Identification of Salt Stress Responding Genes Using Transcriptome Analysis in Green Alga Chlamydomonas reinhardtii
Abstract
1. Introduction
2. Results
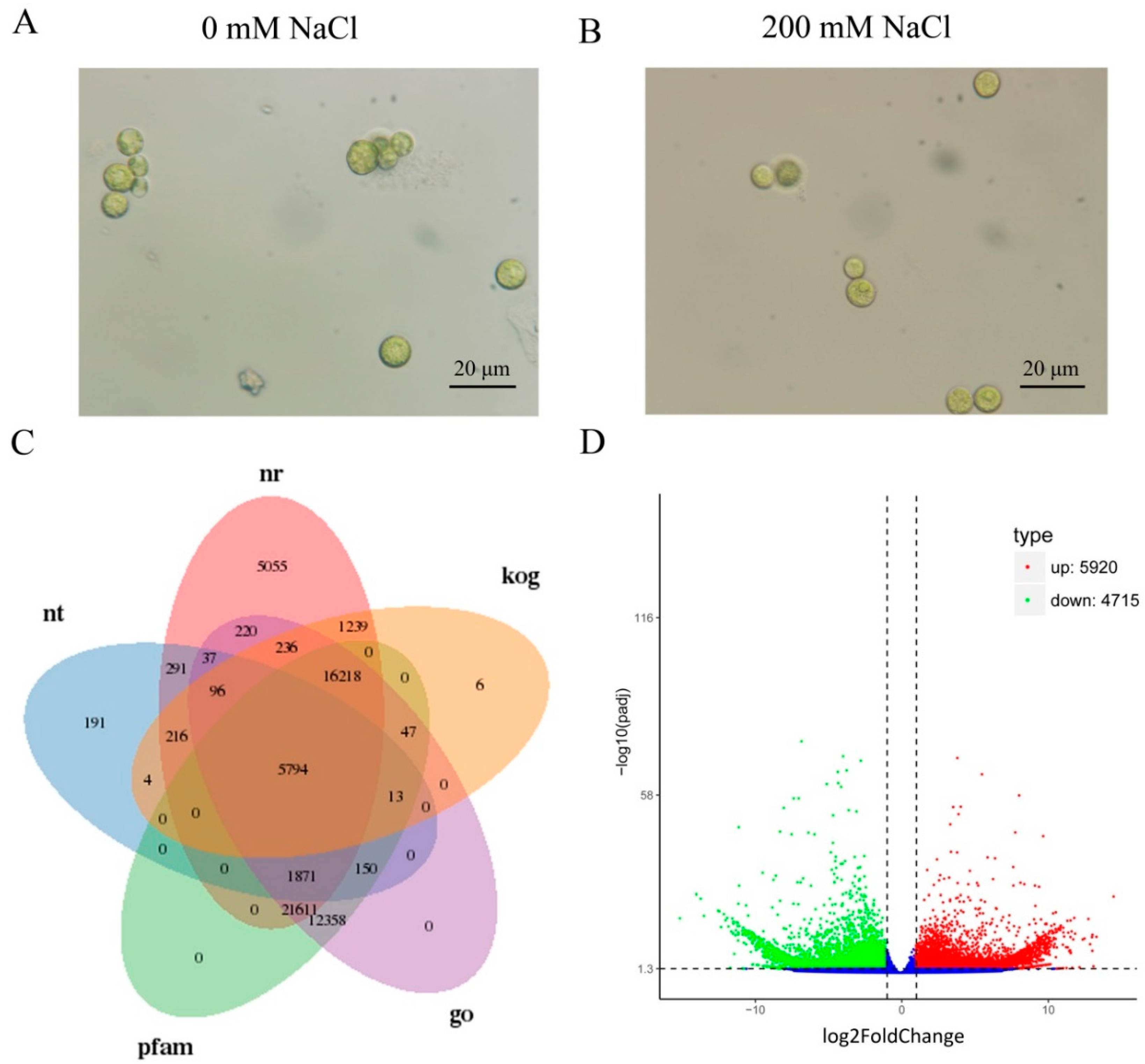
2.1. Transcriptome Profiling of C. reinhardtii
2.2. Functional Annotations of Unigenes
2.3. Differently Expressed Genes (DEGs) Calculation
2.4. GO Enrichment of DEGs
2.5. MapMan Enrichment of DEGs
2.6. KEGG Enrichment of DEGs
2.7. The Differentially Expressed TFs and PKs
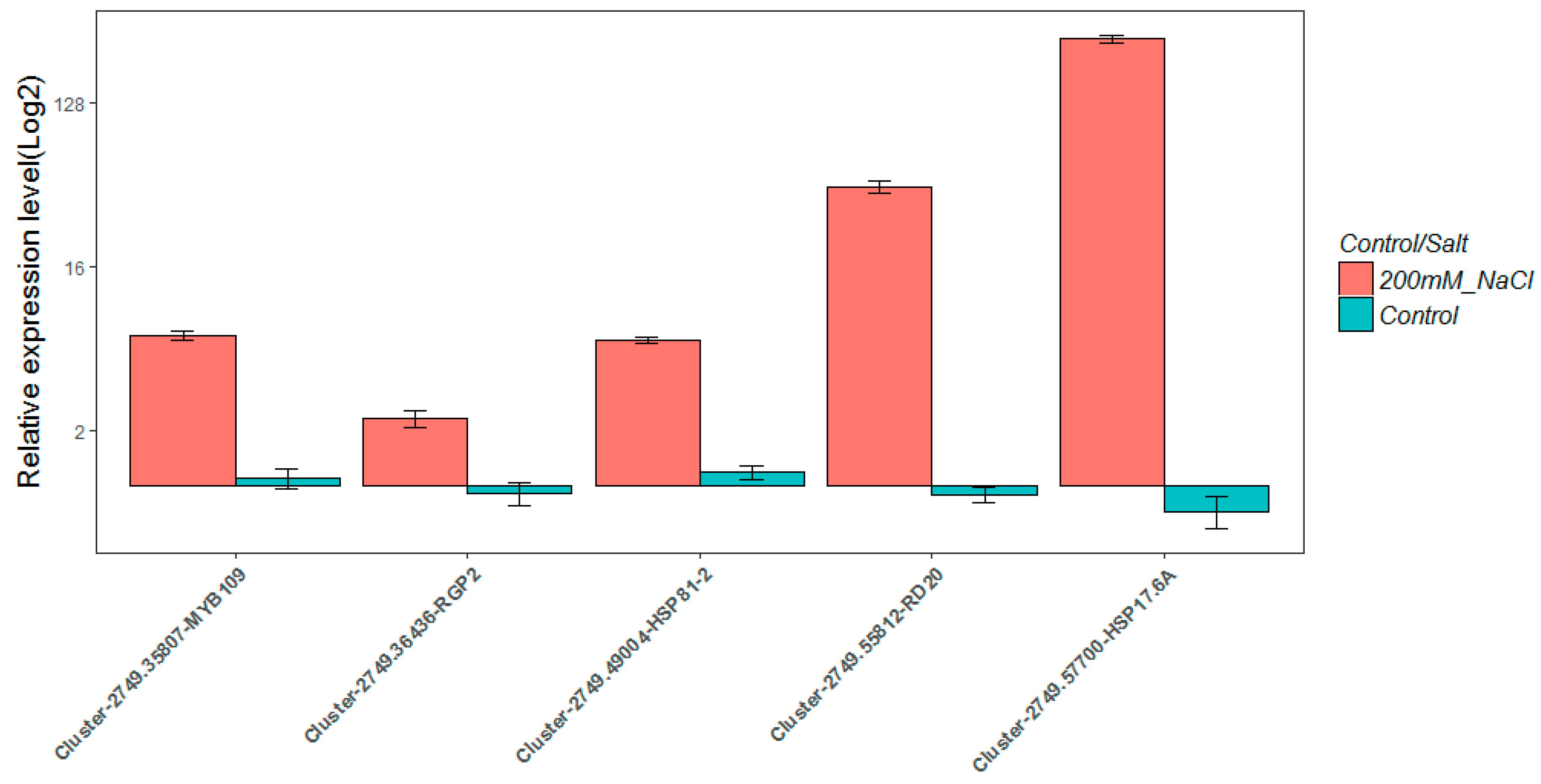
2.8. Real-Time Quantitative PCR Validation
3. Discussion
4. Materials and Methods
4.1. Chlamydomonas Material Preparation, Salt Stress Treatment and RNA Extraction
4.2. Illumina Library Construction and Sequencing
4.3. De Novo Transcriptome Assembling and Unigene Annotation
4.4. Calculation and Comparison of Unigene Expression
4.5. Gene Ontology (GO), Transcription Factors (TFs) and Protein Kinases (PKs) Prediction
4.6. GO, KEGG and MapMan Annotation and Enrichment
4.7. Real-Time Quantitative PCR (qRT-PCR) Verification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DEGs | differentially expressed genes |
| TFs | transcriptional factors |
| PKs | protein kinases |
| GO | gene ontology |
| FPKM | fragment per kilobase of exon model per million mapped reads |
| qRT-PCR | real-time quantitative PCR |
| BP | biological processes |
| PSI | photosystem I |
| PSII | photosystem II |
| PCD | programmed cell death |
| ROS | reactive oxygen species |
| PA | phosphatidic acid |
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Boil. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, B.S. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 2015, 115, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Epstein, E. Salt-tolerant crops: Origins, development, and prospects of the concept. Plant Soil 1985, 89, 187–198. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Zhang, X.J.; Fan, S.J. Meta-analysis of salt-related gene expression profiles identifies common signatures of salt stress responses in Arabidopsis. Plant Syst. Evol. 2017, 303, 757–774. [Google Scholar] [CrossRef]
- Yuan, F.; Leng, B.Y.; Wang, B.S. Progress in Studying Salt Secretion from the Salt Glands in Recretohalophytes: How Do Plants Secrete Salt? Front. Plant Sci. 2016, 7, 977. [Google Scholar] [CrossRef] [PubMed]
- Khona, D.K.; Shirolikar, S.M.; Gawde, K.K.; Hom, E.; Deodhar, M.A.; D’Souza, J.S. Characterization of salt stress-induced palmelloids in the green alga, Chlamydomonas Reinhardtii. Algal Res. 2016, 16, 434–448. [Google Scholar] [CrossRef]
- Shen, X.Y.; Wang, Z.L.; Song, X.F.; Xu, J.J.; Jiang, C.Y.; Zhao, Y.X.; Ma, C.L.; Zhang, H. Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Boil. 2014, 86, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Perrineau, M.M.; Zelzion, E.; Gross, J.; Price, D.C.; Boyd, J.; Bhattacharya, D. Evolution of salt tolerance in a laboratory reared population of Chlamydomonas Reinhardtii. Environ. Microbiol. 2014, 16, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Shabala, S.; Munns, R.; Shabala, S. Salinity stress: Physiological constraints and adaptive mechanisms. In Plant Stress Physiology; CABI: Wallingford, UK, 2012. [Google Scholar]
