Abstract
N-acetyl-5-methoxytryptamine (Melatonin), as a crucial messenger in plants, functions in adjusting biological rhythms, stress tolerance, plant growth and development. Several studies have shown the retardation effect of exogenous melatonin treatment on plant growth and development. However, the in vivo role of melatonin in regulating plant leaf growth and the underlying mechanism are still unclear. In this study, we found that high concentration of melatonin suppressed leaf growth in Arabidopsis by reducing both cell size and cell number. Further kinetic analysis of the fifth leaves showed that melatonin remarkably inhibited cell division rate. Additionally, flow cytometic analysis indicated that melatonin negatively regulated endoreduplication during leaf development. Consistently, the expression analysis revealed that melatonin regulated the transcriptional levels of key genes of cell cycle and ribosome. Taken together, this study suggests that high concentration of melatonin negatively regulated the leaf growth and development in Arabidopsis, through modulation of endoreduplication and the transcripts of cell cycle and ribosomal key genes.
Keywords:
melatonin; leaf growth; cell proliferation; cell expansion; endoreduplication; Arabidopsis 1. Introduction
N-acetyl-5-methoxytryptamine (Melatonin), as an endogenous biomolecule first discovered in the pineal gland of cow [1], was later found to be extensively in almost all living organisms, including plants [2,3,4,5,6]. Melatonin participates in the regulation of many physiological processes in animals, including sleep, circadian rhythms, body temperature regulation, immune responses, etc. [7,8,9,10]. In the most recent 20 years, studies on melatonin have revealed that it functions as an important messenger in regulating the response of plant to both abiotic and biotic stresses, including cold, heat, salt, drought, cadmium, zinc sulfate, and pathogen attacks [11,12,13,14,15,16,17,18,19,20,21,22]. Melatonin also participates in the processes of root growth and architecture, shoot development, flowering, seed germination and fruit ripening [23,24,25,26,27,28,29]. In addition, the auxin-like effects of melatonin are controversial [23,30,31,32,33,34,35,36,37].
Organ size is an important feature of plant morphology. With the determinate growth fate, plant organ size is relative constancy within a given species; however, it varied remarkably among different plant species, indicating the final size of an organ is mainly determined by endogenous generated signals [38,39]. Besides, the motionlessness of plants makes them more susceptible to exogenously generated signals, such as light, temperature, water, nutrients, plant hormones, biotic stress, and so on [40,41,42]. The leaf occurrence and development in Arabidopsis thaliana consists of three important processes: leaf primordia initiation, leaf polarity establishment and leaf growth [43]. Cell proliferation and expansion are two different but interconnected events that are necessary for the process of leaf growth. Cell proliferation generates new cells from primordium with relatively small cell size, while cell expansion causes further growth in size of the new cells [44,45]. During the switching phase, cell proliferation of leaves first stops at leaf-apex, thereafter most cells exit the cell cycle gradually, and then the leaf cells start to expand from the tip to the base [45,46,47]. The final leaf size is the result of strict spatial and temporal genetic control and coordination of these two successive but overlapping phases [45]. However, only changes on level of cell proliferation or cell expansion will not certainly alter the leaf size because of the compensation effect caused by reduced cell proliferation and the DNA ploidy increase resulted from endoreduplication [38,48]. Thus, leaf size is not simply controlled by cell number or cell size [38,48,49].
To date, although numerous studies have shown that melatonin play important roles in plant growth, whether melatonin participates in regulating leaf growth and the underlying mechanism are still unclear in higher plants. In this study, wild-type Arabidopsis (WT, Columbia-0 ecotype) were treated with various concentrations of melatonin, and the results showed that high concentration of melatonin dramatically suppressed leaf growth by reducing the cell number and cell size. Further comprehensive analyses suggested that melatonin might regulate the leaf growth by inhibiting cell proliferation and endoreduplication.
2. Results
2.1. High Concentration of Melatonin Suppresses the Leaf Growth in Arabidopsis by Reduced Cell Size and Cell Number
To investigate the effect of melatonin on leaf growth in Arabidopsis, WT (Col-0) seeds were firstly germinated for one day on 1/2 MS (Murashige & Skoog) media to alleviate the effect of melatonin on seeds germination; then, the germinated seeds were transferred onto new 1/2 MS media with different concentrations of melatonin and cultured for another six days. Cotyledons of the samples were harvested and photographed at indicated time-points. By measuring and statistical analysis, results showed that the melatonin treatment decreased the leaf size in concentration-dependent manner (Figure 1B). After six-day treatment, both 600 and 1000 μM melatonin caused significant decrease of leaf size, with the inhibition level of 27.4% and 40.3%, respectively. The regulation of organ size is mediated by cell proliferation and cell expansion [50]. Many regulatory factors eventually lead to changes in cell number or cell size. Thus, we wonder if melatonin reduces the leaf size by affecting cell size or cell number of cotyledons. To test our hypothesis, cotyledons of seedlings grown under indicated concentrations of melatonin were then bleached and cleared for observation. We found that both average cell area and cell number of palisade cells were significantly suppressed in cotyledons treated with 600 and 1000 μM melatonin (Figure 1A,C,D). Our data showed that low concentration of melatonin had little effect on cotyledon growth (Figure 1), and our previous study indicated that 10–50 μM melatonin had little effect on endogenous melatonin content [18]. Thus, 1000 μM melatonin was used to treat the Arabidopsis seedlings for further analyses in this study.
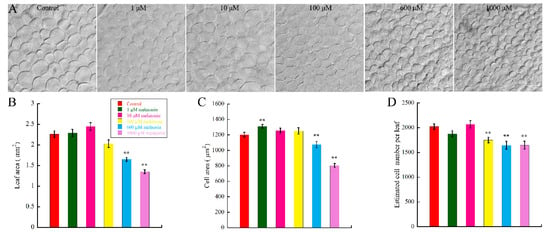
Figure 1.
Effect of high concentration of melatonin on leaf size, cell size and cell number of Arabidopsis cotyledons. After one-day germination, seeds were transferred to 1/2 MS (Murashige & Skoog) medium with indicated concentrations of melatonin for another six days, and the leaf area, cell area and cell number were measured with software Image J. (A) Palisade cells of cotyledons cultured in control and melatonin treated medium. Scale bar = 50 μm; (B) Leaf area; (C) Cell area; and (D) Estimated cell number per leaf of Arabidopsis growing on medium with control and increasing concentration of melatonin. More than 20 cotyledons from 10 seedlings per experiment from three independent experiments were measured for statistic analysis. Values represent mean ± SD, ** p < 0.01 by a Student’s t-test.
