Abstract
Flavonoids, a class of phenolic compounds, are one of the main functional components and have a wide range of molecular structures and biological activities in Polygonatum. A few of them, including homoisoflavonoids, chalcones, isoflavones, and flavones, were identified in Polygonatum and displayed a wide range of powerful biological activities, such as anti-cancer, anti-viral, and blood sugar regulation. However, few studies have systematically been published on the flavonoid biosynthesis pathway in Polygonatum cyrtonema Hua. Therefore, in the present study, a combined transcriptome and metabolome analysis was performed on the leaf, stem, rhizome, and root tissues of P. cyrtonema to uncover the synthesis pathway of flavonoids and to identify key regulatory genes. Flavonoid-targeted metabolomics detected a total of 65 active substances from four different tissues, among which 49 substances were first study to identify in Polygonatum, and 38 substances were flavonoids. A total of 19 differentially accumulated metabolites (DAMs) (five flavonols, three flavones, two dihydrochalcones, two flavanones, one flavanol, five phenylpropanoids, and one coumarin) were finally screened by KEGG enrichment analysis. Transcriptome analysis indicated that a total of 222 unigenes encoding 28 enzymes were annotated into three flavonoid biosynthesis pathways, which were “phenylpropanoid biosynthesis”, “flavonoid biosynthesis”, and “flavone and flavonol biosynthesis”. The combined analysis of the metabolome and transcriptome revealed that 37 differentially expressed genes (DEGs) encoding 11 enzymes (C4H, PAL, 4CL, CHS, CHI, F3H, DFR, LAR, ANR, FNS, FLS) and 19 DAMs were more likely to be regulated in the flavonoid biosynthesis pathway. The expression of 11 DEGs was validated by qRT-PCR, resulting in good agreement with the RNA-Seq. Our studies provide a theoretical basis for further elucidating the flavonoid biosynthesis pathway in Polygonatum.
1. Introduction
Polygonatum is a medicinal food homology perennial plant in the family Asparagaceae [1,2]. Currently, three species of Polygonatum were listed in the Chinese Pharmacopoeia, namely P. sibiricum Delar. ex Redoute, P. kingianum Coll. et Hemsl, and P. cyrtonema Hua [3]. Polygonatum is neutral and sweet and has been used in China for over two thousand years. Modern pharmacological research has shown that it has various pharmacological activities, including anti-oxidant, hypoglycemic, hypolipidemic, anti-tumor, anti-bacterial, and anti-inflammatory and enhances immunity [4]. The chemical composition of Polygonatum includes polysaccharides, saponins, flavonoids, amino acids, inorganic salts, volatile oils, lignans, and alkaloids [5]. Among them, flavonoids are one of the main functional components.
Flavonoids are a large group of polyphenolic compounds in medical plants and exhibit a number of medicinal benefits, including anti-oxidation [6], anti-bacterial [7], and anti-tumor [8]. The flavonoids in Engelhardia roxburghiana Wall. can inhibit myocardial cell autophagy and lipid accumulation in thoracic aortas of high-fat-diet-treated mice [9]. Polygonatum flavonoids also exhibit similar beneficial effects [10,11,12,13]. In addition, different flavonoid compounds might demonstrate different pharmacological effects, such as homoisoflavonoids from Polygonatum odoratum. 3-(2′,4′-dihydroxybenzyl)-5,7-dihydroxy-6-methyl-chroman-4-one could inhibit the proliferation of cancer cells [14]. Tectoridin shows the effects of sensitizing adipocytes for insulin, which promotes the full utilization of insulin and lowers blood glucose concentrations [15]. Odoratumone A exhibits stronger DPPH radical scavenging activities [16]. The homoisoflavonoids 5,7-dihydroxy-3-(2-hydroxy-4-methoxybenzyl)-8-methyl-chroman-4-one isolated from Polygonum verticillatum have a considerable antibacterial action [17]. Considering the differences in the efficacy of flavonoids in Polygonatum, it is necessary to separate and identify the monomer of such compounds, which will provide a more rigorous and systematic basis for clinical applications.
Since 1969, three flavonoids have been isolated from the fresh leaves of Polygonatum multiflorum on the basis of chemical, UV, and H-NMR spectral properties [18]. Using various chromatography and HPLC techniques for extraction and purification, 90 flavonoids were gradually identified from the genus Polygonatum based on their physicochemical properties and spectral characteristics. Metabolomics technology employs multi-reaction monitoring techniques to collect vast amounts of data and establish a metabolite specimen database, achieving high-throughput and accurate qualitative and quantitative analysis of metabolites [19], and providing more comprehensive and precise information on metabolites [20]. In recent years, 53 flavonoids have been isolated from the genus Polygonatum using metabolomics techniques. UPLC-ESI-MS/MS analysis revealed 22 flavonoids in the rhizomes of P. cyrtonema, including chalcones, dihydroflavones, dihydroflavonols, flavones, flavonoids, flavonoid carbonosides, and flavonols [21]. Sharma et al. [22] employed UPLC-MS/MS to identify 10, 15, and 2 flavonoids in Polygonatum verticillatum’s fruits, leaves, and rhizomes, respectively. Research demonstrated that leaf flavonoid content was the highest in all tested tissues of P. sibiricum in the forest [23]. Roots, leaves, flowers, and fruits of Polygonatum were edible, which were recorded in the Collection of Commentaries on the Classic of the Materia Medica (Ben Cao Jing Ji Zhu) compiled by Tao Hongjing in the Southern Liang dynasty (502–557 AD). Most of the Polygonatum flavonoids were isolated from rhizomes [24]. Yet, few studies have been published about flavonoids from the other tissues of Polygonatum. Therefore, it is necessary to study the flavonoids in different tissues of Polygonatum.
Medicinal plants contain an abundance of secondary metabolites, which are controlled by a cluster of functional genes. RNA-Seq technology, which utilizes high-throughput sequencing to analyze the transcriptome of organisms, has been widely applied in new gene identification, molecular markers development, and metabolic pathways elucidation in medicinal plants [25,26,27,28]. As a result, transcriptomics is an effective way to identify genes involved in secondary metabolite synthesis. The biosynthesis of flavonoids is regulated by a series of enzymes related to the phenylpropanoid and flavonoid pathways in medicinal plants [29]. Phenylalanine ammonialyase (PAL) is the first enzyme of the phenylpropanoid pathway, and chalcone synthase (CHS) is the entry-step enzyme that is responsible for the biosynthesis of the flavonoid backbone [30]. Chalcone isomerase (CHI) catalyzes the formation of dihydroflavonoid compounds, which then undergo intramolecular cyclization reactions via the action of dihydroflavonol 4-reductase (DFR) and flavonol synthase (FLS) on different branches, resulting in the evolution of various flavonoid compounds such as flavones, flavonols, isoflavones, and anthocyanins [31]. Transcription analysis obtained a total of 18, 15, and 26 enzymes related to the flavonoid biosynthesis pathway in Ephedra sinica Stapf [32], Scutellaria barbata D. Don [33], and Sophora japonica Linn [34], respectively. In Ginkgo biloba, 13 gene families encoding 111 enzymes involved in flavonoid biosynthesis were identified and characterized [35]. In Polygonatum, a total of 26 key enzymes associated with flavonoid synthesis pathways were identified through transcriptional analysis [36,37]. Therefore, such a pathway in Polygonatum still remains unclear and requires further exploration.
The integration analysis of metabolomics with transcriptomics can elucidate the relationship between genes and metabolites more effectively, so as to provide a more accurate and reliable result [38]. Integrative transcriptional and metabolic analysis of peanut skins with significant differences in anthocyanin types and contents showed that flavonoid biosynthesis was the key pathway for the formation of seed coat color, and delphinidin and cyanidin were the main differential metabolites [39]. Through comprehensive analysis of metabolomics and transcriptomics, Wang et al. [40] found that terpenes were the main volatile compounds, and 17 key genes were significantly related with the terpenoid synthesis pathway, providing references for understanding the aroma biosynthesis and perfume formulations of Dendrobium loddigesii Rolfe. This study aims to identify flavonoid compounds in four tissues of Polygonatum, to determine the flavonoid synthesis pathway and its regulatory mechanisms through integrated metabolomics and transcriptomics techniques. The results of the present study will provide new insights for investigating the transcriptional regulation of flavonoids in Polygonatum.