- Young, M.A.; Rancier, D.G.; Roy, J.L.; Lunn, S.R.; Armstrong, S.A.; Headley, J.V. Seeding conditions of the halophyte Atriplex patula for optimal growth on a salt impacted site. Int. J. Phytoremediat. 2011, 13, 674–680. [Google Scholar] [CrossRef]
- Rao, A.R.; Sarada, R.; Ravishankar, G.A. Enhancement of carotenoids in green alga Botyrocccus braunii in various autotrophic media under stress conditions. Int. J. Biomed. Pharm. Sci. 2010, 4, 87–92. [Google Scholar]
- Stanier, R.Y.; Kunisawa, R.; Mandel, M.; Cohenbazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 1971, 35, 171–205. [Google Scholar] [PubMed]
- Liu, F.; Jin, Z.; Wang, Y.; Bi, Y.; Melton, R.J. Plastid Genome of Dictyopteris divaricata (Dictyotales, Phaeophyceae): Understanding the Evolution of Plastid Genomes in Brown Algae. Mar. Biotechnol. 2017, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sayed, O.H. Chlorophyll Fluorescence as a Tool in Cereal Crop Research. Photosynthetica 2003, 41, 321–330. [Google Scholar] [CrossRef]
- Zuo, Z.; Chen, Z.; Zhu, Y.; Bai, Y.; Wang, Y. Effects of NaCl and Na2CO3 stresses on photosynthetic ability of Chlamydomonas Reinhardtii. Biologia 2014, 69, 1314–1322. [Google Scholar] [CrossRef]
- Fedina, I.S.; Georgieva, K.; Grigorova, I. Response of Barley Seedlings to UV-B Radiation as Affected by Proline and NaCl. J. Plant Physiol. 2003, 47, 549–554. [Google Scholar] [CrossRef]
- Khan, N.A. NaCl-Inhibited Chlorophyll Synthesis and Associated Changes in Ethylene Evolution and Antioxidative Enzyme Activities in Wheat. Boil. Plant. 2003, 47, 437–440. [Google Scholar] [CrossRef]
- Demetriou, G.; Neonaki, C.; Navakoudis, E.; Kotzabasis, K. Salt stress impact on the molecular structure and function of the photosynthetic apparatus—The protective role of polyamines. Biochim. Et Biophys. Acta 2007, 1767, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.D.; Shen, Y.G. Salt shock induces state II transition of the photosynthetic apparatus in dark-adapted Dunaliella salina cells. Environ. Exp. Bot. 2006, 57, 19–24. [Google Scholar] [CrossRef]
- Neelam, S.; Subramanyam, R. Alteration of photochemistry and protein degradation of photosystem II from Chlamydomonas reinhardtii under high salt grown cells. J. Photochem. Photobiol. B Boil. 2013, 124, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Vega, J.M.; Garbayo, I.; Domínguez, M.J.; Vigara, J. Effect of abiotic stress on photosynthesis and respiration in: Induction of oxidative stress. Enzym. Microb. Technol. 2007, 40, 163–167. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Lan, W.J.; He, X.; Shao, Q.; Wang, B.S.; Zhao, X.J. Expression of a Suaeda salsa Vacuolar H+/Ca2+ Transporter Gene in Arabidopsis Contributes to Physiological Changes in Salinity. Plant Mol. Boil. Rep. 2012, 30, 470–477. [Google Scholar] [CrossRef]
- Qi, Y.C.; Liu, W.Q.; Qiu, L.Y.; Zhang, S.M.; Ma, L.; Zhang, H. Overexpression of glutathione S-transferase gene increases salt tolerance of arabidopsis. Russ. J. Plant Physiol. 2010, 57, 233–240. [Google Scholar] [CrossRef]