To deeply reveal the long-time effect of melatonin on the growth and development of Arabidopsis leaves, we performed the kinetic analysis after treatment at 6, 11, 16 and 21 days. The results showed that the treated seedlings exhibited smaller size compared with the control (Figure 2A). Thus, the cotyledons at 6, 11, 16 and 21 days; first pair of true leaves (leaves 1 + 2) at 11, 16 and 21 days; and the fifth leaves (leaf 5) at 16 and 21 days were selected for statistical analysis. We found that the leaf area was inhibited dramatically by melatonin treatment (Figure 2B–D). Both palisade cell area and cell number per leaf were decreased in the melatonin treated plants, except for the palisade cell number in cotyledons harvested at 11, 16 and 21 days (Figure 2E–J). Taken together, these results indicated that melatonin-mediated repression of leaf size might be due to both reduced cell size and cell number per leaf, as evidenced by the inhibition of cell proliferation and expansion during leaf growth in Arabidopsis.
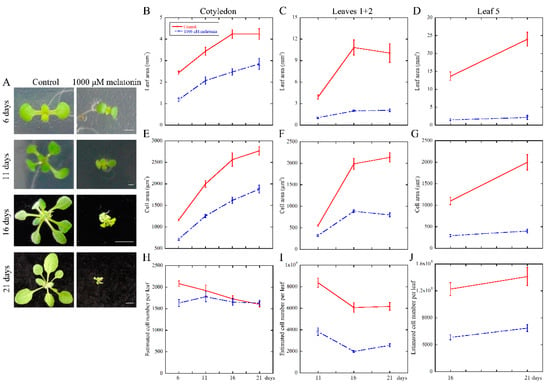
Figure 2.
High concentration of melatonin restricted the development of leaf area by reducing the cell area and cell number of Arabidopsis seedlings. One-day-old seeds were kept growing under control and 1000 μM melatonin for another 6, 11, 16 and 21 days. (A) Digital images of control and 1000 μM melatonin treated Arabidopsis seedlings at a series of distinct developmental stages. Scale bars represented 1, 1, 5 and 5 mm, respectively, from the top to bottom; (B–J) Growth kinetic analysis of leaves in Arabidopsis seedlings grown under control and 1000 μM melatonin. (B) Leaf are; (E) cell area and (H) estimated cell number of cotyledons selected at 6, 11, 16 and 21 days after germination; (C) Leaf area; (F) cell area and (I) estimated cell number of leaves 1 + 2 harvested at 11, 16 and 21 days, and (D) Leaf area; (G) cell area and (J) estimated cell number of the fifth leaves were picked at 16 and 21 days were analyzed. At least 10 seedlings were selected to measure the leaf area, cell area and estimated cell number per leaf. Three independent experiments were measured for statistical analysis. Values represent mean ± SE.
2.2. Melatonin Negatively Affects the Cell Division Rate of Leaves During Leaf Growth
To further illustrate the relationship between melatonin and cell proliferation during leaf development, we continued monitoring the growth kinetics of the fifth leaves of Arabidopsis after treatment for 14 to 22 days. Then, the cell division rate of palisade cells were estimated [51], as shown in Figure 3B,C. We found that 1000 μM melatonin treatment obviously reduced the cell division rate of palisade cells (Figure 3D), suggesting that melatonin treatment caused defective cell proliferation. In the 48-h interval between Day 14 and Day 16, the cell division rate of control and melatonin treated palisade cells was 0.0145 cells cell−1 h−1 and 0.0028 cells cell−1 h−1, respectively. The cell cycle duration of control palisade cells was 1/0.0145 h (68.9 h), while that of the melatonin treated samples was 1/0.0028 h (357.1 h), indicating that the duration of the cell cycle was prolonged by the melatonin treatment. These data suggested that melatonin suppressed the cell number in Arabidopsis leaves by reducing the cell division rate.
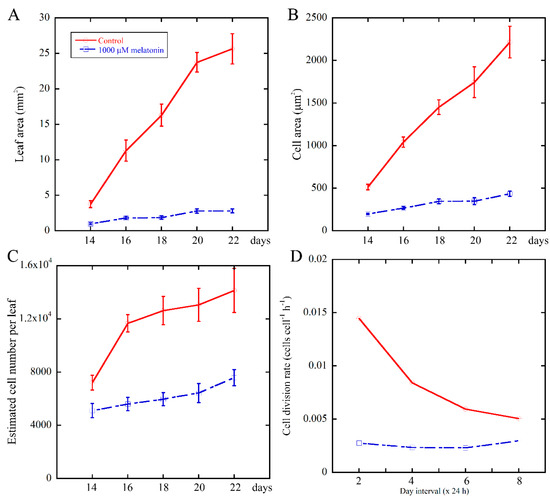
Figure 3.
Growth kinetics of the fifth leaves in Arabidopsis seedlings grown under control and 1000 μM melatonin. Leaf area (A); cell area (B); estimated cell number per leaf (C); and cell division rate (D) were determined from at least 10 leaves for each independent experiments. Three biological replicates were taken for statistical analysis. Values represent mean ± SE.
2.3. Melatonin Regulate Endoreduplication during Leaf Development
A large increase of cell size is mainly achieved by cell expansion, which is closely related with endoreduplication, in which several rounds of DNA replication take place [52,53,54]. The above results suggested that melatonin negatively altered the cell size of Arabidopsis, so we wondered that whether melatonin actually affects DNA ploidy during cell cycle. The cotyledons, leaves 1 + 2 and the fifth leaves in Arabidopsis 21-day seedlings of control and melatonin treated were harvested for flow cytometic analysis. As shown in Figure 4A–C, melatonin caused significant increase of 2C fractions in all three tissues, and corresponding decrease of 8C and 16C fractions in cotyledons and leaves 1 + 2, while the 4C and 8C fractions of the fifth leaves were reduced by melatonin. The endoreduplication index (EI) represents the average number of endocycles performed per cells. As showed in Figure 4D, melatonin significantly reduced the EI values of cotyledons, leaves 1 + 2 and the fifth leaves by 11.42%, 34.2% and 22.7%, respectively. These results showed that melatonin caused reduced DNA content and EI during leaf development.
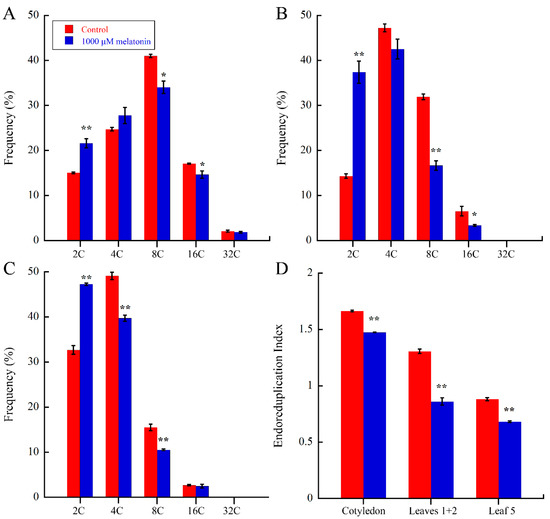
Figure 4.