2. Results
2.1. Metabolic Profiling
The phenolics metabolites of the four tissues from P. cyrtonema were investigated based on UPLC-ESI-MS/MS. A total of 65 active substances were identified, among which 49 substances were first study to identify in Polygonatum (Table S1), and 38 substances were flavonoids, including 14 flavonols, 9 flavones, 7 flavanones, 3 dihydrochalcones, 2 anthocyanins, 2 isoflavones, and 1 flavanol (Table 1). The highest number and content of metabolites were detected in leaves and the lowest in rhizomes. There were 17 metabolites shared across the tissues, among which rutin had the highest proportion of flavonoid monomers in each tissue. The top ten metabolites with the highest content are shown in Table S1, which belong to flavonols, flavones, benzoic acid derivatives, and phenylpropanoids (Table S1).

Table 1.
Quantity and content of metabolites in different tissues of P. cyrtonema.
2.2. Metabolite Analysis Based on PCA and OPLS-DA
Principle component analysis (PCA) indicated that the replicates for each tissue grouped was based on the detected metabolites, suggesting that the results were reliable, and there was less degree of variability within the tissues (Figure 1). In this study, a supervised orthogonal partial least-squares discriminant analysis (OPLS-DA) model was employed to compare the metabolites content of the samples in pairs to evaluate the difference between Leaf and Stem (R2X = 0.987, R2Y = 0.998, Q2 = 0.997), Leaf and Rhizome (R2X = 0.996, R2Y = 1, Q2 = 0.999), Leaf and Root (R2X = 0.995, R2Y = 1, Q2 = 0.999), Stem and Rhizome (R2X = 0.998, R2Y = 1, Q2 = 0.99), Stem and Root (R2X = 0.995, R2Y = 1, Q2 = 0.99), and Rhizome and Root (R2X = 0.965, R2Y = 0.997, Q2 = 0.991) (Figure 2a–f). The Q2 values of all comparison groups exceeded 0.9, demonstrating that such models were stable and reliable and could be used to further screen differential flavonoid metabolites.
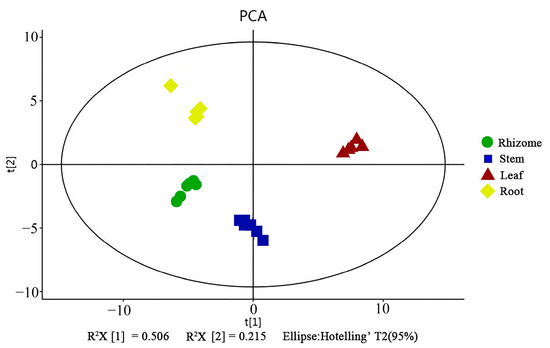
Figure 1.
Metabolite analysis on the basis of principal component analysis (PCA).
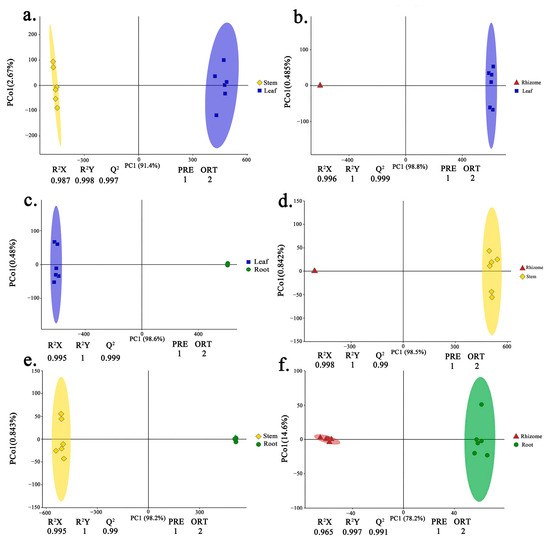
Figure 2.
Metabolite analysis based on orthogonal partial least-squares discriminant analysis (OPLS-DA). (a–f) OPLS-DA model plots for the comparison group Leaf vs. Stem, Leaf vs. Rhizome, Leaf vs. Root, Stem vs. Rhizome, Stem vs. Root, Rhizome vs. Root, respectively. The colorful shading represents the confidence ellipse.
2.3. Differentially Accumulated Metabolite Screening in Leaf, Stem, Rhizome, and Root Tissues
Pairwise comparisons were conducted among the six groups and a total of 62 differentially accumulated metabolites (DAMs) were screened out. Among all comparison groups, the leaf and rhizome had the most DAMs, while the rhizome and root showed the least (Figure 3). Moreover, most metabolites in leaves were upregulated. After taking the intersection of each comparison group, isorhamnetin, isorhamnetin-3-O-glucoside, malvin, vitexin, and phlorizin were common DAMs in the six comparison groups, while myricetin and 4-hydroxycinnamic acid were only screened between rhizomes and leaves, as well as between rhizomes and stems, respectively (Table S2).
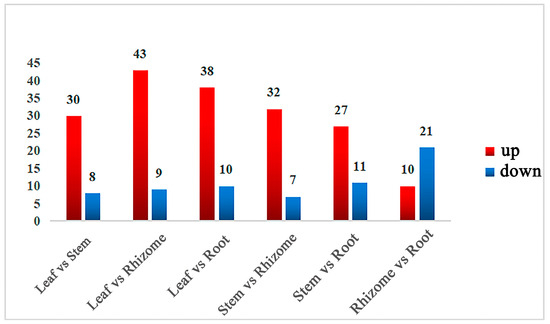
Figure 3.
Statistics of differently expressed metabolites. The statistical analysis of the number of differential metabolites for all different groups is presented, with the red color indicating upregulated metabolites and the blue color indicating downregulated metabolites.
2.4. Functional Annotation and Enrichment Analysis of Differentially Accumulated Metabolites
To obtain detailed pathway information, DAMs of each comparison group were annotated by searching against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Table S3). The results showed that DAMs from each comparison were enriched in flavonoid biosynthesis pathways (Figure 4), including phenylpropanoid biosynthesis (ko00940), flavonoid biosynthesis (ko00941), and flavone and flavonol biosynthesis (ko00944). A total of 19 DAMs were annotated into these pathways, including 5 flavonols, 3 flavones, 2 dihydrochalcones, 2 flavanones, 1 flavanol, 5 phenylpropanoids and 1 coumarin (Table 2).
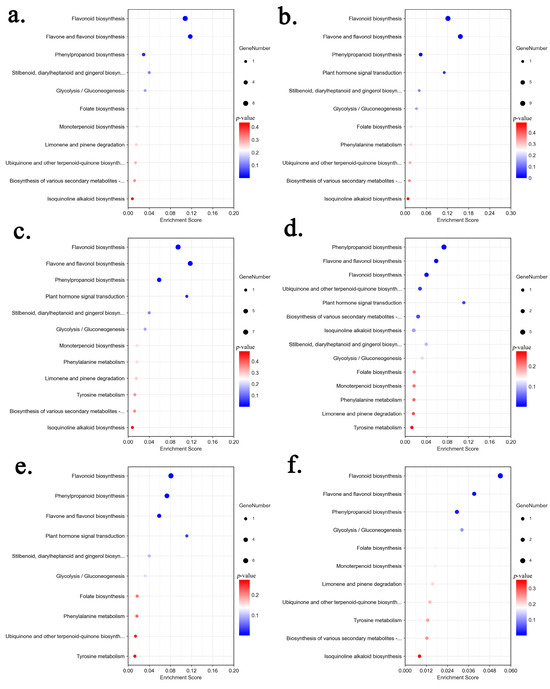
Figure 4.
KEGG pathway enrichment of the differential metabolites in each comparison: (a) Leaf vs. Stem, (b) Leaf vs. Rhizome, (c) Leaf vs. Root, (d) Stem vs. Rhizome, (e) Stem vs. Root, (f) Rhizome vs. Root.

Table 2.
Differential flavonoid metabolites enriched by KEGG.
2.5. Functional Annotation and Enrichment Analysis of Differential Flavonoid Genes
RNA-sequencing analysis was performed on the leaf, stem, rhizome, and root of P. cyrtonema. The differentially expressed genes (DEGs) identified from the six pairs of comparisons were further subjected to KEGG pathway enrichment analyses to screen genes associated with flavonoid biosynthesis (Table S4). The results showed that a total of 222 unigenes encoding 28 enzymes were annotated into flavonoid synthesis pathways, and 7 enzymes were first identified in Polygonatum (Table 3). Most of the DEGs were annotated into the phenylpropanoid biosynthetic pathway (ko00940); 47 DEGs were annotated to the biosynthesis of flavonoid (ko00941); the flavone and flavonol biosynthesis pathway was associated with 6 annotated DEGs (ko00944) (Table 3).