- Zhang, S.R.; Song, J.; Wang, H.; Feng, G. Effect of salinity on seed germination, ion content and photosynthesis of cotyledons in halophytes or xerophyte growing in Central Asia. J. Plant Ecol. 2010, 3, 259–267. [Google Scholar] [CrossRef]
- Lu, T.; Lu, G.; Fan, D.; Zhu, C.; Wei, L.; Qiang, Z.; Qi, F.; Yan, Z.; Guo, Y.; Li, W. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010, 20, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Xiao, Z.; Li, M.W.; Wong, F.L.; Yung, W.S.; Ku, Y.S.; Wang, Q.; Wang, X.; Xie, M.; Yim, A.K. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 2018. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Sui, N.; Duan, G.; Liu, Y.; Han, Y.; Liu, S.; Wan, S.; Li, G. Identification of Metabolites and Transcripts Involved in Salt Stress and Recovery in Peanut. Front. Plant Sci. 2018, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, Y.; Han, G.; Song, J.; Wang, B. NaCl markedly improved the reproductive capacity of the euhalophyte Suaeda Salsa. Funct. Plant Boil. 2018, 45, 350. [Google Scholar] [CrossRef]
- Cao, S.; Du, X.H.; Li, L.H.; Liu, Y.D.; Zhang, L.; Pan, X.; Li, Y.; Li, H.; Lu, H. Overexpression of Populus tomentosa cytosolic ascorbate peroxidase enhances abiotic stress tolerance in tobacco plants. Russ. J. Plant Physiol. 2017, 64, 224–234. [Google Scholar] [CrossRef]
- Sui, N.; Tian, S.S.; Wang, W.Q.; Wang, M.J.; Fan, H. Overexpression of Glycerol-3-Phosphate Acyltransferase from Suaeda salsa Improves Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1337. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Zhang, Q.; Cui, F.; Hou, L.; Zhao, S.Z.; Xia, H.; Qiu, J.J.; Li, T.T.; Zhang, Y.; Wang, X.J.; et al. Genome-Wide Analysis of Gene Expression Provides New Insights into Cold Responses in Thellungiella Salsuginea. Front. Plant Sci. 2017, 8, 713. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Lyu, M.J.A.; Leng, B.Y.; Zhu, X.G.; Wang, B.S. The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Boil. 2016, 91, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Lyu, M.J.A.; Leng, B.Y.; Zheng, G.Y.; Feng, Z.T.; Li, P.H.; Zhu, X.G.; Wang, B.S. Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 2015, 38, 1637–1657. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.T.; Deng, Y.Q.; Fan, H.; Sun, Q.J.; Sui, N.; Wang, B.S. Effects of NaCl stress on the growth and photosynthetic characteristics of Ulmus pumila L. seedlings in sand culture. Photosynthetica 2014, 52, 313–320. [Google Scholar] [CrossRef]
- Yuan, F.; Chen, M.; Yang, J.C.; Leng, B.Y.; Wang, B.S. A system for the transformation and regeneration of the recretohalophyte Limonium bicolor. In Vitro Cell. Dev. Boil. Plant 2014, 50, 610–617. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, C.Z.; Li, M.; Sun, W.; Liu, Y.; Xia, H.; Sun, M.N.; Li, A.Q.; Li, C.S.; Zhao, S.Z.; et al. Genome-wide identification of Thellungiella salsuginea microRNAs with putative roles in the salt stress response. BMC Plant Boil. 2013, 13, 180. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.H.; Jia, W.J.; Song, J.; Wang, D.A.; Chen, M.; Wang, B.S. Thellungilla halophila is more adaptive to salinity than Arabidopsis thaliana at stages of seed germination and seedling establishment. Acta Physiol. Plant. 