Abstract
New techniques are on the horizon for the detection of small leukemic clones in both, acute leukemias and myeloproliferative disorders. A promising approach is based on digital polymerase chain reaction (PCR). Digital PCR (dPCR) is a breakthrough technology designed to provide absolute nucleic acid quantification. It is particularly useful to detect a low amount of target and therefore it represents an alternative method for detecting measurable residual disease (MRD). The main advantages are the high precision, the very reliable quantification, the absolute quantification without the need for a standard curve, and the excellent reproducibility. Nowadays the main disadvantages of this strategy are the costs that are still higher than standard qPCR, the lack of standardized methods, and the limited number of laboratories that are equipped with instruments for dPCR. Several studies describing the possibility and advantages of using digital PCR for the detection of specific leukemic transcripts or mutations have already been published. In this review we summarize the available data on the use of dPCR in acute myeloid leukemia and myeloproliferative disorders.
1. Introduction
Polymerase chain reaction (PCR) is a revolutionary method for DNA amplification set up in 1985 by Kary Mullis, that allows the quantification of nucleic acids by amplification with the enzyme DNA polymerase [1].
The conventional assay is based on the principle that amplification of the nucleic acid is exponential thus it can be quantified by comparing the number of amplification cycles and the amount of PCR end-products to those of a reference sample [2,3,4].
In a conventional PCR, a solution of template DNA, DNA polymerase, deoxyribonucleotide triphosphates (dNTPs), primers, and buffer solution are subjected to a series of thermal cycles to amplify millions of copies of a template DNA [2,3,4].
In the current modified version of the PCR, the first step of a thermal cycle begins with a process called initialization or hot start to activate the polymerase at a temperature ranging from 94 °C to 96 °C. This step is followed by a denaturation step at high temperature between 93 °C and 98 °C. This step breaks the bonds in double stranded DNA (dsDNA) thus generating two single-stranded DNA (ssDNA) molecules. After denaturation, a phase called annealing starts at lower temperature. Primers are annealed to complementary sequences of the single-stranded DNA molecule. Subsequently the PCR mixture is heated to allow incomplete DNA sequence to be extended by the DNA polymerase in the presence of free dNTPs thus generating a new double-stranded DNA.
Despite the high level of sensitivity of this method, the evaluation of the amount of transcript can be hampered by many factors that can lead to inaccuracies. Among these, the low initial concentration of target molecules may not be amplified to detectable levels. In addition, the PCR amplification product can reach a plateau after a certain number of cycles. Finally, PCR amplification efficiency can differ in different samples. All these limits can impact on the accuracy of the quantification of the nucleic acid.
Real Time quantitative PCR (qPCR) represents the evolution of standard PCR. With this technique the products are continuously monitored throughout the reaction cycles using fluorescent dyes [5].
The initial amount of DNA template can be established by comparing the fluorescence output curve of the qPCR with a standard curve generated with different known initial numbers of DNA copies. The cycle threshold (Ct) is defined as the number of cycles required for the fluorescent signal to cross the threshold and to become detectable. Ct levels are inversely proportional to the amount of target nucleic acid in the sample.
The qPCR technique is more accurate than standard PCR and finds application in molecular diagnosis of many hematological malignancies and in the detection of a low amount of residual disease [6].
Nowadays, due to the high level of sensitivity, the qPCR technique, together with multiparameter flow cytometry (MFC) is considered the gold standard for the detection of malignant cells in different types of hematological malignancies [6].
2. Digital PCR
Digital PCR (dPCR) represents an innovative evolution of qPCR with many practical advantages over qPCR. Although initially described in 1999 [7] by Vogelstein and Kinzler, dPCR has become only recently a reliable and applicable tool. The first report [7] described the successful detection of Ras mutations with a high level of sensitivity in a cohort of patients with colorectal cancer [7]. With this method the exponential signal of PCR is converted into a linear digital signal which is most suitable for identification of genetic mutation.
From the first report in 1999, dPCR has been successfully applied to cancer genome studies. In the last few years the interest for this method in the hematological setting has progressively increased as testified by the number of papers in literature reporting the usefulness of this method for the quantification of specific leukemic aberrations. The main applications include evaluations of gene expression (e.g., miRNAs), pathogen quantification, rare allele detection, germline and somatic copy number variation estimation, viral load analysis, and microbial quantification [8].
In dPCR, the polymerase chain reaction mixture along with the necessary fluorophore is compartmentalized into several smaller units, each unit undergoes the same thermal cycles as in the case of a conventional PCR. Usually, dPCR employs the same primer sets, fluorescent labels, and enzymatic reagents as for traditional RT-PCR, unless recommended differently by the manufacturers. The key element of dPCR is the partition of the sample into thousands of individual PCR reactions in essence generating a limiting dilution [9]. As for RT-PCR, dPCR offers a highly precise and sensitive approach with the main advantage over RT-PCR of avoiding the need for a reference or standard curve. Despite this, it is necessary to admit that the same advantage can be obtained by performing a duplex-PCR with the inclusion of a reference gene.
Digital PCR method is based on three main points: the compartmentalization of the target, PCR on each single molecule and Poisson statistics (Figure 1). Following partition and amplification, the negative fraction is used to generate an absolute count of the number of target molecules in the sample, all without reference to standards or controls [10]. Nowadays, different commercialized digital PCR platforms are available. The first is based on Chip in a tube design (BioRad-QX200 digital PCR System, Bio-Rad system, Hercules, CA, USA). The second tool is based on micro-well chip (Life Technologies-QuantStudio3D® Digital PCR, Life Technologies, Carlsbad, CA, USA). An additional platform is based on the microfluidic-chamber (Stilla Technologies-Naica Crystal dPCR, Villejuif, France and Fluidigm-BioMark® HD, Fluidigm Corporation, San Francisco, CA, USA). Micro-well chip-based and microfluidic-chamber-based (cdPCR) technology can contain up to a few thousand individual reactions for each sample. Droplet dPCR (ddPCR) is a method based on emulsion PCR. The sample is fractionated into 20,000 droplets and the amplification of the template molecules occurs in each droplet [11]. The high partition of ddPCR, makes this method very sensitive and potentially useful for both, research and diagnostic purposes. The main advantages of dPCR compared to RT-PCR are the high precision, the very reliable quantification, the absolute quantification without the need for a standard curve, and excellent reproducibility [12].