Nuclear polyploidization analysis of leaves in control and 1000 μM melatonin treated Arabidopsis seedlings at 21 days after one-day germination. Ploidy levels of cotyledons (A); leaves 1 + 2 (B); and the fifth leaves (C) were performed by flow cytometic analysis. Three biological replicates were taken for statistical analysis. Values represent mean ± SD, * p < 0.05; ** p < 0.01 by a Student’s t-test and (D) endoreduplication index (EI) analyzed from flow cytometic data of cotyledons, leaves 1 + 2 and the fifth leaves. EI represents the average number of endocycles undergone by a typical nucleus and was calculated from these percentage values as follows: EI = [(0 × 2C) + (1 × 4C) + (2 × 8C) + (3 × 16C) + (4 × 32C)]/100 [55]. Values represent mean ± SD, ** p < 0.01 by a Student’s t-test.
2.4. Melatonin Regulated Several Cell Cycle and Ribosomal Related Genes
Cell cycle is an essential process in organ growth, driven by a complex series of events and precisely regulated at the level of transcription especially [56]. Several core cell cycle genes have been widely identified [52,57,58,59,60]. To examine whether melatonin treatment affected the transcriptional levels of key genes of cell cycle, the quantitative real-time PCR analysis were performed on cotyledons (6-, 11-, 16- and 21-day treatment), leaves 1 + 2 (11-, 16- and 21-day treatment) and the fifth leaves (16- and 21-day treatment). CYCA2;3, CYCD3;1 and CYCD3;3 are key regulators of ploidy levels in Arabidopsis endoreduplication [60,61,62]. CYCB1;1 and CYCP2;1 are markers in G2-M phase activation in Arabidopsis [63,64]. HTA10 (Histone H2A gene 10), PCNA1 (Proliferating Cell Nuclear Antigen1) and RNR1 (RiboNucleotide Reductase1) are S-phase gene markers [60,65]. As shown in Figure 5, the transcript levels of cyclins significantly increased in most of the development stage of all three tissues after the melatonin treatment, while those of CYCP2;1 and CYCB1;1 were down-regulated by the melatonin treatment. The S-phase key genes (HTA10, PCNA1 and RNR1) showed significantly higher transcript levels at most of the time points after melatonin treatment.
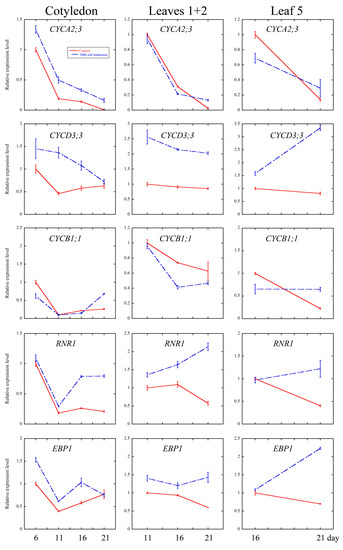
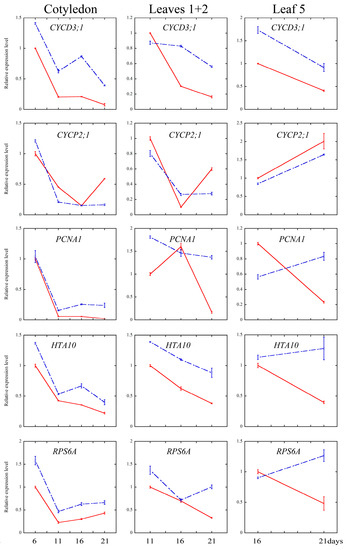
Figure 5.
Quantitative real-time PCR (qRT-PCR) analysis of cell cycle and ribosomal related genes’ expression under control and 1000 μM melatonin treatment. Relative fold changes of the expression of CYCA 2;3, CYCD 3;1, CYCD 3;3, CYCP 2;1, CYCB 1;1, PCNA1, RNR1, HTA10, EBP1 and RPS6A were quantified by real-time PCR, and the expression levels of the indicated genes in control leaves at six days were set to 1. Values represent mean ± SD.
Cell growth is consistent with the rate of protein synthesis [66]. Several ribosomal proteins play important role in morphological and developmental regulation of Arabidopsis [60,67,68]. Here, we found that the transcripts of RPS6A (Ribosomal Protein S6A) and EBP1 (ERBB-3-Binding Protein1) were up-regulated by melatonin, indicating the involvement of protein synthesis in melatonin regulated cell growth.
3. Discussion
Melatonin, a natural product first found in plants less than 30 years ago, has been shown to play various functions in plants. In addition to its functions in regulating stress responses [6,33,35,69], melatonin was also proven to be involved in the regulation of plant growth [31]. We have found that high dose of melatonin inhibited the root growth in Arabidopsis seedlings and suppressed the floral transition in Arabidopsis in our previous studies [23,24]. Consistently, it has been reported that 1 mM melatonin treatment of four-week-old detached Arabidopsis leaves caused increased oxidative stress resistance [15]. However, whether melatonin also has an effect in regulating leaf growth is still unknown. In the present study, the effect of melatonin on the growth and development of Arabidopsis leaf was examined, and the underlying mechanism was also partially investigated.
3.1. Melatonin Represses Cell Proliferation and Cell Expansion in Arabidopsis Leaf Growth
In this study, we first tested the effect of melatonin on leaf growth of Arabidopsis. The results showed that low concentration of melatonin (1, 10 and 100 μM) had no effect on cotyledon size after treatment for six days (Figure 1B). Our preliminary experiments and previous studies indicated that low concentration of melatonin (10–50 μM) had little effect on the content of endogenous melatonin and the development of plant root [18,23]. However, higher concentration of melatonin (600 and 1000 μM) dramatically suppressed the size of Arabidopsis cotyledon in a dose-dependent manner (Figure 1B). After digging deeper into the effects of melatonin, we found that both palisade cell size and cell number were reduced by high concentrations of melatonin, consistent with its effects on cotyledon size (Figure 1A,C,D). It is generally believed that Arabidopsis cotyledon cells are highly differentiated in seeds already, and the growth of cotyledons is attributed to the cell expansion [70,71]. Further studies showed that the cell cycling of cotyledon palisade cells is reactivated after seeds imbibition, which relies on de novo sucrose synthesis [72]. Here, the kinetic analysis of cotyledons, the first pair of true leaves and the fifth leaves in control and 1000 μM melatonin treated seedlings showed coincident results (Figure 2), except the cell number of cotyledons in seedlings treated for 11, 16 and 21 days. Based on these results, melatonin may affect both cell proliferation and cell expansion in leaf growth.
The cell division rate and duration, as important parameters of cell proliferation, have significant impacts on the leaf size [51]. The decline of cell division rate of palisade cells in melatonin-treated seedlings (Figure 3) suggested that melatonin suppressed the cell number in Arabidopsis leaves by reduced cell division rate.