2012, 34, 1287–1294. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, F.; Zhou, J.J.; Chen, F.; Wang, B.S.; Xie, X.Z. Phytochrome B control of total leaf area and stomatal density affects drought tolerance in rice. Plant Mol. Boil. 2012, 78, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.C.; Fu, T.T.; Sui, N.; Guo, J.R.; Feng, G.; Fan, J.L.; Song, J. The role of salinity in seed maturation of the euhalophyte Suaeda Salsa. Plant Biosyst. 2016, 150, 83–90. [Google Scholar] [CrossRef]
- Waśkiewicz, A.; Muzolf-Panek, M.; Goliński, P. Phenolic Content Changes in Plants under Salt Stress; Springer: New York, NY, USA, 2013; pp. 283–314. [Google Scholar]
- Sudhir, P.; Murthy, S.D.S. Effects of salt stress on basic processes of photosynthesis. Photosynthetica 2004, 42, 481–486. [Google Scholar] [CrossRef]
- Maršálek, B.; Zahradníčková, H.; Hronková, M. Extracellular Production of Abscisic Acid by Soil Algae under Salt, Acid or Drought Stress. Z. Für Naturforschung C 1992, 47, 701–704. [Google Scholar] [CrossRef]
- Pineau, B.; Gérard-Hirne, C.; Selve, C. Carotenoid binding to photosystems I and II of Chlamydomonas reinhardtii, cells grown under weak light or exposed to intense light. Plant Physiol. Bioch. 2001, 39, 73–85. [Google Scholar] [CrossRef]
- Ding, F.; Chen, M.; Sui, N.; Wang, B.S. Ca2+ significantly enhanced development and salt-secretion rate of salt glands of Limonium bicolor under NaCl treatment. S. Afr. J. Bot. 2010, 76, 95–101. [Google Scholar] [CrossRef]
- Feng, Z.T.; Deng, Y.Q.; Zhang, S.C.; Liang, X.; Yuan, F.; Hao, J.L.; Zhang, J.C.; Sun, S.F.; Wang, B.S. K+ accumulation in the cytoplasm and nucleus of the salt gland cells of Limonium bicolor accompanies increased rates of salt secretion under NaCl treatment using NanoSIMS. Plant Sci. 2015, 238, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Shao, Q.; Bao, H.Y.; Wang, B.S. Cloning and Characterization of a Ca2+/H+ Antiporter from Halophyte Suaeda salsa L. Plant Mol. Boil. Rep. 2011, 29, 449–457. [Google Scholar] [CrossRef]
- Yang, S.; Li, L.; Zhang, J.; Geng, Y.; Guo, F.; Wang, J.; Meng, J.; Sui, N.; Wan, S.; Li, X. Transcriptome and Differential Expression Profiling Analysis of the Mechanism of Ca(2+) Regulation in Peanut (Arachis hypogaea) Pod Development. Front. Plant Sci. 2017, 8, 1609. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Liao, C.C.; Zhao, S.S.; Wang, C.W.; Guo, Y. The Glycosyltransferase QUA1 Regulates Chloroplast-Associated Calcium Signaling During Salt and Drought Stress in Arabidopsis. Plant Cell Physiol. 2017, 58, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Zhang, Z.; Zhang, X.J.; Yao, Y.; Wang, R.; Duan, B.Y.; Fan, S.J. Comprehensive meta-analysis and co-expression network analysis identify candidate genes for salt stress response in Arabidopsis. Plant Biosyst. 2018. [Google Scholar] [CrossRef]
- Arisz, S.A.; Valianpour, F.; van Gennip, A.H.; Munnik, T. Substrate preference of stress-activated phospholipase D in Chlamydomonas and its contribution to PA formation. Plant J. Cell Mol. Boil. 2003, 34, 595–604. [Google Scholar] [CrossRef]