Table 3.
Genes involved in the synthetic pathway of flavonoids.
2.6. Combined Analysis of Transcriptome and Metabolome Analysis
The Pearson correlation coefficient was used to calculate the relationship between gene expression and metabolite level. The results revealed that 37 DEGs encoding 11 key enzymes were significantly correlated with 19 DAMs (Figure 5a). In addition, the expression profiles of 6 genes (4CL1, 4CL2, CHS2, CHS3, DFR1, LAR) correlate well with those of the flavonoid abundance, which was significantly positively correlated (p < 0.01) with the content of astragalin, catechin, chlorogenic acid, cosmosiin, ferulic acid, kaempferol, naringin, phlorizin, quercetin, and quercitrin. Furthermore, significant positive associations were discovered between luteolin and 4CL2, CHS3, phloretin and 4CL1, 4CL2, CHS3, rutin and 4CL1, 4CL2, CHS3, DFR1, LAR, and between sinapic acid and 4CL1, CHS2, DFR1, LAR. It seemed that these 14 metabolites were more likely to be regulated in the flavonoid biosynthesis pathway, which was in correspondence with the results of the heatmap (Figure 5). Due to the KEGG pathway, transcriptomic and metabolomic data, we outlined potential pathways for flavonoid biosynthesis and accumulation in P. cyrtonema (Figure 5b).
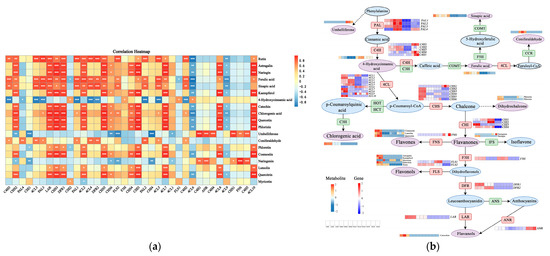
Figure 5.
Transcription metabolism conjoint analysis data plot. (a) Correlation heatmap between differentially accumulated metabolites and differentially expressed genes. * p < 0.05, ** p < 0.01, *** p < 0.001; (b) The flavonoids biosynthesis and accumulation pathway of P. cyrtonema. The circle represents metabolite, and the purple circle indicates metabolite annotated by KEGG enrichment analysis. The box represents genes, and the red box represents the gene selected for joint analysis. The solid line indicates the metabolic reactions in only one step, the dash line presents more than one step of the metabolic reaction.
2.7. Validation of Differentially Expressed Genes by Quantitative Real-Time PCR
To validate the accuracy of the RNA-Seq data, 11 differentially expressed genes encoding 9 key enzymes were selected for a quantitative real-time PCR (qRT-PCR) analysis. As illustrated in Figure 6, the expression patterns of these genes were consistent with the corresponding FPKM values derived from RNA-Seq analysis, which indicated the reliability of the transcriptome. The core genes for flavonoid biosynthesis, such as PAL3, C4H2, CHS1, CHS4, F3H, DFR1, LAR, and ANR, were more highly expressed in the leaves than those in the rhizome.
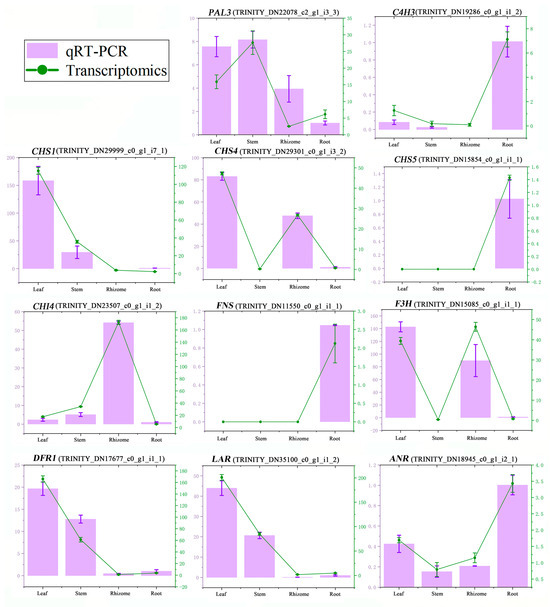
Figure 6.
qRT-PCR validation of 11 DEGs. The X-axis means four tissues of P. cyrtonema. The left Y-axis represents the relative expression of each gene in four tissues by qRT-PCR, and the right Y-axis represents the FPKM value of each gene in four tissues by transcriptomics.
3. Discussion
Mass spectrometry-based metabolomics approaches allow for detection and quantification of many thousands of metabolic products simultaneously, shedding light on the metabolic pathways of effective chemical synthesis in medicinal plants. In the present study, a wide-ranging analysis of phenolic metabolites using targeted metabolomics was conducted in four different tissues of P. cyrtonema, and a total of 65 phenolic metabolites from 12 categories were detected (Table S1). Flavones and flavonols were the main phenolic metabolites. The highest proportion of flavones and flavonols in flavonoids was also reported in Tetratigma hemsleyanum Diels et Gilg [42], Hippophae rhamnoides L. [43], Ziziphi Spinosae Semen [44], Kadsura coccinea [45], and Cannabis sativa L. [46]. Moreover, the content of flavonoids was highest in leaves and lowest in rhizomes (Table 1), which was also reported in P. sibiricum in the forest [23]. The higher content of flavonoids in the leaves was also reported in P. cyrtonema [47], Bupleurum chinense DC. [48], Perilla frutescens (L.) Britt. [49], Hippophae rhamnoides L. [50], and Dendrobium officinale Kimura et Migo [51]. It seemed that leaves were the main source of flavonoids, which could be related to chemical defense in plants. Flavonoids are phenolic compounds that have an undesirable bitter taste, which helps to reduce herbivory [47]. In addition, flavonoids are the primary UV absorbers, preventing or limiting UV-B damage to plants [48]. The most abundant flavonoid in Polygonatum was rutin, which belongs to the flavonols. Such result was also described in Golden Buckwheat and Lonicera japonica Thunb. [52,53]. Among the detected active substances, 49 substances were first study to identify in this species (Table S1). Researchers have extracted a total of 127 Polygonatum flavonoids, which are categorized according to their chemical structures into the classes of homoisoflavones, isoflavones, flavonoids, chalcones, and dihydroflavonoids. Homoisoflavones, a special subclass of flavonoids [54], are rarely found in nature, mainly existing in Fabaceae and Asparagaceae families [55], and accounted for nearly half of the total identified flavonoid compounds. Only 19 substances of the identified Polygonatum flavonoids had biological reference standards and were investigated with UPLC-ESI-MS/MS in the four tissues of P. cyrtonema. Fifteen of them were successfully detected, indicating high detection efficiency of such metabolomics (Table S1). Three of the remaining four compounds, isoliquiritigenin, liquiritigenin, and jaceosidin, were exclusively detected in P. kingianum [24,56,57]. However, apigenin was not detected in this study, possibly due to its low concentration. The top ten metabolites listed in Table S1 had the effects of anti-oxidation and anti-bacterial [58,59,60,61,62,63,64,65,66,67]. In addition, most of them also have anti-tumor (except salicylic acid), and hypoglycemic (except salicylic acid and vanillin) effects.
