulose Synthesis. Curr. Biol. 2007, 17, 922–931. [Google Scholar] [CrossRef]
- Chai, G.; Qi, G.; Cao, Y.; Wang, Z.; Yu, L.; Tang, X.; Yu, Y.; Wang, D.; Kong, Y.; Zhou, G. Poplar PdC3H17 and PdC3H18 are direct targets of PdMYB3 and PdMYB21, and positively regulate secondary wall formation in Arabidopsis and poplar. New Phytol. 2014, 203, 520–534. [Google Scholar] [CrossRef] [PubMed]
- Scheible, W.-R.; Pauly, M. Glycosyltransferases and cell wall biosynthesis: Novel players and insights. Curr. Opin. Plant Biol. 2004, 7, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Sun, J.; Xia, G.-X. Cloning and characterization of a gene for an LRR receptor-like protein kinase associated with cotton fiber development. Mol. Genet. Genom. 2005, 273, 217–224. [Google Scholar] [CrossRef]
- Kobayashi, H.; Fukuda, H. Involvement of calmodulin and calmodulin-binding proteins in the differentiation of tracheary elements in Zinnia cells. Planta 1994, 194, 388–394. [Google Scholar] [CrossRef]
- Barcelo, A.R. Peroxidase and not laccase is the enzyme responsible for cell wall lignification in the secondary thickening of xylem vessels in Lupinus. Protoplasma 1995, 186, 41–44. [Google Scholar]
- Yin, Y.F.; Jiang, X.M. Effect of peroxidase location in cell wall on lignification progress of secondary xylem cell of Populus tomentosa Carr. J. Chin. Electron Microsc. Soc. 2007, 1, 49–54. [Google Scholar]
- Fukuda, H. Programmed cell death during vascular system formation. Cell Death Differ. 1997, 4, 684–688. [Google Scholar] [CrossRef][Green Version]
- Ohashi-lto, K.; Oda, Y.; Fukuda, H. Arabidopsis VASCULAR-RELATED NAG-DOMAIN6 Directly Regulates the Genes That Govern Programmed Cell Death and Secondary Wall Formation during Xylem Differentiation. Plant Cell 2010, 22, 3461–3473. [Google Scholar] [CrossRef] [PubMed]
- Engstrom, E.M.; Andersen, C.M.; Gumulak-Smith, J.; Hu, J.; Orlova, E.; Sozzani, R.; Bowman, J.L. Arabidopsis homologs of the petunia hairy meristem gene are required for maintenance of shoot and root indeterminacy. Plant Physiol. 2010, 155, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Stuurman, J.; Jaggi, F.; Kuhlemeier, C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002, 16, 2213–2218. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; York, W.S.; Darvill, A.G. Important new players in secondary wall synthesis. Trends Plant Sci. 2006, 11, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Richardson, E.A.; Ye, Z. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef]
- Pyo, H.; Demura, T.; Fukuda, H. Vascular cell expression patterns of Arabidopsis bZIP group I genes. Plant Biotechnol. 2006, 23, 497–501. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, X.; Ran, L.; Li, C.; Fan, D.; Luo, K. PtoMYB156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar. Sci. Rep. 2017, 7, 41209. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.L.; Zhong, R.; Fowler, S.; Lyskowski, D.; Piyasena, H.; Carleton, K.; Spicer, C.; Ye, Z. The Poplar MYB Transcription Factors, PtrMYB3 and PtrMYB20, are Involved in the Regulation of Secondary Wall Biosynthesis. Plant Cell Physiol. 2010, 51, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Kotake, T.; Kamihara, K.; Tsuna, K.; Aohara, T.; Kaneko, Y.; Takatsuji, H.; Tsumuraya, Y.; Kawasaki, S. Rice BRITTLE CULM 3 (BC3) encodes a classical dynamin OsDRP2B essential for proper secondary cell wall synthesis. Planta 2010, 232, 95–108. [Google Scholar] [CrossRef]
- Xiong, G.; Li, R.; Qian, Q.; Song, X.; Liu, X.; Yu, Y.; Zeng, D.; Wan, J.; Li, J.; Zhou, Y. The rice dynamin-related protein DRP2B mediates membrane trafficking, and thereby plays a critical role in secondary cell wall cellulose biosynthesis. Plant J. 2010, 64, 56–70. [Google Scholar] [CrossRef]
- Zhang, B.; Su, X.; Zheng, S. Establishment of a highly efficient plant regeneration system of Populus davidiana × P. bolleana and study of its genetic stability. J. Beijing For. Univ. 2008, 30, 68–73. [Google Scholar]
- Brunner, A.M.; Yakovlev, I.; Strauss, S.H. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 2004, 4, 14. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Radonic, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Iida, A.; Seki, M.; Kamada, M.; Yamada, Y.; Morikawa, H. Gene delivery into cultured plant cells by DNA-coated gold particles accelerated by a pneumatic particle gun. Theor. Appl. Genet. 1990, 80, 813–816. [Google Scholar] [CrossRef]
- Franklin, G.L. Preparation of Thin Sections of Synthetic Resins and Wood-Resin Composites, and a New Macerating Method for Wood. Nature 1945, 155, 51. [Google Scholar] [CrossRef]
- Whiting, P.; Favis, B.D.; St-Germain, F.G.T.; Goring, D.A.I. Fractional Separation of Middle Lamella and Secondary Wall Tissue From Spruce Wood. J. Wood Chem. Technol. 1981, 1, 29–42. [Google Scholar] [CrossRef]
- Chen, T. Uncertainty evaluation of determination of nitrogen, carbon and hydrogen in fir with elemental analyzer. Shandong Chem. Ind. 2015, 44, 70–71. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7, Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef]
- Yang, Y.; Li, R.; Qi, M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000, 22, 543–551. [Google Scholar] [CrossRef]
- Jefferson, R.; Kavanagh, T.A.; Bevan, M. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.V.; Hellsten, U.; Putnam, N.H.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).