- Zhou, J.J.; Liu, Q.Q.; Zhang, F.; Wang, Y.Y.; Zhang, S.Y.; Cheng, H.M.; Yan, L.H.; Li, L.; Chen, F.; Xie, X.Z. Overexpression of OsPIL15, a phytochromeinteracting factor- like protein gene, represses etiolated seedling growth in rice. J. Integr. Plant Boil. 2014, 56, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Li, M.; Li, K.; Song, J.; Wang, B.S. Increase in unsaturated fatty acids in membrane lipids of Suaeda salsa L. enhances protection of photosystem II under high salinity. Photosynthetica 2010, 48, 623–629. [Google Scholar] [CrossRef]
- Arisz, S.A.; Munnik, T. The salt stress-induced LPA response in Chlamydomonas is produced via PLA2 hydrolysis of DGK-generated phosphatidic acid. J. Lipid Res. 2011, 52, 2012–2020. [Google Scholar] [CrossRef] [PubMed]
- Meijer, H.J.; Arisz, S.A.; Van Himbergen, J.A.; Musgrave, A.; Munnik, T. Hyperosmotic stress rapidly generates lyso-phosphatidic acid in Chlamydomonas. Plant J. 2010, 25, 541–548. [Google Scholar] [CrossRef]
- Takagi, M.; Karseno; Yoshida, T. Effect of Salt Concentration on Intracellular Accumulation of Lipids and Triacylglyceride in Marine Microalgae Dunaliella Cells. J. Biosci. Bioeng. 2006, 101, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A. Osmoregulation in Dunaliella, Part II: Photosynthesis and starch contribute carbon for glycerol synthesis during a salt stress in Dunaliella tertiolecta. Plant Physiol. Biochem. 2007, 45, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Yokthongwattana, C.; Mahong, B.; Roytrakul, S.; Phaonaklop, N.; Narangajavana, J.; Yokthongwattana, K. Proteomic analysis of salinity-stressed Chlamydomonas reinhardtii revealed differential suppression and induction of a large number of important housekeeping proteins. Planta 2012, 235, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.B.; Qi, X.Y.; Wang, Z.L.; Li, P.H.; Wu, C.X.; Zhang, H.; Zhao, Y.X. Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol. Biochem. 2013, 69, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Pang, C.H.; Li, K.; Wang, B.S. Overexpression of SsCHLAPXs confers protection against oxidative stress induced by high light in transgenic Arabidopsis thaliana. Physiol. Plant. 2011, 143, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Ferjani, A.; Mustardy, L.; Sulpice, R.; Marin, K.; Suzuki, I.; Hagemann, M.; Murata, N. Glucosylglycerol, a compatible solute, sustains cell division under salt stress. Plant Physiol. 2003, 131, 1628–1637. [Google Scholar] [CrossRef] [PubMed]
- Zeid, I.M. Trehalose as osmoprotectant for maize under salinity-induced stress. Res. J. Agric. Boil. Sci. 2009, 5, 613–622. [Google Scholar]
- Xu, J.J.; Li, Y.Y.; Ma, X.L.; Ding, J.F.; Wang, K.; Wang, S.S.; Tian, Y.; Zhang, H.; Zhu, X.G. Whole transcriptome analysis using next-generation sequencing of model species Setaria viridis to support C4 photosynthesis research. Plant Mol. Boil. 2013, 83, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Han, G.L. Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. Acta Physiol. Plant. 2014, 36, 983–992. [Google Scholar] [CrossRef]