In the current study, both DAMs and DEGs annotated by the KEGG database were mainly enriched in phenylpropanoid biosynthesis (ko00940), flavonoid biosynthesis (ko00941), and flavone and flavonol biosynthesis (ko00944) pathways (Table 2 and Table S4). In medicinal plants, differential genes related to flavonoid biosynthesis are subjected to the phenylpropane biosynthesis pathway [68,69,70,71], flavonoid biosynthesis pathway [69,71], flavone and flavonol biosynthesis pathways [69,70,71], and isoflavone biosynthesis pathway [69,71] by KEGG pathway analysis. The first three pathways have also been reported in P. kingianum [41] and P. cyrtonema [36]. After comparing with other results in Polygonatum, our study identified more genes that were 194, 47, and 6 genes in the phenylpropane biosynthesis, flavonoid biosynthesis, flavonoid and flavonol biosynthesis pathways, respectively. Significantly different phenolic compounds contents in the detected tissues helps to identify more DEGs. The transcriptome analysis identified 28 enzymes encoded by flavonoid-related DEGs, of which 21 were reported in Polygonatum, including CHI, CHS, CCOMT, and CYP73A [36,37,41,72] (Table 3). Among the 7 newly identified enzymes, katG, TOGT1, CSE, and PRDX6 were enriched in phenylpropanoid biosynthesis, PGT1 and FNS were annotated to flavonoid biosynthesis, and FG2 was annotated to flavone and flavonol biosynthesis. Flavone synthase (FNS) catalyzes the synthesis of flavones from dihydroflavones. LjFNS was first cloned from Lonicera japonica using Chromosome walking and might be involved in flowers development due to the highest expression in flowers by qRT-PCR analysis [73]. Integrated metabolomic and transcriptomic study revealed that both upregulated DAMs and DGEs were associated with the flavonoid biosynthesis pathway, and the upregulation of the expression of genes phlorizin synthase (PGT1), shikimate O-hydroxycinnamoyltransferase (HCT), and chalcone synthase (CHS) promotes p-coumaroyl-CoA in sugarcane roots transferred into homoeriodictyol chalcone and 5-deoxyleucopelargonidin [74]. The partial cDNA of flavonol-3-O-glucoside L-rhamnosyltransferase (FG2) involved in the rutin biosynthetic pathway was first cloned in C. spinosa plants [75]. Other medicinal plants such as soybeans [76], sugarcane [77], and Populus alba×P. glandulosa also successfully isolated FG2 [78]. The constructed flavonoid synthesis pathway and the related newly identified genes provide a reference for subsequent functional studies of the candidate genes and comparative studies with other species.
The association analysis of metabolomics and transcriptomics confirmed that 19 DAMs regulated in the flavonoid biosynthetic pathway were significantly associated with 37 DEGs encoding 11 enzymes (Figure 5a). Through de novo transcriptome assembly and metabolomic analysis of shell and leaves of Trapa bispinosa Roxb, both the 15 DEGs (CHS, FLS, DFR, ANR…) and 42 metabolites (luteolin, delphinidin…) involving phenylpropanoid and flavonoid biosynthesis were identified to construct flavonoid biosynthesis pathways and related networks (Yin et al.) [79]. Combined transcriptome and metabonomic data of C. paliurus leaves at different developmental stages revealed that the expression trends in differential flavonoids metabolites and genes were significantly related [80]. Among DEGs, six genes (4CL1, 4CL2, CHS2, CHS3, DFR1, LAR)-related flavonoid biosynthesis also showed strong correlations with DAMs (Figure 5a). Coumaric acid generates coumaroyl-CoA in the presence of 4CL, which was the primary rate-limiting enzyme in the phenylalanine pathway [81]. Both 4CL1 and 4CL2 were strongly positively linked with 14 DMFs, including catechin, kaempferol, and caffeic acid, etc., which was consistent with results in Lycium chinense fruits [82]. CHS was the first and major rate-limiting enzyme in the flavonoid biosynthesis pathway and was one of the most explored enzymes in medicinal plants. It catalyzed the formation of chalcone, a substance from which different flavonoids were derived [83]. The overexpression of EaCHS1 in peels of C. reticulata ‘Chachi’ increased the production of downstream flavonoids and the expression of related genes in the phenylpropanoid pathway [84]. There was a substantial positive association between SeCHS1 and five flavonoid compounds in Sechium edulel [85]. Our findings also revealed that CHS2 and CHS3 showed a highly positive relationship with 12 and 13 differential metabolites, respectively (Figure 5a). In addition, a negative association was found between CHS2 and umbelliferone. Flavonoid content was negatively correlated with the expression of CHS during fruit development of Lycium chinense [82]. This may be due to spatiotemporal expression differences or functional redundancy of CHS genes. DFR was an essential enzyme responsible for the diversification of anthocyanins and ellagitannins [86]. The expression of the DFR1 gene was highly positively correlated with the content of 12 flavonoid metabolites in this study. DFR gene expression positively correlated with the accumulation of flavonoids in leaves of field conifers under heavy metal stress [87]. Wang et al. [88] found a strong positive correlation of the accumulation of catechin and eriodictyol with the DFR gene expression in red cell line of Saussurea medusa. LAR was a key enzyme in the plant flavonoid synthesis pathway that catalyzed the conversion of colorless anthocyanins into catechins [89]. The expression of the LAR gene was positively associated with 12 flavonoids in the current investigation, except for umbelliferone. Correlation analysis revealed that the content of total flavonoids, total flavanols, and four flavanol components including catechins and epicatechin showed a significantly strong correlation with the LAR gene in jujube fruits [90].
We also demonstrated that the majority of genes involved in the synthesis pathway of flavonoids exhibited heightened expression levels in the leaves and displayed lower expression levels in rhizomes, which was further confirmed by qRT-PCR (Figure 6). As a result, the most differential metabolites were identified between leaf and rhizome (Figure 3). In general, these preliminary results demonstrated the correlation of metabolites and genes in the flavonoid synthesis pathway and established the foundation for future research on the specific mechanisms regulating the synthesis of secondary metabolites in P. cyrtonema.
4. Materials and Methods
4.1. Plant Materials
Polygonatum cyrtonema Hua, as a wild resource, was collected in Qingyang, Anhui Province (30°34′15″ N, 117°48′22″ E), and then planted in Yuhang, Zhejiang Province (30°48′56″ N, 119°76′04″ E), in November 2018. The samples were identified by Zongsuo Liang, professor at the Zhejiang Sci-Tech University. The root, rhizome, stem, and leaf of P. cyrtonema were collected from three individual plants on the sunny day of 22 April 2022. The biological replicates were conducted three times for each tissue. All samples were cleaned with ultrapure water and were immediately frozen in liquid nitrogen and stored at −80 °C for metabolomic, transcriptomic analyses and quantitative real-time PCR analysis.
4.2. Metabolome Analysis Based on UPLC-ESI-MS/MS
Samples were crushed for 2 min at 60 Hz using a grinder and then dissolved in a methanol–water solution (V (water):V (methanol) = 1:2) before metabolic analysis. To improve the extraction rate, we conducted ultrasound three times and vortexed twice. After centrifugation of the liquid (10,000 rpm, 10 min), the supernatant was aspirated, filtered through a microporous membrane (0.22 μm pore size), and stored in brown injection vials for UPLC-ESI-MS/MS analysis.
Liquid chromatography was performed using a Waters ACQUITY UPLC HSS T3 (Milford, MA, USA, 1.8 μm, 2.1 mm × 100 mm) column. The mobile phase consisted of 0.1% formic acid aqueous solution for phase A and acetonitrile for phase B. The mass spectrometry conditions used in this study involved Ionspray voltage (IS) at 5500 V or −4500 V, curtain gas (CUR) at 35 psi, and a Source Temperature at 500 °C. To perform absolute quantitative analysis on 130 phenolic compounds (Table S5), we employed a phenolic compound analysis method package based on the LC-MS/MS platform developed by LuMing Biotechnology Co., Ltd. (Shanghai, China). Targeted metabolites were analyzed in Schedule multiple reaction monitoring (SRM) mode. The MRM pairs, declustering potentials (DP), and collision energies (CE) were optimized for each analyte. Data acquisitions and further analysis were conducted using Analyst software (https://sciex.com/products/software/analyst-software). SCIEX OS-MQ software (https://sciex.com/products/software/sciex-os-software) was used to quantify all metabolites. The QCs were injected at regular intervals (every 10 samples) throughout the analytical run to provide a set of data from which repeatability can be assessed. The data analysis was based on peak area normalization. The detection parameters of reference substance were deselected for peak retention time alignment with minimum intensity at 15% of base peak intensity, noise elimination level at 10.00, and isotopic peaks were excluded.
4.3. Transcriptomic Analysis
Total RNA was extracted using the mirVana miRNA Isolation Kit, and RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The libraries were constructed using the TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. Then, these libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten), and 125 bp/150 bp paired-end reads were generated.
Raw data (raw reads) were processed using Trimmomatic [91] (http://www.usadellab.org/cms/index.php). The reads containing ploy-N and the low-quality (Q < 20) reads were filtered out to obtain the clean reads. The clean reads were de novo assembled into transcripts by using Trinity [92] (vesion:2.4) in the paired-end method. The longest transcript of each unigene was chosen for subsequent analysis.
4.4. Quantitative Real-Time PCR (qRT-PCR) Analysis
Quantitative real-time PCR (qRT-PCR) was performed using the Taq Pro Universal SYBR qPCR Master Mix Kit (Vazyme, Nanjing, China). Gene primers designed using Primer 6.0 are listed in Table 4. The 10 µL reaction mixture contained 1 µL of cDNA, 0.2 µL of each primer, 5 µL of 2× SYBR qPCR Master Mix, and 3.6 µL of RNase-free water. All qRT-PCR analyses were performed using the following conditions: denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s, and then at 60 °C for 30 s. All reactions were repeated three times in the experiments, and the 2−ΔΔCt method [93] was used to calculate the relative expression of each unigene.