3.2. Melatonin Inhibites Endoreduplication during Leaf Growth
Endoreduplication, also known as endocycle, refers to the fact that cells undergo DNA replication without cell division, and is the mainly cause of cell ploidy change [73,74]. Moreover, the endocycle can lead to a larger nucleus and the enlargement of cell size in certain cells [75,76,77,78,79]. Small organ size in bin4 is primarily caused by suppressed cell expansion associated with defects in endoreduplication [80]. Overexpression of Arabidopsis thaliana homeobox 12 (ATHB12) resulted in increased cell size, along with elevated ploidy [81]. Our results showed that 1000 μM melatonin caused significantly 2C fraction increase in cotyledons, the first pair of true leaves, and the fifth leaves, in accordance with the decrease of higher DNA content (Figure 4A–C). Similar findings were reported by Posmyk [82], that 100 μM pretreatment of melatonin in red cabbage seeds significantly inhibited DNA endoreduplication [82]. Moreover, the endoreduplication index (EI) of melatonin treated leaves was significantly lower than that of control leaves (Figure 4D). Therefore, melatonin affected the DNA ploidy and endoreduplication during the development of leaf. Jasmonate also negatively affects leaf size in Arabidopsis, and causes DNA content change by delaying the onset of endoreduplication [60]. It is well known that gibberellic acid (GA) promote plant growth through promoting the degradation of DELLAs, which were proven to perform the inhibitory activity of plant growth through reducing the rates of both cell proliferation and cell expansion [83]. In our recent studies, we found that melatonin-mediated protein stabilization of DELLAs in Arabidopsis [24]. Thus, DELLAs may also be involved in melatonin-mediated leaf development, and this needs to be further investigated.
3.3. Melatonin Regulates Cell Cycle Progression
On the one hand, it has been reported that both high concentrations (1 mM) and low concentrations (100 pM) of melatonin treatment can result in extensive transcriptional reprogramming in Arabidopsis, including the transcripts of various stress-related genes [15]. The extensive transcriptional reprogramming is consistent with the wide participation of melatonin in plant stress resistance, at least partially [11,12,13,14,15,16,17,18,19,20,21,22]. On the other hand, melatonin is also involved in several developmental processes of plants, including seed germination, shoot development, root growth and architecture, flowering and fruit ripening [23,24,25,26,27,28,29]. To our knowledge, this is the first study to report the involvement of melatonin in leaf development. In this study, we focused on the effect of melatonin on the development of leaf and the underlying mechanism. We found that high concentration of melatonin suppressed the cell proliferation and endoreduplication, and the expression of related key genes in leaf growth might contribute to this issue. Numerous genes were identified and shown to be involved in regulating cell cycle precisely on transcriptional level [52,57,58,59,60], and the mutations of some key genes of cell cycling lead to serious defects of leaf occurrence and development [65,84,85]. CYCA2;3, as a key regulator of DNA contents in Arabidopsis, negatively affects endocycles. Overexpression of CYCA2;3 restrained endocycles of Arabidopsis leaves in a dose-dependent manner [62]. CYCD3 subgroup of D-type cyclins (CYCD) family, composed of CYCD3;1, CYCD3;2 and CYCD3;3, were shown to regulate the balance between cell division and cell expansion [61,86,87]. Overexpression of CYCD3;1 dramatically repressed endoreduplication [61]. Triple mutant cycd3;1-3 showed larger cells and initiated endoreduplication in petals that usually do not undergo endocycles [86]. In our study, the transcriptional level of CYCA2;3 in cotyledons was elevated at all the time points after melatonin treatment, and in leaves 1 + 2 and leaf 5, the transcriptional level of CYCA2;3 was also up-regulated after melatonin treatment for 21 days. Moreover, the expression levels of CYCD3;1 and CYCD3;3 were up-regulated by melatonin in all three tissues at most of the time points (Figure 5). Since CYCA2;3, CYCD3;1, CYCD3;2, and CYCD3;3 are negative regulators of endoreduplication, the higher transcripts of these genes in the melatonin treated leaves might contribute to the inhibition of endoreduplication.
CYCP2;1 was proven to interact with CDKA;1, CDKB2;1 and CDKB2;2 with role in the transition of G2 to M phase during meristem activation, as evidenced by the significant G2 arrest of the cycp2;1 mutants [65,88]. As shown in Figure 5, melatonin treatment down-regulated the expression level of CYCP2;1 in cotyledons and leaves 1 + 2 at 11 and 21 days, and also in the fifth leaves at 16 and 21 days. At CYCB1;1 is a positive regulator and expressed only at the G2-M phase transition [89,90]. Our findings showed that melatonin treatment lead to lower expression of CYCB1;1 in cotyledons and leaves. Similar results were found in the methyl jasmonate (MeJA) treated Arabidopsis CYCB1;1:Dbox-GUS seedlings, which showed down-regulation of CYCB1;1 in the arrest of leaf development [54]. Reduction of transcriptional level of CYCP2;1 and CYCB1;1 indicated that melatonin negatively regulated the phase transition of G2 to M in Arabidopsis leaves, which is consistent with reduced cell number in the melatonin treated leaves (Figure 1, Figure 2 and Figure 3).
PCNA (Proliferating Cell Nuclear Antigen) is a key nuclear protein of eukaryotic dividing cells in the early S phase [91]. PCNA acts as a sliding platform loaded onto the DNA duplex, and involved in DNA synthesis, cell cycle regulation and DNA repair [92,93]. RNRs (Ribonucleotide reductases) are critical for DNA damage checkpoint pathways in yeast, mammals and higher plants like Arabidopsis [94,95]. The elevated expression level of PCNA1 and the induced transcriptional level of RNR1 in melatonin treated cotyledons and leaves were shown in this study. Histon H2A proteins consist of 13 members in Arabidopsis [96]. Several reports have shown that histone genes is coupled to the S phase, and emphasized their roles in DNA replication and cell proliferation [97,98,99,100]. Our experiments showed that the expression of HTA10 was up-regulated by melatonin in all three tissues at all the time points. We also found that the transcript levels of some ribosomal genes, EBP1 and RPS6A, were induced by melatonin treatment in all three tissues at most time points (Figure 5). Noir et al. have showed that keeping high expression levels of histones, ribosomal proteins and pre-RC components by MeJA could help the cells maintaining a potential stand-by mode [60]. The modulation of melatonin on these genes’ transcripts indicated that melatonin treated cells still hold their potency to recover from the exogenous treatment.