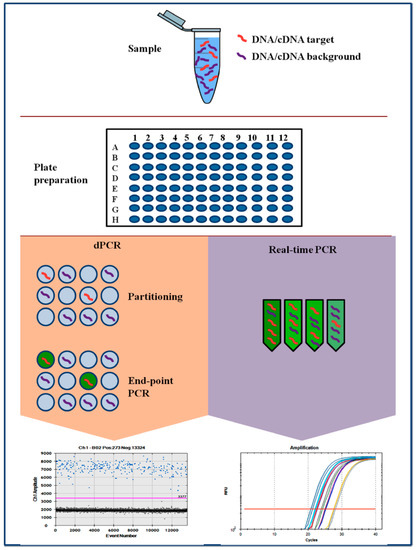
Figure 1.
Comparison of PCR-based techniques. RT-PCR and dPCR using the same amplification reagents and fluorescent labeling system. In dPCR, the sample is first partitioned such that each partition contains either a few or no DNA sequences. Fluorescence is measured at the end of the PCR. In qPCR, the amount of amplified DNA is measured at each cycle during the PCR reaction.
Finally, recent papers have reported the higher tolerance of dPCR as compared to RQ-PCR to different types of inhibitors that can damage DNA or make DNA less accessible, like salts including KCl and NaCl, ionic detergents such as sodium deocycholate, sarkosyl, and SDS, ethanol, isopropanol, and phenol among others. This is mainly due to the compartmentalization of target sequences in smaller volumes [8,13].
An increasing number of manuscripts is published every month on the use of dPCR in hematological diseases. In this review we summarize the current knowledge on dPCR in myeloid neoplasms.
3. dPCR in Chronic Myeloproliferative Disorders: The Example of Chronic Myeloid Leukemia (CML)
In chronic myeloid leukemia (CML), the presence of a specific marker, the Philadelphia chromosome, together with the corresponding molecular marker (BCR-ABL fusion transcripts) provides a unique opportunity for the monitoring of the disease, at diagnosis and during therapy [14]. Lots of data clearly show that in CML both, cytogenetic and molecular responses, are strictly correlated to the final outcome and survival of the patients [15,16]. The use of adequate and standardized methods allows the degree of responses and the depth of disease reduction at specific time points during treatment to be established [17,18,19]. This has become fundamental for a correct clinical management of CML patients. The efforts to standardize the molecular monitoring and the criteria of response led to the definition of “optimal response” that may represent not only the highest probability of survival but also the possibility to discontinue the tyrosine-kinase inhibitor (TKI) treatment and therefore to live in a treatment-free remission (TFR) status [20,21]. Therefore, at least for some patients, deep degrees of molecular response, as MR4 and MR4.5, whose precise definition has been recently introduced and which are prerequisites to try discontinuation, are becoming the target to be achieved even in common clinical practice [15].
Currently, both European Leukemia Net (ELN) [15] and National Comprehensive Cancer Network (NCCN) [21] recommend the use of qPCR to monitor MRD or to establish the depth of molecular response.
With the aim of improving PCR sensitivity to increase the predictive value of molecular response, several studies compared the use of dPCR and qPCR. In the study published by Goh and colleagues, [22] forty-three CML patients were screened by conventional qPCR, by replicate qPCR (rRQ-PCR) and by nanofluidic digital PCR. Interestingly they demonstrated that rRQ-PCR, as well as dPCR with pre-amplification, has a 2–3 log of improvement compared to conventional RQ-PCR. Twenty-four of 32 PCR negative samples as assayed by conventional qPCR showed detectable BCR–ABL in rRQ-PCR and/or dPCR. Furthermore they were able to demonstrate that, using dPCR with a pre-amplification step, the progressive reduction of MRD level could be precisely monitored even when undetectable by conventional qPCR.
In summary, in this study, both rRQ-PCR and ddPCR succeed in the detection of BCR–ABL transcripts not detectable in conventional RQ-PCR.
These data showed the potential feasibility of highly sensitive PCR approaches for molecular monitoring and suggested the clinical relevance for future CML management by allowing further characterization of patients who achieve PCR negativity by qPCR and who are candidates for therapy discontinuation.
In 2018 Wang and colleagues [23] analyzed the peripheral blood samples from 61 CML patients who presented an undetectable level of BCR-ABL by qPCR after tyrosine kinase inhibitor (TKI) treatment in three successive analyses performed once every 3 months. In parallel, they measured the fusion transcript by droplet d-PCR and they found that 18% of the patients tested positive by this analysis. Importantly, during follow up they documented that the few cases who lost MR4 presented a markedly increased transcript amount, detected by dd-PCR, a few months before the loss of response. This increase was not detectable by conventional qPCR. These data led to the initial conclusion that dd-PCR is more sensitive in measuring deep molecular responses.
Different from what is reported above, the study by Alikian and colleagues [24], by analyzing a cohort of 70 CML patients, demonstrated that dPCR has a comparable performance to qPCR for the quantification of BCR-ABL1. Their conclusion is that qPCR already has a high level of sensitivity, close to the single molecule level.
4. Digital PCR in Philadelphia Negative Chronic Myeloproliferative Neoplasms (Ph-MPN)
Philadelphia-negative chronic myeloproliferative neoplasms (MPNs) are clonal disorders that present JAK2V617F mutation in a percentage ranging from 50% to 95% of cases [25]. Calreticulin (CALR) gene mutations have recently been discovered in about 20%–35% of patients affected by essential thrombocythemia (ET) and primary myelofibrosis (PMF) [26]. An additional percentage of ET patients presents with MPL mutation [27].
JAK2, CALR, and MPL “driver” mutations can be targeted for diagnosis and sometimes to monitor the disease response (i.e., after allogeneic stem cell transplantation) although the role of these markers in the evaluation of clinical response to currently available therapies is still questionable.
Due to the lack of effective therapies, able to induce molecular remission in MPN patients, the development of techniques for the detection of MRD has not been heavily pushed. Recently, a new scenario is on the horizon with new targeted therapies available or under development [27].
Furthermore, for myelofibrosis, the allogeneic hematopoietic stem cell transplantation (HSCT) might be curative [28]. In all these settings a precise evaluation of the kinetics of the disease might provide relevant information for treatment choices [29].
The clinical relevance of determining JAK2, CALR, and MPL based MRD has been demonstrated with several therapeutically approaches including interferon alpha, JAK1/2 inhibitors, and allogeneic stem cell transplantation [30].
Recently few studies have been published comparing qPCR and dPCR for the measurement of JAK2V617F [31,32,33].