Table 4.
Primer sequence information of qRT-PCR.
4.5. Data Analysis
Principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA) were used to analyze the trend of metabolites. The Student’s t-test was performed for pairwise comparisons. The differentially accumulated metabolites and expressed genes were screened according to p ≤ 0.05 and |log2 (fold change)| > 1 rules. Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, the related pathways of the DAMs and DEGs were determined. The OECloud tools (https://cloud.oebiotech.com) (accessed on 20 March 2023) application was used for Pearson’s correlation analysis. A significance level of p < 0.05 was considered statistically significant.
5. Conclusions
In the present study, metabolomics and transcriptomics were employed to identify metabolites and genes associated with the flavonoids biosynthesis pathway, respectively. Most of the detected substances were first study to identify in Polygonatum. A total of 222 differentially expressed genes encoding 28 enzymes and 14 different flavonoid metabolites were identified in four tissues of P. cyrtonema. Integrated analysis of transcriptomic and metabolomic data revealed that these 14 different flavonoid metabolites were more likely to be regulated by flavonoid biosynthesis-related genes, especially 4CL1, 4CL2, CHS2, CHS3, DFR1, and LAR. Our results provide valuable information on Polygonatum flavonoid regulation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules29102248/s1, Table S1: 65 phenolic substance information; Table S2: Differentially accumulated metabolites; Table S3: KEGG pathway enrichment of differentially accumulated metabolites; Table S4: KEGG pathway enrichment of differentially expressed genes; Table S5: Standard information. References [94,95,96,97,98,99,100] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, L.Y. and Q.Y.; methodology, L.Z.; software, F.R. and Z.Z.; validation, Q.J. and L.Y.; formal analysis, L.Z. and L.Y.; investigation, L.Y., F.R., and Z.Z.; resources, Q.J.; data curation, L.Y.; writing—original draft preparation, L.Y.; writing—review and editing, Q.J.; visualization, L.Y. and Z.Z.; supervision, Q.J.; project administration, Q.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Key Research and Development Project of Zhejiang Province (2023C02017), Agricultural Major Technology Collaborative Promotion Plan Project of Zhejiang Province (2022XTTGZYC03).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within this article and Supplementary Materials. Raw Illumina sequencing reads have been deposited in the NCBI Sequence Read Archive Database under accession PRJNA1068333.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Liu, R.; Lu, J.; Xing, J.Y.; Du, M.; Wang, M.X.; Zhang, L.; Li, Y.F.; Zhang, C.H.; Wu, Y. Transcriptome and metabolome analyses revealing the potential mechanism of seed germination in Polygonatum cyrtonema. Sci. Rep. 2021, 11, 12161. [Google Scholar] [CrossRef]
- Zhang, W.W.; Xia, L.; Peng, F.; Song, C.; Manzoor, M.A.; Cai, Y.; Jin, Q. Transcriptomics and metabolomics changes triggered by exogenous 6-benzylaminopurine in relieving epicotyl dormancy of Polygonatum cyrtonema Hua seeds. Front. Plant Sci. 2022, 13, 961899. [Google Scholar] [CrossRef]
- Ren, H.M.; Deng, Y.L.; Zhang, J.L.; Ye, X.W.; Xia, L.T.; Liu, M.M.; Liu, Y. Research progress on processing history evolution, chemical components and pharmacological effects of Polygonati Rhizoma. China J. Chin. Mater. Med. 2020, 45, 4163–4182. [Google Scholar] [CrossRef] [PubMed]
- Tao, A.E.; Zhang, X.C.; Du, Z.F.; Zhao, F.Y.; Xia, C.L.; Duan, B.Z. Research progress on flavonoids in plants of Polygonatum Mill. and their pharmacological activities. Chin. Tradi. Herb. Drugs 2018, 49, 2163–2171. [Google Scholar] [CrossRef]
- Jiao, J.; Huang, W.L.; Bai, Z.Q.; Liu, F.; Ma, C.D.; Liang, Z.S. DNA barcoding for the efficient and accurate identification of medicinal polygonati rhizoma in China. PLoS ONE 2018, 13, e0201015. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.F.; Liu, Y.X.; Li, Y.H.; Hao, Q.Q.; He, X.H.; Wang, J.G. Extraction and antioxidant activity of flavonoids from sea-buckthorn. J. Anhui Sci. Technol. Univ. 2024, 38, 88–96. [Google Scholar] [CrossRef]
- Liu, S.J.; Xing, Y.B.; Xing, P.; Chen, X.P. Characterization of the antibacterial activity of Perilla flavone. Food Res. Dev. 2021, 42, 163–168. [Google Scholar] [CrossRef]
- Chen, M.; Wang, J.T.; Gao, W.H.; Lei, S.P.; Zhu, Y.H. Total flavonoids in Scutellaria barbata prevents NLRP3 inflammasome expression in tumor cells by affecting autologous pathway. China J. Chin. Mater. Med. 2017, 42, 4841–4846. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Huang, L.; Li, D.; He, J.H.; Li, Y.J.; He, F.; Fang, W.R.; Wei, G.N. Total flavonoids of Engelhardia roxburghiana Wall. leaves alleviated foam cells formation through AKT/mTOR-Mediated autophagy in the progression of atherosclerosis. Chem. Biodivers. 2021, 18, e2100308. [Google Scholar] [CrossRef]
- Lin, H.R. Two homoisoflavonoids act as peroxisome proliferator-activated receptor agonists. Med. Chem. Res. 2015, 24, 2898–2905. [Google Scholar] [CrossRef]
- Chen, Y.J.; Shi, X.; Qu, R.; Peng, W.S. Study on extraction process of flavonoids from Polygonatum kingianum Coll. et Hemsland antioxidative activities of flavonoids. Sci. Technol. Food Ind. 2013, 34, 222–225. [Google Scholar] [CrossRef]
- Ning, D.L.; Liu, J.; Li, M.; Li, W.J.; Li, l.; Zhang, H.L.; Sun, J.K. Anti-proliferation effect of homoisoflavonoids extracted from Polygonati odorati Rhizoma on A549 Cells. Chin. J. Exp. Tradit. Med. Form. 2017, 23, 174–179. [Google Scholar] [CrossRef]
- Khan, H.; Saeed, M.; Muhammad, N.; Gilani, A.H.; Muhammad, N.; Ur Rehman, N.; Mehmood, M.H.; Ashraf, N. Antispasmodic and antidiarrheal activities of rhizomes of Polygonatum verticillatum maneuvered predominately through activation of K+ channels: Components identification through TLC. Toxicol. Ind. Health 2016, 32, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Li, L.H.; Ren, F.Z.; Zheng, Z.H.; Chen, S.H.; Gao, Y.Q.; Zhu, X.L. Studies on biological activity of homoisoflavanones from Polygonatum odratum (Mill.) Druce. J. Hebei Norm. Univ. 2012, 36, 134–137. [Google Scholar]
- Zhang, H.; Yang, F.; Qi, J.; Song, X.C.; Hu, Z.F.; Zhu, D.N.; Yu, B.Y. Homoisoflavonoids from the fibrous roots of Polygonatum odoratum with glucose uptake-stimulatory activity in 3T3-L1 adipocytes. J. Nat. Prod. 2010, 73, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.L.; Zhang, Y.P.; Zhao, H.D.; Liang, J.S.; Zhang, Y.; Shi, S.Y. Antioxidant homoisoflavonoids from Polygonatum odoratum. Food Chem. 2015, 186, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Patial, V.; Singh, D.; Sharma, U.; Kumar, D. Antimicrobial homoisoflavonoids from the Rhizomes of Polygonatum verticillatum. Chem. Biodivers. 2018, 15, e1800430. [Google Scholar] [CrossRef] [PubMed]