4. Experimental Section
4.1. Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the WT plant. Seeds were sterilized with 70% (v/v) ethanol for 1 min and 1% sodium hypochlorite for 16 min. After washing with distilled water for 3–5 times, seeds were placed at 4 °C for 2 days to break dormancy. Stratified-seeds were then sown on half-strength Murashige and Skoog medium with 1% sucrose and 0.85% agar, and were transferred to a culture room under dark/light cycles of 8/16 h at the temperature of 22 °C for 24 h, to reduce the effect of melatonin treatment on seed germination. Seeds were then transferred to new plates with or without melatonin for another 6–21 day culture.
4.2. Leaf Measurements
Arabidopsis seeds germinating for one day were transferred to new 1/2 MS medium and medium containing different concentrations of melatonin. After another 6-day treatment, cotyledons of control and samples were first rinsed in bleach solution with 75% ethanol and 25% acetic acid for 2 h or overnight at 37 °C to remove chlorophyll, and then mounted in basic solution (7% sodium hydroxide in 70% ethanol) for 15 min at room temperature. After immersed in 60%, 40%, 20% and 10% ethanol for 20 min successively, the cotyledons were mounted with clearing solution (50 g of chloral hydrate, 15 mL of water and 10 mL of glycerol) on glass slides, and the palisade cells at the central region of a half leaf beside the mid-vein were captured with differential interference contrast (DIC) objective of Leica TCS SP8 (Leica, Mannheim, Germany) laser scanning confocal microscope to determine cell size and cell number per unit area [101]. Those slides were then photographed under a Leica M205 FA (Leica, Singapore) stereo microscope to determine the area of the cotyledons. The areas of both cotyledons and cells were measured by software Image J (Wayne Rasband, Bethesda, MD, USA, available online: http://rsbweb.nih.gov/ij/, version 1.47 g). The estimated total palisade cell numbers per cotyledon were calculated by cotyledon and cell areas. Cell division rate (d) was calculated from N1 = N0 2dt [46]. D = (log2N1 − log2N0)/(t1 − t0). N and t represent the cell number and time (h) respectively. Results presented are average values of more than 10 seedlings per treatment from three independent experiments. Statistical analysis was conducted in KaleidaGraph 4.03 (Synergy Software, Reading, PA, USA).
4.3. Flow Cytometry Experiments and Ploidy Measurement
After one-day germination, seeds were transferred onto new 1/2 MS medium and medium supplemented with 1000 μM melatonin for another 21 days. Cotyledons, the first true leaves and the fifth leaves of control and melatonin treated samples were selected and rinsed in 1 mL cold Galbraith’s buffer (45 mM MgCl2, 20 mM MOPS, 30 mM Sodium Citrate, 0.1% Trtion X-100, pH 7.0 with 1 M NaOH) [102]. Cotyledons or leaves were cut by a blade irregularly and quickly, filtered through miracloth (22–25 μm, Millipore, Billerica, MA, USA). Filtrate was treated with RNase A with the final concentration of 50 μg/mL for 20 min, and then stained with Propidium Iodide (PI) (Sigma-Aldrich, St. Louis, MO, USA) with the final concentration of 50 μg/mL for 20 min at 4 °C. All the samples were measured by BD Accuri C6 Flow Cytometer. At least 10,000 nuclei isolated were used for ploidy measurement. Flow Cytometry Experiments were repeated at least three times from independent biological replicates. Statistical analysis was conducted in KaleidaGraph 4.03.
4.4 Quantitative Real-Time Pcr Analysis
One-day-old Arabidopsis seeds were transferred to new 1/2 MS medium and medium containing 1000μM melatonin. After another 6–21 day treatment, cotyledons (6-, 11-, 16-, and 21-day treatment), the first true leaves (11-, 16-, and 21-day treatment), and the fifth rosette leaf (16- and 21-day treatment) of control and samples were collected, and total RNA was isolated from leaves treated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For cDNA synthesis, 2 μg of total RNA from different samples was used for reverse transcription with RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s recommendations. To analyze the transcript levels of cell cycle-related genes in control and treated leaves, quantitative real-time PCR was performed with Roche LightCyler 96® in a 20 μL reaction volume containing SYBR Green dye (FastStart Essential DNA Green Master, Roche, Mannheim, Germany). AtPDF2 (protein phosphatase 2, AT1G13320) was chosen as an internal control [103]. Relative expression levels were estimated using the 2−ΔΔCt method [104]. All primers used in the study are listed in Supplementary Materials, Table S1.
5. Conclusions
In this study, we found that high concentration of melatonin caused smaller leaf size in Arabidopsis by reducing cell size and cell number, as confirmed by the kinetic analysis of leaf growth and flow cytometic analysis of leaf DNA ploidy. The results showed that melatonin suppressed the cell division rate and endoreduplication. Expression analysis of cell cycle genes and ribosomal genes suggested melatonin negatively regulated cell division and endocycle. In summary, this study provides a direct link between melatonin and leaf development, and emphasized the modulation of melatonin in retarded cell proliferation, endoreduplication and cell expansion. We also highlight the involvement of melatonin in the maintenance of a stand-by mode in Arabidopsis leaf cells.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/5/991/s1.
Acknowledgements
This work was supported by the Hainan Natural Science Foundation (No. 20163052), the startup funding and the scientific research foundation of Hainan University (No. kyqd1516 and No. kyqd1531) and the National Natural Science Foundation of China (No. 31570249).