In the study by Fontanelli [31] and colleagues, 99 patients affected by MPN with JAK2V617F were evaluated in parallel by means of qPCR and droplet dPCR. This latter showed a higher sensitivity than qPCR in detecting the mutation. They also confirmed an increased mutant allele burden from ET to PV and PMF.
Link-Lenczowska and colleagues were able to substantially confirm this finding [32]. They analyzed 63 MPN patients, six of them treated with ruxolitinib, by qPCR and dPCR. Basically, they found high concordance between the two methods, both demonstrated to be highly sensitive and both were capable of detecting the JAK2V617F mutation at diagnosis of MPN with a limit of detection of 0.12% for qPCR and 0.01% for ddPCR. The study suggests an advantage of ddPCR in monitoring MRD when the allele burden is below the threshold of detection of qPCR.
Finally, the clinical utility of dPCR for JAK2V617F mutation was investigated by Waterhouse and colleagues [33] in 59 patients with MPN, many of them after hematopoietic stem cell transplantation. The limit of detection was 0.01% for both qPCR and ddPCR. The JAK2V617F mutation was detected in 43 out of 59 patients by both PCR platforms. However, in 14% of the samples, JAK2V617F mutation was detected only with ddPCR. Interestingly, all these patients were analyzed shortly after allogeneic HSCT. Although available data on dPCR for JAK2 detection after HSCT are still immature, this study suggests an intriguing role of dPCR in the context of myeloproliferative disorders. Similar to JAK2V617F mutation, the monitoring of CALR mutation might improve the therapeutic strategies. Mansier and colleagues [34] developed a digital PCR technique that allowed detection of types 1 and 2 CALR allelic burdens. They found that compared with the commonly used fluorescent PCR analysis, digital PCR is more precise, reproducible, and accurate. In their hands this method reached a very high sensitivity being able to detect at least 0.025% CALR mutants. They applied this method to patients with primary myelofibrosis who underwent hematopoietic stem cell transplant and they were able to predict relapse according to the reappearance of CALR mutations after HSCT. After the achievement of dPCR negativity, the reappearance of a low level of mutation, although in a single patient, preceded the hematologic relapse. This study suggests the possibility of using dPCR for CALR mutation measurement for MRD detection.
5. Minimal Residual Disease in Acute Myeloid Leukemias
Currently, the post remission treatment of patients affected by acute myeloid leukemias is based on the genetic profile of leukemic cells at diagnosis and on the level of minimal residual disease after induction and consolidation chemotherapy mainly detected by multiparameter flow cytometry and by q-PCR for the assessment of fusion transcripts levels (i.e., CBFB-MYH11 and RUNX1-RUNX1T1 and PML-RARα) of mutations, mainly NPM1 [35,36]. The issue of MRD monitoring has been recently addressed by the European Leukemia Net (ELN) [6] in an attempt to standardize the methodology and to provide suggestion on when and how to monitor MRD.
A new promising approach for the detection of MRD is based on dPCR. We report below the current knowledge on the use of dPCR for detection of leukemic cells.
6. Digital PCR for Acute Myeloid Leukemia Monitoring: The Case of Acute Promyelocytic Leukemia
Acute promyelocytic leukemia (APL) is a hematological malignancy commonly associated with the chromosomal translocation t(15;17)(q24;q21), which involves the promyelocytic leukemia (PML) and the retinoic acid receptor-α (RARα) genes, resulting in the oncogenic fusion transcript PML-RARα [37].
In acute promyelocytic leukemia, the achievement of PCR negativity for PML-RARα at the end of consolidation treatment is the most informative in terms of prediction of outcome and it is associated with a low risk of relapse and a high probability of long-term survival [38,39]. The achievement of PCR negativity retains its clinical significance independently of the therapeutic strategies, all trans-retinoic acid (ATRA) associated with chemotherapy or ATRA and arsenic trioxide [38,39].
Recently Brunetti and colleagues [40] investigated the role of droplet dPCR for PML-RARα to monitor MRD in a cohort of 21 patients affected by APL. Droplet dPCR exhibited a sensitivity and specificity of 95% and 91% respectively for bcr1 and bcr3 transcripts and showed a good concordance with qPCR. The authors suggested that one of the main advantages of ddPCR-based monitoring of MRD is represented by the absolute quantification. They also believe that ddPCR could potentially provide crucial information for the management of patients whose MRD fluctuates under the level of detection of qPCR.
Similar results have been reported by Yuan and colleagues [41] in 28 APL patients. They confirmed the good concordance between ddPCR and qPCR in the detection of PML-RARα in clinical samples, but showed advantages of dPCR over qPCR in terms of precision, limit of detection, and other basic performance parameters.
In conclusion, based on the available data, at present dPCR could represent a complementary approach to monitor MRD in APL, particularly for those patients at high risk of relapse.
7. Acute Myeloid Leukemia with IDH1/IDH2 Mutations
Since the evidence that IDH1/IDH2 genes can be mutated in about 10%–20% of AML, many groups are investigating the possibility of using IDH1/2 as a marker for MRD detection.
Until now, few data have been generated on the clearance of IDH1/IDH2 after chemotherapy and during remission and contrasting data are reported in the literature [42,43]. Few studies reported the stability and suitability of IDH as a marker of MRD [42,44]. Recently Petrova and colleagues [45] published the evaluation of MRD in 90 patients, 22% of them with IDH1/IDH2 mutations. They based the assessment on NGS and digital droplet PCR. Many patients presented additional mutations such as NPM1 or MLL-PDT. They concluded that the persistence of IDH1/2 correlates with the treatment response, although being less sensitive than NPM1 in predicting relapse.
8. Monitoring C-KIT Exon 17 Mutations by Droplet Digital PCR in Patients with Core-Binding Factor AML
The cytogenetic aberrations involving core binding factor (CBF) include t(8;21)(q22;q22) and inv(16)(p13.1q22)/t(16;16) (p13.1;q22) [43]. CBF-AML are considered leukemias with a highly favorable prognosis although the outcome is highly impacted by the presence of additional genetic mutations, as exemplified by the mutation of FLT3, C-KIT, or NRAS gene [46,47,48,49]. In particular the mutations of C-KIT gene that result in the constitutive activation of tyrosine kinase activity have a significant impact on the prognosis of CBF acute myeloid leukemias. In particular, C-KIT mutations are associated with a higher incidence of relapse, so that CBF AML with C-KIT mutations are classified into the intermediate risk group by the NCCN Guideline [46,47,48,49]. These mutations are most frequently located in exon 17 (54%) encoding the kinase-activation loop, or in exon 8 (28%) affecting the extracellular portion of C-KIT receptor [46]. C-KIT mutations have been described initially as a single point mutation but the detection of double mutations has recently been described in AML patients [48].