- Skrzypczakowa, L. C-glycosyls in Polygonatum multiflorum. Diss Pharm. Pharmacol. 1969, 21, 261–266. [Google Scholar]
- Li, J.; Yan, G.H.; Duan, X.W.; Zhang, K.C.; Zhang, X.M.; Zhou, Y.; Wu, C.B.; Zhang, X.; Tan, S.N.; Hua, X.; et al. Research progress and trends in metabolomics of fruit trees. Front. Plant Sci. 2022, 13, 13881856. [Google Scholar] [CrossRef]
- Lin, C.C.; Zhao, Y.; Liu, R.; Lu, Q. Research progress in the application of proteomics and metabolomics in bee products. Sci. Technol. Food Ind. 2023, 44, 377–386. [Google Scholar] [CrossRef]
- Han, Z.G.; Gong, Q.Q.; Huang, S.Y.; Meng, X.Y.; Xu, Y.; Li, L.G.; Shi, Y.; Lin, J.H.; Chen, X.L.; Li, C.; et al. Machine learning uncovers accumulation mechanism of flavonoid compounds in Polygonatum cyrtonema Hua. Plant Physiol. Biochem. 2023, 201, 107839. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Joshi, R.; Kumar, D. Metabolomics insights and bioprospection of Polygonatum verticillatum: An important dietary medicinal herb of alpine Himalaya. Food Res. Int. 2021, 148, 110619. [Google Scholar] [PubMed]
- Wu, K.J.; Wang, F.F.; Chang, H.; Liang, Z.S.; Yang, Q.W.; Ma, C.D.; Jia, Q.J. Effects of different cultivation methods on the accumulation of main chemical constituents of four-year-old Polygonatum sibiricum. Chin. Tradi. Patent Med. 2021, 43, 2433–2437. [Google Scholar] [CrossRef]
- Li, A.L.; Li, W.J.; Wu, W.; Zhang, T.Y.; Zhang, J.; Wu, Y.; Liu, R. Comparison of flavonoids, amino acids and phenolic acids in different Polygonatum rhizomes. Chin. J. Appl. Environ. Biol. 2023, 1–10. [Google Scholar] [CrossRef]
- Hui, F.; Liu, X.Y.; Li, Z.X.; Liu, F.S.; Yang, S.H. Application of transcriptome sequencing in study of medicinal plants. Chin. Tradi. Herb. Drugs 2019, 50, 6149–6155. [Google Scholar] [CrossRef]
- Liu, H.B.; ShangGuan, Y.N.; Pan, Y.C.; Zhao, Z.Q.; Li, L.; Xu, D.L. Applications of RNA-Seq technology on medicinal plants. Chin. Tradi. Herb. Drugs 2019, 50, 5346–5354. [Google Scholar] [CrossRef]
- Mao, R.J.; Bai, Z.Q.; Wu, J.W.; Han, R.L.; Zhang, X.M.; Chai, W.G.; Liang, Z.S. Transcriptome and HPLC analysis reveal the regulatory mechanisms of aurantio-obtusin in space environment-induced Senna obtusifolia Lines. Int. J. Environ. Res. Public Health 2022, 19, 898. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Y.T.; Guo, J.; Yang, J.; Zhang, X.D.; Wang, Z.X.; Cheng, Y.; Du, Z.W.; Qi, Z.C.; Huang, Y.B.; et al. Integrated transcriptomics and proteomics to reveal regulation mechanism and evolution of SmWRKY61 on tanshinone biosynthesis in Salvia miltiorrhiza and Salvia castanea. Front. Plant Sci. 2022, 12, 820582. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Guo, K.Y.; Liu, L.D.; Tian, W.; Xie, X.L.; Wen, S.Q.; Wen, C.X. Integrated transcriptomic and metabolomic data reveal the flavonoid biosynthesis metabolic pathway in Perilla frutescens (L.) leaves. Sci. Rep. 2020, 10, 16207. [Google Scholar] [CrossRef]
- Zou, L.Q.; Wang, C.X.; Kuang, X.J.; Li, Y.; Sun, C. Advance in flavonoids biosynthetic pathway and synthetic biology. Chin. J. Chin. Mater. Medi. 2016, 41, 4124–4128. [Google Scholar] [CrossRef]
- Guo, J.N. Flavonoids Biosynthesis Pathway and Key Genes Function Analysis in Meconopsis horridula at Different Altitudes. Master’s Thesis, Tibet University, Tibet, China, 20 May 2023. [Google Scholar]
- Ma, J.; Cheng, T.L.; Sun, C.Y.; Deng, N.; Shi, S.Q.; Jiang, Z.P. Characterization of transcriptome reveals pathway of flavonoids in Ephedra sinica Stapf. Acta Agric. Zhejiangensis 2016, 28, 609–617. [Google Scholar]
- Zou, Y.H.; Liu, C.Y.; Lin, Z.Y.; Li, Z. Transcriptome sequencing and flavonoid biosynthesis related genes of Scutellaria barbata D. Don. Fujian J. Agric. Sci. 2018, 33, 1242–1250. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, D.X.; Song, X.H.; Li, L.Y. Transcriptome analysis reveals candidate genes involved in flavonols biosyhthesis in Sophora japonica. Chin. J. Chin. Mater. Med. 2018, 43, 2682–2689. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.A.; Meng, Z.L.; Zhang, H.Y.; Chu, Y.X.; Qiu, Y.Y.; Jin, B.A.; Wang, L. Identification and characterization of thirteen gene families involved in flavonoid biosynthesis in Ginkgo biloba. Ind. Crop. Prod. 2022, 188, 115576. [Google Scholar] [CrossRef]
- Ye, B.H.; Yang, Y.; Zhu, L.J.; Shi, C.G.; Chen, Y.W.; Hu, C.J.; Song, Q.Y.; Li, H.B. Analysis of genes expression involved in flavonoids biosynthesis in Polygonatum cyrtonema based on comparative transcriptome. Food Sci. Biotech. 2022, 41, 84–92. [Google Scholar] [CrossRef]
- Xiao, Y.Z.; Han, S.M.; Qin, Z.; Li, C.Q. Analysis of transcriptome sequencing and related genes of flavonoids biosynthesis from Polygonatum kingianum. J. Henan Agric. Univ. 2020, 54, 931–940. [Google Scholar] [CrossRef]
- Xu, J.Q.; Li, L.; Wang, S.B.; Ma, X.W.; Wu, H.X.; Xu, W.T.; Liang, Q.Z.; Chen, J.Z. Review of integrated metabolome and transcriptome analysis used for disclosing physiological mechanism in fruit crops. J. Fruit Sci. 2020, 37, 1413–1424. [Google Scholar] [CrossRef]
- Li, J.W.; Ma, Y.C.; Yang, X.L.; Wang, M.; Cui, S.L.; Hou, M.Y.; Liu, L.F.; Hu, M.D.; Jiang, X.X.; Mu, G.J. Transcriptomics-metabolomics combined analysis highlight the mechanism of testa pigment formation in peanut (Arachis hypogaea L.). J. Plant Genet. Resour. 2022, 23, 240–254. [Google Scholar] [CrossRef]
- Wang, S.; Du, Z.; Yang, X.; Wang, L.; Xia, K.; Chen, Z. An integrated analysis of metabolomics and transcriptomics reveals significant differences in floral scents and related gene expression between two Varieties of Dendrobium loddigesii. Appl. Sci. 2022, 12, 1262. [Google Scholar] [CrossRef]
- Chai, J.H.; Wang, X.J.; Yi, C.; Zhou, J.B.; Liu, D.; Zhao, N.; He, T.T. Transcriptome analysis of Polygonatum sibiricum and identification of putative genes involved in main secondary metabolism pathway. Mol. Plant Breed. 2023, 1–12. [Google Scholar]
- Xu, F.; Liang, Z.S.; Han, R.L. Research progress of flavonoids in Tetrastigma hemsleyanum Diels et Gilg and their pharmacological activities. Genomics Appl. Biol. 2021, 40, 3704–3716. [Google Scholar] [CrossRef]
- Ren, L.C.C.; Liu, Z.H.; Dong, Q.; Wang, H.L.; Hu, N. Recent progress on the flavonoid components and pharmacological effects of Hippophae rhamnoides L. Chin. J. Med. Chem. 2023, 33, 598–617. [Google Scholar] [CrossRef]