- Heifetz, P.B.; Boynton, J.E. Effects of Acetate on Facultative Autotrophy in Chlamydomonas reinhardtii Assessed by Photosynthetic Measurements and Stable Isotope Analyses. Plant Physiol. 2000, 122, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Soussi, M.; Ocaña, A.; Lluch, C. Effects of salt stress on growth, photosynthesis and nitrogen fixation in chick-pea (Cicer arietinum L.). J. Exp. Bot. 1998, 49, 1329–1337. [Google Scholar] [CrossRef]
- Zhong, M.; Yuan, Y.; Shu, S.; Sun, J.; Guo, S.; Yuan, R.; Tang, Y. Effects of exogenous putrescine on glycolysis and Krebs cycle metabolism in cucumber leaves subjected to salt stress. Plant Growth Regul. 2016, 79, 319–330. [Google Scholar] [CrossRef]
- Huang, J.; Li, Z.Y.; Biener, G.; Xiong, E.H.; Malik, S.; Eaton, N.; Zhao, C.Z.; Raicu, V.; Kong, H.Z.; Zhao, D.Z. Carbonic Anhydrases Function in Anther Cell Differentiation Downstream of the Receptor-Like Kinase EMS1. Plant Cell 2017, 29, 1335–1356. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.X.; Xu, Y.G.; Wang, S.; Shi, W.W.; Liu, R.R.; Feng, G.; Song, J. Salinity affects production and salt tolerance of dimorphic seeds of Suaeda Salsa. Plant Physiol. Biochem. 2015, 95, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Han, N.; Ding, T.L.; Zhou, F.; Wang, B.S. SsHKT1;1 is a potassium transporter of the C3 halophyte Suaeda salsa that is involved in salt tolerance. Funct. Plant Boil. 2014, 41, 790–802. [Google Scholar] [CrossRef]
- Li, K.; Pang, C.H.; Ding, F.; Sui, N.; Feng, Z.T.; Wang, B.S. Overexpression of Suaeda salsa stroma ascorbate peroxidase in Arabidopsis chloroplasts enhances salt tolerance of plants. S. Afr. J. Bot. 2012, 78, 235–245. [Google Scholar] [CrossRef]
- Liu, S.S.; Wang, W.Q.; Li, M.; Wan, S.B.; Sui, N. Antioxidants and unsaturated fatty acids are involved in salt tolerance in peanut. Acta Physiol. Plant. 2017, 39, 207. [Google Scholar] [CrossRef]
- Chen, M.; Song, J.; Wang, B.S. NaCl increases the activity of the plasma membrane H+-ATPase in C3 halophyte Suaeda salsa callus. Acta Physiol. Plant. 2010, 32, 27–36. [Google Scholar] [CrossRef]
- Song, J.; Shi, G.W.; Gao, B.; Fan, H.; Wang, B.S. Waterlogging and salinity effects on two Suaeda salsa populations. Physiol. Plant. 2011, 141, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Yang, D.Y.; Li, X.D.; Zhao, S.Y.; Sui, N.; Meng, Q.W. Physiological changes in fruit ripening caused by overexpression of tomato SlAN2, an R2R3-MYB factor. Plant Physiol. Biochem. 2015, 89, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Ma, X.L.; Zhao, J.L.; Xu, J.J.; Shi, J.F.; Zhu, X.G.; Zhao, Y.X.; Zhang, H. Developmental Genetic Mechanisms of C4 Syndrome Based on Transcriptome Analysis of C3 Cotyledons and C4 Assimilating Shoots in Haloxylon Ammodendron. PLoS ONE 2015, 10, e0117175. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-Seq: Reference generation and analysis with Trinity. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed]