Author Contributions
Haitao Shi conceived and directed this study, and revised the manuscript; Qiannan Wang designed and performed the experiments, analyzed the data, and wrote and revised the manuscript; Bang An performed the experiments, analyzed the data and revised the manuscript; Hongli Luo designed the experiments and revised the manuscript; and Chaozu He designed the experiments and revised the manuscript. All authors approved the manuscript and the version to be published.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lerner, A.B.; Case, J.D.; Takahashi, Y.; Lee, T.H.; Mori, W. Isolation of melatonin, a pineal factor that lightens melanocytes. J. Am. Chem. Soc. 1958, 80, 2587. [Google Scholar] [CrossRef]
- Poeggeler, B.; Balzer, I.; Hardeland, R.; Lerchl, A. Pineal hormone melatonin oscillates also in the dinoflagellate Gonyaulax polyedra. Naturwissenschaften 1991, 78, 268–269. [Google Scholar] [CrossRef]
- Dubbels, R.; Reiter, R.J.; Klenke, E.; Goebel, A.; Schnakenberg, E.; Ehlers, C.; Schiwara, H.W.; Schloot, W. Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography-mass spectrometry. J. Pineal Res. 1995, 18, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Hattori, A.; Migitaka, H.; Iigo, M.; Itoh, M.; Yamamoto, K.; Ohtani-Kaneko, R.; Hara, M.; Suzuki, T.; Reiter, R.J. Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Biochem. Mol. Biol. Int. 1995, 35, 627–634. [Google Scholar] [PubMed]
- Reiter, R.J.; Tan, D.X.; Rosales-Corral, S.; Manchester, L.C. The universal nature, unequal distribution and antioxidant functions of melatonin and its derivatives. Mini. Rev. Med. Chem. 2013, 13, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, R. Melatonin in plants-diversity of levels and multiplicity of functions. Front. Plant Sci. 2016, 7, 198. [Google Scholar] [CrossRef] [PubMed]
- Jan, J.E.; Reiter, R.J.; Wasdell, M.B.; Bax, M. The role of the thalamus in sleep, pineal melatonin production, and circadian rhythm sleep disorders. J. Pineal Res. 2009, 46, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, R.; Madrid, J.A.; Tan, D.X.; Reiter, R.J. Melatonin, the circadian multioscillator system and health: The need for detailed analyses of peripheral melatonin signaling. J. Pineal Res. 2012, 52, 139–166. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Vico, A.; Lardone, P.J.; Álvarez-Sánchez, N.; Rodríguez-Rodríguez, A.; Guerrero, J.M. Melatonin: Buffering the immune system. Int. J. Mol. Sci. 2013, 14, 8638–8683. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tan, D.X.; Galano, A. Melatonin: Exceeding expectations. Physiology 2014, 29, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Lee, K.; Park, S.; Kim, Y.S.; Back, K. Enhanced production of melatonin by ectopic overexpression of human serotonin N-acetyltransferase plays a role in cold resistance in transgenic rice seedlings. J. Pineal Res. 2010, 49, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wang, P.; Li, M.; Ke, X.; Li, C.; Liang, D.; Wu, S.; Ma, X.; Li, C.; Zou, Y.; et al. Exogenous melatonin improves Malus resistance to marssonina apple blotch. J. Pineal Res. 2013, 54, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhao, B.; Zhang, H.J.; Weeda, S.; Yang, C.; Yang, Z.C.; Ren, S.; Guo, Y.D. Melatonin promotes water-stress tolerance, lateral root formation, and seed germination in cucumber (Cucumis sativus L.). J. Pineal Res. 2013, 54, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chan, Z. The Cysteine2/Histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS thaliana 6- activated C-REPEAT-BINDING FACTOR pathway is essential for melatonin-mediated freezing stress resistance in Arabidopsis. J. Pineal Res. 2014, 57, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Weeda, S.; Zhang, N.; Zhao, X.; Ndip, G.; Guo, Y.; Buck, G.A.; Fu, C.; Ren, S. Arabidopsis transcriptome analysis reveals key roles of melatonin in plant defense systems. PLoS ONE 2014, 9, e93462. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chen, Y.; Tan, D.X.; Reiter, R.J.; Chan, Z.; He, C. Melatonin induces nitric oxide and the potential mechanisms relate to innate immunity against bacterial pathogen infection in Arabidopsis. J. Pineal Res. 2015, 59, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Jiang, C.; Ye, T.; Tan, D.X.; Reiter, R.J.; Zhang, H.; Liu, R.; Chan, Z. Comparative physiological, metabolomic, and transcriptomic analyses reveal mechanisms of improved abiotic stress resistance in bermudagrass (Cynodon dactylon (L). Pers.) by exogenous melatonin. J. Exp. Bot. 2015, 66, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Reiter, R.J.; Tan, D.X.; Chan, Z. INDOLE-3-ACETIC ACID INDUCIBLE 17 positively modulates natural leaf senescence through melatonin-mediated pathway in Arabidopsis. J. Pineal Res. 2015, 58, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Tan, D.X.; Reiter, R.J.; Ye, T.; Yang, F.; Chan, Z. Melatonin induces class A1 heat shock factors (HSFA1s) and their possible involvement of thermotolerance in Arabidopsis. J. Pineal Res. 2015, 58, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Tan, D.X.; Reiter, R.J.; Chan, Z. Comparative physiological and proteomic analyses reveal the actions of melatonin in the reduction of oxidative stress in bermudagrass (Cynodon dactylon (L). Pers.). J. Pineal Res. 2015, 59, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.Y.; Zhang, Y.; Xu, Y.P.; Qi, Z.Y.; Li, M.Q.; Ahammed, G.J.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Reiter, R.J.; et al. HsfA1a upregulates melatonin biosynthesis to confer cadmium tolerance in tomato plants. J. Pineal Res. 2017, 62, e12387. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tan, D.X.; Zhou, Z.; Cruz, M.H.; Fuentes-Broto, L.; Galano, A. Phytomelatonin: Assisting plants to survive and thrive. Molecules 2015, 20, 7396–7437. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; An, B.; Wei, Y.; Reiter, R.J.; Shi, H.; Luo, H.; He, C. Melatonin regulates root meristem by repressing auxin synthesis and polar auxin transport in Arabidopsis. Front. Plant Sci. 2016, 15, 1882. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wei, Y.; Wang, Q.; Reiter, R.J.; He, C. Melatonin mediates the stabilization of DELLA proteins to repress the floral transition in Arabidopsis. J. Pineal Res. 2016, 60, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhang, N.; Wang, J.; Zhang, H.; Li, D.; Shi, J.; Li, R.; Weeda, S.; Zhao, B.; Ren, S.; et al. Melatonin promotes ripening and improves quality of tomato fruit during postharvest life. J. Exp. Bot. 2014, 66, 657–658. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhang, N.; Wang, J.; Cao, Y.; Li, X.; Zhang, H.; Zhang, L.; Tan, D.X.; Guo, Y.D. A label-free differential proteomics analysis reveals the effect of melatonin on promoting fruit ripening and anthocyanin accumulation upon postharvest in tomato. J. Pineal Res. 2016, 61, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Gong, B.; Sun, S.; Liu, S.; Wang, X.; Wei, M.; Yang, F.; Li, Y.; Shi, Q. Promoting roles of melatonin in adventitious root development of Solanum lycopersicum L. by regulating auxin and nitric oxide signaling. Front. Plant Sci. 2016, 7, 718. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, H.J.; Sun, Q.Q.; Cao, Y.Y.; Li, X.; Zhao, B.; Wu, P.; Guo, Y.D. Proteomic analysis reveals a role of melatonin in promoting cucumber seed germination under high salinity by regulating energy production. Sci. Rep. 2017, 7, 503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Sun, Y.; Liu, Z.; Sun, Y. Effects of melatonin on seedling growth, mineral nutrition, and nitrogen metabolism in cucumber under nitrate stress. J. Pineal Res. 2017, 62. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: Plant growth regulator and/or biostimulator during stress? Trends Plant Sci. 2014, 19, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Functions of melatonin plants: A review. J. Pineal Res. 2015, 59, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Paredes, S.D.; Korkmaz, A.; Manchester, L.C.; Tan, D.X.; Reiter, R.J. Phytomelatonin: A review. J. Exp. Bot. 2009, 60, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.X.; Hardeland, R.; Manchester, L.C.; Korkmaz, A.; Ma, S.; Rosales-Corral, S.; Reiter, R.J. Functional roles of melatonin in plants and perspectives in nutritional and agricultural science. J. Exp. Bot. 2012, 63, 577–597. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B. Phytomelatonin: Discovery, content, and role in plants. Adv. Bot. 2014, 2014, 815769. [Google Scholar] [CrossRef]
- Hardeland, R. Melatonin in plants and other phototrophs—Advances and gaps concerning the diversity of functions. J. Exp. Bot. 2015, 66, 627–646. [Google Scholar] [CrossRef] [PubMed]
- Pelagio-Flores, R.; Muñoz-Parra, E.; Ortiz-Castro, R.; López-Bucio, J. Melatonin regulates Arabidopsis root system architecture likely acting independently of auxin signaling. J. Pineal Res. 2012, 53, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Seo, H.; Park, C.; Park, W.J. Examination of the auxin hypothesis of phytomelatonin action in classical auxin assay systems in maize. J. Plant Physiol. 2016, 190, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Tsukaya, H. Organ shape and size: A lesson from studies of leaf morphogenesis. Curr. Opin. Plant Biol. 2003, 6, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Mizukami, Y. A matter of size: Developmental control of organ size in plants. Curr. Opin. Plant Biol. 2001, 4, 533–539. [Google Scholar] [CrossRef]
- Walch-Liu, P.; Neumann, G.; Bangerth, F.; Engels, C. Rapid effects of nitrogen form on leaf morphogenesis in tobacco. J. Exp. Bot. 2000, 51, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Thingnaes, E.; Torre, S.; Ernstsen, A.; Moe, R. Day and night temperature responses in Arabidopsis: Effects on gibberellin and auxin content, cell size, morphology and flowering time. Ann. Bot. 2003, 92, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Kozuka, T.; Horiguchi, G.; Kim, G.T.; Ohgishi, M.; Sakai, T.; Tsukaya, H. The different growth responses of the Arabidopsis thaliana leaf blade and the petiole during shade avoidance are regulated by photoreceptors and sugar. Plant Cell Physiol. 2005, 46, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.G.; Hake, S. The initiation and determination of leaves. Plant Cell 1992, 4, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Anastasiou, E.; Lenhard, M. Growing up to one’s standard. Curr. Opin. Plant Biol. 2007, 10, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, N.; Vanhaeren, H.; Inzé, D. Leaf size control: Complex coordination of cell division and expansion. Trends Plant Sci. 2012, 17, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, P.M.; Bonetta, D.; Tsukaya, H.; Dengler, R.E.; Dengler, N.G. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 1999, 215, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Nath, U.; Crawford, B.C.; Carpenter, R.; Coen, E. Genetic control of surface curvature. Science 2003, 299, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Tsukaya, H. Controlling size in multicellular organs: Focus on the leaf. PLoS Biol. 2008, 6, e174. [Google Scholar] [CrossRef] [PubMed]
- Beemster, G.T.; Fiorani, F.; Inzé, D. Cell cycle: The key to plant growth control? Trends Plant Sci. 2003, 8, 154–158. [Google Scholar] [CrossRef]
- Horiguchi, G.; Ferjani, A.; Fujikura, U.; Tsukaya, H. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 2006, 119, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Fiorani, F.; Beemster, G.T. Quantitative analyses of cell division in plants. Plant Mol. Boil. 2006, 60, 963–979. [Google Scholar] [CrossRef] [PubMed]
- Beemster, G.T.; de Veylder, L.; Vercruysse, S.; West, G.; Rombaut, D.; van Hummelen, P.; Galichet, A.; Gruissem, W.; Inzé, D.; Vuylesteke, M. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 2005, 138, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Inzé, D.; de Veylder, L. Cell cycle regulation in plant development. Annu. Rev. Genet. 2006, 40, 77–105. [Google Scholar] [CrossRef] [PubMed]
- Breuer, C.; Ishida, T.; Sugimoto, K. Developmental control of endocycles and cell growth in plants. Curr. Opin. Plant Biol. 2010, 13, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Cookson, S.J.; Radziejwoski, A.; Granier, C. Cell and leaf size plasticity in Arabidopsis: What is the role of endoreduplication? Plant Cell Environ. 2006, 29, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Burssens, S.; Montagu, M.V.; Inzé, D. The cell cycle in Arabidopsis. Plant Physiol. Biochem. 1998, 36, 9–19. [Google Scholar] [CrossRef]
- Vandepoele, K.; Raes, J.; de Veylder, L.; Rouzé, P.; Rombauts, S.; Inzé, D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 2002, 14, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Menges, M.; de Jager, S.M.; Gruissem, W.; Murray, J.A. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005, 41, 546–566. [Google Scholar] [CrossRef] [PubMed]
- López-Juez, E.; Dillon, E.; Magyar, Z.; Khan, S.; Hazeldine, S.; de Jager, S.M.; Murray, J.A.; Beemster, G.T.; Bögre, L.; Shanahan, H. Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 2008, 20, 947–968. [Google Scholar] [CrossRef] [PubMed]
- Noir, S.; Bömer, M.; Takahashi, N.; Ishida, T.; Tsui, T.L.; Balbi, V.; Shanahan, H.; Sugimoto, K.; Devoto, A. Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 2013, 161, 1930–1951. [Google Scholar] [CrossRef] [PubMed]
- Dewitte, W.; Riou-Khamlichi, C.; Scofield, S.; Healy, J.S.; Jacqmard, A.; Kilby, N.J.; Murray, J.A. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 2003, 15, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.K.; Ohashi, Y.; Tsuge, T.; Yoshizumi, T.; Matsui, M.; Oka, A.; Aoyama, T. The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 2006, 18, 382–396. [Google Scholar] [CrossRef] [PubMed]
- González-García, M.P.; Vilarrasa-Blasi, J.; Zhiponova, M.; Divol, F.; Mora-García, S.; Russinova, E.; Caño-Delgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Skylar, A.; Chang, P.L.; Bisova, K.; Wu, X. CYCP2;1 integrates genetic and nutritional information to promote meristem cell division in Arabidopsis. Dev. Boil. 2014, 393, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Francis, D. The plant cell cycle—15 Years on. New Phytol. 2007, 174, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, N.; Kodama, H.; Ando, S.; Komamine, A. Synthesis of protein and mRNA is necessary for the transition of suspension-cultured Catharanthus roseus cells from G1 to the S phase of the cell cycle. Physiol. Plant. 1990, 80, 95–101. [Google Scholar] [CrossRef]