- Li, B.J.; Yang, Y.G.; Song, Z.X.; Tang, Z.S. Research progress of flavonoids in Ziziphi Spinosae Semen. Cent. South Pharm. 2023, 21, 2690–2697. [Google Scholar]
- Gao, J.F.; Zhou, W.; Liu, N.; Yang, Y. Analysis of flavonoids in different tissues of Kadsura coccinea plant by widely-targeted metabolomics. Guihaia 2022, 42, 1193–1203. [Google Scholar] [CrossRef]
- Jiang, S.; Wan, L.; Xu, Z.X.; Yan, J.J.; Zheng, C.N. Research progress on flavonoids of Cannabis sativa L. Chin. Agric. Sci. Bull. 2021, 37, 120–128. [Google Scholar] [CrossRef]
- Zhao, H.Y.; Luo, Y.; Deng, X.K.; Gao, P. The main chemical constituents and antioxidant activities of Polygonatum cyrtonema Hua. J. Anhui Agric. Univ. 2020, 47, 793–797. [Google Scholar] [CrossRef]
- Li, H.M.; Gao, Y.; Shao, X.F.; Qin, J.K.; Wu, Y.C.; Zhao, M. Study on total flavonoids content and comparison of antioxidant activity in different parts of Bupleurum chinense DC. from different provenances. Chin. Food Addit. 2022, 33, 211–217. [Google Scholar] [CrossRef]
- Zhang, C.W.; Guo, J.Q.; Li, W.P.; Luo, Y.J.; Yao, Y.; Xu, C.Q.; Shen, G.A.; Suo, F.M.; Guo, B.L. Content correlation of eight phenolic acids and flavonoids in different medicinal parts of PA-type Perilla germplasms. Chin. J. Chin. Mater. Med. 2022, 47, 3447–3451. [Google Scholar] [CrossRef] [PubMed]
- Tang, K. Comparison of total flavonoids content in different parts of Sea buckthorn. Heilongjiang Agric. Sci. 2022, 3, 64–67. [Google Scholar] [CrossRef]
- Lou, G.G.; Xia, J.; Yang, J.; Wang, H.P.; Liang, Z.S.; Xiao, Y.; Li, Z.D.; Zhang, Y.; Liu, Z.C.; Shi, W.L.; et al. Differences in the chemical composition of Dendrobium officinale Kimura et Migo and Dendrobium crepidatum Lindl based on UPLC-O-TOF-MS/MS and metabolomics. Acta Pharm. Sin. 2021, 56, 3331–3344. [Google Scholar] [CrossRef]
- Huang, J.; Wang, L.Y.; Tang, B.; Ren, R.R.; Shi, T.X.; Zhu, L.W.; Deng, J.; Liang, C.G.; Wang, Y.; Chen, Q.F. Integrated transcriptomics and widely targeted metabolomics analyses provide insights into flavonoid biosynthesis in the rhizomes of Golden Buckwheat (Fagopyrum cymosum). Front. Plant Sci. 2022, 13, 803472. [Google Scholar] [CrossRef]
- Huang, W.J.; Xiong, L.W.; Zhang, L.F.; Zhang, F.; Han, X.Y.; Zhang, Y.Q.; Zhang, L.; Yang, H.B. Study on content variation of flavonoids in different germplasm during development of Lonicerae Japonicae Flos. Chin. Tradi. Herb. Drugs 2022, 53, 3156–3164. [Google Scholar] [CrossRef]
- Wang, Y.C.; Liu, F.; Liang, Z.S.; Peng, L.; Wang, B.Q.; Yu, J.; Su, Y.Y.; Ma, C.D. Homoisoflavonoids and the antioxidant activity of Ophiopogon japonicus root. Iran J. Pharm. Res. 2017, 16, 357–365. [Google Scholar]
- Lin, L.G.; Liu, Q.Y.; Ye, Y. Naturally occurring homoisoflavonoids and their pharmacological activities. Planta Med. 2014, 80, 1053–1066. [Google Scholar] [CrossRef]
- Wang, Y.F.; Mu, T.H.; Chen, J.J.; Luo, S.D. Study on the chemical constituents of Polygonatum kingianum. China J. Chin. Mater. Med. 2003, 28, 524–527. [Google Scholar] [CrossRef]
- Liu, S.; Hu, S.T.; Jia, Q.J.; Liang, Z.S. Advances in chemical constituents and pharmacological effects of Polygonati Rhizoma. Nat. Prod. Res. Dev. 2021, 33, 1783–1796. [Google Scholar] [CrossRef]
- Yang, S.Y.; Song, J.Z.; Yang, S.J.; Li, J.Y.; Jiang, H.M.; Sui, F.Q.; Li, L.J. Research progress on pharmacological action and new dosage forms of rutin. Chin. J. Mod. Appl. Pharm. 2022, 39, 1360–1370. [Google Scholar] [CrossRef]
- Patel, D.K. Medicinal importance, pharmacological activities and analytical aspects of a flavonoid glycoside ‘Nicotiflorin’ in the medicine. Drug Metab. Bioanal. Lett. 2022, 15, 2–11. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, X.; Li, Q.S.; Deng, H.; Li, J.K. Research progress of natural flavone isoorientin. Chin. J. Food Biotech. 2019, 39, 21–27. [Google Scholar] [CrossRef]
- Zheng, B.S.; Yang, W.H.; Xu, Q.X.; Liu, R.H. Antitumor activity of ferulic acid and its colonic metabolites. J. South China Univ. Technol. (Nat. Sci.) 2022, 50, 30–40. [Google Scholar]
- Zhang, H.B. Effects of sinapic acid on oxidative stress and mitochondrial function in skeletal muscles of rats with exercise injury. Mol. Plant Breed. 2023, 21, 8227–8233. [Google Scholar] [CrossRef]
- Shen, Y.J.; Hao, Y.Y.; Yang, J.Q. Observation of efficacy on supramolecular salicylic acid combined with doxycycline in treatment of moderate to severe acne. Int. J. Biomed. Eng. 2023, 38, 188–191. [Google Scholar] [CrossRef]
- He, X.; Fan, R.F.; Su, L.J.; Kang, X. Plant resource distribution and pharmacological action of orientin. J. Liaoning Univ. Tradi. Chin. Med. 2020, 22, 176–181. [Google Scholar] [CrossRef]
- Wang, L.; Yang, W.; Yao, Q.; Chen, J.; Hu, C.; Xia, Y.; Dong, X.P.; Shi, Q.; Chen, Z.B.; Chen, C. Inhibitory effect on PrPsc deposition and cell protection of vanillic acid in prion-infected cells. Chin. J. Virol. 2022, 38, 896–904. [Google Scholar] [CrossRef]
- Han, T.T. Research on the Mechanism about Narcissoside Relieves Insulin Resistance. Master’s Thesis, Liaoning University, Shenyang, China, 1 May 2023. [Google Scholar]
- Guo, T.T. Study on the Mechanism of Vanillin’s Preprotective Effect on LPS Induced Acute Lung Injury. Ph.D. Thesis, Jilin University, Changchun, China, 1 June 2020. [Google Scholar] [CrossRef]
- Glagoleva, A.Y.; Vikhorev, A.V.; Shmakov, N.A.; Morozov, S.V.; Chernyak, E.I.; Vasiliev, G.V.; Shatskaya, N.V.; Khlestkina, E.K.; Shoeva, O.Y. Features of activity of the phenylpropanoid biosynthesis pathway in melanin-accumulating Barley Grains. Front. Plant Sci. 2022, 3, 923717. [Google Scholar] [CrossRef]
- Shan, T.Y.; Xu, J.Y.; Zhong, X.X.; Zhang, J.J.; He, B.; Tao, Y.J.; Wu, J.W. Full-length transcriptome sequencing provides new insights into the complexity of flavonoid biosynthesis in Glechoma longituba. Physiol. Plant 2023, 175, e14104. [Google Scholar] [CrossRef]
- Wu, X.; Wang, C.K.; Zuo, H.Y.; Chen, Z.H.; Wu, S.B.; Zhou, M.Q. Identification of potential genes involved in biosynthesis of flavonoid and analysis of biosynthetic pathway in Fagopyrum dibotrys. Chin. J. Chin. Mater. Med. 2021, 46, 1084–1093. [Google Scholar] [CrossRef]
- Deshmukh, A.B.; Datir, S.S.; Bhonde, Y.; Kelkar, N.; Samdani, P.; Tamhane, V.A. De novo root transcriptome of a medicinally important rare tree Oroxylum indicum for characterization of the flavonoid biosynthesis pathway. Phytochemistry 2018, 156, 201–213. [Google Scholar] [CrossRef]