- Okuda, S.; Yamada, T.; Hamajima, M.; Itoh, M.; Katayama, T.; Bork, P.; Goto, S.; Kanehisa, M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008, 36, W423–W426. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mao, X.; Cai, T.; Luo, J.; Wei, L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Blasing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Kruger, P.; Selbig, J.; Muller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. Cell Mol. Boil. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Sample Name | Sample Description | Total Reads | Total Mapped | Ratio of Mapped Reads |
|---|---|---|---|---|
| C_0_1 | Control replication 1 | 56,438,218 | 51,454,456 | 91.17% |
| C_0_2 | Control replication 2 | 72,853,712 | 66,008,290 | 90.60% |
| C_0_3 | Control replication 3 | 47,551,786 | 43,268,544 | 90.99% |
| S_200_1 | Salt stress replication 1 | 56,962,722 | 51,633,614 | 90.64% |
| S_200_2 | Salt stress replication 2 | 52,926,804 | 47,815,814 | 90.34% |
| S_200_3 | Salt stress replication 3 | 55,998,748 | 50,507,824 | 90.19% |
| Gene_ID | L2fc | pval | BP Description |
|---|---|---|---|
| Up-regulated | |||
| Cluster-2749.47186 | 3.894 | 3.77 × 10−75 | |
| Cluster-2749.64181 | 5.573 | 1.55 × 10−69 | transcription, DNA-templated |
| Cluster-2749.61362 | 8.112 | 1.95 × 10−62 | potassium ion transport |
| Cluster-2749.33332 | 4.129 | 1.19 × 10−58 | signal transduction |
| Cluster-2749.48242 | 3.610 | 1.64 × 10−58 | |
| Cluster-2749.21356 | 3.975 | 4.34 × 10−56 | |
| Cluster-2749.37168 | 3.413 | 1.00 × 10−52 | |
| Cluster-2749.23874 | 7.849 | 5.01 × 10−50 | lipid metabolic process |
| Cluster-2749.57700 | 9.756 | 9.76 × 10−49 | iron-sulfur cluster assembly |
| Cluster-2749.59287 | 3.459 | 1.42 × 10−43 | cell adhesion |
| Cluster-2749.53252 | 3.877 | 2.36 × 10−43 | pathogenesis |
| Cluster-2749.49912 | 5.957 | 1.07 × 10−41 | lipoprotein metabolic process |
| Cluster-2749.84953 | 6.468 | 2.29 × 10−41 | |
| Cluster-2749.82821 | 2.504 | 5.20 × 10−41 | regulation of protein kinase activity |
| Cluster-2749.3203 | 7.706 | 1.83 × 10−38 | |
| Down-regulated | |||
| Cluster-2749.44503 | −8.623 | 4.01 × 10−178 | chlorophyll metabolic process |
| Cluster-2749.61923 | −6.748 | 6.07 × 10−81 | proteolysis |
| Cluster-2749.38883 | −3.906 | 7.54 × 10−76 | |
| Cluster-2749.44595 | −2.699 | 3.50 × 10−74 | metabolic process |
| Cluster-2749.45379 | −3.663 | 6.53 × 10−71 | regulation of transcription, DNA-templated |
| Cluster-2749.49076 | −4.268 | 2.29 × 10−70 | chlorophyll biosynthetic process |
| Cluster-2749.44117 | −4.239 | 1.30 × 10−66 | oxidation-reduction process |
| Cluster-2749.42573 | −5.023 | 3.04 × 10−66 | protein glycosylation |
| Cluster-2749.32226 | −4.043 | 2.67 × 10−65 | proteolysis |
| Cluster-2749.45636 | −7.283 | 1.98 × 10−61 | |
| Cluster-2749.44732 | −6.934 | 2.08 × 10−61 | |
| Cluster-2749.49721 | −7.951 | 3.18 × 10−58 | |
| Cluster-2749.65261 | −3.524 | 1.91 × 10−57 | |
| Cluster-2749.36258 | −2.996 | 5.32 × 10−57 | |
| Cluster-2749.43872 | −4.589 | 1.10 × 10−55 | cell adhesion |
| GO ID | GO Term | Annotated Gene Number | Enriched Gene Number | p-Value |
|---|---|---|---|---|
| Up-Regulated | ||||
| GO:0008150 | biological process | 33682 | 2820 | 1.00 × 10−30 |
| GO:0055114 | oxidation-reduction process | 3653 | 385 | 2.90 × 10−27 |
| GO:0046686 | response to cadmium ion | 1317 | 159 | 3.40 × 10−18 |