Although we still lack solid data generated by qPCR on the prognostic significance of the mutation burden of c-KIT, an attempt to study the dynamic of the mutated clone has been done by Tan and colleagues [50]. They investigated the dynamic evolution of CBF-AML clones with C-KIT mutations by ddPCR combined with sequencing [50]. The study included 75 patients with CBF-AML, 19% of them with double C-KIT mutation.
Droplet digital PCR revealed that these double mutations can occur in either the same or different clones. Interestingly, according to the mutation, the clone was shown to present a different sensitivity to treatment. In particular, C-KITN822 mutation confers to the clone a better response to treatment as compared to C-KITD816 mutation. Moreover, D816 clone was the predominant clone at relapse. In addition, patients with double mutation had a better overall survival and event-free survival than those with a single mutation although the statistical significance was not reached, probably due to the small sample size. The study demonstrated that ddPCR is an effective method for monitoring clonal evolution in AML.
9. WT1 Assessment by Digital PCR as a Marker of MRD
The usefulness of WT1 quantitative assessment q-PCR as a marker for measurable residual disease (MRD) detection in acute myeloid leukemia was demonstrated years ago [51,52,53,54,55]. Many studies clearly showed that WT1 gene is overexpressed in about 80%–90% of patients affected by AML [51,52]. The persistence of WT1 overexpression after chemotherapy is always indicative of persistence of leukemic cells [51,52,53,54,55]. Based on current evidence WT1 can be considered a universal marker of AML. In a European study aimed at standardizing the method used for WT1 measurement, it was shown that the clearance of the transcript to normal values is highly predictive of relapse [55]. Additional studies suggested that the persistence of abnormal WT1 values after induction or consolidation impacts on the probability of relapse. An increase of WT1 levels during follow up always predict leukemia recurrence [51,52,53,54,55]. The main advantages of WT1 assay are that it can be measured in PB and that the method has been standardized [55].
Koizumi and colleagues [56] analyzed WT1 by ddPCR and qPCR in 40 peripheral blood and bone marrow samples obtained from patients affected by acute leukemias and myelodysplastic syndromes. They found a strong correlation between the two methods (R = 0.99) but they demonstrated that dPCR is able to accurately detect lower WT1 levels compared to qPCR. Based on these results the authors concluded that dPCR technology can be utilized to measure WT1 based MRD with high accuracy.
10. dPCR for MRD Detection in the Setting of Allogeneic Stem Cell Transplantation
Allogeneic hematopoietic stem cell transplantation is a consolidated therapy for the cure of patients with AML. Despite the significant number of patients achieving remission after chemotherapy and stem cell transplantation, the relapse rate is still significant. Many studies demonstrated that the presence of MRD both at the time of HSCT or after HSCT is a negative prognostic factor with high impact on survival [57,58]. Thus, nowadays, the detection of MRD to identify patients at high risk of relapse is mandatory. Different published studies addressed the issue of improving the sensitivity of PCR in order to increase the predictive value of the method. Bill and colleagues [59] analyzed by droplet dPCR the pre-transplant samples, both peripheral blood PB and bone marrow BM) of 51 nucleophosmin 1 (NPM1)-mutated AML patients transplanted in complete remission. The authors demonstrated that MRD positive patients have a higher cumulative incidence of relapse and shorter overall survival. In addition, the patients who are still positive for NPM1 mutation by dPCR before allogeneic HSCT have a worse prognosis, independently from other known prognostic markers, or from the conditioning they received. The authors envisage the possibility of using dPCR routinely for patients with mutated NPM1 to guide treatment and improve patients’ outcomes.
A larger study has been published by Valero-Garcia and colleagues [60]. They compared the sensitivity and accuracy of qPCR and dPCR and their capacity to predict relapse. They analyzed 28 adult patients who underwent allogeneic HSCT. They detected an increasing mixed chimerism prior to relapse in 100% of patients who relapsed. Compared with conventional qPCR, dPCR was able to predict relapse with a median anticipation period of 63 days versus 45.5 days by qPCR. Overall, 56% of the relapses were predicted earlier with dPCR whereas 38% of the relapses where detected simultaneously using both techniques. In only one case relapse was predicted earlier with qPCR. The presented data strongly support the notion that dPCR is a powerful tool to predict relapse after HSCT.
A summary of the main targets analyzed by dPCR and qPCR is shown in Table 1.

Table 1.
A summary of the main targets analyzed by dPCR and qPCR.
11. Chimerism Analysis
Finally, another important diagnostic tool in the setting of allogeneic HSCT is represented by chimerism analysis. For this purpose George and colleagues [61] set up a ddPCR assay and analyzed patients who received HSCT. With this method they detected the persistence of recipient cells with a high level of sensitivity. The techniques currently used to assess chimerism after hematopoietic stem cell transplantation are capillary electrophoresis analysis of STRs amplified by PCR and qPCR detection of polymorphic indel loci in the human genome. The main limit of capillary electrophoresis is the sensitivity. This method can detect chimerism fractions >1% of blood or marrow cells with the risk of missing clinically relevant low-level recipient cells. The second is a highly sensitive method with the disadvantage of being characterized by variable efficiency and the need for extensive validation and standards to be run with each assay [62,63,64].
The limit of capillary electrophoresis can be clinically relevant in the setting of cellular therapies that frequently result in microchimerism (donor chimerism <1%) [62].
George and colleagues described a highly sensitive droplet digital PCR assay with good performance throughout the range of clinically relevant chimerism measurements [64]. They validated the assay in serially diluted samples. The levels of detection and quantification of the assay were 0.01%.
12. Innovative Application of Digital PCR: The Case of Methylation Analysis
DNA methylation is an epigenetic modification that plays a key role in genome regulation [65]. Aberrant DNA methylation contributes to the genesis of tumors including hematological malignancies such as acute leukemia and myelodysplastic syndromes [66,67].
The regulation of CpG methylation has been demonstrated to play a role in stem cell maintenance and differentiation [68]. In hematological malignancies, global aberrant DNA methylation has been widely documented and it has been associated with disease progression and response to therapy [69,70]. Therefore, methylation analysis will help to understand the pathogenesis of leukemia and will represent new therapeutic targets. Alu sequences have been demonstrated to contribute to establish the epigenetic landscape of cancer cells, and several papers have been focused on this topic [71,72].