- Wang, Y.; Tan, F.Y.; Zhang, L.P.; He, Y.F.; Fan, J.H.; Li, W.; Peng, S.J.; Qiu, Q.H. Characteristic analysis of Polygonatum Sibiricum transcriptome and study of related functional genes. J. Sichuan Norm. Univ. (Nat. Sci.) 2022, 45, 103–109. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Jiang, X.H. Cloning and sequence analysis of the LjFNS gene of honeysuckle flavonoid synthase. Jiangsu Agric. Sci. 2018, 46, 47–50. [Google Scholar] [CrossRef]
- Nong, Q.; Malviya, M.K.; Solanki, M.K.; Lin, L.; Xie, J.L.; Mo, Z.H.; Wang, Z.P.; Song, X.P.; Huang, X.; Li, C.N.; et al. Integrated metabolomic and transcriptomic study unveils the gene regulatory mechanisms of sugarcane growth promotion during interaction with an endophytic nitrogen-fixing bacteria. Plant Biol. 2023, 23, 54. [Google Scholar] [CrossRef]
- Kianersi, F.; Abdollahi, M.R.; Mirzaie-asl, A.; Dastan, D.; Rasheed, F. Identification and tissue-specific expression of rutin biosynthetic pathway genes in Capparis spinosa elicited with salicylic acid and methyl jasmonate. Sci. Rep. 2020, 10, 8884. [Google Scholar] [CrossRef]
- Rojas Rodas, F.; Rodriguez, T.O.; Murai, Y.; Iwashina, T.; Sugawara, S.; Suzuki, M.; Nakabayashi, R.; Yonekura-Sakakibara, K.; Saito, K.; Kitajima, J.; et al. Linkage mapping, molecular cloning and functional analysis of soybean gene Fg2 encoding flavonol 3-O-glucoside (1→6) rhamnosyltransferase. Plant Mol. Biol. 2014, 84, 287–300. [Google Scholar] [CrossRef]
- Yuan, Z.N.; Dong, F.; Pang, Z.Q.; Fallah, N.; Zhou, Y.M.; Li, Z.; Hu, C.H. Integrated metabolomics and transcriptome analyses unveil pathways involved in sugar content and rind color of two sugarcane varieties. Front. Plant Sci. 2022, 13, 921536. [Google Scholar] [CrossRef]
- Yan, H.L.; Zhang, X.X.; Li, X.; Wang, X.L.; Li, H.X.; Zhao, Q.S.; Yin, P.; Guo, R.X.; Pei, X.N.; Hu, X.Q.; et al. Integrated transcriptome and metabolome analyses reveal the anthocyanin biosynthesis pathway in AmRosea1 overexpression 84K poplar. Front. Bioeng. Biotechnol. 2022, 10, 911701. [Google Scholar] [CrossRef]
- Yin, D.J.; Ye, S.J.; Sun, X.Y.; Chen, Q.Y.; Min, T.; Wang, H.X.; Wang, L.M. Integrative analysis of the transcriptome and metabolome reveals genes involved in phenylpropanoid and flavonoid biosynthesis in the Trapa bispinosa Roxb. Front. Plant Sci. 2022, 13, 913265. [Google Scholar] [CrossRef]
- Sheng, X.L.; Chen, H.W.; Wang, J.M.; Zheng, Y.L.; Li, Y.L.; Jin, Z.X.; Li, J.M. Joint transcriptomic and metabolic analysis of flavonoids in Cyclocarya paliurus leaves. ACS Omega 2021, 6, 9028–9038. [Google Scholar] [CrossRef]
- Liu, C.; Feng, T.T.; Liu, X.W.; Ding, J.X.; Shi, H.; Pan, J.; Zhou, Y. Transcriptome analysis and identification of related genes involved in secondary metabolism biosynthesis in Ardisia crispa. Chin. Tradi. Herb. Drugs 2021, 52, 1434–1447. [Google Scholar]
- Zhang, K.M.; Geng, G.G.; Qiao, F. Correlation analysis of enzyme activities, gene expression and flavonoid accumulation during fruit development in Lycium barbarum. Mol. Plant Breed. 2023, 1–13. [Google Scholar]
- Kang, Y.L.; Pei, J.; Cai, W.L.; Liu, W.; Luo, J.; Wu, Q.H. Research progress on flavonoid metabolic synthesis pathway and related function genes in medicinal plants. Chin. Tradi. Herb. Drugs 2014, 45, 336–1341. [Google Scholar]
- Su, J.M.; Peng, T.H.; Bai, M.; Bai, H.Y.; Li, H.S.; Pan, H.M.; He, H.J.; Liu, H.; Wu, H. Transcriptome and metabolome analyses provide insights into the flavonoid accumulation in peels of Citrus reticulata ‘Chachi’. Molecules 2022, 27, 6476. [Google Scholar] [CrossRef]
- Pu, Y.T.; Wang, C.; Jiang, Y.W.; Wang, X.J.; Ai, Y.J.; Zhuang, W.B. Metabolic profiling and transcriptome analysis provide insights into the accumulation of flavonoids in chayote fruit during storage. Front. Nutr. 2023, 10, 1029745. [Google Scholar] [CrossRef]
- Dick, C.A.; Buenrostro, J.; Butler, T.; Carlson, M.L.; Kliebenstein, D.J.; Whittall, J.B. Arctic mustard flower color polymorphism controlled by petal-specific down regulation at the threshold of the anthocyanin biosynthetic pathway. PLoS ONE 2011, 6, e18230. [Google Scholar] [CrossRef]
- Peng, J. Study on the Sex Differentiation of Flavonoids Content and Key Genes Screening in Wild Gender Broussonetia papyrifera Leaves under Heavy Metal Stress. Ph.D. Thesis, CSUFT, Changsha, China, 1 June 2022. [Google Scholar] [CrossRef]
- Wang, Y.J.; Li, H.H.; Fu, W.Y.; Gao, Y.; Wang, B.J.; Li, L. Flavonoids contents and expression analysis of related genes in red cell line of Saussurea medusa. Chin. J. Biotechnol. 2014, 30, 1225–1234. [Google Scholar] [CrossRef]
- Zhou, P.N.; Wan, Q.Y.; Zhang, X.Q.; Gong, L. Cloning and bioinformatics analysis of LAR gene from Ampelopsis megalophylla. Genomics Appl. Biol. 2019, 38, 3654–3660. [Google Scholar] [CrossRef]
- Li, X. Study on the Dynamic Changes of Flavonoids in Jujube Fruits and the Function of Related Genes. Master’s Thesis, Northwest Sci-Tech University Agriculture, Xianyang, China, 1 May 2020. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xu, S.; Qu, Z.Y.; Bi, J.L.; Li, T.T.; Gan, X.F.; Wei, G.L.; Du, J.F.; He, M.; Fan, B.L. Optimization of extraction by BP-ANN and determination of flavonoids in three varieties of polygonati rhizoma before and after irradiation by HPLC chromatographic fingerprint. Sci. Technol. Food Ind. 2021, 42, 257–264. [Google Scholar] [CrossRef]
- Jiang, L.Y.; Wu, P. Research progress on functional components and deep processing of Rhizoma polygonati. China Fruit Veg. 2022, 42, 14–20. [Google Scholar] [CrossRef]
- Wang, S.; Wang, L.L.; Fang, J.J.; Liu, K.L.; Wang, Y.Z.; Zhang, C. Correlation analysis between color and content changes of five components of wine-processed Polygonatum kingianum Rhizoma during processing. Chin. J. Exp. Tradit. Med. Form. 2022, 28, 56–162. [Google Scholar] [CrossRef]
- Liang, Z.H.; Pan, Y.J.; Qiu, L.Y.; Wu, X.Y.; Xu, X.Q.; Shu, W.Y.; Yuan, Q. Analysis on chemical components changes of Polygonati Rhizoma in processing of nine times steaming and nine times sunning by UPLC-O-TOF-MS/MS. Chin. Tradi. Herb. Drugs 2022, 53, 4948–4957. [Google Scholar] [CrossRef]
- Zhang, K. Comparative Study on Chemical Constituents of Plant from Polygonum. Master’s Thesis, Peking Union Medical College, Beijing, China, 1 June 2022. [Google Scholar] [CrossRef]
- Xu, J.X.; Liu, L.; Yang, S.X.; Kuang, Y. Chemical constituents from aerial part of Polygonatum cyrtonema. Chin. Tradi. Herb. Drugs 2016, 47, 3569–3572. [Google Scholar] [CrossRef]
- Song, Y.J.; Guo, T.; Wang, R.J.; Cui, C.L.; Aili, Y.E.; Qu, J.Q.; Xu, C. Effects of compatibility of different prepared products of Polygonati Rhizoma on the contents of six components in Huangjing pills. West China J. Pharm. Sci. 2019, 38, 75–78. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).