| GO:0042542 | response to hydrogen peroxide | 189 | 41 | 1.10 × 10−15 |
| GO:0009408 | response to heat | 717 | 122 | 1.40 × 10−15 |
| GO:0051259 | protein oligomerization | 109 | 25 | 7.50 × 10−12 |
| GO:0010090 | trichome morphogenesis | 131 | 26 | 4.60 × 10−10 |
| GO:0009414 | response to water deprivation | 668 | 79 | 6.70 × 10−10 |
| GO:0009651 | response to salt stress | 1488 | 143 | 3.90 × 10−09 |
| GO:0043335 | protein unfolding | 39 | 14 | 1.80 × 10−08 |
| GO:0016036 | cellular response to phosphate starvation | 262 | 40 | 2.40 × 10−08 |
| GO:0010030 | positive regulation of seed germination | 85 | 20 | 6.50 × 10−08 |
| GO:0030866 | cortical actin cytoskeleton organization | 31 | 12 | 7.20 × 10−08 |
| GO:0016477 | cell migration | 31 | 12 | 7.20 × 10−08 |
| GO:0045010 | actin nucleation | 31 | 12 | 7.20 × 10−08 |
| Down-Regulated | ||||
| GO:0008150 | biological process | 33682 | 2018 | 1.00 × 10−30 |
| GO:0009768 | photosynthesis, light harvesting in photosystem I | 87 | 46 | 1.00 × 10−30 |
| GO:0009645 | response to low light intensity stimulus | 72 | 37 | 1.00 × 10−30 |
| GO:0015995 | chlorophyll biosynthetic process | 242 | 54 | 4.40 × 10−29 |
| GO:0009644 | response to high light intensity | 393 | 71 | 6.70 × 10−22 |
| GO:0006412 | translation | 1779 | 179 | 3.80 × 10−16 |
| GO:0009409 | response to cold | 978 | 103 | 5.30 × 10−16 |
| GO:0009269 | response to desiccation | 41 | 18 | 1.00 × 10−14 |
| GO:0009769 | photosynthesis, light harvesting in photosystem II | 36 | 17 | 1.30 × 10−14 |
| GO:0010218 | response to far red light | 101 | 25 | 8.70 × 10−14 |
| GO:0006364 | rRNA processing | 742 | 89 | 2.10 × 10−12 |
| GO:0010114 | response to red light | 159 | 28 | 5.90 × 10−11 |
| GO:0015979 | photosynthesis | 853 | 137 | 2.40 × 10−10 |
| GO:0009097 | isoleucine biosynthetic process | 53 | 16 | 2.60 × 10−10 |
| GO:0009099 | valine biosynthetic process | 43 | 14 | 1.10 × 10−09 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Qian, Z.; Luo, M.; Fan, S.; Zhang, X.; Zhang, L. Identification of Salt Stress Responding Genes Using Transcriptome Analysis in Green Alga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2018, 19, 3359. https://doi.org/10.3390/ijms19113359
Wang N, Qian Z, Luo M, Fan S, Zhang X, Zhang L. Identification of Salt Stress Responding Genes Using Transcriptome Analysis in Green Alga Chlamydomonas reinhardtii. International Journal of Molecular Sciences. 2018; 19(11):3359. https://doi.org/10.3390/ijms19113359
Chicago/Turabian StyleWang, Ning, Zhixin Qian, Manwei Luo, Shoujin Fan, Xuejie Zhang, and Luoyan Zhang. 2018. "Identification of Salt Stress Responding Genes Using Transcriptome Analysis in Green Alga Chlamydomonas reinhardtii" International Journal of Molecular Sciences 19, no. 11: 3359. https://doi.org/10.3390/ijms19113359
APA StyleWang, N., Qian, Z., Luo, M., Fan, S., Zhang, X., & Zhang, L. (2018). Identification of Salt Stress Responding Genes Using Transcriptome Analysis in Green Alga Chlamydomonas reinhardtii. International Journal of Molecular Sciences, 19(11), 3359. https://doi.org/10.3390/ijms19113359