Orsini and colleagues [70] developed a new method of investigating Alu differential methylation, based on droplet digital PCR (ddPCR) technology. Although this method has not been applied to hematological malignancies and no data are available in this setting, this approach could be potentially useful to profile patients affected by hematologic disorders including myelodysplastic syndromes (MDS) for diagnostic and prognostic purposes.
13. Conclusions
A great deal of evidence has encouraged the investigation of dPCR for MRD monitoring in many hematological malignancies, especially in those patients who can reach very deep molecular responses with pharmacological or cellular therapies. Examples can be represented by the possibility to discontinue TKI therapy in CML patients or by the possibility of donor lymphocyte infusions in patients with detectable MRD after HSCT.
The real advantage of using dPCR will probably become clearer in the next few years with the improvement of the available strategies and the accumulation of data.
Author Contributions
D.C. wrote the paper, J.P., V.R., G.A, M.D., C.F., and G.S. revised and approved the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| PCR | polymerase chain reaction |
| dNTP | deoxyribonucleotide triphosphates |
| dsDNA | double stranded DNA |
| ssDNA | single stranded DNA |
| qPCR | quantitative polymerase chain reaction |
| MFC | multiparameter flow cytometry |
| Ct | threshold cycle |
| dPCR | digital polymerase chain reaction |
| ddPCR | droplet digital digital polymerase chain reaction |
| CML | chronic myeloid leukemia |
| TKI | tyrosine kinase inhibitor |
| TFR | treatment free remission |
| MR | molecular remission |
| MR4 | molecular remissionwith 4 logs of transcript reduction |
| MR4.5 | Molecular remissionwith 4.5 logs of transcript reduction |
| ELN | European Leukemia Net |
| NCCN | National Comprehensive Cancer Network |
| rRQ-PCR | replicate quantitative polymerase chain reaction |
| MPN | myeloproliferative neoplasms |
| JAK2 | janus kinase 2 |
| CALR | calreticulin |
| ET | essential thrombocythemia |
| PMF | primary myelofibrosis |
| MRD | minimal residual disease |
| WT1 | Wilms tumor gene |
| HSCT | hematopoietic stem cell transplantation |
| APL | acute promyelocytic leukemia |
| PML | promyelocytic leukemia |
| RARa | retinoic acid receptor alpha |
| ATRA | all trans retinoic acid |
| IDH1/2 | Isocitrate dehydrogenase 1/2 |
| NPM1 | nucleophosmin |
| MDS | Myelodysplastic syndromes |
References
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273. [Google Scholar] [CrossRef]
- Boyle, J. Molecular Biology, 2nd Edition David P. Clark and Nanette J. Pazdernik, Academic Press-Cell, 2012, 928 pp., ISBN 978-0-1237–8594-7, $135.00 (hardback). Biochem. Mol. Biol. Educ. 2014, 42, 100–101. [Google Scholar] [CrossRef]
- Butler, J.M. Chapter 4—PCR Amplification: Capabilities and Cautions. In Advanced Topics in Forensic DNA Typing: Methodology; Butler, J.M., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 69–97. [Google Scholar] [CrossRef]
- Najafov, A.; Hoxhaj, G. Chapter 1—Introduction. In PCR Guru; Najafov, A., Hoxhaj, G., Eds.; Academic Press, 2017; pp. 1–6. Available online: https://doi.org/10.1016/B978-0-12-804231-1.00001-8 (accessed on 2 December 2016).
- DiNardo, C.D.; Luger, S.M. Beyond morphology: minimal residual disease detection in acute myeloid leukemia. Curr. Opin. Hematol. 2012, 19, 82–88. [Google Scholar] [CrossRef]
- Schuurhuis, G.J.; Heuser, M.; Freeman, S.; Bene, M.C.; Buccisano, F.; Cloos, J.; Grimwade, D.; Haferlach, T.; Hills, R.K.; Hourigan, C.S.; et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood 2018, 131, 1275–1291. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Debski, P.R.; Gewartowski, K.; Bajer, S.; Garstecki, P. Calibration-free assays on standard real-time PCR devices. Sci. Rep. 2017, 7, 44854. [Google Scholar] [CrossRef] [PubMed]
- Dodd, D.W.; Gagnon, K.T.; Corey, D.R. Digital quantitation of potential therapeutic target RNAs. Nucleic Acid Ther. 2013, 23, 188–194. [Google Scholar] [CrossRef]
- Dong, L.; Meng, Y.; Sui, Z.; Wang, J.; Wu, L.; Fu, B. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci. Rep. 2015, 5, 13174. [Google Scholar] [CrossRef]
- Quan, P.L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18. [Google Scholar] [CrossRef]
- Huggett, J.F.; Cowen, S.; Foy, C.A. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin. Chem. 2015, 61, 79–88. [Google Scholar] [CrossRef]
- Shanmuganathan, N.; Hughes, T.P. Molecular monitoring in CML: how deep? How often? How should it influence therapy? Hematol. Am. Soc. Hematol. Educ. Program 2018, 2018, 168–176. [Google Scholar] [CrossRef]
- Marin, D.; Milojkovic, D.; Olavarria, E.; Khorashad, J.S.; de Lavallade, H.; Reid, A.G.; Foroni, L.; Rezvani, K.; Bua, M.; Dazzi, F.; et al. European LeukemiaNet criteria for failure or suboptimal response reliably identify patients with CML in early chronic phase treated with imatinib whose eventual outcome is poor. Blood 2008, 112, 4437–4444. [Google Scholar] [CrossRef]
- Baccarani, M.; Deininger, M.W.; Rosti, G.; Hochhaus, A.; Soverini, S.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Guilhot, F.; et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 2013, 122, 872–884. [Google Scholar] [CrossRef]