- Ito, T.; Kim, G.T.; Shinozaki, K. Disruption of an Arabidopsis S-13 homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000, 22, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; van Dijk, M.F.; Venken, R.J.; Quint, A.; Hooykaas, P.; Offringa, R. An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 2001, 128, 4289–4299. [Google Scholar] [PubMed]
- Shi, H.; Chen, K.; Wei, Y.; He, C. Fundamental issues of melatonin-mediated stress signaling in plants. Front. Plant Sci. 2016, 7, 1124. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, S.G.; Briarty, L.G. Cotyledon cell development in Arabidopsis thaliana during reserve deposition. Can. J. Bot. 1992, 70, 151–164. [Google Scholar] [CrossRef]
- Tsukaya, H.; Tsuge, T.; Uchimiya, H. The cotyledon: A superior system for studies of leaf development. Planta 1994, 195, 309–312. [Google Scholar] [CrossRef]
- Ferjani, A.; Segami, S.; Horiguchi, G.; Muto, Y.; Maeshima, M.; Tsukaya, H. Keep an eye on PPi: The vacuolar-type H+-pyrophosphatase regulates postgerminative development in Arabidopsis. Plant Cell 2011, 23, 2895–2908. [Google Scholar] [CrossRef] [PubMed]
- Sabelli, P.A.; Larkins, B.A. The endoreduplication cell cycle: Regulation and function. Plant Cell Monogr. 2008, 9, 75–100. [Google Scholar]
- Yoshizumi, T.; Breuer, C.; Matsui, M.; Sugimoto-Shirasu, K. Plant cell growth signaling and its link to ploidy. Plant Cell Monogr. 2008, 10, 107–125. [Google Scholar]
- Edgar, B.A.; Orr-Weaver, T.L. Endoreplication cell cycles: More for less. Cell 2001, 105, 297–306. [Google Scholar] [CrossRef]
- Sugimoto-Shirasu, K.; Roberts, K. “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 2003, 6, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Bramsiepe, J.; Wester, K.; Weinl, C.; Roodbarkelari, F.; Kasili, R.; Larkin, J.C.; Hülskamp, M.; Schnittger, A. Endoreplication controls cell fate maintenance. PLoS Genet. 2010, 6, e1000996. [Google Scholar] [CrossRef] [PubMed]
- Orr-Weaver, T.L. When bigger is better: The role of polyploidy in organogenesis. Trends Genet. 2015, 31, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, Y.; Hasegawa, J.; Fujikura, U.; Hoshino, R.; Matsunaga, S.; Tsukaya, H. The coordination of ploidy and cell size differs between cell layers in leaves. Development 2016, 143, 1120–1125. [Google Scholar] [CrossRef] [PubMed]
- Breuer, C.; Stacey, N.J.; West, C.E.; Zhao, Y.; Chory, J.; Tsukaya, H.; Azumi, Y.; Maxwell, A.; Roberts, K.; Sugimoto-Shirasu, K. BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell 2007, 19, 3655–3668. [Google Scholar] [CrossRef] [PubMed]
- Hur, Y.S.; Um, J.H.; Kim, S.; Kim, K.; Park, H.J.; Lim, J.S.; Kim, W.Y.; Jun, S.E.; Yoon, E.K.; Lim, J.; et al. Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytol. 2015, 205, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Posmyk, M.M.; Kuran, H.; Marciniak, K.; Janas, K.M. Presowing seed treatment with melatonin protects red cabbage seedlings against toxic copper ion concentrations. J Pineal Res. 2008, 45, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Gusti, A.; Cheminant, S.; Alioua, M.; Dhondt, S.; Coppens, F.; Beemster, G.T.; Genschik, P. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Boil. 2009, 19, 1188–1193. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.U.; Buechel, S.; Zhao, Z.; Ljung, K.; Novák, O.; Busch, W.; Schuster, C.; Lohmann, J.U. Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 2008, 20, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, H.; Ohno, R.; Umeda, M. The Arabidopsis cyclin-dependent kinase-activating kinase CDKF;1 is a major regulator of cell proliferation and cell expansion but is dispensable for CDKA activation. Plant J. 2009, 59, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Menges, M.; Samland, A.K.; Planchais, S.; Murray, J.A. The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 2006, 18, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Dewitte, W.; Scofield, S.; Alcasabas, A.A.; Maughan, S.C.; Menges, M.; Braun, N.; Collins, C.; Nieuwland, J.; Prinsen, E.; Sundaresan, V.; et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14537–14542. [Google Scholar] [CrossRef] [PubMed]
- Torres Acosta, J.A.; de Almeida Engler, J.; Raes, J.; Magyar, Z.; de Groodt, R.; Inzé, D.; de Veylder, L. Molecular characterization of Arabidopsis PHO80-like proteins, a novel class of CDKA;1-interacting cyclins. Cell Mol. Life Sci. 2004, 61, 1485–1497. [Google Scholar] [CrossRef] [PubMed]
- Doerner, P.; Jørgensen, J.E.; You, R.; Steppuhn, J.; Lamb, C. Control of root growth and development by cyclin expression. Nature 1996, 380, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Shaul, O.; Mironov, V.; Burssens, S.; van Montagu, M.; Inze’, D. Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 1996, 93, 4868–4872. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.F.; Mathews, M.B. Regulation of proliferating cell nuclear antigen during the cell cycle. J. Biol. Chem. 1989, 264, 13856–13864. [Google Scholar] [PubMed]
- Strzalka, W.; Ziemienowicz, A. Proliferating cell nuclear antigen (PCNA): A key factor in DNA replication and cell cycle regulation. Ann. Bot. 2011, 107, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, A.; Concia, L.; Maggio, C.; Raynaud, C.; Bergounioux, C.; Crespan, E.; Cella, R.; Maga, G. Oxidative DNA damage bypass in Arabidopsis thaliana requires DNA polymerase λ and proliferating cell nuclear antigen 2. Plant Cell 2011, 23, 806–822. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhou, Z.; Elledge, S.J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 1998, 94, 595–605. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Z. Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 2006, 18, 350–365. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Sardesai, N.; Fujinuma, T.; Chan, C.W.; Gelvin, S.B. Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell 2006, 18, 1575–1589. [Google Scholar] [CrossRef] [PubMed]
- Sundas, A.; Tandre, K.; Kvarnheden, A.; Engstrom, P. cDNA sequence and expression of an intron-containing histone H2A gene from Norway spruce, Picea abies. Plant Mol. Biol. 1993, 21, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Reichheld, J.P.; Gigot, C.; Gigot, N.C. Multilevel regulation of histone gene expression during the cell cycle in tobacco cells. Nucleic Acids Res. 1998, 26, 3255–3262. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Kennedy, B.K.; Lawrence, B.D.; Barbie, D.A.; Matera, G.; Fletcher, J.A.; Harlow, E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000, 14, 2283–2297. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.M.; Ye, X.; Hall, C.; Santos, H.; Ma, T.; Kao, G.D.; Yen, T.J.; Harper, J.W.; Adams, P.D. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 2002, 22, 7459–7472. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Qin, Z.; Yan, J.; Zhang, X.; Hu, Y. Arabidopsis ORGAN SIZE RELATED1 regulates organ growth and final organ size in orchestration with ARGOS and ARL. New Phytol. 2011, 191, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).