- White, H.; Deprez, L.; Corbisier, P.; Hall, V.; Lin, F.; Mazoua, S.; Trapmann, S.; Aggerholm, A.; Andrikovics, H.; Akiki, S.; et al. A certified plasmid reference material for the standardisation of BCR-ABL1 mRNA quantification by real-time quantitative PCR. Leukemia 2015, 29, 369–376. [Google Scholar] [CrossRef]
- Cross, N.C.; Hochhaus, A.; Muller, M.C. Molecular monitoring of chronic myeloid leukemia: principles and interlaboratory standardization. Ann. Hematol. 2015, 94, S219–S225. [Google Scholar] [CrossRef]
- Cross, N.C.; White, H.E.; Ernst, T.; Welden, L.; Dietz, C.; Saglio, G.; Mahon, F.X.; Wong, C.C.; Zheng, D.; Wong, S.; et al. Development and evaluation of a secondary reference panel for BCR-ABL1 quantification on the International Scale. Leukemia 2016, 30, 1844–1852. [Google Scholar] [CrossRef] [PubMed]
- Saussele, S.; Richter, J.; Guilhot, J.; Gruber, F.X.; Hjorth-Hansen, H.; Almeida, A.; Janssen, J.; Mayer, J.; Koskenvesa, P.; Panayiotidis, P.; et al. Discontinuation of tyrosine kinase inhibitor therapy in chronic myeloid leukaemia (EURO-SKI): a prespecified interim analysis of a prospective, multicentre, non-randomised, trial. Lancet. Oncol. 2018, 19, 747–757. [Google Scholar] [CrossRef]
- Chamoun, K.; Kantarjian, H.; Atallah, R.; Gonzalez, G.N.; Issa, G.C.; Rios, M.B.; Garcia-Manero, G.; Borthakur, G.; Ravandi, F.; Jain, N.; et al. Tyrosine kinase inhibitor discontinuation in patients with chronic myeloid leukemia: a single-institution experience. J. Hematol. Oncol. 2019, 12, 1. [Google Scholar] [CrossRef]
- Goh, H.G.; Lin, M.; Fukushima, T.; Saglio, G.; Kim, D.; Choi, S.Y.; Kim, S.H.; Lee, J.; Lee, Y.S.; Oh, S.M.; et al. Sensitive quantitation of minimal residual disease in chronic myeloid leukemia using nanofluidic digital polymerase chain reaction assay. Leuk. Lymphoma 2011, 52, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Zheng, C.F.; Liu, Z.; Tan, Y.H.; Chen, X.H.; Zhao, B.L.; Li, G.X.; Xu, Z.F.; Ren, F.G.; Zhang, Y.F.; et al. Droplet digital PCR for BCR/ABL(P210) detection of chronic myeloid leukemia: A high sensitive method of the minimal residual disease and disease progression. Eur. J. Haematol. 2018, 101, 291–296. [Google Scholar] [CrossRef]
- Alikian, M.; Whale, A.S.; Akiki, S.; Piechocki, K.; Torrado, C.; Myint, T.; Cowen, S.; Griffiths, M.; Reid, A.G.; Apperley, J.; et al. RT-qPCR and RT-Digital PCR: A Comparison of Different Platforms for the Evaluation of Residual Disease in Chronic Myeloid Leukemia. Clin. Chem. 2017, 63, 525–531. [Google Scholar] [CrossRef]
- Kralovics, R.; Passamonti, F.; Buser, A.S.; Teo, S.S.; Tiedt, R.; Passweg, J.R.; Tichelli, A.; Cazzola, M.; Skoda, R.C. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 2005, 352, 1779–1790. [Google Scholar] [CrossRef]
- Nangalia, J.; Massie, C.E.; Baxter, E.J.; Nice, F.L.; Gundem, G.; Wedge, D.C.; Avezov, E.; Li, J.; Kollmann, K.; Kent, D.G.; et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 2013, 369, 2391–2405. [Google Scholar] [CrossRef]
- Alvarez-Larran, A.; Martinez, D.; Arenillas, L.; Rubio, A.; Arellano-Rodrigo, E.; Hernandez Boluda, J.C.; Papaleo, N.; Caballero, G.; Martinez, C.; Ferrer-Marin, F.; et al. Essential thrombocythaemia with mutation in MPL: clinicopathological correlation and comparison with JAK2V617F-mutated and CALR-mutated genotypes. J. Clin. Pathol. 2018, 71, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Vannucchi, A.M.; Harrison, C.N. Emerging treatments for classical myeloproliferative neoplasms. Blood 2017, 129, 693–703. [Google Scholar] [CrossRef]
- Gagelmann, N.; Ditschkowski, M.; Bogdanov, R.; Bredin, S.; Robin, M.; Cassinat, B.; Shahswar, R.; Thol, F.; Heuser, M.; Socie, G.; et al. Comprehensive clinical-molecular transplant scoring system for myelofibrosis undergoing stem cell transplantation. Blood 2019. [Google Scholar] [CrossRef] [PubMed]
- Guglielmelli, P.; Pietra, D.; Pane, F.; Pancrazzi, A.; Cazzola, M.; Vannucchi, A.M.; Tura, S.; Barosi, G. Recommendations for molecular testing in classical Ph1-neg myeloproliferative disorders-A consensus project of the Italian Society of Hematology. Leuk. Res. 2017, 58, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Fontanelli, G.; Barate, C.; Ciabatti, E.; Guerrini, F.; Grassi, S.; Del Re, M.; Morganti, R.; Petrini, I.; Arici, R.; Barsotti, S.; et al. Real-Time PCR and Droplet Digital PCR: two techniques for detection of the JAK2(V617F) mutation in Philadelphia-negative chronic myeloproliferative neoplasms. Int. J. Lab. Hematol. 2015, 37, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Link-Lenczowska, D.; Pallisgaard, N.; Cordua, S.; Zawada, M.; Czekalska, S.; Krochmalczyk, D.; Kandula, Z.; Sacha, T. A comparison of qPCR and ddPCR used for quantification of the JAK2 V617F allele burden in Ph negative MPNs. Ann. Hematol. 2018, 97, 2299–2308. [Google Scholar] [CrossRef]
- Waterhouse, M.; Follo, M.; Pfeifer, D.; von Bubnoff, N.; Duyster, J.; Bertz, H.; Finke, J. Sensitive and accurate quantification of JAK2 V617F mutation in chronic myeloproliferative neoplasms by droplet digital PCR. Ann. Hematol. 2016, 95, 739–744. [Google Scholar] [CrossRef]
- Mansier, O.; Migeon, M.; Saint-Lezer, A.; James, C.; Verger, E.; Robin, M.; Socie, G.; Bidet, A.; Mahon, F.X.; Cassinat, B.; et al. Quantification of the Mutant CALR Allelic Burden by Digital PCR: Application to Minimal Residual Disease Evaluation after Bone Marrow Transplantation. J. Mol. Diagn. 2016, 18, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Hourigan, C.S.; Gale, R.P.; Gormley, N.J.; Ossenkoppele, G.J.; Walter, R.B. Measurable residual disease testing in acute myeloid leukaemia. Leukemia 2017, 31, 1482–1490. [Google Scholar] [CrossRef] [PubMed]
- Dohner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Buchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Lo-Coco, F.; Hasan, S.K. Understanding the molecular pathogenesis of acute promyelocytic leukemia. Best Pract. Res. Clin. Haematol. 2014, 27, 3–9. [Google Scholar] [CrossRef]
- Grimwade, D.; Jovanovic, J.V.; Hills, R.K.; Nugent, E.A.; Patel, Y.; Flora, R.; Diverio, D.; Jones, K.; Aslett, H.; Batson, E.; et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 3650–3658. [Google Scholar] [CrossRef]
- Platzbecker, U.; Avvisati, G.; Cicconi, L.; Thiede, C.; Paoloni, F.; Vignetti, M.; Ferrara, F.; Divona, M.; Albano, F.; Efficace, F.; et al. Improved Outcomes With Retinoic Acid and Arsenic Trioxide Compared With Retinoic Acid and Chemotherapy in Non-High-Risk Acute Promyelocytic Leukemia: Final Results of the Randomized Italian-German APL0406 Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 605–612. [Google Scholar] [CrossRef]
- Brunetti, C.; Anelli, L.; Zagaria, A.; Minervini, A.; Minervini, C.F.; Casieri, P.; Coccaro, N.; Cumbo, C.; Tota, G.; Impera, L.; et al. Droplet Digital PCR Is a Reliable Tool for Monitoring Minimal Residual Disease in Acute Promyelocytic Leukemia. J. Mol. Diagn. 2017, 19, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Cui, M.; Yu, S.; Wang, H.; Jing, R. Droplet digital PCR for quantification of PML-RARalpha in acute promyelocytic leukemia: a comprehensive comparison with real-time PCR. Anal. Bioanal. Chem. 2019, 411, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Jeziskova, I.; Razga, F.; Toskova, M.; Dvorakova, D.; Timilsina, S.; Mayer, J.; Racil, Z. Quantitative detection of IDH2 mutation for minimal residual disease monitoring in patients with acute myeloid leukemia and its comparison with mutations in NPM1 gene. Leuk. Lymphoma 2013, 54, 867–870. [Google Scholar] [CrossRef] [PubMed]
- Chou, W.C.; Peng, K.Y.; Lei, W.C.; Ko, B.S.; Tsay, W.; Kuo, C.H.; Tien, H.F. Persistence of mutant isocitrate dehydrogenase in patients with acute myeloid leukemia in remission. Leukemia 2012, 26, 527–529. [Google Scholar] [CrossRef][Green Version]
- Brambati, C.; Galbiati, S.; Xue, E.; Toffalori, C.; Crucitti, L.; Greco, R.; Sala, E.; Crippa, A.; Chiesa, L.; Soriani, N.; et al. Droplet digital polymerase chain reaction for DNMT3A and IDH1/2 mutations to improve early detection of acute myeloid leukemia relapse after allogeneic hematopoietic stem cell transplantation. Haematologica 2016, 101, e157–e161. [Google Scholar] [CrossRef]
- Petrova, L.; Vrbacky, F.; Lanska, M.; Zavrelova, A.; Zak, P.; Hrochova, K. IDH1 and IDH2 mutations in patients with acute myeloid leukemia: Suitable targets for minimal residual disease monitoring? Clin. Biochem. 2018, 61, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Beghini, A.; Peterlongo, P.; Ripamonti, C.B.; Larizza, L.; Cairoli, R.; Morra, E.; Mecucci, C. C-kit mutations in core binding factor leukemias. Blood 2000, 95, 726–727. [Google Scholar] [PubMed]
- Pollard, J.A.; Alonzo, T.A.; Gerbing, R.B.; Ho, P.A.; Zeng, R.; Ravindranath, Y.; Dahl, G.; Lacayo, N.J.; Becton, D.; Chang, M.; et al. Prevalence and prognostic significance of KIT mutations in pediatric patients with core binding factor AML enrolled on serial pediatric cooperative trials for de novo AML. Blood 2010, 115, 2372–2379. [Google Scholar] [CrossRef]
- Allen, C.; Hills, R.K.; Lamb, K.; Evans, C.; Tinsley, S.; Sellar, R.; O’Brien, M.; Yin, J.L.; Burnett, A.K.; Linch, D.C.; et al. The importance of relative mutant level for evaluating impact on outcome of KIT, FLT3 and CBL mutations in core-binding factor acute myeloid leukemia. Leukemia 2013, 27, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, M.R.; Abboud, C.N.; Altman, J.; Appelbaum, F.R.; Arber, D.A.; Attar, E.; Borate, U.; Coutre, S.E.; Damon, L.E.; Goorha, S.; et al. NCCN Clinical Practice Guidelines Acute myeloid leukemia. J. Natl. Compr. Cancer Netw. 2012, 10, 984–1021. [Google Scholar] [CrossRef]
- Tan, Y.; Liu, Z.; Wang, W.; Zhu, G.; Guo, J.; Chen, X.; Zheng, C.; Xu, Z.; Chang, J.; Ren, F.; et al. Monitoring of clonal evolution of double C-KIT exon 17 mutations by Droplet Digital PCR in patients with core-binding factor acute myeloid leukemia. Leuk. Res. 2018, 69, 89–93. [Google Scholar] [CrossRef]
- Inoue, K.; Ogawa, H.; Yamagami, T.; Soma, T.; Tani, Y.; Tatekawa, T.; Oji, Y.; Tamaki, H.; Kyo, T.; Dohy, H.; et al. Long-term follow-up of minimal residual disease in leukemia patients by monitoring WT1 (Wilms tumor gene) expression levels. Blood 1996, 88, 2267–2278. [Google Scholar]
- Cilloni, D.; Gottardi, E.; De Micheli, D.; Serra, A.; Volpe, G.; Messa, F.; Rege-Cambrin, G.; Guerrasio, A.; Divona, M.; Lo Coco, F.; et al. Quantitative assessment of WT1 expression by real time quantitative PCR may be a useful tool for monitoring minimal residual disease in acute leukemia patients. Leukemia 2002, 16, 2115–2121. [Google Scholar] [CrossRef]
- Ostergaard, M.; Olesen, L.H.; Hasle, H.; Kjeldsen, E.; Hokland, P. WT1 gene expression: an excellent tool for monitoring minimal residual disease in 70% of acute myeloid leukaemia patients—results from a single-centre study. Br. J. Haematol. 2004, 125, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Lapillonne, H.; Renneville, A.; Auvrignon, A.; Flamant, C.; Blaise, A.; Perot, C.; Lai, J.L.; Ballerini, P.; Mazingue, F.; Fasola, S.; et al. High WT1 expression after induction therapy predicts high risk of relapse and death in pediatric acute myeloid leukemia. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2006, 24, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Cilloni, D.; Renneville, A.; Hermitte, F.; Hills, R.K.; Daly, S.; Jovanovic, J.V.; Gottardi, E.; Fava, M.; Schnittger, S.; Weiss, T.; et al. Real-time quantitative polymerase chain reaction detection of minimal residual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: a European LeukemiaNet study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 5195–5201. [Google Scholar] [CrossRef]
- Koizumi, Y.; Furuya, D.; Endo, T.; Asanuma, K.; Yanagihara, N.; Takahashi, S. Quantification of Wilms’ tumor 1 mRNA by digital polymerase chain reaction. Int. J. Hematol. 2018, 107, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Othus, M.; Araki, D.; Wood, B.L.; Radich, J.P.; Halpern, A.B.; Mielcarek, M.; Estey, E.H.; Appelbaum, F.R.; Walter, R.B. Pre- and post-transplant quantification of measurable (‘minimal’) residual disease via multiparameter flow cytometry in adult acute myeloid leukemia. Leukemia 2016, 30, 1456–1464. [Google Scholar] [CrossRef]
- Balsat, M.; Renneville, A.; Thomas, X.; de Botton, S.; Caillot, D.; Marceau, A.; Lemasle, E.; Marolleau, J.P.; Nibourel, O.; Berthon, C.; et al. Postinduction Minimal Residual Disease Predicts Outcome and Benefit From Allogeneic Stem Cell Transplantation in Acute Myeloid Leukemia With NPM1 Mutation: A Study by the Acute Leukemia French Association Group. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 185–193. [Google Scholar] [CrossRef]
- Bill, M.; Grimm, J.; Jentzsch, M.; Kloss, L.; Goldmann, K.; Schulz, J.; Beinicke, S.; Hantschel, J.; Cross, M.; Vucinic, V.; et al. Digital droplet PCR-based absolute quantification of pre-transplant NPM1 mutation burden predicts relapse in acute myeloid leukemia patients. Ann. Hematol. 2018, 97, 1757–1765. [Google Scholar] [CrossRef]
- Valero-Garcia, J.; Gonzalez-Espinosa, M.D.C.; Barrios, M.; Carmona-Antonanzas, G.; Garcia-Planells, J.; Ruiz-Lafora, C.; Fuentes-Galvez, A.; Jimenez-Velasco, A. Earlier relapse detection after allogeneic haematopoietic stem cell transplantation by chimerism assays: Digital PCR versus quantitative real-time PCR of insertion/deletion polymorphisms. PLoS ONE 2019, 14, e0212708. [Google Scholar] [CrossRef]
- Kliman, D.; Castellano-Gonzalez, G.; Withers, B.; Street, J.; Tegg, E.; Mirochnik, O.; Lai, J.; Clancy, L.; Gottlieb, D.; Blyth, E. Ultra-Sensitive Droplet Digital PCR for the Assessment of Microchimerism in Cellular Therapies. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2018, 24, 1069–1078. [Google Scholar] [CrossRef]
- Kim, S.Y.; Jeong, M.H.; Park, N.; Ra, E.; Park, H.; Seo, S.H.; Kim, J.Y.; Seong, M.W.; Park, S.S. Chimerism monitoring after allogeneic hematopoietic stem cell transplantation using quantitative real-time PCR of biallelic insertion/deletion polymorphisms. J. Mol. Diagn. 2014, 16, 679–688. [Google Scholar] [CrossRef]
- Stahl, T.; Rothe, C.; Bohme, M.U.; Kohl, A.; Kroger, N.; Fehse, B. Digital PCR Panel for Sensitive Hematopoietic Chimerism Quantification after Allogeneic Stem Cell Transplantation. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef]
- George, D.; Czech, J.; John, B.; Yu, M.; Jennings, L.J. Detection and quantification of chimerism by droplet digital PCR. Chimerism 2013, 4, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Shilatifard, A. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016, 30, 2021–2041. [Google Scholar] [CrossRef]
- Goyama, S.; Kitamura, T. Epigenetics in normal and malignant hematopoiesis: An overview and update 2017. Cancer Sci. 2017, 108, 553–562. [Google Scholar] [CrossRef]
- Saied, M.H.; Marzec, J.; Khalid, S.; Smith, P.; Down, T.A.; Rakyan, V.K.; Molloy, G.; Raghavan, M.; Debernardi, S.; Young, B.D. Genome wide analysis of acute myeloid leukemia reveal leukemia specific methylome and subtype specific hypomethylation of repeats. PLoS ONE 2012, 7, e33213. [Google Scholar] [CrossRef]
- Yang, J.; Ryan, D.J.; Wang, W.; Tsang, J.C.; Lan, G.; Masaki, H.; Gao, X.; Antunes, L.; Yu, Y.; Zhu, Z.; et al. Establishment of mouse expanded potential stem cells. Nature 2017, 550, 393–397. [Google Scholar] [CrossRef]
- Nouzova, M.; Holtan, N.; Oshiro, M.M.; Isett, R.B.; Munoz-Rodriguez, J.L.; List, A.F.; Narro, M.L.; Miller, S.J.; Merchant, N.C.; Futscher, B.W. Epigenomic changes during leukemia cell differentiation: analysis of histone acetylation and cytosine methylation using CpG island microarrays. J. Pharmacol. Exp. Ther. 2004, 311, 968–981. [Google Scholar] [CrossRef]
- Yang, A.S.; Doshi, K.D.; Choi, S.W.; Mason, J.B.; Mannari, R.K.; Gharybian, V.; Luna, R.; Rashid, A.; Shen, L.; Estecio, M.R.; et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006, 66, 5495–5503. [Google Scholar] [CrossRef] [PubMed]
- Jorda, M.; Diez-Villanueva, A.; Mallona, I.; Martin, B.; Lois, S.; Barrera, V.; Esteller, M.; Vavouri, T.; Peinado, M.A. The epigenetic landscape of Alu repeats delineates the structural and functional genomic architecture of colon cancer cells. Genome Res. 2017, 27, 118–132. [Google Scholar] [CrossRef]
- Chen, J.; Huan, W.; Zuo, H.; Zhao, L.; Huang, C.; Liu, X.; Hou, S.; Qi, J.; Shi, W. Alu methylation serves as a biomarker for non-invasive diagnosis of glioma. Oncotarget 2016, 7, 26099–26106. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).